Abstract
Candida albicans is known to form polymicrobial biofilms with various Streptococcus spp., including mitis and mutans group streptococci. Streptococcus gordonii (mitis group) has been shown to bind avidly to C. albicans hyphae via direct cell-to-cell interaction, while the cariogenic pathogen Streptococcus mutans (mutans group) interacts with the fungal cells via extracellular glucans. However, the biophysical properties of these cross-kingdom interactions at the single-cell level during the early stage of biofilm formation remain understudied. Here, we examined the binding forces between S. mutans (or S. gordonii) and C. albicans in the presence and absence of in situ glucans on the fungal surface using single-cell atomic force microscopy and their influence on biofilm initiation and subsequent development under cariogenic conditions. The data show that S. gordonii binding force to the C. albicans surface is significantly higher than that ofS. mutans to the fungal surface (~2-fold). However, S. mutans binding forces are dramatically enhanced when the C. albicans cell surface is locally coated with extracellular glucans (~6-fold vs. uncoated C. albicans), which vastly exceeds the forces between S. gordonii andC. albicans. The enhanced binding affinity of S. mutans to glucan-coated C. albicans resulted in a larger structure during early biofilm initiation compared to S. gordonii–C. albicans biofilms. Ultimately, this resulted in S. mutans dominance composition in the 3-species biofilm model under cariogenic conditions. This study provides a novel biophysical aspect of Candida-streptococcal interaction whereby extracellular glucans may selectively favor S. mutans binding interactions with C. albicans during cariogenic biofilm development.
Keywords: glucans, Streptococcus mutans, Streptococcus gordonii, Candida albicans, biophysics, caries
Introduction
Candida albicans is a commonly detected fungal organism in the oral cavity (Ghannoum et al. 2010; Peleg et al. 2010; Koo et al. 2018). C. albicans forms polymicrobial biofilms with a diverse spectrum of bacterial species (Nobile and Johnson 2015). Particularly, interactions between C. albicans and streptococci have been known to play important roles in the pathogenesis of biofilm-derived oral diseases (Falsetta et al. 2014; Koo and Bowen 2014; Nobile and Johnson 2015; Hwang et al. 2017). These cross-kingdom interactions provide mutual benefit for both C. albicans and streptococci, resulting in biofilms of greater microbial carriage and infectivity (Falsetta et al. 2014; Xu et al. 2014; Hwang et al. 2017; Koo et al. 2018), as well as enhanced antimicrobial resistance (Montelongo-Jauregui et al. 2016; Kim et al. 2018).
Mitis group streptococci include early colonizers of oral cavity surfaces, such as Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii (Zheng et al. 2016).C. albicans has been extensively shown to interact with these mitis group streptococci, particularly with S. gordonii, through well-characterized cell-wall surface proteins/receptors on both organisms, resulting in increased biofilm biomass (Ricker et al. 2014; Xu et al. 2014; Jack et al. 2015; O’Donnell et al. 2015). Their interaction is largely mediated by an interaction between C. albicans surface proteins (Als3p, Eap1p, and Hwp1p) and S. gordonii cell-wall associated polypeptides (SspA, SspB, and CshA) (Bamford et al. 2009; Nobbs et al. 2009; Nobbs et al. 2010; Xu et al. 2014).
C. albicans also shows synergistic interactions with mutans group streptococci (Streptococcus mutans) but through a completely different interaction mechanism from the one with S. gordonii (Gregoire et al. 2011; Falsetta et al. 2014; Hwang et al. 2017). Interestingly, the cariogenic pathogen S. mutans has limited direct cell-to-cell binding interaction with C. albicans in the absence of sucrose. Instead, their coadhesion is greatly enhanced by S. mutans–derived glucosyltransferases (Gtfs) that adhere tightly to the C. albicans cell surface, resulting in a large production of glucans in situ in the presence of sucrose (Gregoire et al. 2011; Hwang et al. 2015; Hwang et al. 2017). The glucans on the fungal surface provide an enhanced binding site for S. mutans and also allow C. albicans to colonize dental surfaces, thereby completing an ideal mechanism for cariogenic cross-kingdom biofilm formation.
Although biofilm studies point to synergistic interkingdom associations between C. albicans and S. mutans (or S. gordonii), the influences of in situ glucans on Candida-streptococcal interactions at the single-cell level in the early stage of biofilm formation remain largely unknown. Here, we hypothesized that the binding forces between C. albicans and S. mutans (or S. gordonii) would be altered by glucans on the fungal-cell surface, thereby influencing biofilm initiation and further development. We employed single-cell atomic force microscopy (sc-AFM) to directly measure the binding forces of S. mutans/S. gordonii to C. albicans in the presence/absence of glucans on C. albicans. The data showed that the glucan coating on C. albicans significantly increases adhesive interactions between S. mutans and C. albicans. Notably, the binding force of S. mutans to glucan-coated Candida was substantially higher (~6-fold) than the one from S. gordonii–C. albicans (either uncoated or glucan coated). By employing static/dynamic polymicrobial biofilm models, we confirmed that the enhanced interaction between S. mutans and C. albicans via extracellular glucans resulted in a higher population of S. mutans (vs. S. gordonii) when these 3 species were cocultured under a sucrose-rich condition. Altogether, we demonstrated that extracellular glucans may selectively favor S. mutans binding interactions with C. albicans (vs. S. gordonii) at a single-cell level, which aided the establishment of the pathogen in 3-species polymicrobial biofilm under cariogenic (sucrose-rich) conditions.
Materials and Methods
Microorganisms and Culture Conditions
C. albicans SC5314 or C. albicans SN250 tagged with a red fluorescent protein (tdTomato; gift from Damian J. Krysan, University of Iowa), S. mutans UA159 or a green fluorescent protein (GFP)–expressing S. mutans (LDH-gfp; gift from Justin Merritt, Oregon Health & Science University), and S. gordonii DL1 were used. See the Appendix for more details.
Immobilization of C. albicans and Functionalization of AFM Probes with Streptococci
Prior to sc-AFM analysis, C. albicans cells were chemically immobilized on a glass slide, and AFM probes were functionalized with streptococci as detailed elsewhere (Hwang et al. 2015). Briefly, a coverslip was cleaned and treated with poly-L-lysine (pLL) solution (Sigma-Aldrich). Washed C. albicans were immobilized on a pLL-coated coverslip. Then, the coverslip was gently washed with demineralized (DI) water and kept hydrated until the AFM analysis. For in situ glucan formation on the C. albicans surface, 15 U of glucosyltransferase B (GtfB) enzyme was applied to the immobilized C. albicans on a coverslip and incubated. The coverslip was gently washed with DI water, followed by incubating with a sucrose substrate as described previously (Hwang et al. 2015).
To functionalize the AFM probe with streptococci, we used the tipless AFM cantilever (PNP-TR-TL-50; NanoWorld). The AFM probes were cleaned and immersed in pLL solution. The pLL-coated AFM cantilever was then incubated with washed bacterial cells. After binding, the functionalized probes were washed using phosphate-buffered saline (PBS). See the Appendix for more details.
sc-AFM Methodology
All force measurements were obtained using an MFP-3D AFM (Asylum Research) in fluid phase at room temperature under PBS. Silicon-nitride probes were calibrated using the thermal-tune method (Hutter and Bechhoefer 1993) and examined optically. Cells were imaged in contact mode at randomly selected locations. Force-distance curves were obtained from a minimum of 10 individual microbial cells from at least 3 distinct culture preparations per strain. See the Appendix for more details.
Time-Lapse Imaging of Cross-Kingdom Biofilm Initiation
To investigate the role of glucan coating on C. albicans in the cross-kingdom biofilm initiation, 15 U of GtfB was added to incubate with C. albicans. GtfB-coated C. albicans were washed with PBS and centrifuged, followed by incubating with sucrose substrate. Biofilms were formed using our saliva-coated hydroxyapatite (sHA) model as described elsewhere (Paula et al. 2020). Briefly, each sHA disc was inoculated with microbial cells in UFYTE broth containing 1% (w/v) sucrose at 37°C under 5% CO2. The inoculum concentrations for streptococci and C. albicans were approximately 2 × 108 colony-forming units (CFU)·mL−1 of S. mutans (or S. gordonii) and 2 × 106 CFU·mL−1 of C. albicans. Microbe-attached sHA discs were transferred to the flow-cell device (FC310; BioSurface Technologies) for real-time time-lapse imaging using an upright microscope (LSM 800; Zeiss) with a 40× (numerical aperture, 1.2) water objective. Taken images at 60 and 180 min were subject to the quantification of the number of coadhesion and biofilm biovolume/thickness, respectively. See the Appendix for more details.
Dual and 3-Species Cross-Kingdom Biofilm Model
Biofilms were formed using our sHA model with modification. For dual-species biofilms, each disc was inoculated with ~2 × 106 CFU·mL−1 of S. mutans (or S. gordonii) and ~2 × 104 CFU·mL−1 of uncoated C. albicans and incubated in UFYTE broth containing either 1% sucrose or 1% glucose at 37°C under 5% CO2 for 18 h. For 3 species, ~2 × 106 CFU·mL−1 of S. mutans, ~2 × 106 CFU·mL−1 of S. gordonii, and ~2 × 104 CFU·mL−1 of uncoated C. albicans were inoculated under the same condition. Then, the biofilms were subjected to CFU, total biomass, and imaging analyses. Imaging was performed using a Zeiss LSM 800 upright confocal laser scanning microscope with a 20× (numerical aperture, 1.0) water-immersion objective. Taken images were subject to the quantification of the biovolume of each strain. See the Appendix for more details.
Statistical Analyses
Data analysis was conducted using pairwise comparisons of multiple groups with regression models based on the ranked values. For 2-group comparisons, Kruskal-Wallis tests were used (nonparametric and based on ranks). A significance level set at 5% was used.
Results
To investigate the physical interactions between bacterial cells (S. mutans [Sm] or S. gordonii [Sg]) and the C. albicans surface (uncoated [Ca] or coated with glucans [gCa]), AFM probes functionalized with the bacteria were used (Fig. 1a). As a binding counterpart of streptococci, a layer of C. albicans was immobilized on a coverslip. For reliable detection of intercellular forces, we verified the functionalization of bacteria on the AFM probes using fluorescent imaging, showing that a few Streptococcus cells are located on AFM probes (Fig. 1b1, b2). Quantitative analysis showed that there were no significant differences in the number of bacteria on AFM probes between Sm and Sg (Appendix Fig. 1). Immobilization of C. albicans was also verified via confocal imaging (Fig. 1c1). Finally, glucan coating on C. albicans was confirmed by detecting fluorescently labeled glucans on fungal surfaces (Fig. 1c2, Appendix Fig. 2).
Figure 1.

Single-cell atomic force microscopy (sc-AFM) analysis of the adhesive interactions between Streptococcus and Candida albicans. (a) Schematic diagram of sc-AFM. AFM tipless cantilever probes were functionalized with Streptococcus mutans (or Streptococcus gordonii), and C. albicans cells were immobilized on a coverslip. Fluorescent image of S. mutans (b1) or S. gordonii (b2) functionalized AFM probes. (c1) Confocal image of immobilized C. albicans on the poly-L-lysine-coated coverslip. (c2) Total Internal Reflection Fluorescence-AFM image of glucans formed on immobilized C. albicans.
With sc-AFM, we measured the binding forces and rupture lengths of bacteria to fungus to quantify their adhesive interactions. We tested the binding strength of the nonfunctionalized AFM probe to C. albicans as a control, which showed mostly low binding forces and short rupture lengths (Appendix Fig. 3). When looking at the adhesion forces and rupture lengths, we noticed striking differences between Sm-Ca and Sg-Ca (Fig. 2). The Sm-Ca force distribution showed scattered low binding frequencies with high binding failure (~50%) (Fig. 2a, light-red columns), whereas the binding forces of Sg-Ca were more concentrated between 0.4 and 1.2 nN with low failure (<10%) (Fig. 2c, light-green columns). Thus, the average adhesion force of Sg-Ca was significantly higher (0.62 nN) compared with the Sm-Ca binding force (0.35 nN) (Appendix Table 1). Oppositely, the rupture lengths of Sm-Ca were substantially shorter (0.30 µm) than Sg-Ca (0.68 µm) (Fig. 2b, d; Appendix Table 1).
Figure 2.
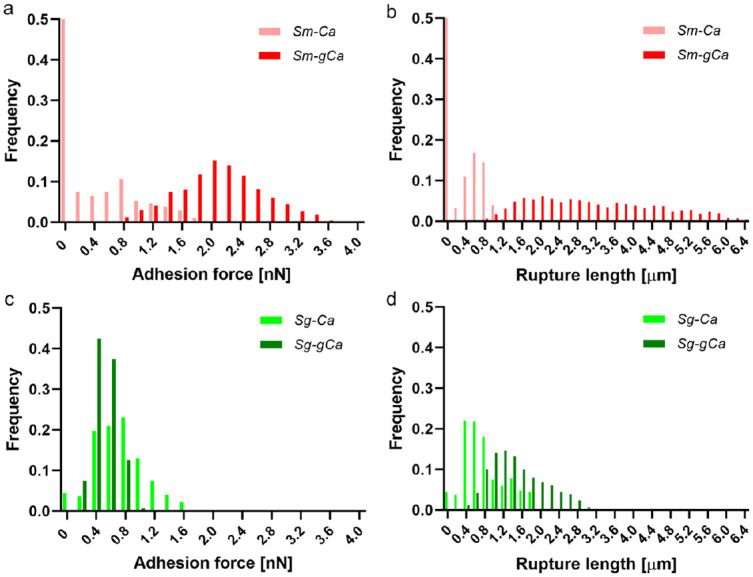
Binding forces and rupture lengths of Streptococcus to yeast Candida albicans surfaces. (a) Adhesion force histogram and (b) rupture length histogram of Streptococcus mutans to uncoated (light-red columns) or glucan-coated yeast C. albicans (dark-red columns). (c) Adhesion force histogram and (d) rupture length histogram of Streptococcus gordonii to uncoated (light-green columns) or glucan-coated yeast C. albicans (dark-green columns).
Next, we tested how the in situ glucans affect the binding forces between the streptococci and C. albicans. As expected, the Sm-Ca interaction was dramatically enhanced by glucan coating on the C. albicans surface, resulting in the elevation of the average adhesion force to 2.00 nN (~6-fold) (Fig. 2a, dark-red columns; Appendix Table 1). Furthermore, the frequency of binding failure (0 nN) was greatly diminished. We concomitantly observed a significant increase (~10-fold) of the rupture length of Sm-gCa, showing around 3.06 µm (Fig. 2c, dark-green columns; Appendix Table 1). Unexpectedly, glucan coating on C. albicans did not increase adhesive interaction between Sg and Ca; rather, Sg-gCa showed a slight reduction of average adhesion force to 0.41 nN (Fig. 2c). Although the average rupture length of Sg-gCa increased to 1.34 µm (Fig. 2d), that of the Sm-gCa condition was still significantly longer (>2-fold). Altogether, the data revealed that the glucan coating on C. albicans dramatically altered the binding behavior of streptococci to C. albicans.
As C. albicans exhibits different morphotypes, we also quantified the binding forces of Sm/Sg to uncoated hyphalC. albicans (hCa) or glucan-coated hyphal C. albicans (ghCa). Similar to Sm-Ca and Sm-gCa data, binding forces and rupture lengths of Sm-ghCa were significantly higher (>4-fold vs. Sm-hCa; Fig. 3a, b; Appendix Table 1). Interestingly, we observed a significant increase in binding forces and rupture lengths of Sg to hyphae (>2-fold) compared with Sg to yeast (Fig. 2c, d; Fig. 3c, d; Appendix Table 1). However, glucan coating on hyphal C. albicans significantly reduced adhesive forces and rupture lengths of Sg (Fig. 3c, d; Appendix Table 1). The highest binding forces of Sm-gCa than any other measures (regardless of C. albicans morphotypes) suggested the critical role of glucans in mediating the S. mutans–C. albicans interaction rather than S. gordonii–C. albicans.
Figure 3.

Binding force and rupture length of Streptococcus to hyphal Candida albicans surfaces. (a) Adhesion force histogram and (b) rupture length histogram of Streptococcus mutans to uncoated (light-red columns) or glucan-coated hyphal C. albicans (dark-red columns). (c) Adhesion force histogram and (d) rupture length histogram of Streptococcus gordonii to uncoated (light-green columns) or glucan-coated hyphal C. albicans (dark-green columns).
The presence of glucans on C. albicans may also influence the cospecies biofilm initiation and development. Thus, we explored the dynamics of cross-kingdom biofilm development using a time-lapse confocal imaging methodology. Excitingly, findings from sc-AFM were further confirmed whereby in situ glucans enhanced S. mutans–C. albicans coadhesion in the early stage of biofilm colonization and subsequent development. When Sm and gCa were cocultured, their interaction was facilitated (Fig. 4a), possibly due to their high binding strengths. However, there was no visible initial coadhesion from Sg-gCa (Fig. 4b). We compared the number of coadhesions between Sm-gCa and Sg-gCa after 60 min of incubation; Sm-gCa exhibited ~6-fold higher frequency of the initial cross-kingdom interaction (vs. Sg-gCa; Fig. 4c). An amalgamated structure consisting of Sm and gCa grew together on the sHA disc in the presence of sucrose, while Sg and gCa developed their structure remotely up to 180 min (Fig. 4a, b). Cross-sectional images and quantified biovolumes/thickness of these biofilms at 180 min clearly showed that a thick and multilayered structure was developed from Sm-gCa, while only a flat and thin-layered structure was visible from Sg-gCa (Fig. 4d, e). Taken together, the data revealed that initial cross-kingdom interaction could modulate the later phases of biofilm development.
Figure 4.
Cross-kingdom interactions of Streptococcus mutans–glucan-coated Candida albicans and Streptococcus gordonii–glucan-coated C. albicans. Time-lapse images of (a) S. mutans–glucan-coated C. albicans (Sm-gCa) and (b) S. gordonii–glucan-coated C. albicans (Sg-gCa) biofilm initiation in the presence of 1% sucrose. Red color on the far-left panels of (a) and (b) showed glucans were formed on C. albicans surfaces. Extracellular polysaccharides channel was removed in time-lapse images (0 to 180 min) to clearly show Streptococcus–C. albicans direct cell-to-cell interactions. (c) The number of coadhesions between S. mutans (or S. gordonii) and glucan-coated C. albicans per area (319.45 × 319.45 µm) in each biofilm after 60 min of incubation (n = 20). (d) Orthogonal views of each biofilm at 180 min (left: Sm-gCa; right: Sg-gCa). (e) Biovolumes and the maximum thickness of each biofilm at 180 min (n = 3). Scale bars indicate 10 µm. Asterisk indicates that the values are significantly different between Sm-gCa and Sg-gCa. (P < 0.05).
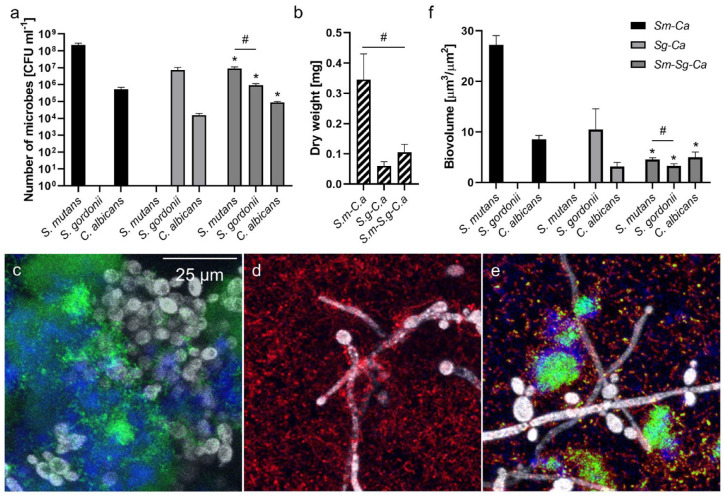
Therefore, we determined the properties (CFU and dry weight [DW]) of 18-h-old dual-species (Sm-Ca or Sg-Ca) and 3-species (Sm-Sg-Ca) biofilms to assess the important role of glucans in cross-kingdom biofilm development. As shown in Figure 5a, we observed that the population of C. albicans was substantially higher (~35-fold) when they were cocultured with S. mutans (vs. S. gordonii). Furthermore, the CFU of S. mutans was significantly higher (~30-fold) than the CFU of S. gordonii. Interestingly, when 3 species were cocultured, all populations were reduced possibly due to competition between S. mutans and S. gordonii as well as the acceleration of nutrient depletion or accumulation of toxic compounds. Nevertheless, the CFU of S. mutans was still a log higher than S. gordonii in the 3-species biofilm (Fig. 5a). C. albicans population in 3 species was also ~6-fold higher (vs. Sg-Ca), while it was ~6-fold lower (vs. Sm-Ca). This trend was also observed from biofilm biomass; Sm-Ca exhibited the highest biomass, whereas it decreased whenS. gordonii was involved (Fig. 5b). In addition, biofilm morphologies were in line with binding force data; lots of yeastC. albicans (gray) interacted with S. mutans (green) via in situ extracellular polysaccharides (EPS) (blue) (Fig. 5c), while abundant S. gordonii (red) bound to the hyphal C. albicans (Fig. 5d). In a 3-species biofilm, we observed a similar pattern that S. mutans microcolonies interacted with C. albicans through EPS, and numerous S. gordonii cells were bound to hyphal C. albicans (Fig. 5e). Quantified biovolumes of each component also showed a similar trend to CFU data (Fig. 5a, f). Finally, we assessed biofilm properties cultured in 1% glucose to see whether the removal of competitive advantages for S. mutans might shift microbial composition. As expected, the numbers of both S. mutans and C. albicans as well as DW in Sm-Ca were reduced significantly when cultured under 1% glucose (vs. 1% sucrose; Appendix Fig. 4). Moreover, the S. mutans population was significantly lower than the S. gordonii population in Sm-Sg-Ca under the glucose condition (Appendix Fig. 4), which showed a reversed trend to the sucrose condition (Fig. 5a). Collectively, our data reveal how “glucan surface-priming” enhances S. mutans–C. albicans coadhesion, thereby influencing the cross-kingdom biofilm initiation and development.
Figure 5.
Properties of dual- and 3-species biofilms cultured in 1% sucrose. (a) Colony-forming units (CFU) and (b) dry weight of Streptococcus mutans–Candida albicans, Streptococcus gordonii–C. albicans, and S. mutans–S. gordonii–C. albicans biofilms. Representative confocal images of (c) S. mutans–C. albicans, (d) S. gordonii–C. albicans, and (e) S. mutans–S. gordonii–C. albicans biofilms. Green, red, blue, and gray colors indicate S. mutans, S. gordonii, Extracellular polysaccharides-matrix, and C. albicans, respectively. (f) Biovolumes of each strain in each biofilm. Asterisk indicates that the values are significantly different from dual-species biofilms. Hash indicates that the values are significantly different among groups. (P < 0.05) (n = 8).
Discussion
AFM is an advanced tool for imaging and measuring forces at the nanoscale (Helenius et al. 2008; Dufrêne 2014). By functionalizing the AFM probe with bacterial organisms and immobilizing fungal cells on the surface, biophysical properties of the binding interactions between these key players can be unveiled (Müller et al. 2009; Dufrêne 2015). The results of this study showed how species-specific cross-kingdom adhesive behaviors affected the cariogenic biofilm formation under sugar-rich diets.
Previous studies revealed that S. mutans–derived GtfB avidly bound to the C. albicans surface, thereby producing large amounts of extracellular α-glucans on the fungal surface (Gregoire et al. 2011; Hwang et al. 2015). Here, the S. mutans–C. albicans interaction in the cariogenic conditions (+glucans) yielded the highest binding forces of all conditions, confirming the important role of in situ glucans on C. albicans in mediating their cross-kingdom interaction. Interestingly, we observed increased rupture lengths of both streptococci to gCa, possibly due to the long chain of polysaccharides (glucans) attributed to an extension of rupture length when unfolded (Francius et al. 2009). As glucans on the C. albicans surface did not favor the S. gordonii attachment, it is plausible that the glucan binding proteins (Gbp) on the S. mutans membrane contribute to S. mutans binding to the Candida surface through locally produced glucans (Banas and Vickerman 2003). However, additional studies using isogenic Gbp-defective mutant strains are required to further confirm the strong binding affinity between glucans and Gbp.
An avid interaction between S. gordonii and hyphal C. albicans has been well documented (Bamford et al. 2009; Ricker et al. 2014; Jack et al. 2015). In this study, we observed a substantial increase in binding affinity of Sg-hCa (vs. Sg-Ca). It may confirm the important role of cell surface adhesin of S. gordonii (SspA/SspB) (Bamford et al. 2009) and hyphal wall protein of C. albicans (ALS3) (Silverman et al. 2010; Bamford et al. 2015) in their interaction. However, the binding forces of Sg-hCa were still significantly lower than Sm-gCa (or Sm-ghCa) (Appendix Table 1), suggesting the critical role of glucans in favoring the S. mutans–C. albicans interaction amid cariogenic biofilm initiation and subsequent development (vs. S. gordonii–C. albicans). Interestingly, the binding force of Pseudomonas aeruginosa to C. albicans showed a similar pattern to the S. gordonii to C. albicans interaction: increased binding forces of P. aeruginosa to hyphal C. albicans (vs. yeast C. albicans) (Ovchinnikova et al. 2012). Also, Staphylococcus aureus bound strongly to hyphal C. albicans through surface adhesin (Als3p) (Peters et al. 2012), which showed a similar binding mechanism of S. gordonii to hyphal C. albicans. Collectively, our data reveal a distinct binding characteristic of the S. mutans–C. albicans interaction mediated by surface-adsorbed insoluble α-glucans.
Specifically, those high binding forces of Sm-gCa under cariogenic conditions may help S. mutans compete with other bacteria while building oral biofilms. For example, S. gordonii is known to be capable of antagonizing and competing with S. mutans (Kreth et al. 2008; Abranches et al. 2018; Huang et al. 2018). However, data from the 3-species biofilm experiment showed that enhanced interaction between S. mutans and C. albicans resulted in a significantly higher population of S. mutans (vs. S. gordonii) in the early stage of biofilms (Fig. 5). Intriguingly, the S. mutans dominance in Sm-Sg-Ca was not maintained when cultured in the glucose condition (Appendix Fig. 4). It suggested that enhanced S. mutans–C. albicans interaction via in situ glucans on Candida under a sucrose-rich condition could boost S. mutans competitiveness against S. gordonii.
Although we observed the distinct role of S. mutans–derived α-glucans in modulating Streptococcus-Candida interactions, various glucosyltransferases from S. mutans, as well as other streptococci (e.g., S. gordonii or S. oralis), appeared to play different roles in forming biofilms with C. albicans. For example, S. gordonii GtfG promoted dual-species biofilm development with C. albicans (Ricker et al. 2014), while binding of S. oralis GtfR to fungal cells did not play a major role in their biofilm formation (Souza et al. 2020). Furthermore, it revealed that the presence of sucrose in each model adversely affected the cross-kingdom biofilm interaction; sucrose further augmented S. gordonii–C. albicans biofilm formation (Ricker et al. 2014), while it negatively impacted S. oralis–C. albicans biofilms (Souza et al. 2020). Finally, it is noteworthy that bacteria-fungus interaction in human saliva could be distinctly modulated by various salivary components. Therefore, further investigation of the role of various glucosyltransferases on the streptococci–C. albicans polymicrobial interaction in human saliva supplemented with different types of sugars is required to shed light on an enhanced understanding of the pathogenesis of severe tooth decay.
In summary, the data reveal that in situ glucans on the surface of C. albicans selectively favor the S. mutans binding to C. albicans over S. gordonii. The early alliance between S. mutans and glucan-coated C. albicans assists the establishment of cariogenic S. mutans against S. gordonii in a 3-species mixed biofilm model under cariogenic (sucrose-rich) conditions while developing biofilm structures. Our findings provide a biophysical aspect of the distinct streptococci–C. albicans interactions that are largely governed by bacterially produced extracellular polysaccharides.
Author Contributions
S.X. Wan, contributed to data acquisition, analysis, and interpretation, drafted the manuscript; J. Tian, Y. Liu, A. Dhall, contributed to data acquisition and analysis, drafted the manuscript; H. Koo, contributed to conception and design, critically revised the manuscript; G. Hwang, contributed to conception, design, and data interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520950286 for Cross-Kingdom Cell-to-Cell Interactions in Cariogenic Biofilm Initiation by S.X. Wan, J. Tian, Y. Liu, A. Dhall, H. Koo and G. Hwang in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported in part by the National Institutes for Dental and Craniofacial Research (NIDCR) grants DE027970 (GH) and DE025220 (HK) and was carried out in part at the Singh Center for Nanotechnology, which is supported by the NSF National Nanotechnology Coordinated Infrastructure Program under grant NNCI-1542153.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iDs: A. Dhall  https://orcid.org/0000-0001-8950-9994
https://orcid.org/0000-0001-8950-9994
References
- Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA. 2018. Biology of oral streptococci. Microbiol Spectr. 6(5):GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. 2015. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 161(Pt 1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CV, D’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 77(9):3696–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA, Vickerman MM. 2003. Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med. 14(2):89–99. [DOI] [PubMed] [Google Scholar]
- Dufrêne YF. 2014. Atomic force microscopy in microbiology: new structural and functional insights into the microbial cell surface. mBio. 5(4):e01363-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF. 2015. Sticky microbes: forces in microbial cell adhesion. Trends Microbiol. 23(6):376–382. [DOI] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 82(5):1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francius G, Alsteens D, Dupres V, Lebeer S, De Keersmaecker S, Vanderleyden J, Gruber HJ, Dufrêne YF. 2009. Stretching polysaccharides on live cells using single molecule force spectroscopy. Nat Protoc. 4(6):939–946. [DOI] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 8(6):e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, et al. 2011. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 77(18):6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J, Heisenberg CP, Gaub HE, Muller DJ. 2008. Single-cell force spectroscopy. J Cell Sci. 121(11):1785–1791. [DOI] [PubMed] [Google Scholar]
- Huang X, Browngardt CM, Jiang M, Ahn SJ, Burne RA, Nascimento MM. 2018. Diversity in antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Caries Res. 52(1–2):88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter JL, Bechhoefer J. 1993. Calibration of atomic-force microscope tips. Rev Sci Instrum. 64(7):1868–1873. [Google Scholar]
- Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. 2017. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13(6):e1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Marsh G, Gao L, Waugh R, Koo H. 2015. Binding force dynamics of Streptococcus mutans–glucosyltransferase B to Candida albicans. J Dent Res. 94(9):1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack AA, Daniels DE, Jepson MA, Margaret Vickerman M, Lamont RJ, Jenkinson HF, Nobbs AH. 2015. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 161(Pt2):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Liu Y, Benhamou RI, Sanchez H, Simon-Soro A, Li Y, Hwang G, Fridman M, Andes DR, Koo H. 2018. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 12(6):1427–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Andes DR, Krysan DJ. 2018. Candida–streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 14(12):e1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Bowen WH. 2014. Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Future Microbiol. 9(12):1295–1297. [DOI] [PubMed] [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 190(13):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelongo-Jauregui D, Srinivasan A, Ramasubramanian AK, Lopez-Ribot JL. 2016. An in vitro model for oral mixed biofilms of Candida albicans and Streptococcus gordonii in synthetic saliva. Front Microbiol. 7:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DJ, Helenius J, Alsteens D, Dufrêne YF. 2009. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol. 5(6):383–390. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 73(3):407–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. 2010. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell. 9(10):1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD. 2015. Candida albicans biofilms and human disease. Annu Rev Microbiol. 69(1):71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LE, Millhouse E, Sherry L, Kean R, Malcolm J, Nile CJ, Ramage G. 2015. Polymicrobial Candida biofilms: friends and foe in the oral cavity. FEMS Yeast Res. 15(7):fov077. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova ES, Krom BP, Van Der Mei HC, Busscher HJ. 2012. Force microscopic and thermodynamic analysis of the adhesion between Pseudomonas aeruginosa and Candida albicans. Soft Matter. 8(24):6454–6461. [Google Scholar]
- Paula AJ, Hwang G, Koo H. 2020. Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nat Commun. 11(1):1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Hogan DA, Mylonakis E. 2010. Medically important bacterial–fungal interactions. Nat Rev Microbiol. 8(5):340–349. [DOI] [PubMed] [Google Scholar]
- Peters BM, Ovchinnikova ES, Krom BP, Schlecht LM, Zhou H, Hoyer LL, Busscher HJ, van der Mei HC, Jabra-Rizk MA, Shirtliff ME. 2012. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology. 158(12):2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker A, Vickerman M, Dongari-Bagtzoglou A. 2014. Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans.J Oral Microbiol. 6. doi: 10.3402/jom.v6.23419. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 78(11):4644–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza JGS, Bertolini M, Thompson A, Mansfield JM, Grassmann AA, Maas K, Caimano MJ, Barao VAR, Vickerman MM, Dongari-Bagtzoglou A. 2020. Role of glucosyltransferase R in biofilm interactions between Streptococcus oralis and Candida albicans. ISME J. 14(5):1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou A. 2014. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 29(3):99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Tan TK, Paterson IC, Mutha NVR, Siow CC, Tan SY, Old LA, Jakubovics NS, Choo SW. 2016. Streptobase: an oral Streptococcus mitis group genomic resource and analysis platform. PLoS One. 11(5): e0151908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520950286 for Cross-Kingdom Cell-to-Cell Interactions in Cariogenic Biofilm Initiation by S.X. Wan, J. Tian, Y. Liu, A. Dhall, H. Koo and G. Hwang in Journal of Dental Research