Abstract
Objective
Human mesenchymal stem cells (hMSCs) are a promising source for regenerative medicine, especially mesodermal lineages. Clinical applications require an understanding of the mechanisms for transcriptional control to maintain the desired cell type. The aim of this study was to identify novel markers for differentiation of hMSCs into bone or cartilage with the use of Kartogenin, by RNA analysis using microarray technology, and explore the role of RhoA-Rho associated protein kinase (ROCK) inhibition in these.
Methods
Commercial human bone marrow derived primary mesenchymal stem cells were purchased from ATCC. Cells were differentiated in vitro in 2-dimensional cultures using Kartogenin as the main cartilage inducer and bone morphogenetic protein 2 for bone differentiation; cells were cultured with and without ROCK inhibitor Y-27632. After 21 days of culture, whole RNA was extracted and analyzed via Affimetrix microarrays. The most significant hits were validated by quantitative polymerase chain reaction.
Results
We found a total of 1,757 genes that were either up- or downregulated on differentiation, when compared to P1 hMSC (control) at day 0 of differentiation. Two members of the Serpin superfamily, SERPINA9 and SERPINB2, were significantly upregulated in the cartilage groups, whereas they were unchanged in the bone groups with and without ROCK inhibition.
Conclusions
SERPINA9 and SERPINB2 are novel differentiation markers, and molecular regulator candidates for hMSC lineage commitment toward bone and cartilage, providing a new tool for regenerative medicine. Our study highlights the roles of these 2 genes, with significant upregulation of both in cell cultures stimulated with Kartogenin.
Keywords: mesenchymal stem cells, SERPINA9, SERPINB2, chondrocytes, osteoblasts
Introduction
Human mesenchymal stem cells (hMSCs), multipotent stem cells able to differentiate into 4 main lineages including osteoblasts and chondrocytes, are a very promising cell source for regenerative medicine.1 Clinical applications require an understanding of the mechanisms for transcriptional control to maintain the desired cell type. Cartilage regeneration techniques rely primarily in cell therapy, with the first technique describing autologous chondrocyte implantation.2 The best results have been accomplished with the use of chondrocytes for cartilage restoration, since the ultimate goal is hyaline cartilage.2 Despite progress been made, new cell sources are being researched, namely, bone marrow–derived hMSCs. These must express the following cell surface receptors in at least 95%: CD105, CD73, and CD90; and be negative for CD45, CD14, CD34, CD19, and Class II HLA.3 Use of these cells has given mixed results over the years, the main concern being that quality of regenerated cartilage is not as good as the hyaline variety found in articular cartilage.4 Also, several studies have seen deterioration of this neocartilage after a median of 18 months.2,5
In the past years another approach to regenerate hyaline cartilage for articular purposes has focused on the use of stem cells from several origins, and commitment for having a developmental recapitulation of cartilage in vitro. In order to do so, developmental gene regulation must be taken into account. The master regulator of cartilage development is the gene SOX9.6 In early studies, it was demonstrated that both cartilage and bone lineages share a common osteochondro progenitor and that the RUNX2 gene, which is master regulator for bone formation, is downstream of SOX9, at least in the case of endochondral ossification, whereas for intramembranous bone formation mesenchymal stem cells can differentiate directly into bone cells.6
Differentiation of mesenchymal stem cells into cartilage has previously been achieved using members of the transforming growth factor (TGF) β family, especially TGF-β3.4 Differentiation to bone has been extensively described in the past, also using other members of the superfamily named bone morphogenetic proteins (BMP), with the best one characterized being BMP2.7 The new strategy to induce certain genetic expression patterns is to bypass cell surface receptors using small chemical molecules that are able to target the cytosolic or nuclear proteins that are transcription factors.8 Kartogenin has recently been reported to induce chondrogenesis directly by dissociating CBFβ from filamin A, to later translocate to the nucleus and activate RUNX1.9 On the other hand, inhibition of BMP type I receptor kinase has recently demonstrated to commit hMSCs to articular cartilage.10
The RhoA-Rho associated protein kinase (ROCK) branch of the small Rho GTPases has been the most researched pathway in lineage commitment, due to easy manipulation with chemical ROCK inhibitors, such as Fasudil and Y-27632.11 This pathway has been implicated in lineage commitment of mesenchymal stem cells to many cell types.11 Interestingly, ROCK inhibition can promote or inhibit chondrogenesis and/or osteogenesis, depending on the developmental stage of the cell type used for the in vitro assays.11
The aim of this study was to identify novel markers for differentiation of hMSCs into bone and cartilage. In order to do this, a commercial bone marrow–derived hMSC line previously validated for differentiation to adipocytes, chondrocytes, and osteocytes was used, differentiating them in vitro into cartilage and bone. Cells were cultured with and without ROCK inhibitor Y-27632. RNA was extracted and analyzed via Affimetrix microarrays, with the most significant hits validated by quantitative polymerase chain reaction (qPCR).
Methods
Cell Culture
Multipotent mesenchymal stem cells with the ability to differentiate to both cartilage and bone lineages PCS-500-012 from ATCC were used.9 This cell line has been validated for differentiation to adipocytes, chondrocytes, and osteocytes, being positive for CD29, CD44, CD73, CD90, CD105, and CD166 and negative for CD14, CD19, CD34, and CD45, which are markers considered as International Society for Cellular Therapy (ISCT) criteria for characterizing MCSs.12 Cells were cultured with DMEM:F12 (Dulbecco’s modified Eagle medium), 10% FCS (fetal calf serum), Pen/Strep 1:100, and Glutamine 1:100 (Gibco-Invitrogen, Carlsbad, CA).
In Vitro Chondrogenic Differentiation
To set up the differentiation assay, 3 technical replicates were cultured in parallel in 24-well plates; 5 × 104 cells per 1 cm2 were seeded. Basal media was supplemented with Kartogenin (KGN) 100 nM (Sigma) and ascorbic acid 50 µM (Sigma).9
In Vitro Osteogenic Differentiation
To set up the differentiation assay, 3 technical replicates were cultured in parallel in 24-well plates; 5 × 104 cells per 1 cm2 were seeded. Basal media was supplemented with BMP-2 at 50 ng/mL (Peprotech), ascorbic acid 50 µM (Sigma), dexamethasone at 1 × 10−6, and β-glycerol phosphate at a final concentration of 10 mM.
ROCK Inhibition
In order to inhibit RhoA downstream effector ROCK, the chemical inhibitor Y-27632 (Sigma) was used at a final concentration of 10 µM/mL in both groups differentiated to chondrocyte and osteoblast lineage.
RNA Isolation
After 21 days of culture, differentiation medium was removed and the cell monolayer was washed twice with phosphate-buffered saline. QIAGEN RNEasy mini kit was used as recommended by the manufacturer. Briefly, monolayers were removed from the tissue culture plates with a 10-minute incubation time at 37°C with Accutase (Sigma); cells were dislodged mechanically with a P1000 tip and transferred to a clean Eppendorf tube. Cells were pelleted and the supernatant removed. They were then lysed with the RLT buffer and processed until the end of the protocol, eluting with 50 µL of RNase-free water. Integrity and purity of RNA concentration was assessed using ND-1000 Spectrophotometer (Nano-Drop Technologies, Wilmington, DE). The quality of each RNA sample was validated on an Agilent BioAnalyzer 2100 (Agilent Technologies, Germany), and 150 ng of the best-quality samples (RIN values between 8.0 and 9.7) was processed using Affymetrix Whole Transcript Sense Target Labelling Kit (Affymetrix, Santa Clara, CA).
DNA Microarray Global Gene Expression Profiling
For microarray experiments, target cDNA was prepared according to the Whole-Transcript PLUS (WT) Sense Target Labelling Protocol (Affymetrix, Inc., Santa Clara, CA). Briefly, pooled RNA (200 ng) from 3 biological replicates was converted to first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer. Second strand cDNA synthesis was followed by an in vitro transcription to generate cRNA. The cRNA products were used as templates for a second cycle cDNA synthesis where dUTPs are incorporated to the new strand. The cDNA was fragmented using uracil-DNA glycosilase and apurin apirymidin endonuclease. The fragments (40-70 mers) where then labelled by means of a biotin-labelled deoxynucleotide terminal addition reaction. Fragmented and labelled cDNA was hybridized using Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Santa Clara, CA). The arrays were washed, stained for biotinylated cDNA, and scanned according to the manufacturer’s recommendations. The array represents 47,000 total RefSeq transcripts covered with a median of 22 probes per gene, which assures an accurate detection for genome-wide transcript expression changes (http://www.affymetrix.com).
Gene Expression Profiling
Samples were classified into 5 groups with 3 replicates each: (1) Control hMSCs; (2) samples from chondrogenic media (Chondro); (3) samples from chondrogenic media ROCK inhibited with Y-27632 (Chondro Y); (4) osteogenic samples (Osteo); and (5) samples from osteogenic media ROCK inhibited with Y-27632 (Osteo Y). Hierarchical cluster analysis was performed as described previously.13
Quantitative Reverse Transcription-Polymerase Chain Reaction Analysis
In order to validate microarray findings, master transcription factors for each type of tissue (SOX9 for cartilage and RUNX2 for bone) along with those genes with highest differential expression and biological significance (SERPINB2 and SERPINA9) were measured in total RNA from the in vitro differentiation assays. qPCR assays for each gene were carried out in triplicate and normalized to the most stably expressed reference gene in hMSC: peptidyl-prolyl isomerase A (PPIA).14
cDNA Synthesis for qPCR Validation
Complementary DNA (cDNA) synthesis was carried out with QuantiTect Rev. Transcription Kit (QIAGEN) starting from 900 ng of total RNA of each sample. We checked for genomic DNA (gDNA) contamination before the cDNA synthesis by agarose electrophoresis and by real-time PCR after the DNA Wipeout step of the cDNA synthesis procedure. All samples were free of gDNA contamination.
The PCR reaction was performed with RT2 qPCR Primer Assays (QIAGEN) using the following conditions: 1 µL of cDNA, 1 µL of the primer assay, 12.5 1 µL of RT2 SYBR Green ROX Mastermix (QIAGEN), and 10.5 µL of nuclease-free water. Samples were amplified using the following steps: 1 hold of 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds, and finally a melting curve ramping from 50°C to 99°C.
We performed the gene expression assays by triplicate in a Rotor-Gene Q instrument (QIAGEN) obtaining a global mean variation coefficient of 1.37%. The information of the primers used in the expression assays is shown in Supplemental Table 1. The manufacturer (QIAGEN) experimentally verified all primers to amplify a single amplicon of the correct size with uniform PCR efficiency.
Cell Culture Protocol with TGF-β1
In order to further strengthen our results, a standard 21-day differentiation protocol using TGF-β1 was performed in the same primary cells. Cells were cultured with DMEM:F12, 10% FCS, Pen/Strep 1:100, and glutamine 1:100 (Gibco-Invitrogen, Carlsbad, CA). To set up the differentiation assay, 3 technical replicates were cultured in parallel in 24-well plates; 5 × 104 cells per 1 cm2 were seeded. Basal media was supplemented with Transforming Growth Factor β1 (TGF-β1) 10 ng/mL (Peprotech) and ascorbic acid 50 µM (Sigma).15 RNA isolation and qPCR analyses were performed from 3 independent experiments (n = 3) in the same manner as previously described.
Statistical Analysis
For microarray analyses, all possible pairwise comparisons between the 5 groups generated some contrasts of interest. Raw data were background-corrected using Robust Multiarray Average (RMA)16 and normalized using Quantile Normalization.17 Differential expression was determined using statistical linear models with arbitrary coefficients; contrasts of interest were analyzed using the bioconductor library “limma.”18,19 Correction for multiple hypotheses was applied using false discovery rate, and genes were selected as differentially expressed based on a fold-change (FC) <−2 or >2 and considered significant when P value was below 0.05.
For qPCR, relative expression units were calculated with REST software (QIAGEN) using the reference gene PPIA and normalizing with the control group. We compared the expression levels among the 5 groups of each gene using the Kruskal-Wallis test adjusted by multiple comparisons. Statistical analysis and the graphs were performed in the IBM SPSS Statistics 21 statistical package. P values below 0.05 were considered significant.
Results
Molecular Phenotype of Chondrocytes and Osteoblasts with and without ROCK Inhibition
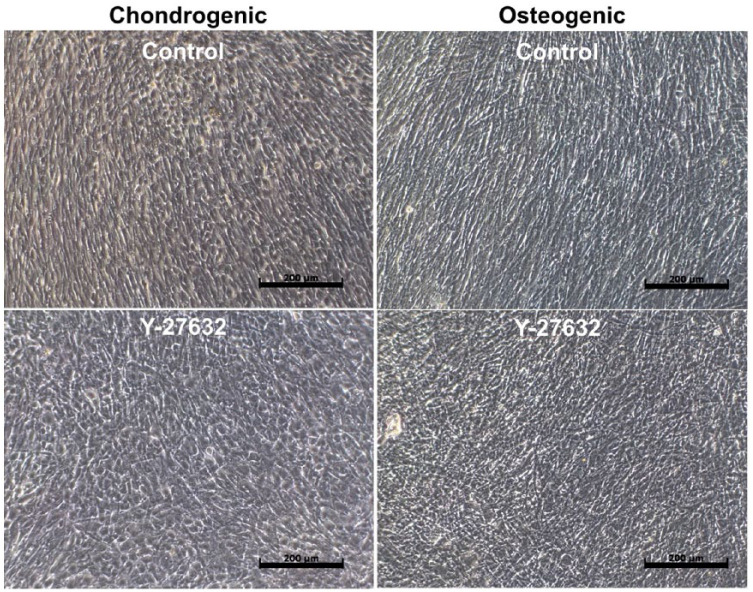
Global gene expression profiling was conducted on hMSCs treated with or without Y-27632 (10 µM/mL) in the presence of osteogenic or chondrogenic media ( Fig. 1 ). Hierarchical clustering based on differentially expressed genes revealed clear separation of control cells treated with the chondrogenic media or osteogenic media regardless of ROCK inhibition. We identified 1,757 genes differentially expressed within 2.0 FC, P < 0.05, 635 were upregulated with KGN treatment and 768 genes upregulated in the BMP2 treatment group. The most representative 100 genes, 51 upregulated with KGN treatment and 49 upregulated in the BMP2 treatment group, are shown in Figure 2 and Table 1 . The complete list of genes and their differential expression between groups are shown in Supplemental Tables S2 to S5.
Figure 1.
High-density monolayer culture in chondrogenic and osteogenic conditions. Phase contrast images of 7-day high-density monolayer cultures in chondrogenic (first column) and osteogenic (second column) conditions, with and without ROCK inhibitor Y-27632. Scale bar: 200 µm.
Figure 2.
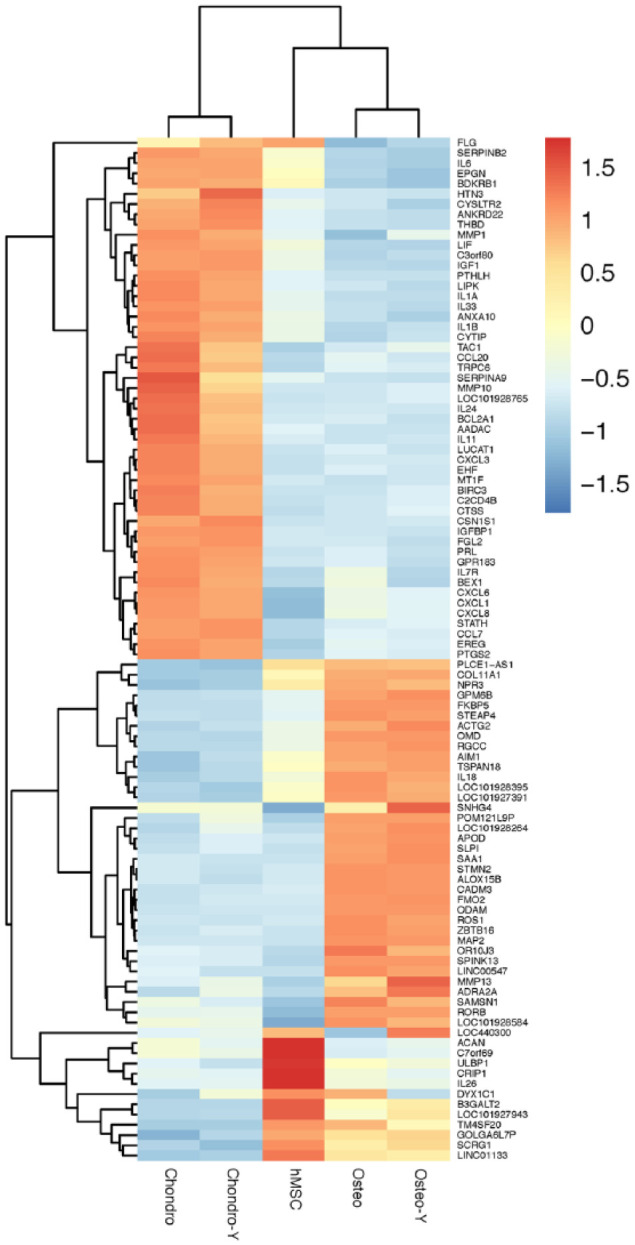
Hierarchical cluster analysis of genes involved in chondrocyte and bone differentiation. Chondro = chondrocytes; Chondro-Y = chondrocytes with ROCK inhibitor Y27632; hMSC = human bone marrow mesenchymal stem cells; Osteo = osteoblasts; Osteo-Y = osteoblasts with ROCK inhibitor Y27632; ROCK = Rho-associated coiled-coil containing protein kinase.
Table 1.
Hundred Most Representative Genes Revealed in Microarray Analysis.
| Upregulation with Kartogenin | Upregulation with BMP2 | ||
|---|---|---|---|
| Gene Symbol | Gene Name | Gene Symbol | Gene Name |
| AADAC | Arylacetamide deacetylase | ACAN | Aggrecan |
| ANKRD22 | Ankyrin repeat domain 22 | ACTG2 | Actin, gamma 2, smooth muscle, enteric |
| ANXA10 | Annexin A10 | ADRA2A | Adrenoceptor alpha 2A |
| BCL2A1 | B-cell CLL/lymphoma 2–related protein A1 | AIM1 | Absent in melanoma 1 |
| BDKRB1 | Bradykinin receptor B1 | ALOX15B | Arachidonate 15-lipoxygenase, type B |
| BEX1 | Brain expressed, X-linked 1 | APOD | Apolipoprotein D |
| BIRC3 | Baculoviral IAP repeat containing 3 | B3GALT2 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| C2CD4B | C2 calcium-dependent domain containing 4B | C7orf69 | Chromosome 7 open reading frame 69 |
| C3orf80 | Chromosome 3 open reading frame 80 | CADM3 | Cell adhesion molecule 3 |
| CCL20 | Chemokine (C-C motif) ligand 20 | COL11A1 | Collagen, type XI, alpha 1 |
| CCL7 | Chemokine (C-C motif) ligand 7 | CRIP1 | Cysteine-rich protein 1 (intestinal) |
| CSN1S1 | Casein alpha s1 | DYX1C1 | Dyslexia susceptibility 1 candidate 1 |
| CTSS | Cathepsin S | FKBP5 | FK506 binding protein 5 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | FMO2 | Flavin containing monooxygenase 2 (nonfunctional) |
| CXCL3 | Chemokine (C-X-C motif) ligand 3 | GOLGA6L7P | Golgin A6 family-like 6 |
| CXCL6 | Chemokine (C-X-C motif) ligand 6 | GPM6B | Glycoprotein M6B |
| CXCL8 | Chemokine (C-X-C motif) ligand 8 | IL18 | Interleukin 18 |
| CYSLTR2 | Cysteinyl leukotriene receptor 2 | IL26 | Interleukin 26 |
| CYTIP | Cytohesin 1 interacting protein | LINC00547 | Long intergenic non-protein coding RNA 547 |
| EHF | Ets homologous factor | LINC01133 | Long intergenic non-protein coding RNA 1133 |
| EPGN | Epithelial mitogen | LOC101927391 | Uncharacterized LOC101927391 |
| EREG | Epiregulin | LOC101927943 | Uncharacterized LOC101927943 |
| FGL2 | Fibrinogen-like 2 | LOC101928264 | Uncharacterized LOC101928264 |
| FLG | Filaggrin | LOC101928395 | Uncharacterized LOC101928395 |
| GPR183 | G protein-coupled receptor 183 | LOC101928584 | Uncharacterized LOC101928584 |
| HTN3 | Histatin 3 | LOC440300 | Chondroitin sulfate proteoglycan 4 pseudogene |
| IGF1 | Insulin-like growth factor-like family member 1 | MAP2 | Microtubule-associated protein 2 |
| IGFBP1 | Insulin-like growth factor binding protein 1 | MMP13 | Matrix metallopeptidase 13 |
| IL11 | Interleukin 11 | NPR3 | Natriuretic peptide receptor 3 |
| IL1A | Interleukin 1, alpha | ODAM | Odontogenic, ameloblast associated |
| IL1B | Interleukin 1, beta | OMD | Osteomodulin |
| IL24 | Interleukin 24 | OR10J3 | Olfactory receptor, family 10, subfamily J, member 3 |
| IL33 | Interleukin 33 | PLCE1-AS1 | Phospholipase C, epsilon 1 antisense RNA 1 |
| IL6 | Interleukin 6 | POM121L9P | POM121 transmembrane nucleoporin-like 9, pseudogene |
| IL7R | Interleukin 7 receptor | RGCC | Regulator of cell cycle |
| LIF | Leukemia inhibitory factor | RORB | RAR-related orphan receptor B |
| LIPK | Long intergenic non-protein coding RNA 1133 | ROS1 | ROS proto-oncogene 1, receptor tyrosine kinase |
| LOC101928765 | Uncharacterized LOC101928765 | SAA1 | Serum amyloid A-like 1 |
| LUCAT1 | Lung cancer associated transcript 1 (non-protein coding) | SAMSN1 | SAM domain, SH3 domain and nuclear localization signals 1 |
| MMP1 | Matrix metallopeptidase 1 | SCRG1 | Stimulator of chondrogenesis 1 |
| MMP10 | Matrix metallopeptidase 10 | SLPI | Secretory leukocyte peptidase inhibitor |
| MT1F | Metallothionein 1F | SNHG4 | Small nucleolar RNA host gene 4 |
| PRL | Prolactin | SPINK13 | Serine peptidase inhibitor, Kazal type 13 (putative) |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | STEAP4 | STEAP family member 4 |
| PTHLH | Parathyroid hormone-like hormone | STMN2 | Stathmin 2 |
| SERPINA9 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 | TM4SF20 | Transmembrane 4 L six family member 20 |
| SERPINB2 | Serpin peptidase inhibitor, clade B (ovalbumin), member 2 | TSPAN18 | Tetraspanin 18 |
| STATH | Statherin | ULBP1 | UL16 binding protein 1 |
| TAC1 | Tachykinin, precursor 1 | ZBTB16 | Zinc finger and BTB domain containing 16 |
| THBD | Thrombomodulin | ||
| TRPC6 | Transient receptor potential cation channel, subfamily C, member 6 | ||
BMP2 = bone morphogenetic protein 2. Genes shown in alphabetical order.
Master Transcription Factors for Cartilage and Bone Expression
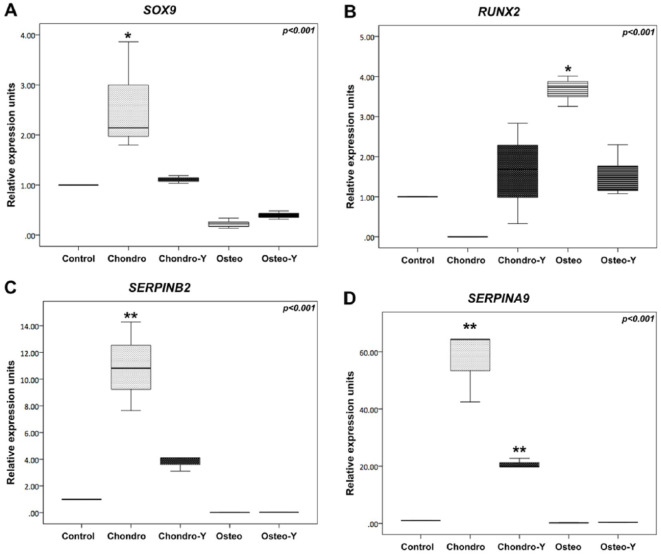
SOX9 and RUNX2 did not show statistically significant differential expression in microarray analysis; nevertheless, these results were further validated by qPCR since they are considered the master transcription factors for each type of tissue. Results of qPCR analysis are shown in Table 2 and Figure 3 . SOX9 expression was significantly upregulated in the chondrogenic group without ROCK inhibition and downregulated in both osteogenic differentiation groups (P < 0.001; Fig. 3A ). Relative expression was maintained at basal level in the ROCK inhibited chondrogenic group. RUNX2 expression was upregulated in the osteogenic group without ROCK inhibition when compared with both chondrogenic groups (P < 0.001; Fig. 3B ).
Table 2.
qPCR Analysis of the Different Culture Conditions. Gene Expression Analyses of SOX9, RUNX2, SERPINB2, and SERPINA9 in hMSCs, and After Cell Differentiation Into Chondrocytes and Osteoblasts, with and without ROCK Inhibition.
| Gene | Relative Gene Expression Units | P Value | |||
|---|---|---|---|---|---|
| Chondro | Chondro-Y | Osteo | Osteo-Y | ||
| SOX9 | 2.14 (2.06) | 1.11 (0.15) | 0.22 (0.20) | 0.39 (0.16) | <0.001 |
| RUNX2 | 0 (0.0) | 1.69 (2.50) | 3.74 (0.75) | 1.23 (1.23) | <0.001 |
| SERPINB2 | 10.82 (6.62) | 4.10 (0.99) | 0.02 (0.01) | 0.03 (0.01) | <0.001 |
| SERPINA9 | 64.32 (21.88) | 19.79 (2.94) | 0.23 (0.07) | 0.38 (0.08) | <0.001 |
Data shown as median and interquartile range, normalized by the control group (hMSCs). hMSCs = human bone marrow mesenchymal stem cells; Chondro = chondrocytes; Chondro-Y = chondrocytes with ROCK inhibitor Y27632; Osteo = osteoblasts; Osteo-Y = osteoblasts with ROCK inhibitor Y27632; ROCK = Rho-associated coiled-coil containing protein kinase.
Figure 3.
Real-time PCR analysis of the different culture conditions. Gene expression analyses of (A) SOX9, (B) RUNX2, (C) SERPINB2, and (D) SERPINA9 in hMSCs, and after cell differentiation into chondrocytes, osteoblasts with and without ROCK inhibition. Box-plots normalized by the control group (hMSCs). Chondro = chondrocytes; Chondro-Y = chondrocytes with ROCK inhibitor Y27632; hMSCs = human bone marrow mesenchymal stem cells; Osteo = osteoblasts; Osteo-Y = osteoblasts with ROCK inhibitor Y27632; ROCK = Rho-associated coiled-coil containing protein kinase. *P < 0.05 as compared to Control; **P < 0.05 as compared to all other study groups.
Identification of SERPINB2 and SERPINA9 as Cartilage Differentiation Genes
Among genes highly regulated in the cartilage differentiation group, and without change in the osteogenic differentiation group, were 2 members of the Serpin peptidase inhibitor superfamily: SERPINB2 and SERPINA9. SERPINB2 was 10-fold overexpressed between control and cartilage groups (P = 0.003) and highly significant when comparing both bone groups against cartilage without ROCK inhibition (P < 0.001; Fig. 3C ). SERPINA9 was more than 60-fold overexpressed between control and cartilage groups (P = 0.003) and also highly significant when comparing both groups without ROCK inhibition (P < 0.001; Fig. 3D ).
Characterization of SERPINB2 and SERPINA9 in TGF-β1 Cultures
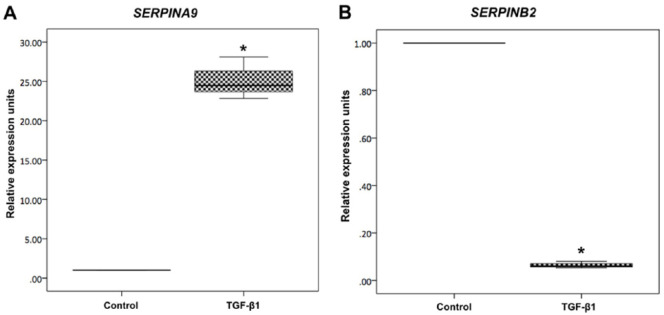
Both Serpins were evaluated in TGF-β1 cultures in order to confirm previous results. SERPINA9 was more than 20-fold upregulated in the TGF-β1 group (P < 0.001; Fig. 4A ); however, SERPINB2 was found downregulated in the TGF-β1 group (P < 0.001; Fig. 4B ), as compared to controls (data shown are representative from 3 independent experiments, n = 3).
Figure 4.
Real-time PCR analysis of culture with TGF-β1. Gene expression analyses of (A) SERPINA9 and (B) SERPINB2 in hMSCs and after cell culture with transforming growth factor beta 1 (TGF-β1). Box-plots normalized by the control group (hMSCs). *P < 0.001 as compared to Control.
Discussion
Serpins (serine proteinase inhibitors) are a superfamily of proteins (350-500 amino acids in size) that fold into a conserved structure and employ a substrate-like inhibitory mechanism.20 To our knowledge, this is the first study to demonstrate SERPINB2 and SERPINA9 are highly overexpressed in hMSCs when stimulated with KGN, and to describe a role of this protein family in chondrogenesis.
In recent years, it has been acknowledged that not only growth factors or chemical molecules drive differentiation of a certain progenitor to a desired lineage; external physical cues can also influence differentiation to the point of restricting, directing or favoring certain lineages.7 Members of the Rho family of small GTPases, specially the downstream effectors ROCKs, have been implicated in multiple functions such as differentiation, maturation, and remodeling of the extracellular matrix.11 ROCK inhibition has been previously reported, via the small chemical inhibitor Y-27632, as promoting differentiation and maturation of osteoblasts in rat primary calvarial cell culture.21 In contrast, ROCK inhibition has been reported to interfere with osteogenesis in hMSCs with BMP2 via downstream effectors SMADs.11 In the present work, we have shown how ROCK inhibition affected gene expression of cells differentiated to bone and cartilage in high-density monolayer cultures from hMSCs. High-density monolayer cultures are considered an appropriate model to explore cartilage differentiation, when at the end of 21 days of culture the so-called monolayer is composed of several layers of cells and in reality is a 3-dimensional model.22 It is also considered an ideal experimental model that prevents the formation of a local hypoxic microenvironment, since pellet cultures may present variations between conditions at the periphery and center of the cultures.23
Microarray results gave no significant upregulation of either SOX9 or RUNX2, which are the master regulators for cartilage and bone, respectively. When validation by qPCR was performed, we identified that Serpins, in particular SERPINB2 and SERPINA9, are highly upregulated when hMSCs are differentiated to cartilage using KGN, suggesting their expression is downstream of the RUNX1 transcription factor.8 In contrast, gene expression was not affected in cells differentiated toward osteoblasts, regardless of ROCK inhibition. These findings could indicate that bone cells might not require these 2 protease inhibitors while differentiating, or that they serve as angiogenic inhibitors, since articular cartilage is avascular.24 In contrast, SERPINF1 was previously reported as a positive regulator for osteoblasts, and mutations of the gene have been implicated in osteogenesis imperfecta and otosclerosis.25
The SERPINA9 gene has been previously described as implicated in B cell development and maturation,26 confined to germinal centers in secondary lymphoid organs.27 SERPINB2 has been characterized as implicated in intracellular stress response.28 Only recently have members of this superfamily been studied in lineage specification of 2 tissues typically arising from mesenchymal stem cells. SERPINB2 was found as a novel negative regulator of osteogenesis and adipogenesis; the authors stimulated hMSCs via TGF-β1 and found a 3-fold downregulation of the SERPINB2 gene. On siRNA SERPINB2 gene inhibition, osteogenic and adipogenic differentiation was stimulated, suggesting TGF-β1 suppresses SERPINB2, promoting osteogenesis and adipogenesis.29 Our findings also confirm downregulation of SERPINB2 on TGF-β1 cultures; however, our study demonstrates upregulation of SERPINA9 in hMSCs when stimulated with either KGN and TGF-β1, which had not previously been described. Furthermore, KGN showed upregulation of SERPINB2, which suggests these Serpins may be differentiation markers of lineage commitment toward cartilage, which has been recently published as the default pathway for these cells.30
Another member of the Serpin superfamily, SERPINH1 or heat shock protein 47, had previously been described as an essential molecular chaperone for cartilage and endochondral bone formation, at collagen last stage assembly.31 Its role was characterized downstream of genes controlling chondrocyte and osteoblast commitment and differentiation. SERPINE2 has been recently reported to be upregulated in cartilage on IL-1α stimulation, inhibiting metalloproteinase activity at the cartilage level to decrease collagen breakdown.32 SERPINA1 and SERPINA3 were also previously explored as markers of chondrogenic differentiation and dedifferentiation; however, bone marrow aspirates were not characterized with ISCT criteria and lacked information for SOX9 expression.33 Finally, SERPINI2 has also been described in MSCs but not in articular cartilage, in a previous study with bone marrow aspirates that were not also not fully characterized or evaluated during differentiation.34 Taking into account what has been previously reported, Serpin superfamily members have antagonist roles for cell differentiation from mesoderm precursors toward cartilage, bone, and adipose tissue.
Our study has various strengths. Our study model was a cell line previously validated for differentiation to adipocytes, chondrocytes, and osteocytes, with complete criteria for characterizing MCSs, which we found is a limitation for most studies previously reported. We decided on the use of this model based on the study publishing the discovery of KGN, which also used a commercial cell line.9 Initially, our findings performed by microarray analysis with regard to Serpins were unexpected; however, we decided to further explore and validate by qPCR with novel constitutive genes specific for hMSC and validated commercial primers (QIAGEN). Upregulation of these genes was very significant, with very stringent methodology. Results were very marked, particularly with SERPINA9 upregulation showing a 60-fold increase with KGN, and a 20-fold increase with TGF-β1, which identifies it as a novel differentiation marker for lineage commitment and potential regulator. We acknowledge that Serpins may provide a new tool for regenerative medicine. However, regenerative efforts must take into account that a single target (i.e., gene, protein) is not sufficient to obtain cell differentiation, and that the extracellular environment is as important.2 Especially when cells are cultured in vitro ex vivo, this hurdle might be tackled using chemical compounds that mimic physical forces, by altering signaling pathways such as the Rho GTPases that translate external stimuli.11 We also acknowledge that functional studies were not performed, that data from this study will require external validation, and that future research is required to check for basal expression of Serpins in native articular cartilage, as well as hypertrophic chondrocytes and subchondral bone.
Conclusions
In conclusion, we describe SERPINA9 and SERPINB2 of the Serpin superfamily of proteins as novel differentiation markers, and molecular regulator candidates for hMSC lineage commitment toward bone and cartilage, providing a new tool for regenerative medicine. Our study highlights the roles of these 2 genes, with significant upregulation of both in cell cultures stimulated with Kartogenin.
Supplemental Material
Supplemental material, Suppl_SERPIN for SERPINA9 and SERPINB2: Novel Cartilage Lineage Differentiation Markers of Human Mesenchymal Stem Cells with Kartogenin by Julio Granados-Montiel, Monica Cruz-Lemini, Claudia Rangel-Escareño, Gabriela Martinez-Nava, Carlos Landa-Solis, Ricardo Gomez-Garcia, Alberto Lopez-Reyes, Alejandro Espinosa-Gutierrez and Clemente Ibarra in CARTILAGE
Footnotes
Acknowledgments and Funding: The authors would like to thank our technician Xochitl Guerrero Alva, Juanjo Lozano, PhD, of the Bioinformatics Platform, CIBEREHD, Hospital Clínic Barcelona, and Dan Jerzain Gutierrez Fuentes, of the Microarray Facility, Instituto Nacional de Medicina Genomica, for their assistance. M. Cruz-Lemini was supported by the National System of Researchers (SNI), of CONACyT. This work was funded by grants received from the Mexican National Council for Science and Technology (CONACyT), PDCPN-2013-216779 and PDCPN-2013-215138, and the Coordinating Commission of National Institutes of Health and High Specialty Hospitals in Mexico (C.C.I.N.S.H.A.E.) special grant internal number 41/14.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iDs: Julio Granados-Montiel  https://orcid.org/0000-0002-0611-6421
https://orcid.org/0000-0002-0611-6421
Monica Cruz-Lemini  https://orcid.org/0000-0002-6807-3578
https://orcid.org/0000-0002-6807-3578
Alberto Lopez-Reyes  https://orcid.org/0000-0002-2575-2589
https://orcid.org/0000-0002-2575-2589
Supplemental material: Supplemental material for this article is available online.
References
- 1. Huang X, Zhong L, Hendriks J, Post JN, Karperien M. The effects of the WNT-signaling modulators BIO and PKF118-310 on the chondrogenic differentiation of human mesenchymal stem cells. Int J Mol Sci. 2018;19:E561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brittberg M. Cellular and acellular approaches for cartilage repair: a philosophical analysis. Cartilage. 2015;6:4S-12S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghaneialvar H, Soltani L, Rahmani HR, Lotfi AS, Soleimani M. Characterization and classification of mesenchymal stem cells in several species using surface markers for cell therapy purposes. Indian J Clin Biochem. 2018;33:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berebichez-Fridman R, Gomez-Garcia R, Granados-Montiel J, Berebichez-Fastlicht E, Olivos-Meza A, Granados J, et al. The holy grail of orthopedic surgery: mesenchymal stem cells-their current uses and potential applications. Stem Cells Int. 2017;2017:2638305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mithoefer K, McAdams T, Williams RJ, Kreuz PC, Mandelbaum BR. Clinical efficacy of the microfracture technique for articular cartilage repair in the knee: an evidence-based systematic analysis. Am J Sports Med. 2009;37:2053-63. [DOI] [PubMed] [Google Scholar]
- 6. Akiyama H. Control of chondrogenesis by the transcription factor Sox9. Mod Rheumatol. 2008;18:213-9. [DOI] [PubMed] [Google Scholar]
- 7. Wang YK, Yu X, Cohen DM, Wozniak MA, Yang MT, Gao L, et al. Bone morphogenetic protein-2-induced signaling and osteogenesis is regulated by cell shape, RhoA/ROCK, and cytoskeletal tension. Stem Cells Dev. 2012;21:1176-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marini JC, Forlino A. Replenishing cartilage from endogenous stem cells. N Engl J Med. 2012;366:2522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, et al. A stem cell-based approach to cartilage repair. Science. 2012;336:717-21. [DOI] [PubMed] [Google Scholar]
- 10. Occhetta P, Pigeot S, Rasponi M, Dasen B, Mehrkens A, Ullrich T, et al. Developmentally inspired programming of adult human mesenchymal stromal cells toward stable chondrogenesis. Proc Natl Acad Sci U S A. 2018;115:4625-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strzelecka-Kiliszek A, Mebarek S, Roszkowska M, Buchet R, Magne D, Pikula S. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochim Biophys Acta Gen Subj. 2017;1861:1009-23. [DOI] [PubMed] [Google Scholar]
- 12. Mennan C, Garcia J, McCarthy H, Owen S, Perry J, Wright K, et al. Human articular chondrocytes retain their phenotype in sustained hypoxia while normoxia promotes their immunomodulatory potential. Cartilage. Epub 2018 Apr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18:581-92. [DOI] [PubMed] [Google Scholar]
- 14. Li X, Yang Q, Bai J, Xuan Y, Wang Y. Identification of appropriate reference genes for human mesenchymal stem cell analysis by quantitative real-time PCR. Biotechnol Lett. 2015;37:67-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy MK, Huey DJ, Hu JC, Athanasiou KA. TGF-β1, GDF-5, and BMP-2 stimulation induces chondrogenesis in expanded human articular chondrocytes and marrow-derived stromal cells. Stem Cells. 2015;33:762-73. [DOI] [PubMed] [Google Scholar]
- 16. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185-93. [DOI] [PubMed] [Google Scholar]
- 18. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. [DOI] [PubMed] [Google Scholar]
- 20. Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293-6. [DOI] [PubMed] [Google Scholar]
- 21. Harmey D, Stenbeck G, Nobes CD, Lax AJ, Grigoriadis AE. Regulation of osteoblast differentiation by Pasteurella multocida toxin (PMT): a role for Rho GTPase in bone formation. J Bone Miner Res. 2004;19:661-70. [DOI] [PubMed] [Google Scholar]
- 22. Craft AM, Rockel JS, Nartiss Y, Kandel RA, Alman BA, Keller GM. Generation of articular chondrocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:638-45. [DOI] [PubMed] [Google Scholar]
- 23. Taheem DK, Foyt DA, Loaiza S, Ferreira SA, Ilic D, Auner HW, et al. Differential regulation of human bone marrow mesenchymal stromal cell chondrogenesis by hypoxia inducible factor-1α hydroxylase inhibitors. Stem Cells. 2018;36:1380-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu L, Yan M, Simkin J, Ketcham PD, Leininger E, Han M, et al. Angiogenesis is inhibitory for mammalian digit regeneration. Regeneration (Oxf). 2014;1:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ziff JL, Crompton M, Powell HR, Lavy JA, Aldren CP, Steel KP, et al. Mutations and altered expression of SERPINF1 in patients with familial otosclerosis. Hum Mol Genet. 2016;25:2393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frazer JK, Jackson DG, Gaillard JP, Lutter M, Liu YJ, Banchereau J, et al. Identification of centerin: a novel human germinal center B cell-restricted serpin. Eur J Immunol. 2000;30:3039-48. [DOI] [PubMed] [Google Scholar]
- 27. Olsson M, Tengvall K, Frankowiack M, Kierczak M, Bergvall K, Axelsson E, et al. Genome-wide analyses suggest mechanisms involving early B-cell development in canine IgA deficiency. PLoS One. 2015;10:e0133844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee JA, Yerbury JJ, Farrawell N, Shearer RF, Constantinescu P, Hatters DM, et al. SerpinB2 (PAI-2) modulates proteostasis via binding misfolded proteins and promotion of cytoprotective inclusion formation. PLoS One. 2015;10:e0130136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elsafadi M, Manikandan M, Atteya M, Abu Dawud R, Almalki S, Ali Kaimkhani Z, et al. SERPINB2 is a novel TGFβ-responsive lineage fate determinant of human bone marrow stromal cells. Sci Rep. 2017;7:10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jing Y, Jing J, Ye L, Liu X, Harris SE, Hinton RJ, et al. Chondrogenesis and osteogenesis are one continuous developmental and lineage defined biological process. Sci Rep. 2017;7:10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Masago Y, Hosoya A, Kawasaki K, Kawano S, Nasu A, Toguchida J, et al. The molecular chaperone Hsp47 is essential for cartilage and endochondral bone formation. J Cell Sci. 2012;125:1118-28. [DOI] [PubMed] [Google Scholar]
- 32. Santoro A, Conde J, Scotece M, Abella V, Lois A, Lopez V, et al. SERPINE2 inhibits IL-1α-induced MMP-13 expression in human chondrocytes: involvement of ERK/NF-κB/AP-1 pathways. PLoS One. 2015;10:e0135979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boeuf S, Steck E, Pelttari K, Hennig T, Buneb A, Benz K, et al. Subtractive gene expression profiling of articular cartilage and mesenchymal stem cells: serpins as cartilage-relevant differentiation markers. Osteoarthritis Cartilage. 2008;16:48-60. [DOI] [PubMed] [Google Scholar]
- 34. Polacek M, Bruun JA, Elvenes J, Figenschau Y, Martinez I. The secretory profiles of cultured human articular chondrocytes and mesenchymal stem cells: implications for autologous cell transplantation strategies. Cell Transplant. 2011;20:1381-93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Suppl_SERPIN for SERPINA9 and SERPINB2: Novel Cartilage Lineage Differentiation Markers of Human Mesenchymal Stem Cells with Kartogenin by Julio Granados-Montiel, Monica Cruz-Lemini, Claudia Rangel-Escareño, Gabriela Martinez-Nava, Carlos Landa-Solis, Ricardo Gomez-Garcia, Alberto Lopez-Reyes, Alejandro Espinosa-Gutierrez and Clemente Ibarra in CARTILAGE