Abstract
In December 2019, an outbreak of a novel coronavirus (SARS-CoV-2) in Wuhan, China resulted in the current COVID-19 global pandemic. The human immune system has not previously encountered this virus, raising the important question as to whether or not protective immunity is generated by infection. Growing evidence suggests that protective immunity can indeed be acquired post-infection—although a handful of reinfection cases have been reported. However, it is still unknown whether the immune response to SARS-CoV-2 leads to some degree of long-lasting protection against the disease or the infection. This review draws insights from previous knowledge regarding the nature and longevity of immunity to the related virus, SARS-CoV, to fill the gaps in our understanding of the immune response to SARS-CoV-2. Deciphering the immunological characteristics that give rise to protective immunity against SARS-CoV-2 is critical to guiding vaccine development and also predicting the course of the pandemic. Here we discuss the recent evidence that characterises the adaptive immune response against SARS-CoV-2 and its potential implications for the generation of memory responses and long-term protection.
Keywords: SARS-CoV-2, COVID-19, protective, immunity, vaccine, immunogenicity
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a novel coronavirus that appeared in Wuhan at the end of 2019 and rapidly escalated into the global pandemic of COVID-19, the disease that results from infection. Similar to previous recently-emerged coronaviruses, SARS and Middle East Respiratory Syndrome (MERS), SARS-CoV-2 is likely to have originated from a zoonotic transmission from bats (1). A recently discovered bat-derived CoV, RmYN02, was found to share 93.3% whole genome identity with SARS-CoV-2 and 97.2% identity in 1ab—the longest coding gene (1). Typically, bats are the natural reservoir for coronaviruses, although transmission to humans often occurs via an intermediate host, which is still under debate for SARS-CoV-2. The pangolin, an animal used in traditional Chinese medicine, has been proposed as intermediate host due to a strong similarity in the receptor binding domain (RBD) between SARS-CoV-2 and pangolin coronavirus (2).
Genome sequencing studies of SARS-CoV-2 showed high levels of whole genome conservation (>99%) across 739 sequences reported on GISAID (3), which suggests that major mutations may be detrimental for viral fitness. A comprehensive analysis of the mutations found in SARS-CoV-2 has been published by Li et al. (4). A single mutation, D614G, affecting the viral spike protein, emerged in Europe and became the dominant circulating virus: this variant has been reported to increase viral infectivity but not affect disease severity (5). SARS-CoV-2 also shares 79%–82% of its genome with SARS-CoV, which was responsible for the 2003 SARS outbreak and is the most closely related coronavirus known to infect humans (6).
SARS-CoV-2 is an enveloped virus with a 30 kb single positive stranded RNA genome (7). It contains 12 canonical open reading frames (ORFs) that are translated either from genomic or subgenomic RNAs by the host cell upon entry (8). Interestingly, recent high-resolution map of coding regions has identified 23 other ORFs. ORFs 2, 4, 5 and 9a encode structural proteins (8). These proteins are the spike (S), the envelope (E), the membrane (M), and the nucleocapsid (N). The rest of the genome encodes non-structural proteins (NSP), such as the RNA dependent RNA polymerase, protease, and helicase, as well as other ORFs that act as accessory proteins, the functions of which are less well understood but assist in the completion of the viral cycle. For example, the NSP1 protein enables immune evasion by supressing host gene expression (9) and ORF7a counteracts host restriction factor Bone Marrow Stromal Antigen 2 (BST2) (10), similar to what has been described for SARS-CoV (11).
Both SARS-CoV and SARS-CoV-2 target the same receptor to infect target cells, ACE2 (Angiotensin-converting enzyme 2), through the highly conserved RBD in the S protein (12, 13). The S protein is composed of two functionally distinct domains; subunit S1, containing the RBD, engages with the ACE2 host cell receptor and the S2 subunit mediates fusion between the viral and the host cell membrane (14, 15). For fusion to occur following ACE2 binding the S protein is cleaved by the TMPRSS2 protease between the S1 and S2 subunits, which triggers fusion into the cell (16). This furin-like cleavage site (FCS) is uniquely present in the S protein of SARS-CoV-2, which might contribute to the significantly greater infectivity of SARS-CoV-2 compared to other known beta-coronaviruses (17). Alternatively, the viral particle can be endocytosed and enter the endosome/lysosomal pathway, where cathepsin L has been found to activate S protein and trigger fusion (15). Other host factors have been suggested to facilitate SARS-CoV-2 cell entry (18–24). Interestingly, Neuropilin-1 (NRP1), highly present in human respiratory and olfactory epithelium, has been shown to potentiate SARS-CoV-2 infectivity in the presence of ACE2 and TMPRSS2 by interacting with the furin-cleaved spike (25).
SARS-CoV-2 has been shown to bind ACE2 with 10-20 fold higher affinity than SARS-CoV, which may explain its greater transmissibility (14). The aforementioned D614G mutation in Spike appears to increase the proportion of Spike trimer components in the “open” conformation, which facilitates ACE2 binding and potentially confers greater infectivity (26). The S viral protein is the main focus for vaccine design as it is known to be highly immunogenic, particularly the RBD region. Moreover, the S protein is indispensable for viral entry and therefore its targeting may reduce or prevent infection. For SARS-CoV, vaccination with the S protein alone was shown to induce an immune response that was likely to be protective (27, 28): for this reason most current vaccine strategies against SARS-CoV-2 are based on the immunogenicity of S. Other structural viral proteins, like the nucleocapsid, have also generated interest, as they were studied in SARS-CoV infection and showed some immunogenicity (29).
The majority of individuals infected with SARS-CoV-2 experience mild-to-moderate disease. Although exact numbers are still debated, a systematic review analysis of 79 published studies found asymptomatic cases to be 20% of PCR-confirmed cases (30). Severe cases, defined as those requiring hospitalisation, are estimated to be around 14% of all confirmed cases. COVID-19 symptoms are diverse, but common manifestations include fever, dry cough, fatigue, loss of taste and/or smell, diarrhoea, and breathlessness (31). Age, sex, and underlying comorbidities, such as diabetes and hypertension, have been associated with disease severity (32). Patients over 65 years old have an odds ratio of 3.4 for requiring hospitalisation compared to 18–44 year olds (33). Critical COVID-19 patients often undergo a sudden clinical deterioration 7–10 days after symptom onset and present with features of acute respiratory distress syndrome (ARDS), along with lymphopaenia and an elevation of inflammatory markers (34, 35). It is important to note that some patients continue to suffer from symptoms for months after the infection has resolved.
The reasons behind the different outcomes are unclear. The main hypothesis is that a dysregulated immune response, probably at the early stages of infection, leads to systemic hyperinflammation (cytokine storm) that may be driving the ARDS and multi-organ damage observed in severe disease (34, 35). However, the reasons why there is loss of control of what is usually a tightly-regulated inflammatory response are still being investigated. Inborn defects in the Type I Interferon (IFN) pathway and anti-IFN auto antibodies have been found in life-threatening COVID-19 (36, 37).
Differences in the viral load of the initial inoculum, along with differences in the genetic sequence of the founder virus, have been proposed as potential factors that may influence overall patient outcome, given that a higher viral load at infection may trigger a stronger immune response. So far, the D614G mutant, associated with higher viral loads and greater viral dissemination, has not been found to correlate with disease severity (5).
A key question yet to be addressed is whether SARS-CoV-2 infection induces long-lasting protective immunity, and if so whether it will simply protect from severe disease or provide sterilising immunity. Our understanding of the immune correlates of protection for SARS-CoV-2 and their durability is limited to very recent data and depends mainly on knowledge gained from SARS-CoV, the most closely related virus known to affect humans. Long-lasting memory T cells to the genetically similar SARS-CoV have been revealed to be still detectable in convalescent patients 17 years after the SARS epidemic (38).
To date, few large-scale studies have characterised the immune response in recovered SARS-CoV-2 patients and it is too soon to evaluate the longevity of the response and its characteristics. The first challenge studies in macaques showed that SARS-CoV-2 infection may result in the development of protective immunity, when the animals were challenged soon after resolution of the primary infection (39, 40). However, a number of cases of reinfection in humans have now been reported a few months after initial infection, challenging the idea of long-lasting protective immunity (41–47).
In this review, we explore the current evidence for long-lasting immunity in SARS-CoV and the immunological parallels with SARS-CoV-2. We also examine the current literature characterising the immune response to SARS-CoV-2 and discuss its potential implications for current and future vaccine strategies. We aim to propose a perspective on whether protective immunity is likely to develop in individuals who have recovered from COVID-19 infection and for how long we might expect it to last. Some of the papers we have included were available on pre-print servers at the time of publication and therefore have not yet been peer-reviewed.
Humoral Immune Response
Antibodies Induced in SARS-CoV and SARS-CoV-2 Infection
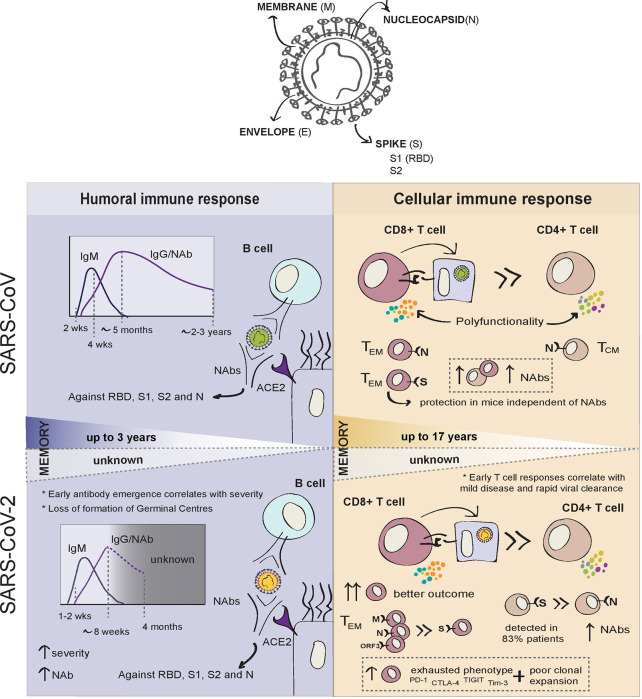
In the previous SARS epidemic, seroconversion was documented at 2 weeks post-onset of symptoms in the majority of patients with SARS-CoV infection ( Figure 1 ). IgM antibodies appeared during acute infection, then, with progressive class-switching to IgG, IgM, and IgG titres increased in parallel during the first weeks after infection. Coinciding with the resolution of infection, IgM titres were shown to wane gradually 4 weeks after symptom onset. IgG levels peaked 4 months later, and both neutralising and non-neutralising antibody titres were shown to remain detectable for at least 2 years. Non-neutralising IgG antibodies became undetectable after 24 months in a small proportion of patients (48).
Figure 1.
Summary of adaptive immune responses to SARS-CoV and SARS-CoV-2.
Recovered SARS patients were found to have high titres of neutralising antibodies (NAbs) directed against the receptor binding domain (RBD) of the S protein (defined as residues 318–510). These specific antibodies maintained high titres up to 5 months post-infection and remained measurable in sera at 24 months follow-up, although notably decreased after 16 months (48).
The immune correlates of the humoral response in SARS-CoV are likely to provide a good basis on which to predict antibody responses for SARS-CoV-2, especially given that the S protein is highly conserved between the two viruses (76% amino acid sequence homology) and antibodies targeting the RBD region appear to be crucial for neutralisation (15). S-based neutralising antibodies generated in humans against SARS-CoV-2 are likely to be similar in structure and function, and to follow homologous kinetics to those produced in response to SARS-CoV.
To date, similarly to SARS-CoV, people infected with SARS-CoV-2 are reported usually to develop IgA and IgM antibodies 7–14 days after symptoms develop and, together with later IgGs, antibodies are present before the resolution of infection ( Figure 1 ) (49). However, COVID-19 seroconversion can occur as early as 2 days after symptom onset (50). In one of the largest studies conducted 44%–56% of patients had developed IgG on day 7 after the onset of symptoms and IgG antibodies were present in 95% of patients by day 20 (51). More recent studies have observed a small decrease in IgG titres 8 weeks after symptom onset (49, 52), despite maintenance of high titres of antibodies against the Spike protein (53, 54). The longevity of the antibody response may depend on their targets. N and S specific IgG antibody titres were found to remain stable for at least 4 months post-diagnosis, while anti-N IgM increased markedly early in infection, before declining to undetectable levels at 2 months (55). For now, only one report has revealed the dynamics of the antibody responses 6 months after convalescence and found anti-S1 IgG and IgA to decrease over time, while anti-N antibodies remained stable. Interestingly, higher anti-N antibody titres correlated with the prevalence of post-infection symptoms (56).
Neutralising Antibodies and Cross-Neutralisation
Neutralising antibodies are key to provide sterilising immunity against a second infection, in particular those targeting the S-ACE2 binding to prevent infection. Many in vitro and in vivo studies have shown that NAbs appear to control SARS-CoV-2 infection (57–59).
In SARS-CoV-2, NAbs have been reported from day 10–15 after symptom onset and correlate with S-targeting antibodies (54) with 90% of seroconverters having detectable NAbs up to 5 months after infection (55). There is a clear association between antibody titres, and NAbs levels, with disease severity (60). NAb titres show an inverse correlation with the patient’s lymphocyte counts, where low lymphocyte counts constitute a strong indicator of poor prognosis (61). Robust antibody responses have been found in mild-to-moderate COVID-19 (55), but others could not detect NAbs in 30% of patients (54), suggesting that, for some patients with mild or asymptomatic infection, NAb titres are either very low or non-existent. The titres of NAbs therefore appear to correlate directly with disease severity, but paradoxically, may provide higher protection against infection on subsequent exposure to SARS-CoV-2.
Most antibody responses in COVID-19 patients appear to target the S viral protein, in particular the S1 subunit and RBD region, which are thought to elicit the most potent neutralising effect (62). Isolated potent NAbs against the RBD were shown to reduce viral burden and to protect against infection in mice and macaques (63, 64).
Given that the RBD sequence is highly conserved between the two viruses, we would expect SARS-CoV-2 NAbs directed against RBD to persist for a similar length of time to that observed for SARS-CoV. Structures of S-specific antibodies bound to the SARS-CoV-2 spike reveal common epitopes and conserved features (65). Murine SARS-CoV S antibodies can inhibit SARS-CoV-2 cell entry, consistent with the similarities of the S protein between the two viruses (66). Serum from SARS-CoV recovered individuals during the 2003 outbreak has been shown to cross-react with the S ectodomain, S, RBD and S2 proteins of SARS-CoV-2 and neutralise SARS-CoV-2 in vitro (67, 68). More recently, RBD-targeting antibodies have been isolated in convalescent SARS-CoV plasma and were shown to cross-neutralise SARS-CoV-2 by targeting multiple conserved epitopes on the S protein (69). However, other reports in humans suggest that convalescent sera from SARS-CoV and SARS-CoV-2 recovered patients show limited cross-neutralisation between the two viruses. These differences were ascribed to seven critical residues that differ in SARS-CoV and SARS-CoV-2 S RBDs, despite 76% amino acid identity (15).
Despite the general capacity of SARS-CoV and SARS-CoV-2 antibodies to cross-react, their ability for cross-neutralisation may be very specific. Such specificity was demonstrated in plasma from subjects with either SARS-CoV and SARS-CoV-2 that contained antibodies able to cross-react in their binding of spike protein but not to cross-neutralise (70). The isolation of SARS-CoV-2 RBD NAbs showed that their neutralising capacity is directly correlated with the ability to compete with ACE2 for RBD binding, but they were unable to neutralise SARS or MERS RBD, despite showing cross-reactivity against their S proteins trimers (71). Moreover, antibodies against seasonal coronaviruses (HCoV) from pre-pandemic sera, showed no cross-neutralising activity for SARS-CoV-2 (72, 73).
The S2 domain of SARS-CoV-2 also appears to be highly conserved (88% sequence homology with SARS-CoV). Similarly to S1, antibodies generated against S2 in SARS-CoV infection were found to be cross-reactive to S2 in SARS-CoV-2 but none were neutralising (69).
COVID-19 patients produce a spectrum of antibodies to both structural and non-structural proteins of SARS-CoV-2. These antibodies, targeting NSP1, ORF3b, ORF7a, and ORF8, may mediate immune functions other than neutralisation that may be beneficial or harmful to the patient (74). Whether antibodies to non-structural proteins add to protection is not yet known.
Humoral Protection
In humans, plasma memory B cells and monoclonal NAbs have been isolated from SARS-CoV-2 convalescent individuals (63, 71). Memory B cells may persist in plasma in the absence of detectable antibody titres and can then be recalled in the event of subsequent infection and confer protection (69). Based on the findings from SARS-CoV studies, memory B cells and antibody-mediated protection may last for up to 3 years, although the efficacy of such protection will likely fall with time as circulating antibody titres decay (69). Many trials have highlighted the potential protective effect of antibodies generated naturally during the course of SARS-CoV-2 infection by using convalescent plasma (CP) from recovered COVID-19 patients to treat critical cases (75–77). However, a recent multicentre, randomised and controlled trial found no difference in mortality or disease progression for those treated with CP (78). Although the efficacy of convalescent plasma is still under debate, this could suggest that the contribution of the humoral immune response may be limited for patients who are already severely ill.
Evidence that SARS-CoV-2 infection can induce protective immunity was first shown by Chandrashekar et al. in rhesus macaques studied soon after initial infection (39). Nine macaques infected with SARS-CoV-2 were re-challenged 35 days after primary infection had been cleared and showed significantly lower viral RNA measured in bronchoalveolar lavage (BAL) compared to challenge-naïve animals. This was accompanied by the effective induction of neutralising antibodies and cell-mediated virus-specific responses (39). The demonstration of protective immunity developing after SARS-CoV-2 infection in macaques gives reason to postulate that protective immunity can also be generated in humans following recovery or vaccination. However, the longevity of such protection is yet to be evaluated.
To date, among the few reinfection cases reported, one study found the humoral immune response to be defective during the second infection, suggesting that a failure of the humoral response during the first infection may account for susceptibility to reinfection (42). In addition, Chen et al. demonstrate a positive correlation between the magnitude of NAb titres and disease severity but also report immense heterogeneity between the NAbs generated—with 80.7% patients producing S1 specific NAbs and only 40% patients producing NAbs to both S1 and S2. Notably, asymptomatic patients failed to generate adequate titres of NAbs, and sera from these patients were unable to neutralise SARS-CoV-2 pseudovirions in vitro (79).
A matter of concern is that, in fatal COVID-19 cases, SARS-CoV-2 appears to impair the generation of germinal centres (GC), where, with the help of T Follicular Helper (TFH) cells, B cells can differentiate into memory B cells or long-lived plasma cells (80). Interestingly, extrafollicular B cell activation has been detected in severely ill patients and correlated with early antibody responses, multi-organ failure and death (81). Lack of GC formation during acute SARS-CoV-2 infection would seriously undermine the generation of long-lived antibody responses. It has been proposed that GC are blocked from forming due to excessive Tumour Necrosis Factor alpha (TNF-α) production in severe disease. Therefore, it is possible that the formation of a memory compartment is compromised for humoral responses because of an excessive inflammatory response.
It is important to understand the correlates of humoral protection and to consider the immunological implications for those individuals with undetectable NAb titres. T cell responses may potentially provide protection against subsequent infection in the absence of NAbs. Early studies appear to show that virus-specific T cell responses are robust and develop in asymptomatic or mild SARS-CoV-2 infections, even in the absence of an antibody response (82).
Evidence so far shows that humoral responses to both SARS and COVID-19 share a number of similarities. However, it will be crucial to characterise the SARS-CoV-2 humoral immune response in relation to disease severity and determine whether the kinetics and the breadth of the antibody response play a role in the resolution of primary infection and protection against reinfection. In COVID-19, the antibody response appears to be stronger in those with more severe symptoms, with higher NAbs titres correlating with disease severity (37), while NAb titres appear to be low or undetectable in young people and asymptomatic cases (62, 82), with 40% of asymptomatic patients becoming seronegative during convalescence (83, 84). This may suggest that protective immunity against COVID-19 is short-lasting in asymptomatic or mild cases, although NAbs have been detected irrespective of symptoms in a cohort of health care workers 4 months after infection (85). However, if protective immunity is short-lasting in asymptomatic cases, it may well be a reason for concern about any public health policies that rely on the generation of herd immunity following natural infection.
Data from SARS-CoV studies indicate that neutralising antibodies may be able to persist for up to 3 years (69). If the same holds true for SARS-CoV-2, regardless of whether the neutralising antibodies are naturally or vaccine-induced, this could provide a reasonable window of protection but not necessarily confer lifelong humoral immunity.
Early Antibody Responses and Antibody-Dependent Enhancement
It has been a matter of debate as to whether certain pre-existing or early-generated antibodies may facilitate disease progression in both SARS and COVID-19 (86). The proposed mechanism is known as antibody-dependent enhancement (ADE), which enables the virus to gain cellular entry via Fc-receptor-mediated internalisation of non-neutralising antibodies bound to virus. ADE has been observed during infection with feline coronaviruses and a number of flaviviruses, which has raised concerns that it may also contribute to the immunopathology of both SARS and COVID-19 (87, 88). A number of animal studies during SARS-CoV vaccine development demonstrated the possibility of deleterious effects caused by ADE as a result of particular non-neutralising antibodies induced by vaccination (89, 90). However, these studies should be taken with caution, as complement and FcR-IgG engagement mechanisms are species-specific.
The first hypothesis for ADE is that pre-existing immunity to endemic strains of HCoV (HCoV-183 229E, HCoV-OC43, 184 HCoV-NL63, and HCoV-HKU1) that cause mild seasonal illness and, despite relatively limited homology to SARS viruses, may potentially share antigens with SARS-CoV-2 (53) and facilitate disease progression mediated by potentially cross-reactive antibodies (62). The rationale for antigenic cross-reactivity (or “original antigenic sin”) contributing to severe disease was based on the observation that a relatively small percentage of severe cases occur in children and young people. Given that fewer pathogenic HCoV are abundant, a previous infection with a closely-related endemic strain of coronavirus with similar immunological epitopes could be sufficient to trigger the production of cross-reactive antibodies in response to SARS-CoV-2 infection. If true, the induction of memory antibodies which fail to neutralise SARS-CoV-2 would not only fail to protect but could also facilitate viral invasion and worsen prognosis.
The second hypothesis for ADE is the early emergence of antibodies against SARS-CoV-2, as there is an increased mortality risk in patients with early emergence of IgG that could implicate a recall immune response (91). In one study, NAbs appeared to emerge early in a small proportion of SARS-CoV patients (17%), during the first 2 weeks of illness, but this group of early responders were shown to have an increased mortality rate (29.6%) (92). The authors suggested that the early emergence of neutralising antibodies correlated with the age of the patients and proposed that the early response implied priming effects from existing humoral memory against endemic strains of HCoV, which then led to higher mortality.
Antibody titres have been observed to correlate with disease severity for SARS-CoV-2, but some have suggested that this is likely to reflect a response to high viral loads rather than being the cause of pathology (62, 79, 93). However, a recent report has found not only an association between the rapid induction of humoral responses and disease severity, but also with an early N-biased antibody response in severe disease and a more balanced or S-dominant response in mild and moderate disease (94). This finding suggests that antibodies induced early against SARS-CoV-2 N could play a pathogenic role in the severity of COVID-19, so the ADE hypotheses should not be completely disregarded at this stage.
To our knowledge, recent results have provided the first direct evidence that anti-S IgG antibodies may correlate with disease severity in COVID-19 patients. Hoepel et al. showed in vitro that serum from severely ill patients formed complexes with SARS-CoV-2 Spike protein and triggered macrophages to release pro-inflammatory cytokines in an antibody-dependent manner. This induced long-lasting endothelial barrier disruption in vitro. The mechanism was found to be linked to an aberrant Fc glycosylation pattern, but the authors also showed that downstream Syk inhibitors could prevent macrophage activation (95).
The current evidence favours the argument that ADE is not the main driver in the pathological immune response against SARS-CoV-2, although it cannot be completely ruled out as a mechanism explaining disease severity in some patients. Arvin et al. have published a review where this is discussed in depth (96). Preliminary studies trialling treatment with convalescent plasma have so far shown no adverse effects in COVID-19 patients (75–77).
A recent pre-clinical study in a mouse model for SARS-CoV-2 failed to demonstrate ADE using recovered patient sera containing anti-SARS-CoV-2 RBD antibodies, although under the same conditions ADE was observed with Zika virus antibodies (97). In contrast to many of the small animal studies for SARS-CoV, no ADE was observed in the three rhesus macaque studies involving challenge with SARS-CoV-2 (39, 98, 99). These studies all demonstrate the induction of Th1 responses rather than Th2 responses following infection with SARS-CoV-2, immunisation with a S protein based viral vector (ChAdOx1) vaccine or with a DNA vaccine (39, 98–100). It is important to note that there are limitations to non-human primate models of ARDS and lung injury, due to differences between human and animal innate immune systems, risk factors, and co-morbidities (101).
To date, two case studies, in Nevada (United States) and another in the Netherlands, have reported patients with more severe clinical symptoms after SARS-CoV-2 reinfection—with one case leading to death (44, 46). This may re-ignite the debate about the role that ADE could play in poor patient outcomes. Strong humoral responses and B cell activity in severely ill patients may also suggest a pathogenic role for antibodies (102), but the use of convalescent plasma has so far not been shown to worsen disease outcome, with some evidence for a slightly better outcome in elderly patients (103). Based on current evidence, it is therefore unlikely that ADE is driving the severity of disease in SARS-CoV-2 infection. Nevertheless, the results from Hoepel et al. suggest that the possibility that some patients may develop a form of ADE should not be completely discounted as a mechanism for disease exacerbation in COVID-19, and therefore the outcomes of spike-based vaccines in human trials should be carefully evaluated.
Cellular Immune Response
Cellular immune responses are critical for both the resolution of viral infections and the development of long-lasting immunity, which will usually last longer than humoral responses ( Figure 1 ). Memory B cells can be recalled into antibody-producing plasmablasts in the event of infection. During the first SARS epidemic, both memory B cells and neutralising antibodies were found to persist in SARS-CoV patients up to 3 years post-infection (69). However, SARS-CoV-specific antibody levels in recovered patients were undetectable at 6 years post-infection while N-specific memory T cells could be detected at the same time point (104). Similarly, a later study showed that SARS-specific memory T cells persisted in blood at 11 years post-infection in three recovered patients (105). Recently, T cell responses to SARS have been detected 17 years after the epidemic and were shown to potently cross-react against SARS-CoV-2 (38). These data support the belief that memory T cells may critically contribute to long-term responses to SARS-CoV-2. However, it is not clear whether the level at which T-cell responses are maintained would be sufficient to provide protective immunity against reinfection, and this was not addressed in these studies.
Lymphopaenia is one of the defining clinical characteristics of severe COVID-19. During SARS-CoV-2 infection, both CD4+ and CD8+ T-cell depletion strongly correlate with the patient’s clinical course (38, 53, 106). The decrease of circulating T cells is probably due to recruitment into the lungs, as well as cell death. High numbers of infiltrating CD8+ T cells and macrophages have been found in the lung tissue in autopsies of patients with significant pulmonary damage (107). Interestingly, preferential depletion of Natural Killer (NK) cells and mucosal-associated innate T (MAIT) cells compared to other T cell subsets has been identified in severe COVID (108–111), also suggesting recruitment to sites of infection, and highlighting the potential role of unconventional T cells in disease pathogenesis. The breadth and magnitude of SARS-CoV-2 specific T cell response in convalescent patients has been correlated with disease severity (112). However in one acute infection study, the early appearance of T cells responses was linked with rapid viral clearance and less severe disease (94).
SARS-CoV T Cell Responses
When assessing T cell responses to the whole SARS-CoV proteome in the previous SARS epidemic, the majority of immunogenic epitopes were found within (but not limited to) the structural proteins, particularly in the S and N proteins, with the S protein inducing the greatest response (113). Among 70% of T cell responses induced against all structural proteins, 41% of those were elicited by the S protein for SARS-CoV (104, 113). In the recent study reporting long-lasting T cell responses in former SARS-CoV patients, two specific regions of the N protein (1-215aa and 206-419aa) were shown to elicit T cell responses in the majority of the individuals studied. In this same report, other non-structural proteins from ORF1 (NSP7 and NSP13) were also shown to elicit a response, but not in the majority of individuals (38). The role of these long-lasting T cell responses as correlates of protection against re-infection is yet to be determined.
From the few studies that identified antigen-specific CD4+ T cells from SARS-CoV recovered patients, responses were generated predominantly against the S and N proteins (114, 115). Interestingly, while N -specific CD4+ T cells exhibited a central memory (CD45RA- CCR7+ CD62L-), N-specific CD8+ T cells showed effector memory (CD45RA+ CCR7- CD62L-) phenotypes (115, 116). SARS-CoV-specific CD8+ T cell responses were far greater in magnitude than CD4+ responses and both CD4+ and CD8+ exhibited a memory phenotype characterised by higher proportions of polyfunctional T cells (113). High levels of CD4+ T cell responses and S-specific NAbs correlated with disease severity, but total and CD8+ T cell responses did not correlate with clinical outcome (113).
SARS-CoV-2 T Cell Responses
In SARS-CoV-2, depletion of both circulating CD4+ and CD8+ T-cells is strongly associated with the patient’s clinical course, but the magnitude of the virus-specific T cell response was found to be proportional to disease severity in convalescent patients (112). The level of T cell responses strongly correlates with NAb titres, consistent with a T-cell-dependent NAb response, whilst higher serum Th2 cytokines (IL-4, IL-5, and IL-10) correlated with mortality rate (117, 118). T cell responses during acute infection to SARS-CoV-2 waned at 1 month post-infection, as expected after the resolution of infection (94). Interestingly, a recent longitudinal analysis found that although the magnitude of T cell responses wanes over time, the number of convalescent individuals with detectable T cell responses had actually increased 6 months post-infection. The authors also found that those still having post-infection symptoms had the most pronounced decrease in T cell responses, reinforcing the idea that T cell responses are critical to the resolution of SARS-CoV-2 infection (56).
Severe COVID-19 correlates with preferential CD8+ lymphopaenia as compared to loss of CD4+ T-cells (119). The CD8+ T-cell response has been shown to be critical for resolving SARS-CoV-2 infection, although virus-specific CD4+ T-cells have been detected more often than specific CD8+ T-cells in recovered COVID-19 patients (38, 53, 106). The total CD8+ T cell count, including the numbers of effector memory CD8+ T cells, negatively correlates with stage of disease progression in COVID-19 patients (93, 120). The cytotoxic phenotype of SARS-CoV-2-specific T cells also appears to correlate with disease severity during acute infection, and virus-specific T cells from recovered COVID-19 individuals are polyfunctional and exhibit a stem-like phenotype (82).
There is some controversy about the phenotype of CD8+ T cells across the spectrum of disease, with some reports finding them exhausted with decreased polyfunctionality and cytotoxicity (121–123).
The severity of disease has been associated with a marked T cell exhaustion phenotype together with poor clonal expansion (102), while moderately-ill patients show robust T cell expansion and higher proportions of highly cytotoxic effector T cell subsets, as well as CD4+ and CD8+ T cells expressing granulysin and CD160+ Natural Killer T (NKT) cells during convalescence (102). Exhausted COVID-19 CD8+ T cells express elevated levels of PD-1, CTLA-4, TIGIT, and Tim-3 (122, 123), but Kusnadi et al. showed that the exhausted phenotype in memory CD8+ T cells was more pronounced in mild disease, as compared to severe COVID-19, where CD8+ T cells appeared to be more polyfunctional and exhibit a pro-survival signature (124). The authors suggest that the failure to establish an exhaustion imprint may lead to the dysregulation of the T cell response and greater disease pathogenesis, which may imply that a distinct phenotype of CTL response is required for durable protection (124). SARS-CoV-2-specific CD8+ T cells in recovered patients were found to predominantly be long-lived TEMRA cells, characterised by the re-expression of the naïve marker CD45RA and biased towards the CD27+CD28+ subset. For this reason, it has been suggested that SARS-CoV-2 specific CD8+ TEMRA cells are likely to contribute to long-lasting protective immunity.
However, when analysed using functional assays, antigen-specific CD8+ T cells presented similar polyfunctional profiles in both severe and mild cases (112). In general, most of the single-cell studies performed find an effector phenotype for CD8+ T cells, with effector memory and terminal effector CD8+ T cells showing increased clonal expansion in peripheral blood, along with upregulated inflammatory markers consistent with their antiviral activity (120).
Broader and stronger SARS-CoV-2-specific T cell responses were found in patients recovered from severe disease, when compared to those who experienced only mild symptoms. In patients with mild infection, the ratio of CD8+:CD4+ SARS-CoV-2-specific T cells appears to be much higher than in severe disease (112). Strikingly, the pattern of virus-specific CD8+ T cell responses to immunodominant epitopes in SARS-CoV-2 is notably different to SARS-CoV, with a large proportion of the total SARS-CoV-2 CD8+ T cell responses targeting M, N and ORF3a (53). During acute infection, early T cell responses against ORF7/8 were detected in mild patients, but were virtually absent during convalescence (94). ORF7a has been recently reported to counteract the restriction factor BST2, which tethers newly-produced viruses to the inner plasma membrane impairing viral release (11). These findings taken together could suggest that immune responses against ORF7 are involved in viral control. Also, ORF8 appears to downregulate MHC-I, by targeting it to the lysosomal compartment for degradation, a classic mechanism of viral immune evasion (125). In contrast, responses against structural proteins (S, M, N, and ORF3a) remained unchanged or increased 1 month after infection (94).
Even though the total CD4+ T cell count is also reduced in COVID-19 patients, the proportion of CD4+ T cells (amongst lymphocytes) appears to remain constant, with numbers of central memory CD4+ T-cells found to be markedly increased in severe disease (120). Naïve CD4+ T cells, T regs, and effector memory CD4+ T cell counts are all lower in acute infection. SARS-CoV-2 infection induces a strong CD4+ T cell response that favours Th1 polarisation, but Th2 and Th17 responses have also been detected (106). As virus-specific CD4+ T cells are usually key to the production of neutralising antibodies, it is unsurprising to observe a significant correlation between NAb titres and the frequency of virus-specific CD4+ T cells (59).
Upregulated expression of inflammatory genes have been highlighted in COVID-19 CD4+ T cells, including IL-1ß, FOS, JUN and KLF6 (120). Notably, Th1 cells that express both Interferon gamma (IFN-γ) and GM-CSF were found only in ICU patients with severe disease, with very few found in mild cases and healthy controls, indicating that a subset of T cells induced by SARS-CoV-2 may potentially be pathogenic (126).
COVID-19 T cell responses showed notably higher frequency of S-specific CD4+ T cell responses, whilst the majority of CD8+ responses were to M, N and ORF proteins (112). SARS-CoV-2 S-reactive CD4+ T cells were found in 83% of COVID-19 patients, which targeted both C and N terminal epitopes and expressed markers of recent activation (CD38 and HLA-DR) (59). In a single donor, about 50% of the CD4-specific responses were found to be directed against the spike protein (53).
Cellular Protection and Cross-Reactivity
T cell responses, and CD8+ responses in particular, may provide durable and robust protection against SARS-CoV-2 infection. The potential for memory T cell responses to clear viruses independent of humoral immunity has been clearly demonstrated for SARS-CoV, as virus-specific memory CD8+ T cells were shown to confer protection against lethal SARS-CoV challenge in mice without virus-specific memory CD4+ T or B cells (127).
Recall of memory cytotoxic T lymphocyte (CTL) responses against S epitopes could be observed 1 year post-infection with SARS-CoV (128). These epitopes were HLA-A*02:01-restricted (S1203–1211, S978–986 and S1167-1175) and were able to elicit high magnitude IFN-γ T cell responses in recovered patients (129). The S-specific CD8+ T cells, targeting S436 or S525 domains, had an effector memory phenotype (CD45RA+ CCR7- CD62L-) (115, 128) Interestingly, CTL responses could be elicited in a small minority of healthy patients without any history of SARS-CoV infection (129), which suggests that there may be cross-reactive memory T cells naturally present in the T cell repertoire that can elicit recall-like responses following SARS-CoV infection.
Regarding T cell cross-protection for SARS-CoV-2, several groups have demonstrated cross-reactivity for T cell responses, but whether this cross-reaction can provide some level of protection has not yet been addressed (38, 53, 130). Grifoni et al. proposed that immunity generated by HCoV exposure could potentially confer protection against SARS-CoV-2 infection, as they found that 40-60% of unexposed individuals generated CD4+ T cell responses against SARS-CoV-2 (53). Similarly, Braun et al. found 34% of healthy individuals to have S-reactive CD4+ T cells (130). Interestingly, these SARS-CoV-2 seronegative healthy donors have S-reactive CD4+ T cells that exclusively respond to C-terminal epitopes—most likely due to overlapping MHC-II epitopes found on the C terminus of endemic HCoV strains that account for 20% of common colds (130, 131). Le Bert et al. also described SARS-CoV-2-reactive T cells in uninfected individuals, which showed different immunodominance patterns to those found in COVID patients. However, the authors ascribe the epitope recognition to conserved fragments amongst animal CoVs, which show low homology with HCoV (38). In addition, some common immunodominant epitope clusters for S, M, and N proteins were previously identified by Peng et al., but dominant epitopes were shown to have little structural resemblance to common HCoV (112). Another group performed single cell (sc)-RNA sequencing in virus-reactive memory CD8+ T cells from convalescent COVID-19 individuals and found similar clusters to pre-pandemic samples when stimulated with SARS-CoV-2 peptides (124).
It will be important to fully elucidate where these T cell epitopes overlap and to determine whether these seronegative individuals show any degree of protection from SARS-CoV-2 infections. Stervbo et al. performed an extensive in silico characterisation of the similarity between SARS-CoV-2 antigens and epitopes for the most commonly found pathogens. Several identical epitopes were found for common HCoV HKU1 and OC43 and were predicted to bind HLA-I and -II (132). Despite similar and cross-reacting epitopes, whether there is any contribution to COVID-19 protection from pre-existing cross-reactive T-cell responses is yet to be determined.
Overall, T cell responses appear to be important for good outcomes with COVID-19. SARS-CoV T cell responses have been shown to be able to persist until now, 17 years after the SARS epidemic (38). Evidence points to a lack of a pre-existing immunity offering cross-protection against SARS-CoV-2. However, if T cells from pre-existing immunity to common HCoV are able to cross-react with SARS-CoV-2 there is the possibility that pre-existing immunity might have some effect in mitigating the spread of the pandemic.
Can Immunopathology in COVID-19 Impair Long-Lasting Protective Immunity?
The current view is that COVID-19 pathophysiology is exacerbated by immunological dysregulation. In severe cases, lymphopenia and dysregulated inflammatory cytokine production account for an excessive immune response leading to a cytokine storm, which in turn can provoke tissue damage, ARDS and multi-organ failure (35, 132). Single-cell RNA-seq from PBMCs from mild and severe COVID patients found a hyper-inflammatory signature in all cell types linked to TNF/IL-1ß-driven inflammation, which co-existed with a strong type I IFN-driven inflammatory response in severe patients (134). In a longitudinal study of patients admitted to UK hospitals, the degree of hyper-inflammation on admission [defined as COV-HI (hyperinflammatory), assessed using plasma C-reactive protein >150 mg/ml or doubling within 24 h and Ferritin >1500 mcg/ml] strongly predicted poor outcomes (135).
Does this hyperinflammatory response impair the formation of a memory compartment and subsequent protective immunity? There is no conclusive evidence yet to suggest an impaired memory compartment in patients with severe COVID-19 compared to those with mild disease. However, the lack of GC formation in severe COVID-19 patients may suggest that despite the high antibody titres linked with severe disease, the humoral response for these patients may be short-lived (80). The significant T cell lymphopaenia of severe cases may also influence whether a successful antibody- and T cell-mediated response can be mounted during this phase of inflammatory drive.
Also, T cell memory formation is known to be impaired in the context of chronic inflammation (136). Whether this holds true in the context of the cytokine storm in response to SARS-CoV-2 infection, and whether this leads to some level of impaired long-lasting T cell immunity is still unknown. There is an urgent need to understand whether the immune response generated after severe COVID-19 with excessive immune activation (COV-HI) differs from that which develops in mild cases. If a primary SARS-CoV-2 immune response cannot fully develop in critically ill patients, the formation of memory immune cells may also fail. Therefore, the differences in the adaptive immune response generated in mild compared to very sick patients, such as antibody titres and epitope targets of antibodies and T-cells, and the adequate formation of the memory compartment are likely to influence the level of protection against SARS-CoV-2 reinfection.
Variability of The Immune Landscape
Prior immune status has been known to play a role in many disease outcomes. Previous infections or co-morbidities can additionally modify the immune landscape. Co-infection with persistent viruses such as human cytomegalovirus (CMV) are known to facilitate disease progression in AIDS by driving immune activation (137), whilst co-morbidities such as obesity, associated with severe COVID-19, lead to a state of chronic inflammation and impaired immune function (138). The immune status generated by co-infections and pre-morbid conditions may correlate with distinct COVID-19 outcomes and warrants further investigation.
It is also possible that genetic differences can influence different immunophenotypes with distinct disease outcomes following SARS-CoV-2 infection. Two recent publications show a clear relationship between life-threatening COVID-19 and defects in the Type I IFN response, either from inborn errors in the IFN signalling pathway (36) or through the acquisition of auto-antibodies against IFN (37). Different immunotypes have been proposed for COVID-19 patients based on integration of B and T cell responses. Differential activation for either CD8+ and CD4+ T cells was observed in subgroups of patients. The same study segregates COVID-19 patients into two distinct patterns for activated B cell responses and identifies a third group with little evidence of an active humoral response (119).
Recent genome-wide association studies (GWAS) and large-scale immunophenotyping studies have compared immunological and genetic profiles between patient outcomes in sepsis (139). Similar functional genomic studies are warranted for COVID-19 to elucidate the patient-to-patient variability that will help us understand if genetics play a role in why a few individuals undergo severe immunopathological responses whilst most do not (140). To date, the largest GWAS of COVID-19 genetics published has identified a six-gene cluster on chromosome 3p21.31 which shows a significant association with severe disease (141). The risk genotype was found to be present at significantly higher frequency in patients who required mechanical ventilation. The locus contains genes implicated in immune regulation, including SLC6A20 which interacts with ACE2 and CXCR6—a receptor known to be expressed in lung-resident T cells and plays a role in the regulation of the inflammatory response to airway infections (142, 143). In addition, genetic variability in Human Leucocyte Antigen (HLA) class I has been shown to affect susceptibility to severe disease in COVID-19, with patients with the HLA-B*46:01 allele being particularly vulnerable (144). Further genetic studies will shed more light on why patient outcomes for Covid-19 can be so dramatically different.
Addressing whether the differences in the host immune response to SARS-CoV-2 is owed to the individual’s previous immunological history, or the result of genetic predisposition, and whether this affects the generation of memory responses will be an important consideration in the development of vaccine strategies.
Vaccine-Induced Immunity
As of 29th October 2020, a total of 45 candidate vaccines against COVID-19 are in clinical trials worldwide, with a further 156 other candidates in pre-clinical evaluation (145). Of those, six vaccines have been approved for early or limited use: one by CanSino, Sputnik V by the Gamaleya Research Institute, EpiVacCorona by the Vector Institute, CoronaVac by Sinovac Biotech, and two by Sinopharm together with the Wuhan Institute and the Beijing Institute of Biological Products. Among the approved vaccines, three contain inactivated virus, two use non-replicating adenoviral vectors and one is protein-based. A total of 10 vaccines are in Phase III clinical trials—including the aforementioned approved vaccines and others which are the first of their kind to enter Phase III—like the mRNA based vaccine by Moderna and the ChAdOx1-vectored vaccine from Oxford/AstraZeneca (145).
The S and the N proteins have proved to be powerful immunogens for vaccine development, as they elicit both humoral and cell-mediated responses, for both SARS-CoV and SARS-CoV-2, as well as and MERS-CoV (27, 105, 146). The general consensus from human studies concludes that the S protein from SARS-CoV is capable of inducing neutralising antibodies as well as CD4+ and CD8+ T cell responses, whilst the N protein mostly elicits T cell responses (146, 147).
Following the first SARS epidemic, S protein-expressing vaccines against SARS-CoV were shown to elicit protective immunity mediated by neutralising antibody production (28, 148, 149). S protein-based DNA vaccines induced high titres of potent neutralising antibodies that were able to inhibit viral entry but lacked cell-mediated protective efficacy against SARS-CoV (28). However, later animal studies showed that in mice, S-specific memory CD8+ T cells could also elicit protective immunity, even in the absence of SARS-CoV-specific memory CD4+ T cells or B cells, after a challenge with a lethal dose of the virus (127). Subsequent trials of N protein encoding DNA vaccines and synthetic N peptide vaccines elicited potent CD8+ T cell responses that conferred protection in SARS-CoV challenge models (150–152). In early preclinical studies of SARS-CoV vaccines conducted in mice, immunisation with N protein-based DNA vaccines have demonstrated induction of potent CTL responses (150). This preclinical evidence, combined with the observations of high neutralising antibody titres in recovered patients, provided good reason to believe that anti-S protein antibodies and CTL responses against N proteins were likely to be required to provide immunity against SARS-CoV. However, the disappearance of SARS-CoV infection prevented efficacy testing of these vaccines.
In the race to develop a SARS-CoV-2 vaccine, the first results have shown that an S-based DNA vaccine in a prime-boost regime in macaques induces NAb titres similar to those found in recovered humans and macaques (32). These antibodies showed better neutralising capacity following vaccination using the full prefusion-stabilised S protein. When challenged with SARS-CoV-2, vaccine-elicited NAbs appeared to be the major immune correlate of protection, together with some innate effector functions directly related to antibody efficacy (39). The ChAdOx1 nCoV-19 vaccine, another ongoing vaccine trial based on a ChAd (Chimpanzee Adenovirus) viral vector expressing the S protein, demonstrated some efficacy in preventing pulmonary pathology following SARS-CoV-2 challenge in non-human primates (98). All immunised macaques (6/6) were protected from developing pneumonia, which was seen in the control group (2/3). The immunised group had significant reductions in viral load in BAL fluid, but no differences were seen in viral load in nasal swabs (NS) compared to controls. Immunisation appears to reduce the severity of pathogenesis without any significant reduction in viral shedding. The authors found no sign of ADE and ascribe the persistence of virus in nasal swabs to the high titres of virus used for the challenge (98). These results are encouraging and could suggest that these first vaccines may provide protection against severe illness, even if they fall short of providing sterile immunity.
In humans, two vaccinations of the mRNA-1273 vaccine, known as the Moderna vaccine and based on a lipid nanoparticle that encapsulates RNA encoding S glycoprotein, have so far proven to elicit antibody and CD4+ T-cell responses but low CD8+ T-cell responses (153). Published Phase I/II data from ChAdOx1 nCoV-19 demonstrates the immunogenicity of the S protein, eliciting both neutralising antibodies and virus-specific effector T cell responses, with no major adverse effects (154). Although neutralising antibodies were induced by a single dose, a prime-boost regimen was shown to induce significantly higher titres of neutralising antibodies—a magnitude that, in a previous macaque study, appeared to be adequate for conferring protection against reinfection (40, 154). At least 10 of the vaccines now in Phase III trials report the induction of NAb titres in response to vaccination (155–157). Though limited data have been reported on cellular immune responses, all 10 appear to generate a detectable virus-specific T cell response in IFN-γ assays, although these vary in magnitude. At this point, it is not certain if vaccine-induced immune responses are durable when compared with natural infection. But results suggest that the induction of a robust virus-specific T cell responses with Th1 type polarisation is certainly possible.
From the published results from phase I/II trials across a range of vaccine subtypes, we find that adjuvanted protein based vaccines are the most immunogenic, followed by mRNA vaccines, followed by ChAdOx1 and inactivated vaccines, with AdV5-based vaccines being the least immunogenic in terms of NAb titres, although it is difficult to directly compare these studies due to differences in the assays being used (155). The AdV5-based vaccines (CanSino) were shown to be reactogenic, especially at higher doses and in older patients who are likely to have pre-existing immunity to AdV5 (158). Certain phase III candidates including the inactivated and mRNA vaccines (from Sinovac and Pfizer respectively) have already shown significantly lower immunogenicity in the older population, and may require booster regimens or higher doses in order to generate a durable protective efficacy—this is expected, as it has also been the case for the influenza vaccine (159–161).
Although these studies note the generation of T cell responses against the S protein, the extent to which protection is owed to the presence of S-specific CD8+ T cells is not addressed. The mRNA-1273 (Moderna) vaccine, for example, showed good CD4+ responses and detectable, but low, CD8+ T cell responses against the S protein (153). It was recently shown that patients who recovered from mild disease possess a higher frequency of CD8+ T cells specific for M or N proteins rather than to the S protein when compared to severe cases (112). The same study points out that though the S protein generates a substantial CD8+ T cell response, it does not appear to be immunodominant (112). This potentially has major implications for candidate vaccines using solely the S antigen, as these might elicit a narrower CD8+ T cell response than natural SARS-CoV-2 infection. An effective CD8+ T cell-inducing vaccine may therefore require additional antigens beyond the spike protein. Emerging studies are identifying epitopes across the SARS-CoV-2 genome predicted to activate both CD4+ and CD8+ responses that may induce long-lasting immunity (125). The combination of different epitopes may help build a robust memory response to provide protection through a powerful and effective vaccine.
Additionally, the majority of the current vaccines in clinical trials are administered through the skin or muscle. This route of administration may fail to generate a protective immune response in the upper respiratory tract that would otherwise be generated in natural infection (98). Hassan et al. demonstrated that a single-dose of intranasal administration of a ChAd-vectored S vaccine induced a local immune response that was sufficient to provide sterilising immunity in mice (163). There is reason to believe that local immunisation with a vaccine that can generate an IgA mucosal antibody response in the upper respiratory tract may provide a robust strategy for effective early clearance, as shown in successful trials of measles and BCG vaccines administered through the aerosol route (164, 165).
Discussion
Emerging evidence from recent SARS-CoV-2 reports, combined with literature from nearly two decades of SARS-CoV research, provide good reason to believe that it should be possible to generate protective immunity against SARS-CoV-2 in humans, either following natural infection or with a vaccine. Long-lasting protective immunity is likely to require both a sufficient titre of circulating NAbs and a strong T cell response, consisting of virus-specific central memory CD4+ and effector memory CD8+ T cells. Whether natural infection will induce a humoral and cell-mediated immune response that provides long-lasting protection against reinfection, and how this compares with the immune response generated by vaccination, and whether the inflammatory response will impair the proper formation of a memory compartment currently remains unknown.
The development of a safe SARS-CoV-2 vaccine is made more complex because of evidence of an immunopathological response underpinning disease severity. Antibody-dependent enhancement, along with direct tissue damage to alveolar cells, have been noted amongst studies conducted in SARS-CoV, and therefore warrants caution in future SARS-CoV-2 vaccine studies. Vaccine candidates using full-length S protein sequences may be counterproductive for certain individuals who may possess a predisposition for generating an immunopathological response to this antigen: the biological basis of adverse responses to S proteins needs to be elucidated.
The recent studies in macaques seem to suggest that counterproductive responses are not observed with the SARS-CoV-2 S protein, and induction of high titres of anti-RBD neutralising antibodies are likely to confer protection with minimal risk of causing an immunopathogenic response, provide a promising avenue for vaccine development. One caveat for vaccines that solely utilise the S protein is that this may well lead to a narrower cellular immune response and reduce the spectrum of antibodies induced – potentially reducing protective efficacy. However, the data from the vaccines currently in clinical trials appear promising, with evidence of robust neutralising antibody titres and various degrees of virus-specific T cell responses against the S protein. Whether or not strong immune responses to the S protein alone will provide adequate protection is still unclear, but the limited evidence from the animal challenge studies previously discussed seem to indicate that this may be sufficient to protect against severe disease. More recently, trials deploying intranasal immunisation using the S protein have been shown to elicit a broad spectrum of antibodies and T cell responses, including local mucosal responses, which may be a promising avenue for improving vaccine efficacy (163).
We are beginning to decipher the differences in immunological profiles between individuals who suffer from severe illness and those with asymptomatic or mild disease. In a similar manner to autoimmune diseases, this dysregulated response may occur only in a particular group of the general population that possess a predisposition to severe disease, based perhaps on initial infectious dose, viral and host genetics, and the individual profile of previous antigenic exposures. For this reason, large-scale immunophenotyping of COVID-19 patients has an important place on the global research roadmap for vaccine development.
Importantly, based on the preclinical studies conducted in SARS-CoV, there still remains a small possibility of generating an immunopathological response through vaccination that causes disease enhancement (86). So far, animal studies for vaccine candidates for SARS-CoV-2 have shown no evidence of immune-enhanced pathology or a Th2 biased response (39, 98), and all 10 candidates in phase III clinical trials have, to date, shown no adverse events directly linked to the vaccine immunisation that warrants concern about disease enhancement (145)
Overall, the evidence from observational studies in both SARS-CoV and SARS-CoV-2 infection, along with the promising data from clinical trials across 42 vaccine candidates, provide sufficient reason to speculate that a vaccine for SARS-CoV-2 will be safe and could provide lasting protective immunity that even if not lifelong might persist for years. Future studies, in due course, are expected to provide conclusive evidence about whether NAbs and/or virus-specific T cell responses will provide protection against infection, transmission and severe disease. Understanding the magnitude and characteristics of virus-specific cellular responses or titres of antibody responses required to provide protection will guide both vaccine design and public health policies to limit spread.
Author Contributions
All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr Biol (2020) 30:2186–203.e3. 10.1016/j.cub.2020.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lam TTY, Jia N, Zhang YW, Shum MHH, Jiang JF, Zhu HC, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature (2020) 583:282–5. 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- 3. Rangan R, Zheludev IN, Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA (2020) 26:937–59. 10.1261/rna.076141.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell (2020) 182:1284–94.e9. 10.1016/j.cell.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell (2020) 182:812–27.e19. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (2020) 395:565–74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol (2020) 92:522–8. 10.1002/jmv.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, et al. The coding capacity of SARS-CoV-2. Nature (2020). 10.1038/s41586-020-2739-1 [DOI] [PubMed] [Google Scholar]
- 9. Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Sci (80- ) (2020) 369:1249–56. 10.1126/science.abc8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor JK, Coleman CM, Postel S, Sisk JM, Bernbaum JG, Venkataraman T, et al. Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference. J Virol (2015) 89:11820–33. 10.1128/jvi.02274-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin-Sancho L, Lewinski MK, Pache L, Stoneham CA, Yin X, Pratt D, et al. Functional Landscape of SARS-CoV-2 Cellular Restriction. bioRxiv (2020). 10.1101/2020.09.29.319566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W, Moore MJ, Vasllieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature (2003) 426:450–4. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Milne S, Yang CX, Timens W, Bossé Y, Sin DD. SARS-CoV-2 receptor ACE2 gene expression and RAAS inhibitors. Lancet Respir Med (2020) 8:E50–1. 10.1016/s2213-2600(20)30224-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Sci (80- ) (2020) 367:1264–9. 10.1126/science.aax0902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun (2020) 11:1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell (2020) 181:271–80.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res (2020) 176:104742. 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palau V, Riera M, Soler MJ. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol Dial Transplant (2020) 35:1071–2. 10.1093/ndt/gfaa093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lambert DW, Yarski M, Warner FJ, Thornhill P, Parkin ET, Smith AI, et al. Tumor necrosis factor-α convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J Biol Chem (2005) 280:30113–9. 10.1074/jbc.M505111200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, et al. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc Natl Acad Sci USA (2008) 105:7809–14. 10.1073/pnas.0711241105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ichimura T, Mori Y, Aschauer P, Padmanabha Das KM, Padera RF, Weins A, et al. KIM-1/TIM-1 is a Receptor for SARS-CoV-2 in Lung and Kidney. medRxiv (2020). 10.1101/2020.09.16.20190694 [DOI] [Google Scholar]
- 22. Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv (2020). 10.1101/2020.03.14.988345 [DOI] [Google Scholar]
- 23. Davanzo GG, Codo AC, Brunetti NS, Boldrini VO, Knittel TL, Monterio LB, et al. SARS-CoV-2 Uses CD4 to Infect T Helper Lymphocytes. medRxiv (2020) 2020.09.25.20200329. 10.1101/2020.09.25.20200329 [DOI] [Google Scholar]
- 24. Radzikowska U, Ding M, Tan G, Zhakparov D, Peng Y, Wawrzyniak P, et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy Eur J Allergy Clin Immunol (2020) 75:2828–44. 10.1111/all.14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (2020) 370:856–60. 10.1126/science.abd2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell (2020) 183:739–51.e8. 10.1016/j.cell.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV - A target for vaccine and therapeutic development. Nat Rev Microbiol (2009) 7:226–36. 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin JE, Louder MK, Holman LSA, Gordon IJ, Enama ME, Larkin BD, et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine (2008) 26:6338–43. 10.1016/j.vaccine.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He Y, Zhou Y, Siddiqui P, Niu J, Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol (2005) 43:3718–26. 10.1128/JCM.43.8.3718-3726.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buitrago-Garcia D, Egli-Gany D, Counotte MJ, Hossmann S, Imeri H, Ipekci AM, et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARSCoV-2 infections: A living systematic review and meta-analysis. PloS Med (2020) 17:e1003346. 10.1371/journal.pmed.1003346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (2020) 395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guan W, Liang W, Zhao Y, Liang H, Chen Z, Li Y, et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J (2020) 55:2000547. 10.1183/13993003.00547-2020.Supp1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Killerby ME, Link-Gelles R, Haight SC, Schrodt CA, England L, Gomes DJ, et al. Characteristics Associated with Hospitalization Among Patients with COVID-19 - Metropolitan Atlanta, Georgia, March-April 2020. MMWR Morb Mortal Wkly Rep (2020) 69:790–4. 10.15585/mmwr.mm6925e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet (2020) 395:1054–62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int J Antimicrob Agents (2020) 55:105954. 10.1016/j.ijantimicag.2020.105954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Sci (80- ) (2020) 370:eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bastard P, Rosen LB, Zhang Q, Zhang Y, Dorgham K, Béziat V, et al. Auto-antibodies against type I IFNs in patients with life- threatening COVID-19. Sci (80- ) (2020) 370:eabd4585. 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature (2020) 584:457–62. 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 39. Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Sci (80- ) (2020) 369:812–7. 10.1126/science.abc4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng W, Bao L, Liu J, Xiao C, Liu J, Xue J, et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Sci (80- ) (2020) 369:818–23. 10.1126/science.abc5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gupta V, Bhoyar RC, Jain A, Srivastava S, Upadhayay R, Imran M, et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis (2020) ciaa1451. 10.1093/cid/ciaa1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldman JD, Wang K, Roltgen K, Nielsen SCA, Roach JC, Naccache SN, et al. Reinfection with SARS-CoV-2 and Failure of Humoral Immunity: a case report. medRxiv (2020). 10.1101/2020.09.22.20192443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. To KK-W, Hung IF-N, Ip JD, Chu AW-H, Chan W-M, Tam AR, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis (2020) ciaa1275. 10.1093/cid/ciaa1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tillett R, Sevinsky J, Hartley P, Kerwin H, Crawford N, Gorzalski A, et al. Genomic Evidence for a Case of Reinfection with SARS-CoV-2. SSRN Electron J (2020). 10.2139/ssrn.3680955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, et al. A Case of Early Re-infection with SARS-CoV-2. Clin Infect Dis (2020) ciaa1436. 10.1093/cid/ciaa1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis (2020) ciaa1330. 10.1093/cid/ciaa1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, Marquez S, Gutierrez B, Rojas-Silva P, et al. COVID-19 Re-Infection by a Phylogenetically Distinct SARS-CoV-2 Variant, First Confirmed Event in South America. SSRN Electron J (2020). 10.2139/ssrn.3686174 [DOI] [Google Scholar]
- 48. Liu W, Fontanet A, Zhang P, Zhan L, Xin Z, Baril L, et al. Two-Year Prospective Study of the Humoral Immune Response of Patients with Severe Acute Respiratory Syndrome. J Infect Dis (2006) 193:792–5. 10.1086/500469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma H, Zeng W, He H, Zhao D, Jiang D, Zhou P, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol (2020) 17:773–5. 10.1038/s41423-020-0474-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med (2020) 26:1033–6. 10.1038/s41591-020-0913-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu X, Sun J, Nie S, Li H, Kong Y, Liang M, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med (2020) 26:1193–5. 10.1038/s41591-020-0949-6 [DOI] [PubMed] [Google Scholar]
- 52. Adams ER, Ainsworth M, Anand R, Andersson MI, Auckland K, Baillie JK, et al. Antibody testing for COVID-19: A report from the National COVID Scientific Advisory Panel. Wellcome Open Res (2020) 5:139. 10.12688/wellcomeopenres.15927.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell (2020) 181:1489–01.e15. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S, et al. Neutralizing Antibody Responses to SARS-CoV-2 in a COVID-19 Recovered Patient Cohort and Their Implications. SSRN Electron J (2020). 10.2139/ssrn.3566211 [DOI] [Google Scholar]
- 55. Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Sci (80- ) (2020) eabd7728. 10.1126/science.abd7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bilich T, Nelde A, Heitmann JS, Maringer Y, Roerden M, Bauer J, et al. Differential Kinetics of T Cell and Antibody Responses Delineate Dominant T Cell Epitopes in Long-Term Immunity after COVID-19. Cell (2020). 10.2139/ssrn.3713528 [DOI] [Google Scholar]
- 57. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: Current State of the Science. Immunity (2020) 52:910–41. 10.1016/j.immuni.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jiang S, Hillyer C, Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol (2020) 41:355–9. 10.1016/j.it.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity (2020) 52:971–7.e3. 10.1016/j.immuni.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen W, Zhang J, Qin X, Wang W, Xu M, Wang L-F, et al. SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. BioMed Pharmacother (2020) 130:110629. 10.1016/j.biopha.2020.110629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ (2020) 368:m1091. 10.1136/bmj.m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick L, Rattigan SM, et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. Nat Commun (2020) 11:4704.e16. 10.1038/s41467-020-18450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell (2020) 82:73–84.e16. 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature (2020) 584:443–9. 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barnes CO, West AP, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell (2020) 182:828–42.e16. 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell (2020) 181:281–92.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pinto D, Park Y, Beltramello M, Walls AC, Tortorici MA, Bianchi S, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature (2020) 583:290–5. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 68. Zhu Y, Yu D, Han Y, Yan H, Chong H, Ren L, et al. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci Adv (2020) 6:eabc9999. 10.1126/sciadv.abc9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Sci (80- ) (2020) 369:731–6. 10.1126/science.abc7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lv H, Wu NC, Tsang OTY, Yuan M, Perera RAPM, Leung WS, et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep (2020) 31:107725. 10.1016/j.celrep.2020.107725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature (2020) 584:115–9. 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- 72. Poston D, Weisblum Y, Wise H, Templeton K, Jenks S, Hatziioannou T, et al. Absence of SARS-CoV-2 neutralizing activity in pre-pandemic sera from individuals with recent seasonal coronavirus infection. medRxiv (2020). 10.1101/2020.10.08.20209650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Edridge AWD, Kaczorowska JM, Hoste ACR, Bakker M, Klein M, Jebbink MF, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med (2020) 26:1691–3. 10.1038/s41591-020-1083-1 [DOI] [PubMed] [Google Scholar]
- 74. Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, et al. Beyond the Spike: identification of viral targets of the antibody response to SARS-CoV-2 in COVID-19 patients. medRxiv (2020). 10.1101/2020.04.30.20085670 [DOI] [Google Scholar]
- 75. Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 Critically Ill Patients with COVID-19 with Convalescent Plasma. JAMA - J Am Med Assoc (2020) 323:1582–9. 10.1001/jama.2020.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eckhardt CM, Cummings MJ, Rajagopalan KN, Borden S, Bitan ZC, Wolf A, et al. Evaluating the efficacy and safety of human anti-SARS-CoV-2 convalescent plasma in severely ill adults with COVID-19: A structured summary of a study protocol for a randomized controlled trial. Trials (2020) 21:499. 10.1186/s13063-020-04422-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest (2020) 130:2757–65. 10.1172/JCI138745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mascuch SJ, Fakhretaha-Aval S, Bowman JC, Ma MTH, Thomas G, Bommarius B, et al. Buzz about RT-qPCR: An RT-qPCR formulation for SARS-CoV-2 detection using reagents produced at Georgia Institute of Technology. medRxiv (2020). 10.1101/2020.07.29.20163949 [DOI] [Google Scholar]
- 79. Chen X, Pan Z, Yue S, Yu F, Zhang J, Yang Y, et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther (2020) 5:180. 10.1038/s41392-020-00301-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, et al. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell (2020) 183:143–57. 10.1016/j.cell.2020.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol (2020) 21:1506–16. 10.1038/s41590-020-00814-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Straling K, Gorin J-B, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell (2020) 183:p158–68.e14. 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ibarrondo FJ, Fulcher JA, Goodman-Meza D, Elliott J, Hofmann C, Hausner MA, et al. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N Engl J Med (2020) 383:1085–7. 10.1056/NEJMc2025179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med (2020) 26:1200–4. 10.1038/s41591-020-0965-6 [DOI] [PubMed] [Google Scholar]
- 85. Reynolds CJ, Swadling L, Gibbons JM, Pade C, Jensen M, Diniz MO, et al. Healthcare workers with mild asymptomatic SARS-CoV-2 infection show T cell responses and neutralising antibodies after the first wave. medRxiv (2020). [Google Scholar]
- 86. Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect (2020) 22:72–3. 10.1016/j.micinf.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weiss RC, Scott FW. Antibody-mediated enhancement of disease in feline infectious peritonitis: Comparisons with dengue hemorrhagic fever. Comp Immunol Microbiol Infect Dis (1981) 4:175–89. 10.1016/0147-9571(81)90003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yip MS, Leung HL, Li PH, Cheung CY, Dutry I, Li D, et al. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J (2016) 22:25–31. [PubMed] [Google Scholar]
- 89. Jaume M, Yip MS, Kam YW, Cheung CY, Kien F, Roberts A, et al. SARS CoV subunit vaccine: Antibodymediated neutralisation and enhancement. Hong Kong Med J (2012) 18:31–6. [PubMed] [Google Scholar]
- 90. Luo F, Liao FL, Wang H, Tang HB, Yang ZQ, Hou W. Evaluation of Antibody-Dependent Enhancement of SARS-CoV Infection in Rhesus Macaques Immunized with an Inactivated SARS-CoV Vaccine. Virol Sin (2018) 33:201–4. 10.1007/s12250-018-0009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J Virol (2019) 94:e02015–9. 10.1128/jvi.02015-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ho MS, Chen WJ, Chen HY, Lin SF, Wang WC, Di J, et al. Neutralizing antibody response and SARS severity. Emerg Infect Dis (2005) 11:1730–7. 10.3201/eid1111.040659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. He R, Lu Z, Zhang L, Fan T, Xiong R, Shen X, et al. The clinical course and its correlated immune status in COVID-19 pneumonia. J Clin Virol (2020) 127:104361. 10.1016/j.jcv.2020.104361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tan AT, Linster M, Wah Tan C, Le Bert N, Ni Chia W, Zhuang Y, et al. Early induction of SARS-CoV-2 specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. bioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hoepel W, Chen H-J, Allahverdiyeva S, Manz X, Aman J, Bonta P, et al. Anti-SARS-CoV-2 IgG from severely ill COVID-19 patients promotes macrophage hyper-inflammatory responses. bioRxiv (2020) 2020.07.13.190140. 10.1101/2020.07.13.190140 [DOI] [Google Scholar]
- 96. Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, et al. Virgin HW. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature (2020) 584:353–63. 10.1038/s41586-020-2538-8 [DOI] [PubMed] [Google Scholar]
- 97. Quinlan BD, Mou H, Zhang L, Guo Y, He W, Ojha A, et al. The SARS-CoV-2 Receptor-Binding Domain Elicits a Potent Neutralizing Response Without Antibody-Dependent Enhancement. SSRN Electron J (2020). 10.2139/ssrn.3575134 [DOI] [Google Scholar]
- 98. van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature (2020) 586:578–82. 10.1038/s41586-020-2608-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Sci (80- ) (2020) 369:806–11. 10.1126/science.abc6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T-cell responses. Nature (2020) 586:594–9. 10.1038/s41586-020-2814-7 [DOI] [PubMed] [Google Scholar]
- 101. Matute-Bello G, Frevert CW, Martin TR. Animal models of acute lung injury. Am J Physiol Lung Cell Mol Physiol (2008) 295:L379–99. 10.1152/ajplung.00010.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang J-Y, Wang X-M, Xing X, Xu Z, Zhang C, Song J-W, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol (2020) 21:1107–18. 10.1038/s41590-020-0762-x [DOI] [PubMed] [Google Scholar]
- 103. Rogers R, Shehadeh F, Mylona EK, Rich J, Neill M, Touzard-Romo F, et al. Convalescent plasma for patients with severe COVID-19: a matched cohort study. Clin Infect Dis (2020) ciaa1548. 10.1093/cid/ciaa1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Oh H-LJ, Chia A, Chang CXL, Leong HN, Ling KL, Grotenbreg GM, et al. Engineering T Cells Specific for a Dominant Severe Acute Respiratory Syndrome Coronavirus CD8 T Cell Epitope. J Virol (2011) 20:10464–71. 10.1128/jvi.05039-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine (2016) 34:2008–14. 10.1016/j.vaccine.2016.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol (2020) 5:eabd2071. 10.1126/sciimmunol.abd2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nienhold R, Ciani Y, Koelzer VH, Tzankov A, Haslbauer JD, Menter T, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun (2020) 11:5086. 10.1038/s41467-020-18854-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J Exp Med (2020) 217:e20200872. 10.1084/jem.20200872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Parrot T, Gorin JB, Ponzetta A, Maleki KT, Kammann T, Emgård J, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol (2020) 5:eabe1670. 10.1126/sciimmunol.abe1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Flament H Beaudoin L, Toubal A, Bertrand L, Lebourgeois S, Gouda Z, Rousseau C, et al. Outcome of SARS-CoV-2 infection linked to MAIT cell activation and cytotoxicity: evidence for an IL-18 dependent mechanism. Medrxiv (2020). 10.1101/2020.08.31.20185082 [DOI] [PubMed] [Google Scholar]
- 111. Maucourant C, Filipovic I, Ponzetta A, Aleman S, Cornillet M, Hertwig L, et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol (2020) 5:eabd6832. 10.1126/SCIIMMUNOL.ABD6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol (2020) 21:1336–45. 10.1038/s41590-020-0782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T Cell Responses to Whole SARS Coronavirus in Humans. J Immunol (2008) 181:5490–500. 10.4049/jimmunol.181.8.5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Libraty DH, O’Neil KM, Baker LM, Acosta LP, Olveda RM. Human CD4+ memory T-lymphocyte responses to SARS coronavirus infection. Virology (2007) 368:317–21. 10.1016/j.virol.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Peng H, Yang Lt, Wang Ly, Li J, Huang J, Lu Zq, et al. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology (2006) 351:466–75. 10.1016/j.virol.2006.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yang LT, Peng H, Zhu ZL, Li G, Huang ZT, Zhao ZX, et al. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol (2006) 120:171–8. 10.1016/j.clim.2006.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest (2020) 130:2620–9. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wan S, Yi Q, Fan S, Lv J, Zhang X, Guo L, et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv (2020). 10.1101/2020.02.10.20021832 [DOI] [Google Scholar]
- 119. Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Sci (80- ) (2020) eabc8511. 10.1126/science.abc8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery (2020). 10.1038/s41421-020-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol (2020) 17:533–5. 10.1038/s41423-020-0402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zheng HY, Zhang M, Yang CX, Zhang N, Wang XC, Yang XP, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol (2020) 17:541–3. 10.1038/s41423-020-0401-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol (2020) 11:827. 10.3389/fimmu.2020.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kusnadi A, Ramírez-Suástegui C, Fajardo V, Chee SJ, Meckiff BJ, Simon H, et al. Severely ill COVID-19 patients display augmented functional properties in SARS-CoV-2-reactive CD8+ T cells. bioRxiv (2020). 10.1101/2020.07.09.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhang Y, Zhang J, Chen Y, Luo B, Yuan Y, Huang F, et al. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I. bioRxiv (2020). 10.1101/2020.05.24.111823. 2020.05.24.111823. [DOI] [Google Scholar]
- 126. Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci Rev (2020) nwaa041. 10.1093/nsr/nwaa041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-Specific Memory CD8 T Cells Provide Substantial Protection from Lethal Severe Acute Respiratory Syndrome Coronavirus Infection. J Virol (2014) 88:11034–44. 10.1128/jvi.01505-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chen H, Hou J, Jiang X, Ma S, Meng M, Wang B, et al. Response of Memory CD8 + T Cells to Severe Acute Respiratory Syndrome (SARS) Coronavirus in Recovered SARS Patients and Healthy Individuals. J Immunol (2005) 175:591–8. 10.4049/jimmunol.175.1.591 [DOI] [PubMed] [Google Scholar]
- 129. Wang B, Chen H, Jiang X, Zhang M, Wan T, Li N, et al. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood (2004) 104:200–6. 10.1182/blood-2003-11-4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature (2020) 587:270–4. 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- 131. Gaunt ER, Hardie A, Claas ECJ, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol (2010) 48:2940–7. 10.1128/JCM.00636-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Stervbo U, Rahmann S, Roch T, Westhof TH, Babel N. SARS-CoV-2 reactive T cells in uninfected individuals are likely expanded by beta-coronaviruses. bioRxiv (2020). 10.1101/2020.07.01.182741 [DOI] [Google Scholar]
- 133. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA - J Am Med Assoc (2020) 323:1061–9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lee JS, Park S, Jeong HW, Ahn JY, Choi SJ, Lee H, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol (2020) 5:eabd1554. 10.1126/sciimmunol.abd1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Manson JJ, Crooks C, Naja M, Ledlie A, Goulden B, Liddle T, et al. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol (2020) 2:e594–602. 10.1016/S2665-9913(20)30275-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stelekati E, Shin H, Doering TA, Dolfi DV, Ziegler CG, Beiting DP, et al. Bystander chronic infection negatively impacts development of CD8+ T cell memory. Immunity (2014) 40:801–13. 10.1016/j.immuni.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Gianella S, Letendre S. Cytomegalovirus and HIV: A Dangerous Pas de Deux. J Infect Dis (2016) 214 Suppl:S67–74. 10.1093/infdis/jiw217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Milner JJ, Beck MA. The impact of obesity on the immune response to infection. Proc Nutr Soc (2012) 71:298–306. 10.1017/S0029665112000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Elhabyan A, Elyaacoub S, Sanad E, Abukhadra A, Elhabyan A, Dinu V. The role of host genetics in susceptibility to severe viral infections in humans and insights into host genetics of severe COVID-19: A systematic review. Virus Res (2020) 289:198163. 10.1016/j.virusres.2020.198163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. GAINS-2 - Sepsis Immunomics trial (UK). Available at: https://www.hra.nhs.uk/covid-19-research/approved-covid-19-research/260007/.
- 141. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med (2020) 383:1522–34. 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wein AN, McMaster SR, Takamura S, Dunbar PR, Cartwright EK, Hayward SL, et al. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J Exp Med (2019) 216:248–2762. 10.1084/jem.20181308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ashhurst AS, Flórido M, Lin LCW, Quan D, Armitage E, Stifter SA, et al. CXCR6-deficiency improves the control of pulmonary mycobacterium tuberculosis and influenza infection independent of T-lymphocyte recruitment to the lungs. Front Immunol (2019) 10:339. 10.3389/fimmu.2019.00339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, et al. Human leukocyte antigen susceptibility map for SARS-CoV-2. J Virol (2020) 94:e00510–20. 10.1128/JVI.00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. WHO Vaccines Landscape (2020). Available at: https://www.who.int/who-documents-detail/draft-landscape-of-covid-19-candidate-vaccines (Accessed October 29, 2020).
- 146. Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, et al. Contributions of the structural proteins of severe respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA (2004) 101:9804–9. 10.1073/pnas.0403492101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Oh HLJ, Gan SKE, Bertoletti A, Tan YJ. Understanding the T cell immune response in SARS coronavirus infection. Emerg Microbes Infect (2012) 1:e23. 10.1038/emi.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, Murphy BR, et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA (2004) 101:6641–6. 10.1073/pnas.0401939101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K. Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature (2004) 428:561–4. 10.1038/nature02463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zhu MS, Pan Y, Chen HQ, Shen Y, Wang XC, Sun YJ, et al. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol Lett (2004) 92:237–43. 10.1016/j.imlet.2004.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhao P, Cao J, Zhao LJ, Qin ZL, Ke JS, Pan W, et al. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology (2005) 331:128–35. 10.1016/j.virol.2004.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ohno S, Kohyama S, Taneichi M, Moriya O, Hayashi H, Oda H, et al. Synthetic peptides coupled to the surface of liposomes effectively induce SARS coronavirus-specific cytotoxic T lymphocytes and viral clearance in HLA-A*0201 transgenic mice. Vaccine (2009) 27:3912–20. 10.1016/j.vaccine.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. N Engl J Med (2020) 383:1920–31. 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet (2020) 396:467–78. 10.1016/S0140-6736(20)31604-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Krammer F. SARS-CoV-2 vaccines in development. Nature (2020) 586:516–27. 10.1038/s41586-020-2798-3 [DOI] [PubMed] [Google Scholar]
- 156. Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatullin AI, Shcheblyakov DV, Dzharullaeva AS, et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet (2020) 396:887–97. 10.1016/S0140-6736(20)31866-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature (2020) 586:583–8. 10.1038/s41586-020-2607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhu F-C, Guan X-H, Li Y-H, Huang J-Y, Jiang T, Hou L-H, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (2020) 396:479–88. 10.1016/S0140-6736(20)31605-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Phase 1/2 study of COVID-19 RNA vaccine BNT162b1 in adults. Nature (2020) 586:589–93. 10.1038/s41586-020-2639-4 [DOI] [PubMed] [Google Scholar]
- 160. Zhang Y, Zeng G. Immunogenicity and Safety of a SARS-CoV-2 Inactivated Vaccine in Healthy Adults Aged 18-59 years: Report of the Randomized, Double-blind, and Placebo-controlled Phase 2 Clinical Trial. medRxiv (2020). 10.1101/2020.07.31.20161216 [DOI] [Google Scholar]
- 161. Dunning AJ, DiazGranados CA, Voloshen T, Hu B, Landolfi VA, Talbot HK. Correlates of Protection against Influenza in the Elderly: Results from an Influenza Vaccine Efficacy Trial. Clin Vaccine Immunol (2016) 23:228–35. 10.1128/CVI.00604-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yarmarkovich M, Warrington JM, Farrel A, Maris JM. Identification of SARS-CoV-2 Vaccine Epitopes Predicted to Induce Long-Term Population-Scale Immunity. Cell Rep Med (2020) 1:100036. 10.1016/j.xcrm.2020.100036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith B, Harvey IB, et al. A single intranasal dose of chimpanzee adenovirus-vectored vaccine confers sterilizing immunity against SARS-CoV-2 infection. bioRxiv (2020) 2020.07.16.205088. 10.1101/2020.07.16.205088 [DOI] [Google Scholar]
- 164. Low N, Bavdekar A, Jeyaseelan L, Hirvez S, Ramanathan K, Andrews N, et al. A randomized, controlled trial of an aerosolized vaccine against measles. N Engl J Med (2015) 372:1519–29. 10.1056/NEJMoa1407417 [DOI] [PubMed] [Google Scholar]
- 165. Satti I, Meyer J, Harris SA, Thomas ZRM, Griffiths K, Antrobus RD, et al. Safety and immunogenicity of a candidate tuberculosis vaccine MVA85A delivered by aerosol in BCG-vaccinated healthy adults: A phase 1, double-blind, randomised controlled trial. Lancet Infect Dis (2014) 14:939–46. 10.1016/S1473-3099(14)70845-X [DOI] [PMC free article] [PubMed] [Google Scholar]