Abstract
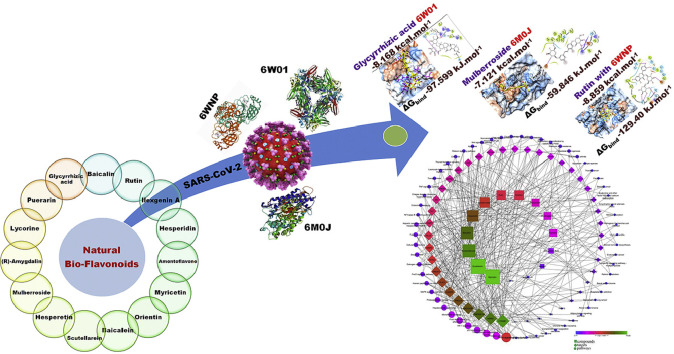
Bioflavonoids are the largest group of plant-derived polyphenolic compounds with diverse biological potential and have also been proven efficacious in the treatment of Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS). The present investigation validates molecular docking, simulation, and MM-PBSA studies of fifteen bioactive bioflavonoids derived from plants as a plausible potential antiviral in the treatment of COVID-19. Molecular docking studies for 15 flavonoids on the three SARS CoV-2 proteins, non-structural protein-15 Endoribonuclease (NSP15), the receptor-binding domain of spike protein (RBD of S protein), and main protease (Mpro/3CLpro) were performed and selected protein-ligand complexes were subjected to Molecular Dynamics simulations. The molecular dynamics trajectories were subjected to free energy calculation by the MM-PBSA method. All flavonoids were further assessed for their effectiveness as adjuvant therapy by network pharmacology analysis on the target proteins. The network pharmacology analysis suggests the involvement of selected bioflavonoids in the modulation of multiple signaling pathways like p53, FoxO, MAPK, Wnt, Rap1, TNF, adipocytokine, and leukocyte transendothelial migration which plays a significant role in immunomodulation, minimizing the oxidative stress and inflammation. Molecular docking and molecular dynamics simulation studies illustrated the potential of glycyrrhizic acid, amentoflavone, and mulberroside in inhibiting key SARS-CoV-2 proteins and these results could be exploited further in designing future ligands from natural sources.
Keywords: Amentoflavone, Bioflavonoids, In-silico study, Glycyrrhizic acid, Novel Coronavirus-2, Mulberroside
Abbreviations: 2019-nCoV, 2019 Novel Coronavirus; CoV, Corona Virus; Å, Angstrom; COVID-19, Coronavirus Disease-2019; MD, Molecular Dynamics; MM-PBSA, Molecular Mechanics Poisson-Boltzmann Surface Area; NSP, Non-structural Protein; ORF, Open Reading Frame; OPLS, Optimized Potentials for Liquid Simulations; RBD, Receptor Binding Domain; RMSD, Root Mean Square Deviation; SARS, Severe Acute Respiratory syndrome; SARS-CoV-2, Severe Acute Respiratory syndrome Coronavirus-2; SDF, Structure Data File; WHO, World Health Organization
Graphical abstract
Highlights
-
•
Bioflavonoids are potential antiviral secondary metabolites.
-
•
Docking, MD and network pharmacology demonstrated antiviral & adjuvant potential.
-
•
MM-PBSA calculations and network pharmacology identified key bioflavonoids.
-
•
Mulberroside, Glycyrrhizic acid and Amentoflavone may hold promise in COVID-19.
1. Introduction
During December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as a global pandemic originating from the Wuhan city of China, and still it is a major global threat due to high mortality and morbidity rates. The coronaviruses contain highly enveloped single stranded RNA with the genome size of 26–32 kilobases and requires the initial RNA synthesis, replication and transcription for its life cycle [[1], [2], [3]]. Two-thirds of the genome of all coronoviruses encodes pp1ab, a replicase polyprotein which comprises of two overlapping open reading frames (ORF1a and ORF1b), cleaved into 16 different non-structural proteins (NSPs) by viral proteases [4]. Amongst these NSPs, NSP-15 is ascribed with nidoviral uridylate-specific endoribonuclease (NendoU) activity involving uridylate-specific cleavage of RNAs. NSP-15/NendoU activity is important in the innate immune responses and the viral life cycle [5]. The structural protein protruding on viral surface called spike (S) glycoprotein which exist as a metastable prefusion homotrimeric form enables the entry of 2019-nCoV into the host cells through structural rearrangement. The binding of the S1 subunit of spike S glycoprotein to the angiotensin-converting enzyme 2 (ACE2) on the host cell leads to the transition of the S2 subunit to a highly stable post-fusion conformation which accelerates the viral entry process. Hence, the drugs targeting the viral proteases, nonstructural proteins and structural protein that block various stages of viral life cycle such as the entry, replication, and proliferation may exhibit a wide spectrum of activity [[6], [7], [8], [9]].
The natural compounds, especially from plant resources, remained the choice in the lead identification programs due to their abundance, safety and broad spectrum of ensuing activities. The flavonoids, bioflavonoids or polyphenolic compounds are a group of secondary metabolites present in fruits and vegetables and are well known for their health benefits. Flavonoids, a group of compounds containing flavan nucleus or a 15-carbon skeleton with two benezene rings linked through pyran ring, are well studied natural compounds comprising more than 6000 structurally well characterized molecules [10]. They are reported to possess health benefits in infectious, oncogenic, inflammatory and degenerative diseases [[11], [12], [13], [14]]. It is noteworthy that flavonoids have been extensively studied against the plethora of viruses to overcome the limitations of existing therapies [[15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27]]. The recent in-silico studies have demonstrated the antiviral potential of natural compounds in COVID-19 [[1], [2], [3],28].
Flavonoids have been proven to have a multidimensional therapeutic effect such as anti-inflammatory, antiviral, antioxidant, cardioprotective, and have also shown their significant role in the treatment of respiratory associated problems, which are a key requirement for a drug to be effective against COVID-19 [[1], [2], [3],28]. In the present investigation, an attempt was made to evaluate the potency of some well-known bioflavonoids having multiple therapeutic properties against COVID-19, which can contribute to the scientific community exploring and identifying treatments for COVID-19. In the present investigation, molecular docking was performed on fifteen potential flavonoids derived from plants against three SARS- CoV-2 proteins namely non-structural protein-15 Endoribonuclease (NSP15), the receptor-binding domain of spike protein (RBD of S protein), and main protease (Mpro, also called 3CLpro) as targeted proteins. Further, the top ranking compounds on each protein were subjected to molecular dynamics simulation and Molecular Mechanics Poisson Boltzmann surface area continuum solvation (MM-PBSA) calculations to gain the deeper insights of binding affinity and possible mode of inhibition.
2. Material and methods
2.1. Molecular docking
The molecular docking analysis of 15 bioflavonoids (Supplementary file) were performed on the crystal structures of NSP-15 (PDB ID: 6W01), RBD of spike protein (PDB ID: 6M0J), and Mpro (PDB ID: 6WNP) with the resolutions 1.9, 2.45 and 1.44 A° respectively using Schrödinger maestro 2018-1 MM. The details of the molecular docking protocol are given in the supplementary file.
2.2. Molecular dynamics (MD) simulation and molecular mechanics- Poisson Boltzmann surface area continuum solvation (MM-PBSA) calculations
MD simulations were performed with the Gromacs 4.5.6 software package [29] and the ligand topologies were parameterized with the Gromos54a7 force field [30,31] on the ATB server [[32], [33], [34]]. The solvated and equilibrated protein-ligand complexes were subjected to 25 ns production phase MD simulations on the remote server of the Bioinformatics Resources and Applications Facility (BRAF), C-DAC, Pune. Further, the 25 ns trajectories were subjected to MM-PBSA calculations [35,36]. The details of the MD and MM-PBSA calculations are given in the supplementary file.
2.3. Network pharmacology of bioflavonoids
For the assessment of network pharmacology three major steps were performed: (a) prediction of bioactive targets, (b) enrichment analysis of regulated targets, and (c) construction of network between bioactives, targets, and pathways and its analysis. Briefly, targets of bio-actives were predicted using DIGEP-Pred [37] at the pharmacological activity (Pa) of 0.5 and the modulated proteins were enriched using STRING [38] ver 11.0 for their cellular components, biological processes, molecular function, and KEGG (https://www.genome.jp/kegg/) pathway database. Similarly, the network between bioactives, their targets, and modulated pathways was constructed using Cytoscape [39] ver 3.5.1; any duplicates were removed and the whole network was analyzed based on edge count for color map and node size.
2.4. Prediction of probable anti-viral activity
The biological spectrum of all the flavonoids was queried using PASS [40] at the pharmacological activity (Pa) > pharmacological inactivity (Pi) and a complete dataset was constructed. Then a complete dataset was queried for the keyword “viral” to identify the probable anti-viral activities against multiple viruses.
3. Results and discussion
3.1. Molecular docking studies
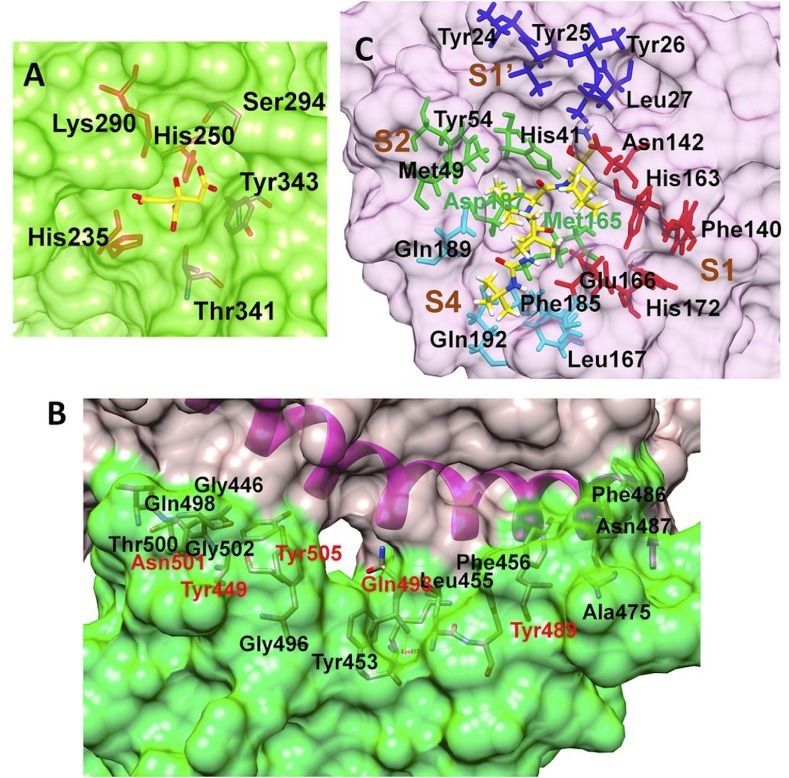
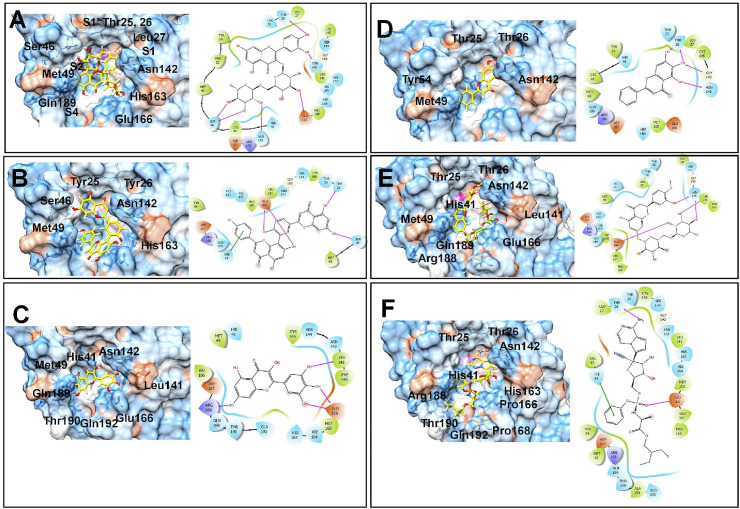
The open reading frame 1 ab (ORF1ab) located at 5’ terminus of the SARS-CoV-2 genome encodes various non-structural proteins (NSP1-16) [41]. The NSP15 protein, a NendoU, is a monomer assemble into hexamer, generated by a dimer of trimers [42]. The crystal structure of NSP15 (PDB ID: 6W01) has been solved with the resolution of 1.90 Å and was found suitable for docking studies. Each monomer has 348 amino acid units and constitutes a catalytic C-terminal nidoviral RNA uridylate-specific endoribonuclease (NendoU) domain, a middle domain, and an N-terminal domain. However, 23 residues are missing in the crystal structure in each monomeric chain and these residues were modeled through the Prime module. The NendoU domain has the endoribonuclease activity which is responsible for cutting the double-stranded (ds) RNA substrates. The C-terminal catalytic domain comprises of two antiparallel β-sheets containing six key amino acids His235, His250, Lys290, Thr341, Tyr343, and Ser294 (Fig. 1 ). The residues His235, His250, Lys290 form a catalytic triad where His235 serves as an acidic residue while His250 and Lys290 serve as a basic residue. The residues Ser294 and Tyr343 govern the uridylate-specific endoribonuclease specificity. A citrate ion is bound at this site and this binding site is exploited in the docking studies.
Fig. 1.
Binding site residues (A) SARS-CoV-2 NSP15 protein (PDB ID: 6W01); (B) SARS-CoV-2 spike RBD (PDB ID: 6M0J) (Spike RBD and ACE-2 surfaces shown in Green and rosy red color respectively); (C) SARS-CoV-2 main protease (Mpro) (PDB ID: 6WNP) (Color scheme for pocket residues: S1 red stick, S1′ blue stick, S2, green stick and S4 pocket cyan stick). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Another target protein explored in this computational study is the SARS-CoV-2 spike RBD. SARS-CoV-2 uses this fusion spike glycoprotein to make entry into the host cells through the ACE-2 receptor [43,44]. The spike glycoprotein has two subunits S1 and S2 which mediate attachment and membrane fusion respectively. The binding of the S1 subunit to the ACE-2 receptor results in the transition of the S2 subunit to a highly stable post-fusion conformation [45,46]. The RBD S1 subunit has 5 twisted β sheets antiparallel to each other designated as β1, β2, β3, β4, and β7 sheets. The region between β4 and β7 strands has extended insertions of short β5 and β6 strands, α4 and α5 helices, and some loops. This extended insertion is called a receptor-binding motif (RBM) and it has most of the residues which bind to ACE-2 [43]. Our recent report [4] confirmed that the residues Lys417, Gly446, Tyr449, Tyr453, Leu455, Phe456, Ala475, Phe486, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501, Gly502, Tyr505 are crucial for binding with N-terminal peptidase domain of ACE-2. However, from the cluster of these residues Gln493, Asn501, Tyr449, Tyr489, and Tyr505 could form the hydrogen bonding interaction, whereas Lys417 forms a salt bridge interaction.
The two overlapping poly-proteins (pp1a, pp1ab) encoded by ORF 1 ab are cleaved into NSP1-16 by the main protease (Mpro) and the papain-like protease (PLpro) [[47], [48], [49]]. Amongst these two proteases, Mpro has gained much attention in drug development due to its key function in viral replication [47]. Numerous crystal structures of SARS-CoV-2 Mpro are available, amongst which the crystal structure (PDB ID: 6WNP) having the resolution 1.44 Å was found most suitable. This crystal structure also has a peptide inhibitor Boceprevir covalently bound to Cys145 residue at its binding site. The main protease forms a dimer of two protomers and each protomer has three domains i.e. domain I, II, and III comprising the residues 8–101, 102–184, and 185–306 respectively. The binding site is located at the cleft between domains I and II. The residues Cys145 and His41 form the catalytic dyad. The binding site has four pockets S1, S1′, S2, and S4. The S1 pocket has the residues Phe140, Asn142, Glu166, His163, Leu141, and His172, while the adjacent S1’ pocket is surrounded by hydrophobic residues Tyr24, Thr25, Thr26, and Leu27. The S2 pocket is surrounded by residues His41, Met49, Tyr54, Met165, and Asp187, while the S4 pocket is surrounded by residues Leu167, Phe185, and Gln192.
The docking results of five top-ranked flavonoids based on the lowest docking scores for each protein are discussed in the case of each protein. The docking results for SARS-CoV-2 NSP15 Endoribonuclease are given in Table 1 and supplementary file- Table S1.
Table 1.
Molecular docking score and binding free energy for the protein-ligand complexes calculated by MM-PBSA analysis, all energies are in kJ.mol−1 with standard deviation in parenthesis.
| Compounds | Dock Score | MM-PBSA (ΔGbind) |
|---|---|---|
|
SARS-CoV-2 NSP-15 endoribonuclease (PDB ID: 6W01) | ||
| Glycyrrhizic acid | −8.168 | −97.599 (±32.621) |
| Baicalin | −7.564 | −47.332 (±17.273) |
| Rutin | −5.575 | −52.561 (±18.521) |
| Remdesivir |
−5.636 |
−62.637 (±20.132) |
|
SARS-CoV-2 spike RBD (PDB ID: 6M0J) | ||
| Mulberroside | −7.121 | −59.846 (±21.624) |
| (R)-Amygdalin | −6.978 | −14.521 (±19.927) |
| Remdesivir |
−4.652 |
−65.198 (±19.800) |
|
SARS-CoV-2 main protease (Mpro) (PDB ID: 6WNP) | ||
| Rutin | −8.859 | −129.402 (±23.245) |
| Amentoflavone | −7.589 | −134.358 (±19.769) |
| Remdesivir | −7.766 | −111.488 (±14.177) |
ΔGbind = Binding free energy (kJ.mol−1), Dock Score = Glide Score.
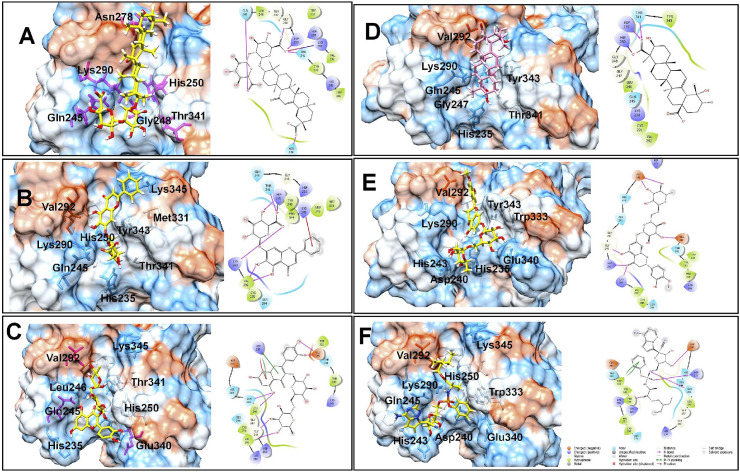
The docking results on the NSP15 endoribonuclease of SARS-CoV-2 revealed that the compounds glycyrrhizic acid, baicalin, rutin, ilexgenin A, hesperidin have the lower docking scores in the range −8.168 to −5.29; while the docking score for the standard drug remdesivir is −5.636. The key residues at the binding site of NSP-25 belong to the C-terminal catalytic domain catalytic triad His235, His250, Lys290, and Thr341, Tyr343, and Ser294. It would be worthwhile to investigate how the compounds interact with the catalytic triad residues and other residues. Glycyrrhizic acid was found most active with the lowest docking score of −8.168 and the carboxylate anion on the sugar moiety forms a hydrogen bond interaction with protonated His250 residue and also forms a salt bridge interaction with basic Lys290 residue (Fig. 2 ). Another carboxylate anion forms a hydrogen bond with Gln245 residue, while the carboxylate anion on the aglycone part forms a hydrogen bond with Asn278. The hydrophobic pentacyclic aglycone moiety of glycyrrhizic acid binds at the hydrophobic pocket surrounded by hydrophobic residues Leu346, Tyr343, Val292 and Trp333 and some charged residues such as Lys345, Hip235, and polar residue Thr341. The Thr341 and Gly248 also form hydrogen bond interactions. Baicalin has the docking score −7.564 and it forms the π-cation interaction between the phenyl ring substituted on chromone ring and positively charged Lys345 residue. The salt bridge interaction is formed between the anionic carboxylate group on sugar moiety and protonated His235 residue. Other residues such as polar Thr341, positively charged Lys290, and hydrophobic Val292 also form hydrogen bonds with the hydroxyl groups. The chromone ring of rutin forms a π-π stacking as well as π-cation interaction with the protonated His235. The hydroxyl groups on the phenyl ring form hydrogen bond interaction with Glu340; while the hydroxyl group at the 5th position of the chromone ring forms hydrogen bond interaction with polar Gln245 and hydrophobic Leu246 residues. The hydroxyl groups of sugar moiety form hydrogen bonds with Gly248 residue. Ilexgenin A, a hydrophobic pentacyclic triterpenoid core ring containing compound, has two carboxylic acid groups both of which are deprotonated at pH 7.4 as predicted by the Epik module. One of the carboxylate ions forms an ionic salt bridge interaction with protonated His250 residue and hydrogen bond interaction with polar Thr341 and protonated His241 residues. The carbonyl oxygen and a hydroxyl group at the 5th position of the chromone ring in hesperidin form hydrogen bonds with Lys290 and protonated His250 residues respectively; while the hydroxyl groups on sugar moieties form hydrogen bonds with Glu340 and Asp240 residues. Interestingly, it was found that the reference drug remdesivir forms the π-π stacking interaction with hydrophobic Trp333 residue. The hydroxyl group on ribose moiety forms a hydrogen bond with negatively charged Asp240 residue. The phosphate oxygen atoms form hydrogen bonds with Gln245 and protonated His235, while the ester carbonyl group forms a hydrogen bond with Pro344 residue. These docking results suggest that the hydrogen bond interactions with the residues forming a catalytic triad His235, His250, Lys290 are important for the binding affinity.
Fig. 2.
Binding poses of five top-ranked compounds and remdesivir at the binding site of SARS-CoV-2 NSP15 Endoribonuclease and 2D interaction diagrams. (A) Glycyrrhizic Acid, (B) Baicalin, (C) Rutin, (D) Ilexgenin A, (E) Hesperidin, and (F) Remdesivir.
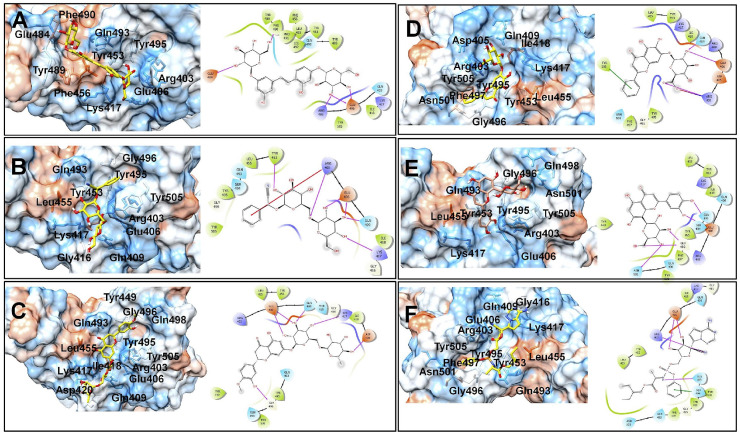
In the case of SARS-CoV-2 spike RBD, the docking scores for five top-ranked compounds from lower to higher values were in the order mulberroside < (R)-amygdalin < hesperidin < baicalin < orientin. This suggests the most favorable binding affinity of SARS-CoV-2 spike RBD is for the compound mulberroside (Supplementary file- Table S2). The hydroxyl groups on the sugar moieties in mulberroside form hydrogen bonds with negatively charged Glu484 and Glu406 residues and positively charged Arg403 residue (Fig. 3 ). The central phenyl chalcone core accommodates in the hydrophobic pocket surrounded by residues Pro491, Leu492, Gln493, Tyr495, Tyr453, and Leu455. In the case of (R)-amygdalin, the phenyl ring forms π-cation interaction with positively charged Arg403 residue. Arg403 also forms a hydrogen bond with the glycosidic linkage oxygen atom. The hydroxyl groups on sugar moiety form hydrogen bonds with Lys417, Gln409, Glu406, and Tyr453 residues. The hydroxyl group on the phenyl ring in hesperidin forms a hydrogen bond with Gly496; while the hydroxyl groups on sugar moieties form a hydrogen bond with negatively charged Glu406, Asp420, and positively charged Lys417. In the case of baicalin the phenyl ring substituted on chromone ring forms π- π stacking interaction with Tyr505; while the hydroxyl group on sugar moiety forms a hydrogen bond with Gln409 and negatively charged Glu406 residue. The deprotonated anionic carboxylate forms a salt bridge with positively charged Arg403 residue. Only hydroxyl groups on sugar moieties in the case of Orientin forms hydrogen bond interaction with Asn501, Gly496, Ser494, Gln493 residues.
Fig. 3.
Binding poses of five top-ranked compounds and remdesivir at the binding site of SARS-CoV-2 Spike RBD. (A) Mulberroside, (B) (R)-Amygdalin, (C) Hesperidin, (D) Baicalin, (E) Orientin, and (F) Remdesivir.
The phenyl ring in remdesivir also forms π- π stacking interaction with Ser494 residue, while the hydroxyl groups on ribose form hydrogen bonds with negatively charged Glu406 residue. The nitrogen from the cyano group forms a hydrogen bond with positively charged Arg403 residue, while the nitrogen atom from the phosphamidon group forms a hydrogen bond with Gln493 residue. Most of these interactions were with the residue clusters from the interface of spike RBD and ACE-2 as mentioned earlier. Many of the favorable interactions were found with some important residues such as Gln493, Asn501, Tyr449, Tyr489, Tyr505, and Lys417.
The docking results for SARS-CoV-2 main protease (Mpro) showed that rutin has the best docking score (−8.859) amongst all flavonoids (Supplementary file- Table S3). Other flavonoids namely amentoflavone, myricetin, baicalein, hesperidin have the docking scores in the range −7.589 to −7.137. The reference drug remdesivir has a docking score of −7.766 which points out the stronger binding affinity of rutin than the reference drug. The binding pose of best docked rutin conformer was found to form the hydrogen bond with two residues deprotonated Glu166 and polar Asn142 at S1 pocket, while at S1′ pocket it forms hydrogen bond interaction with Thr26 (Fig. 4 ). The 3,4 dihydroxyl groups on phenyl substituent form these hydrogen bonds at both S1 and S1′ pockets with Asn142 and Thr26 residue. The hydroxyl groups on α-L-rhamnopyranose moiety form hydrogen bonds with polar Ser46 residue at S2 pocket and polar Gln189 residue at S4 pocket. Thus, rutin was found to occupy the binding site and forming important hydrogen bond interactions at all the pockets of SARS-CoV-2 main protease (Mpro). Remdesivir, on the other hand also occupies all the pockets, but it forms hydrogen bond interactions with S1 pocket residue Glu166 and S1′ pocket residue Thr26. The phenyl ring substituted on phosphoramide forms a π- π stacking interaction with S2 pocket protonated residue His41. Amentoflavone forms hydrogen bond interactions with S1 pocket residue Glu166, S1′ pocket residue Thr25, S2 pockets residues His41, and Ser46. It also forms a π- π stacking interaction with protonated His41 residue. Myricetin forms the hydrogen bond with Glu166 and Leu141 residues at S1 pocket and Arg188 residue at S2 pocket. Baicalein, with hydroxyl groups on the core chromone ring, form hydrogen bond interaction with S1 and S1’ pocket residues Asn142, Gly143, and Thr26. Hesperidin, a close analog of rutin, forms a hydrogen bond with S1 pocket residues Asn142, Leu141, and Glu166. These results suggest that the flavonoid rutin has a better binding affinity due to interactions with key residues at all the pockets of the binding site of SARS-CoV-2 main protease (Mpro).
Fig. 4.
Binding poses of five top-ranked compounds and remdesivir at the binding site of SARS-CoV-2 Main protease (Mpro). (A) Rutin, (B) Amentoflavone, (C) Myricetin, (D) Baicalein, (E) Hesperidin, (F) Remdesivir.
3.2. Molecular dynamics studies and MM-PBSA calculations
Molecular dynamics simulation (MDS) is known to provide binding mode of ligand accurately than the molecular docking. MDS of sufficiently long duration offers the conformational sampling of ligand as well as protein backbone and side-chain atoms. On the other hand, it also provides the essential information in terms of the various non-bonded interactions and consequent energetic of solvated system. Further, it captures the protein folding events, the influence of loop flexibility, and biding site adaptation. This eliminates the limitations of docking studies and provides accurate estimates of the binding free energy and binding affinity. In the present study, 25 ns MDS studies were performed to evaluate the conformational stability, binding free energy, and binding affinity of the compounds with the best docking scores, and the results were compared with reference drug remdesivir. In the case of SARS-CoV-2 NSP-15 endoribonuclease, the top-ranked flavonoids glycyrrhizic acid, baicalin, rutin were selected for MDS; while with the SARS-CoV-2 spike, RBD the flavonoids mulberroside and (R)-amygdalin were selected. In the case of SARS-CoV-2 main protease, the flavonoids rutin and amentoflavone were selected. The MDS of the reference drug remdesivir was also performed. During the MDS stearic clashes were relieved by subjecting the system to the initial steps of energy minimization by steepest descent criteria. The minimized system was subjected to equilibration steps at NPT and NVT conditions. The equilibrated system was subjected to 25 ns production phase MDS and the post MDS analysis which includes analysis of various non-bonded interactions such as H-bond, π stacking and hydrophobic interactions. In order to understand the stability of the system during the MDS timescale, the parameters such as root mean square deviation (RMSD) and root mean square fluctuation (RMSF) for ligands as well as protein were measured. The post MDS trajectories were used in Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) calculations to estimate the binding free energies.
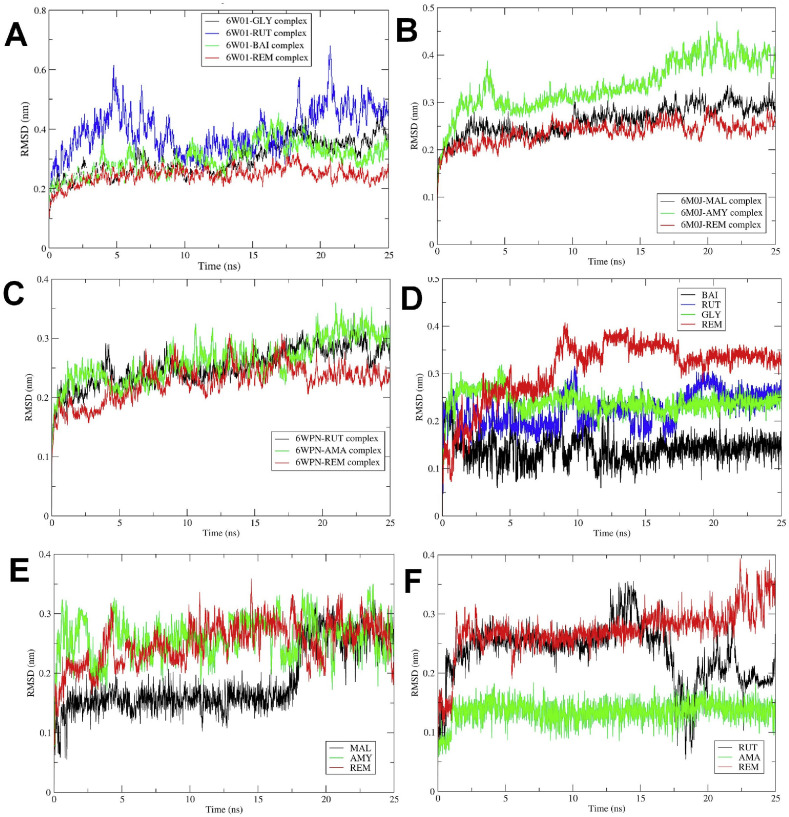
The RMSD is a good measure of conformational stability of protein and ligands and its a measure of extent of deviation in the position of atoms from the starting position. The lower the deviation better is the conformational stability [50]. The RMSD analysis for protein atoms showed that the complex of SARS-CoV-2 NSP-15 endoribonuclease with remdesivir has the lowest RMSD with an average of 0.24 nm (Fig. 5 ). The complex with glycyrrhizic acid has almost equal RMSD as that of remdesivir till 15 ns, but thereafter it increases and the average RMSD is 0.29 nm. The complex of baicalin stabilizes quickly with an average RMSD of 0.299 nm. The complex of rutin has the highest deviations with an average RMSD of 0.39 nm. It was found that though the remdesivir docking score is higher, its complex is stabilized quickly as compared to other complexes, and glycyrrhizic acid and baicalin has almost similar deviations in protein atoms. In the case of SARS-CoV-2 spike RBD, mulberroside and (R)-amygdalin has the best docking scores. The RMSD in protein atoms for these two compounds and the reference drug remdesivir showed that the mulberroside and remdesivir have almost similar deviations in protein atoms with average values of 0.26 and 0.23 respectively suggesting better stabilization in both the cases [51]. While the RMSD for a complex of (R)-amygdalin is having a slightly higher average of 0.33 suggesting lower stability as compared to the other two complexes. In the case of SARS-CoV-2 main protease, the compounds i.e. rutin and amentoflavone, showed better docking scores, and remdesivir has very close RMSD values of 0.25, 0.26, and 0.22 respectively. Moreover, these results are supporting the docking scores.
Fig. 5.
Root Mean Square Deviations. RMSD in protein backbone atoms of - (A) NSP-15 endoribonuclease; (B) Spike RBD; and (C) Main protease (Mpro). RMSD in ligand atoms - (D) NSP-15 endoribonuclease; (E) Spike RBD; and (F) Main protease (Mpro) (MAL: Mulberroside, AMY: (R)-Amygdalin, REM: Remdesivir, RUT: Rutin, AMA: Amentoflavone).
The measurement of RMSD in ligand atoms is also important in judging the overall stability of the protein-ligand complexes. In the case of the complexes of SARS-CoV-2 NSP-15 endoribonuclease with ligands, baicalin atoms have the least deviations with an average RMSD value of 0.14 nm, while the remdesivir atoms have the higher deviations with an average RMSD of 0.30 nm (Fig. 5D). The flavonoids glycyrrhizic acid and rutin have an average RMSD of 0.24 and 0.22 nm respectively. It was found that the RMSD is proportional to the number of rotatable bonds in the molecules. For instance, baicalin with 4 rotatable bonds has the lowest RMSD value, while the remdesivir with 14 rotatable bonds has the highest RMSD value. For glycyrrhizic acid and rutin with 7 and 6 rotatable bonds respectively, the RMSD values vary proportionally. In the case of the complexes of SARS-CoV-2 spike RBD with ligands, all the ligands were found adopting stable conformation with RMSD of around 0.25 nm at the end of 25 ns MDS. Interestingly, in the case of mulberroside, till around 17.5 ns the RMSD is almost stable with a lower RMSD value of around 0.15 nm, but thereafter it quickly raises to around 0.25 nm and remains stable till the end of the simulation. In the case of the complexes of SARS-CoV-2 main protease (Mpro), it can be seen that the flavonoid amentoflavone has the least deviations from the starting structure with an average RMSD of 0.13 nm, suggesting the stability of its complex with the main protease. While, the reference drug remdesivir has a higher but constant deviation with RMSD of around 0.25 nm till 20 ns, but thereafter it rises to around 0.35 nm towards the end of the simulation. This suggests larger deviations in the starting structure of remdesivir. Interestingly, rutin also has a higher initial RMSD of around 0.25 nm, but the RMSD drops to a stable conformation with RMSD around 0.2 nm soon after 20 ns simulation.
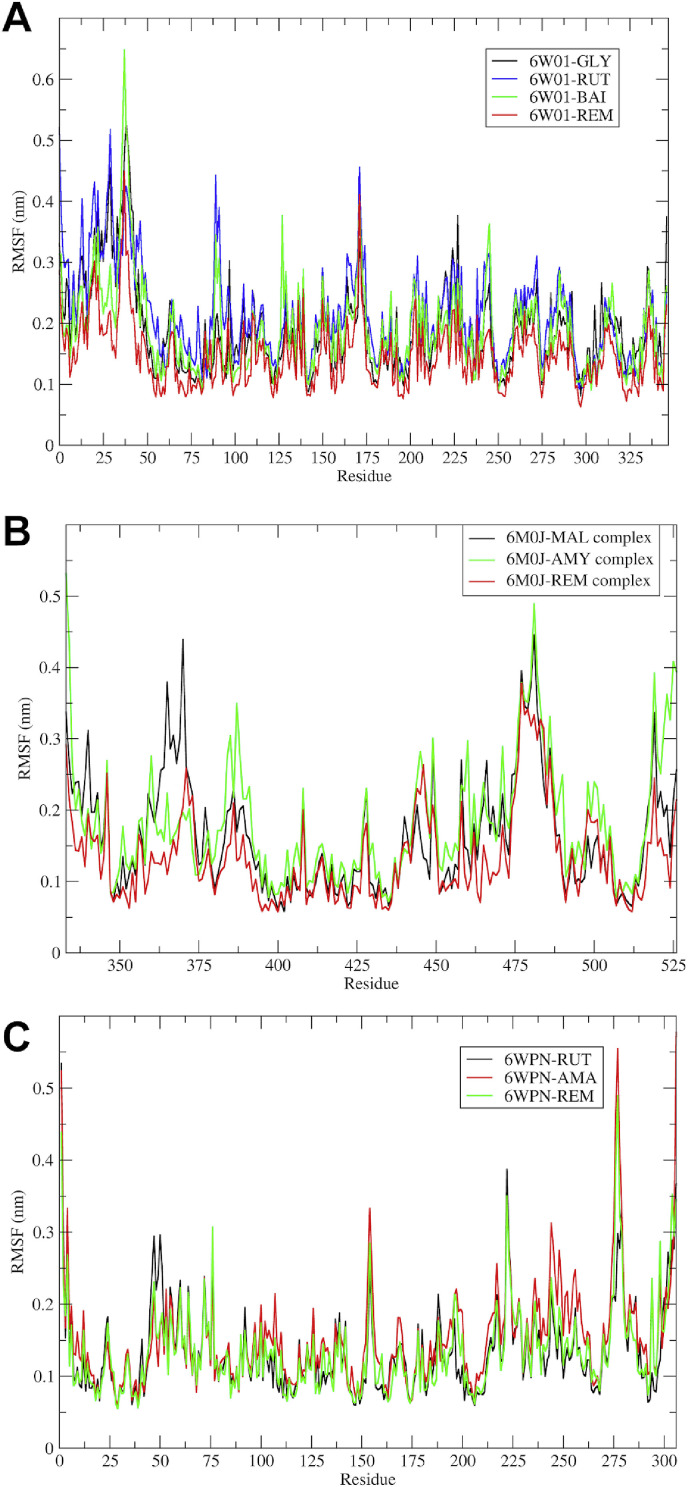
The stability of protein-ligand complexes can be evaluated in terms of Root Mean Square Fluctuations (RMSF) in the atoms of protein residues [52]. It is a good measure of elasticity of the protein residues and points out the binding site adaptation and other phenomena. In the case of the SARS-CoV-2 NSP-15 endoribonuclease, the fluctuations in a cluster of residues were evident with rutin and baicalin; while these were minimal with glycyrrhizic acid and remdesivir (Fig. 6 ).
Fig. 6.
RMSF in residues of proteins. (A) Complexes with SARS-CoV-2 NSP-15 endoribonuclease; (B) complexes with SARS-CoV-2 spike RBD (MAL: Mulberroside, AMY: (R)-Amygdalin, and REM: Remdesivir); and (C) Complexes with SARS-CoV-2 main protease (Mpro) (RUT: Rutin, AMA: Amentoflavone).
Maximum fluctuations were seen amongst the cluster of N-terminal residues ranging from 1 to 50 and to some extent minor fluctuations were seen in the binding site residues in the range 225–346. In the SARS-CoV-2 spike RBD-ligand complexes, the cluster of residues from 350 to 375 from the N-terminal region and binding site residues from 375 to 500 had higher fluctuations. The fluctuations in the N-terminal residues were minimal as observed with remdesivir and maximal with (R)-amygdalin, while the fluctuations at the binding site residues were almost similar to all the compounds. In the case of the complexes of SARS-CoV-2 main protease (Mpro), the fluctuations in the binding site residues 25–50, 150–200, and C-terminal loop region residues 275–306 were observed with all the three compounds.
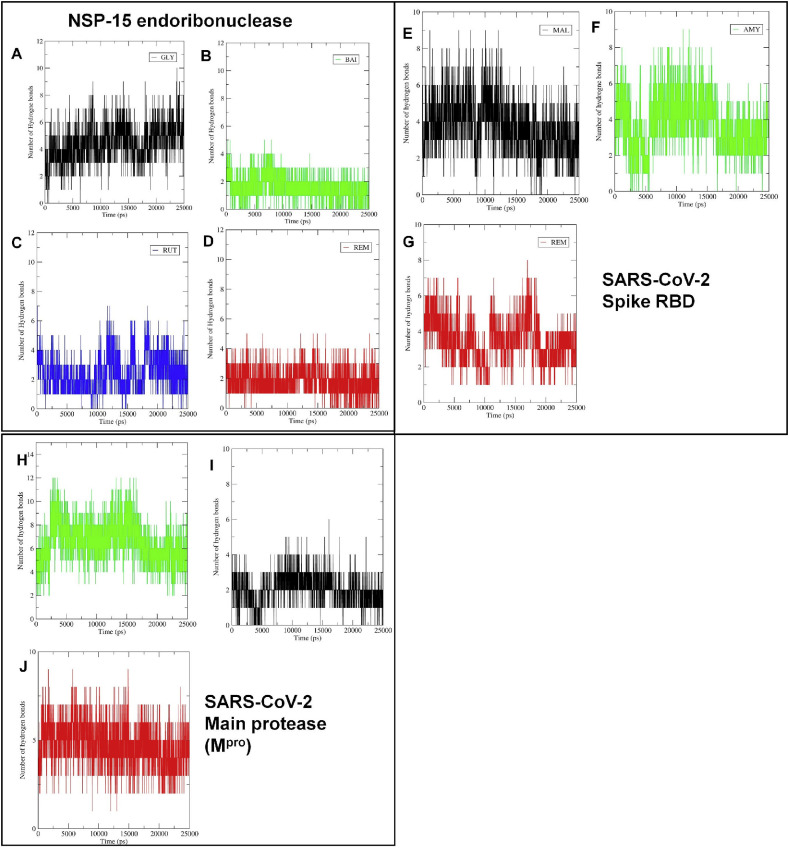
The binding affinity of ligands depends on the non bonded interactions such as hydrogen bond interactions, hydrophobic and ionic interactions [53]. The number of hydrogen bonds formed, and their lifetime indicates the strength and binding affinity of the complexes. The hydrogen bond formation was critically evaluated for all protein-ligand complexes. In the case of SARS-CoV-2 NSP-15 endoribonuclease, the maximum 10 hydrogen bonds were formed with glycyrrhizic acid, while with rutin 7 hydrogen bonds were formed (Fig. 7 ). In the case of baicalin and remdesivir, 5 hydrogen bonds were formed. The estimate of the average number of hydrogen bonds formed during the entire simulation period may be crucial in judging which molecule is constantly forming more number of hydrogen bonds [54]. Glycyrrhizic acid, rutin, baicalin, and remdesivir form an average of 4.45, 2.41, 1.57, and 1.85 number of hydrogen bonds respectively. Thus, glycyrrhizic acid is forming approximately 4 hydrogen bonds constantly throughout the MDS, while other compounds form approximately 1–2 hydrogen bonds constantly. Further, the residues involved in hydrogen bond formation and the lifetime of hydrogen bonds were studied. In this case, a program PyContact [55], a GUI-based tool for analysis of non-covalent interactions was used. The hydrogen bond formation was analyzed with a cut off the radius of 0.35 nm and an angle cut off of 120° criteria [56]. In the case of glycyrrhizic acid, the residues Val292 and Tyr343 form the hydrogen bonds with the longest lifetime, while in the case of baicalin the residue Lys290 forms a hydrogen bond with the longest lifetime (Supplementary file S4- Figure S2). The flavonoid rutin forms the hydrogen bonds with the residues Gln245, Lys290, and Tyr343 with the longest lifetime.
Fig. 7.
Number of hydrogen bonds formed between the molecules and the residues at the binding site. SARS-CoV-2 NSP-15 endoribonuclease (A) Glycyrrhizic acid, (B) Baicalin, (C) Rutin, (D) Remdesivir; SARS-CoV-2 Spike RBD (E) Mulberroside, (F) (R)-Amygdalin, (G) Remdesivir; SARS-CoV-2 Main protease (Mpro). (H) Rutin, (I) Amentoflavone, (J) Remdesivir.
The reference drug remdesivir forms hydrogen bonds with residues Gly248, His235, Lys290, Gln245, Tyr243, His250, and Gly247 with the longest lifetime. These results suggest that remdesivir could produce hydrogen bonds with many key residues at the binding site. It was also observed that the residues Lys290, Tyr343, and Gln245 are important in hydrogen bond formation.
In the case of SARS-CoV-2 spike RBD, the maximum 8 hydrogen bonds were formed with remdesivir; while mulberroside and (R)-amygdalin forms 7 hydrogen bonds each. However, the average number of hydrogen bonds formed with remdesivir, mulberroside, and (R)-amygdalin were 3.52, 3.81, and 3.70 respectively, which suggest that hydrogen bond formation events happen more frequently with mulberroside. The analysis of residue wise hydrogen bond lifetime suggests that the Tyr120, Leu122, Tyr156, Tyr162, and Arg70 residues form hydrogen bonds consistently with longer lifetime (Supplementary file- Figure S3). (R)-Amygdalin residues Arg70 and Glu73 form such hydrogen bonds with longer lifetime. Interestingly, remdesivir forms several such hydrogen bonds with residues Glu160, Tyr120, Tyr162, Ser161, Arg70, and Gly163. These results point to the importance of Tyr120, Tyr162, and Arg70 residues in these compounds. Depending on the consistency of hydrogen bonds formed, remdesivir may have a stronger binding affinity than mulberroside.
In the case of the complexes of SARS-CoV-2 main protease (Mpro), rutin, amentoflavone, and remdesivir form 12, 7, and 9 maximum number of hydrogen bonds respectively. The average number of hydrogen bonds formed during the period of simulation for rutin, amentoflavone, and remdesivir was 5.95, 2.05, and 4.75 respectively. Thus, it is evident that rutin forming a greater number of hydrogen bonds consistently compared to amentoflavone and remdesivir. Rutin may have a more favorable binding affinity due to such hydrogen bond formation than the other two compounds. The hydrogen bond formation pattern at the binding site in terms of hydrogen bond lifetime was investigated which suggests that rutin could form hydrogen bonds with the number of residues at the binding site of SARS-CoV-2 main protease (Mpro) (Supplementary file- Figure S4). Interestingly, the residues from all the four pockets were found involved in such key hydrogen formation. Especially, the S1 pocket residues Glu166, Gly143 and Cys145, S1’ pocket residues Leu27, Thr26, Thr25, S2 pocket residues Cys44, His41, Met165, and Met49 and S4 pocket residue Gln189 were found forming hydrogen bonds consistently. In the case of amentoflavone, these residues and S1 pocket residue His163 were found involved in hydrogen bond formation. Remdesivir forms hydrogen bonds with most of these residues and Thr190, His164, and Pro168 residues. These results suggest that the residues Glu166, His41, Gln189 are important in forming key hydrogen bond interactions.
MM-PBSA calculations carried out MD trajectories can provide more accurate estimates of binding affinity in terms of binding free energy. The g_mmpbsa program [35,36] is useful in calculating the non-bonded interaction energies such as van der Waal energy, electrostatic energy, polar solvation energy, Solvent Accessible Surface Area (SASA) energy, and these energy values are used to calculate the binding free energy (ΔGbinding) (Supplementary file- Table S4). The non-bonded interaction energies such as van der Waal energy and electrostatic energy in terms of Coulomb and Lennard-Jones (LJ) potential functions respectively, has a major influence on the binding free energy (ΔGbinding) estimate. In the case of MDS of SARS-CoV-2 NSP-15 endoribonuclease protein-ligand complexes, the MM-PBSA results showed that glycyrrhizic acid has the lowest van der Waal energy and has the lowest binding free energy of −97.599 kJ/mol. In comparison to the reference drug remdesivir, the binding free energy of glycyrrhizic acid is much lower. This may be in part due to the higher number of hydrogen bonds formed and consequently most favorable interactions at the binding site. This proves that the glycyrrhizic acid, amongst all the flavonoids, interacts most favorably interacts at the binding site of SARS-CoV-2 NSP-15 endoribonuclease. Baicalin and rutin having comparably lower ΔGbinding may have less favorable interactions than the reference drug. The interactions formed by glycyrrhizic acid and remdesivir when compared, it was found that glycyrrhizic acid forms polar interactions with the residues Asn278, Ser294, Cys293, Glu340, Leu246, Leu346, Lys346, Lys345, His243, and Trp333; while the reference drug remdesivir was observed not forming interactions with these residues (Supplementary file- Figure S5).
In the case of MDS of SARS-CoV-2 Spike protein-ligand complexes, the results of MM-PBSA calculations showed that remdesivir has the lowest binding free energy as compared to the other two flavonoids. However, mulberroside was found to have better van der Waals interactions than remdesivir. But the results are not so conclusive in deciding the better binding affinities of flavonoids to this target protein.
In the case of MDS of SARS-CoV-2 Main protease (Mpro) protein-ligand complexes, Amentoflavone was found to have better lower binding free energy than the remdesivir. But only polar solvation energy contributions differ considerably in the final binding free energies.
3.3. Network pharmacology
3.3.1. Targets of bio-actives
A total of 199 proteins were modulated by 15 flavonoids in which myricetin and scutellarein were major protein regulators i.e. 28 (Table 2 ). Similarly, PLAU and TIMP1 proteins were most regulated proteins by the maximum number of bio-actives i.e. 11 (Table 3 ).
Table 2.
Bioactives and their count to regulated targets.
| Bioactives | Count of targeted proteins | Bioactives | Count of targeted proteins |
|---|---|---|---|
| (R)-Amygdalin | 8 | Lycorine | 1 |
| Amentoflavone | 24 | Mulberroside | 15 |
| Baicalein | 22 | Myricetin | 28 |
| Baicalin | 4 | Orientin | 13 |
| Glycyrrhizic Acid | 5 | Puerarin | 8 |
| Hesperetin | 19 | Rutin | 13 |
| Hesperidin | 9 | Scutellarein | 28 |
| Ilexgenin A |
2 |
||
| Total (Count of targeted proteins): 199 | |||
Table 3.
Targets and their count to regulators.
| Proteins | Count of Bioactives | Proteins | Count of Bioactives | Proteins | Count of Bioactives | Proteins | Count of Bioactives |
|---|---|---|---|---|---|---|---|
| AR | 9 | EPAS1 | 4 | MMP2 | 5 | PRDX4 | 5 |
| CASP8 | 4 | ESR2 | 1 | MMP3 | 3 | PRKCA | 1 |
| CAT | 6 | FKBP5 | 1 | MMP7 | 7 | SIRT1 | 4 |
| CBR1 | 1 | FLT1 | 2 | NFE2L2 | 6 | SMN2 | 10 |
| CD83 | 3 | GADD45B | 1 | NOS2 | 7 | TIMP1 | 11 |
| CD86 | 1 | GSS | 1 | NPPB | 5 | TNFRSF1A | 8 |
| CHEK1 | 6 | GYPA | 1 | PGR | 5 | TOP2A | 1 |
| CTNNB1 | 7 | HMOX1 | 9 | PLAT | 9 | TP53I3 | 2 |
| CYP1A1 | 4 | KLK3 | 5 | PLAU | 11 | TP63 | 1 |
| CYP1A2 | 1 | KRT1 | 6 | PPARA | 2 | TP73 | 5 |
| EGLN1 |
2 |
MDM2 |
5 |
PRDX2 |
4 |
VDR |
7 |
| Total: 199 | |||||||
3.3.2. GO enrichment and network analysis
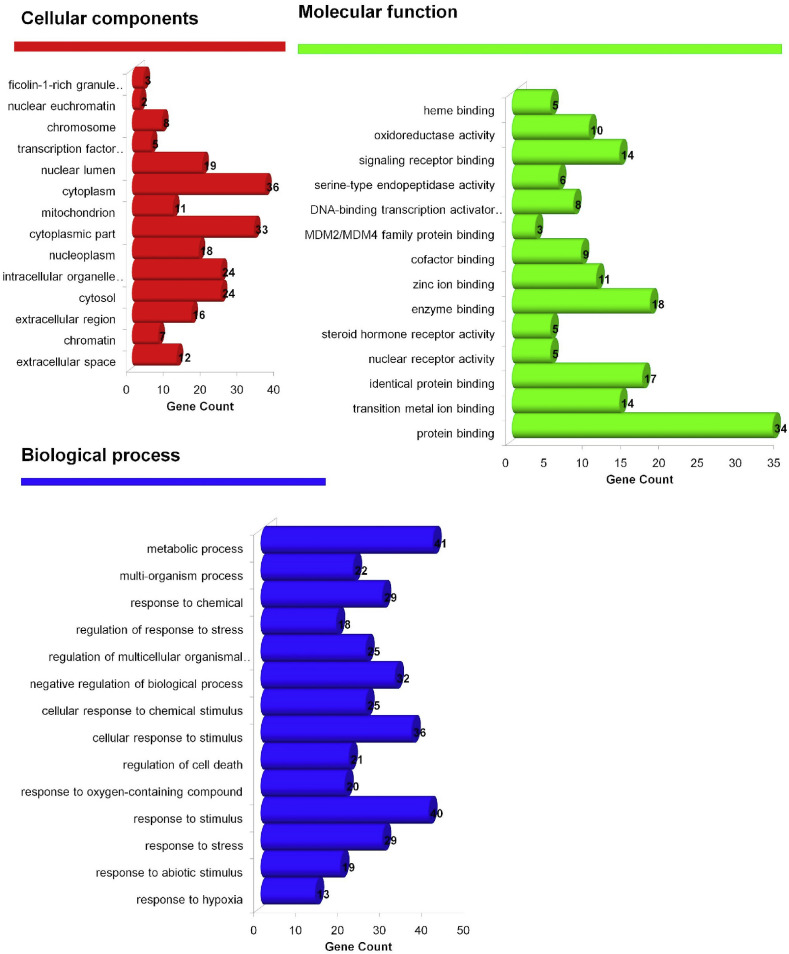
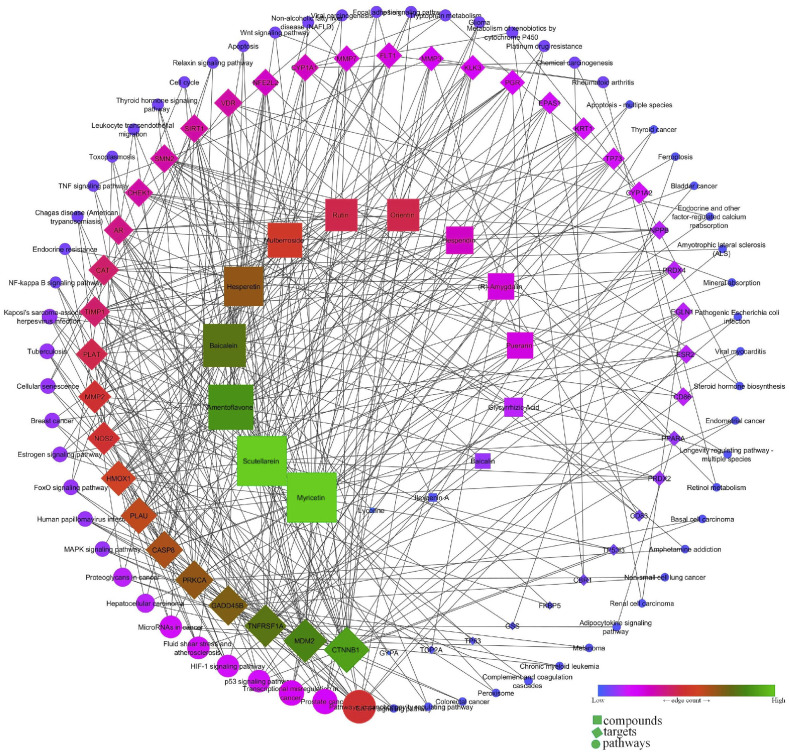
Gene ontology analysis identified 565 biological processes in which response to hypoxia (GO:0001666) scored the lowest false discovery rate via the modulation of 13 genes (CAT, CYP1A1, EGLN1, EPAS1, HMOX1, MDM2, MMP2, NFE2L2, NOS2, PLAT, PLAU, PPARA, SIRT1) against 288 background proteins. Similarly, 69 molecular functions were identified in which protein binding (GO:0005515) was majorly modulated with the lowest false discovery rate via the modulation of 34 genes (AR, CASP8, CAT, CHEK1, CTNNB1, CYP1A1, CYP1A2, EGLN1, EPAS1, ESR2, FKBP5, FLT1, GSS, GYPA, HMOX1, KRT1, MDM2, NFE2L2, NOS2, NPPB, PGR, PLAT, PPARA, PRDX4, PRKCA, SIRT1, SMN2, TIMP1, TNFRSF1A, TOP2A, TP53I3, TP63, TP73, VDR) against 6605 background proteins. Likewise, 14 cellular components were majorly modulated in which extracellular space (GO:0005615) scored minimum false discovery rate via the modulation of 12 genes (CD86, FLT1, HMOX1, KLK3, MMP2, MMP3, MMP7, NPPB, PLAT, PLAU, TIMP1, TNFRSF1A) against 1134 background proteins (Fig. 8 ). Similarly, KEGG analysis identified the modulation of 63 pathways in which pathways in cancer (hsa05200) was primarily modulated by regulating 14 proteins (AR, CASP8, CTNNB1, EGLN1, EPAS1, ESR2, GADD45B, HMOX1, KLK3, MDM2, MMP2, NFE2L2, NOS2, PRKCA) against 515 background proteins with the lowest false discovery rate (Supplementary file- Table S5). Similarly, the network interaction of bio-actives, modulated proteins, and regulated pathways reflected the myricetin and scutellarein with the highest number of proteins in which CTNNB1 was primarily modulated in a maximum of pathways chiefly modulating pathways in cancer (Fig. 9 ).
Fig. 8.
GO Analysis of flavonoid regulated genes.
Fig. 9.
Network interaction of flavonoids, their targets, and regulated pathways.
3.3.3. Prediction of probable anti-viral activity
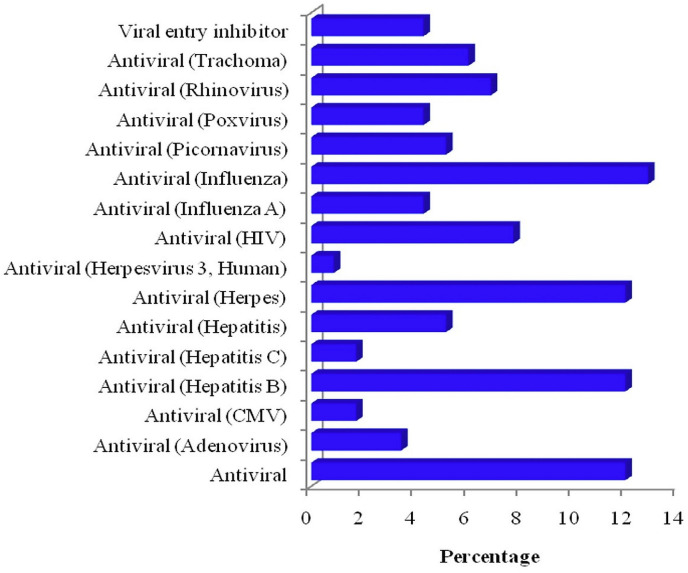
Prediction of the biological spectrum of bio-actives identified 16 different anti-viral activities for the keyword “viral” as anti-adenovirus, CMV, hepatitis B and C, hepatitis, herpes, HIV, influenza, picornavirus, poxvirus, rhinovirus, trachoma, viral entry inhibitor in which the combined action was integrated against ‘antiviral’ (Influenza). The antiviral activity of the combined action of selected flavonoids is presented in Fig. 10 .
Fig. 10.
Predicted anti-viral activity of selected flavonoids against multiple viruses.
Apart from the anti-viral spectrum of the selected bio-actives against the novel coronavirus, the study also aimed to investigate the probable regulation of multiple proteins via the compounds. This is because, during COVID-19 infection, it has been proposed for the deregulation of nutrient and oxygen supply in the affected tissue leading to necrosis which could be the basic pathogenesis observed in autopsy [57]. This process is complex and involved the up-/down-regulation of multiple proteins and regulation of numerous pathways by affecting various cellular components, molecular functioning, and biological process which can be traced via the assessment of gene ontology enrichment analysis. Similarly, the present study enriched the bio-actives regulated proteins to evaluate their synergistic or additive probability for multiple pathways in regulating immunity, inflammation, and oxidative stress.
The risk of COVID-19 infection is reported to be more in subjects with co-morbid multiple infectious diseases (tuberculosis, malaria, HIV, etc) and non-infectious diseases (diabetes, obesity, cancer, etc) due to compromised immunity [58,59]. Similarly, inflammation in the lungs and increased oxidative stress are also contributors to the progression of COVID-19 pathogenesis [60]. The selected bio-actives are the flavonoids, an important class of secondary metabolites with multiple beneficial values including immune booster, anti-inflammatory, and antioxidant activities. Hence, the present study utilized the GO analysis followed by network interaction of flavonoids to evaluate the above triplex action, which would be beneficial in the subjects with compromised immunity.
GO analysis identified the modulation of 565 biological processes in which response to hypoxia was majorly modulated. As explained previously, in COVID-19 infection, hypoxia in one of the major problems due to inadequate O2/CO2 exchanges [61]. Hence, regulation of this pathway could be beneficial in ameliorating the exchange of gas and nutritional supply in infected cells/tissue and could synergist other biological processes like a response to stress and stimulus and minimize reactive oxygen species (ROS) system and inflammation. During drug action, efficacy may be affected via the change in effective concentration at the site of action or via the alteration of the rate of drug elimination [62]. Similarly, in the present study, the combined synergistic/additive also regulated the protein binding which would contribute to the regulation of pharmacokinetic/pharmacodynamic parameters. The previous report also reports the role of extracellular vesicles in viral infection and transmission [63]. In the present study, proteins from extracellular vesicles were predicted to be most affected which could get activated in response to novel coronavirus infection. Similarly, the present KEGG analysis identified the modulation of multiple signaling pathways like p53, FoxO, MAPK, Wnt, Rap1, TNF, Adipocytokine, and Leukocyte transendothelial migration that are involved in immune boost, minimizing the oxidative stress and inflammation [[64], [65], [66], [67], [68], [69], [70], [71]]. Further, the selected bio-actives were also identified to modulate the multiple pathways which are involved in the multiple infectious and non-infectious pathogenesis and would add a beneficial effect in treating the subjects suffering from the same.
4. Conclusion
Bioflavonoids or polyphenolics are well known secondary metabolites possessing diverse activities including antiviral activity. In the present work, the possible antiviral properties of 15 bioflavonoids were investigated through in-silico approaches against SARS-CoV-2- proteins i.e. NSP15, RBD of S protein, and Mpro/3CLpro. The docking studies and MDS confirmed the promising potential of glycyrrhizic acid, baicalin, and rutin in inhibiting the n-CoV-2 key viral proteins. The network pharmacology analysis suggested the contribution of selected bioflavonoids in the modulation of multiple signaling pathways that could play a significant role in immunomodulation, minimizing the oxidative stress and inflammation. This computational assessment of bioflavonoids and network pharmacology work can lead researchers to take up and plan further experimental work on investigation of the inhibition of SARS-CoV-2 vital proteins with the potential secondary metabolite found in this study.
Ethical statement
This study doesn't include any animal or human study.
Funding support
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
SSG and MMW: Conceptualization, Supervision, Investigation;
RBP and RVC: Molecular Docking and Dynamics Studies;
PK and YDD: Network Pharmacology and Analysis;
RBP, RVC and SKS:Methodology, Software Data analysis;
SSG and NSG: Writing- Original draft preparation;
SKP and MA: Formal analysis, Visualization, Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Authors are highly grateful to Dr. B. M. Patil, Professor, Department of Pharmacology and Toxicology, KLE College of Pharmacy, Belagavi and Dr. P.B. Khedekar, Professor, Department of Pharmaceutical Sciences, R. T. M. University, Nagpur, Maharashtra, India for proofreading the article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100504.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sinha S.K., Prasad S.K., Islam M.A., Gurav S.S., Patil R.B., AlFaris N.A. Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J Biomol Struct Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chikhale R.V., Sinha S.K., Patil R.B., Prasad S.K., Shakya A., Gurav N., Prasad R., Dhaswadikar S.R., Wanjari M., Gurav S.S. In-silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J Biomol Struct Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1784289. [DOI] [PubMed] [Google Scholar]
- 3.Chikhale R.V., Gurav S.S., Patil R.B., Sinha S.K., Prasad S.K., Shakya A. SARS-CoV-2 host entry and replication inhibitors from Indian ginseng: an in-silico approach. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1778539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha S.K., Shakya A., Prasad S.K., Singh S., Gurav N.S., Prasad R.S., Gurav S.S. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhardwaj K., Palaninathan S., Alcantara J.M.O., Li Yi L., Guarino L., Sacchettini J.C., Kao C.C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adeoye A.O., Oso B.J., Olaoye I.F., Tijjani H., Adebayo A.I. Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1765876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug for corona virus: a systematic review. Indian J Pharmacol. 2020;52:56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: an overview. J Nutr Sci. 2016;5 doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia E.-Q., Deng G.-F., Guo Y.-J., Li H.-B. Biological activities of polyphenols from grapes. Int J Mol Sci. 2010;11:622–646. doi: 10.3390/ijms11020622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorlach S., Fichna J., Lewandowska U. Polyphenols as mitochondria-targeted anticancer drugs. Canc Lett. 2015;366:141–149. doi: 10.1016/j.canlet.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Iranshahi M., Rezaee R., Parhiz H., Roohbakhsh A., Soltani F. Protective effects of flavonoids against microbes and toxins: the cases of hesperidin and hesperetin. Life Sci. 2015;137:125–132. doi: 10.1016/j.lfs.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Nabavi S.F., Braidy N., Habtemariam S., Orhan I.E., Daglia M., Manayi A., Gortzi O., Nabavi S.M. Neuroprotective effects of chrysin: from chemistry to medicine. Neurochem Int. 2015;90:224–231. doi: 10.1016/j.neuint.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Parvez M.K., Tabish Rehman Md, Alam P., Al-Dosari M.S., Alqasoumi S.I., Alajmi M.F. Plant-derived antiviral drugs as novel hepatitis B virus inhibitors: cell culture and molecular docking study. Saudi Pharmaceut J. 2019;27:389–400. doi: 10.1016/j.jsps.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lani R., Hassandarvish P., Shu M.-H., Phoon W.H., Chu J.J.H., Higgs S., Vanlandingham D., Abu Bakar S., Zandi K. Antiviral activity of selected flavonoids against Chikungunya virus. Antivir Res. 2016;133:50–61. doi: 10.1016/j.antiviral.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S., Pandey A.K. Chemistry and biological activities of flavonoids: an overview. Sci World J. 2013;2013:1–16. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan W., Qian S., Qian P., Li X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112–116. doi: 10.1016/j.virusres.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Bae E.-A., Han M.J., Lee M., Kim D.-H. In vitro inhibitory effect of some flavonoids on rotavirus infectivity. Biol Pharm Bull. 2000;23:1122–1124. doi: 10.1248/bpb.23.1122. [DOI] [PubMed] [Google Scholar]
- 20.Khalid M., Saeed-ur-Rahman, Bilal M., Huang D. Role of flavonoids in plant interactions with the environment and against human pathogens — a review. J. Integr. Agric. 2019;18:211–230. doi: 10.1016/S2095-3119(19)62555-4. [DOI] [Google Scholar]
- 21.Ahmad A., Kaleem M., Ahmed Z., Shafiq H. Therapeutic potential of flavonoids and their mechanism of action against microbial and viral infections—a review. Food Res Int. 2015;77:221–235. doi: 10.1016/j.foodres.2015.06.021. [DOI] [Google Scholar]
- 22.Goh V.S.L., Mok C.-K., Chu J.J.H. Antiviral natural products for arbovirus infections. Molecules. 2020;25:2796. doi: 10.3390/molecules25122796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C.H., Kim J.-E., Song Y.-J. Antiviral activities of quercetin and isoquercitrin against human herpesviruses. Molecules. 2020;25:2379. doi: 10.3390/molecules25102379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Özçelik B., Orhan I., Toker G. Antiviral and antimicrobial assessment of some selected flavonoids. Z Naturforsch C Biosci. 2006;61:632–638. doi: 10.1515/znc-2006-9-1003. [DOI] [PubMed] [Google Scholar]
- 25.Zakaryan H., Arabyan E., Oo A., Zandi K. Flavonoids: promising natural compounds against viral infections. Arch Virol. 2017;162:2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evers D.L., Chao C.-F., Wang X., Zhang Z., Huong S.-M., Huang E.-S. Human cytomegalovirus-inhibitory flavonoids: studies on antiviral activity and mechanism of action. Antivir Res. 2005;68:124–134. doi: 10.1016/j.antiviral.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaul T.N., Middleton E., Ogra P.L. Antiviral effect of flavonoids on human viruses. J Med Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 28.Dey Y.N., Khanal P., Patil B.M., Wanjari M.M., Srivastava B., Gurav S.S. The role of andrographolide and its derivative in COVID-19 associated proteins and immune system. Preprint. 2020:1–24. doi: 10.21203/rs.3.rs-35800/v1. [DOI] [Google Scholar]
- 29.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 30.Schmid N., Eichenberger A.P., Choutko A., Riniker S., Winger M., Mark A.E., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 31.Huang W., Lin Z., van Gunsteren W.F. Validation of the GROMOS 54A7 force field with respect to β-peptide folding. J Chem Theor Comput. 2011;7:1237–1243. doi: 10.1021/ct100747y. [DOI] [PubMed] [Google Scholar]
- 32.Malde A.K., Zuo L., Breeze M., Stroet M., Poger D., Nair P.C., Oostenbrink C., Mark A.E. An automated force field topology builder (ATB) and repository: version 1.0. J Chem Theor Comput. 2011;7:4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- 33.Canzar S., El-Kebir M., Pool R., Elbassioni K., Malde A.K., Mark A.E., Geerke D.P., Stougie L., Klau G.W. Charge group partitioning in biomolecular simulation. J Comput Biol. 2013;20:188–198. doi: 10.1089/cmb.2012.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koziara K.B., Stroet M., Malde A.K., Mark A.E. Testing and validation of the Automated Topology Builder (ATB) version 2.0: prediction of hydration free enthalpies. J Comput Aided Mol Des. 2014;28:221–233. doi: 10.1007/s10822-014-9713-7. [DOI] [PubMed] [Google Scholar]
- 35.Kumari R., Kumar R. Open source drug discovery consortium, A. Lynn, g_mmpbsa - a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 36.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci Unit States Am. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagunin A., Ivanov S., Rudik A., Filimonov D., Poroikov V. DIGEP-Pred: web service for in silico prediction of drug-induced gene expression profiles based on structural formula. Bioinformatics. 2013;29:2062–2063. doi: 10.1093/bioinformatics/btt322. [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poroikov V.V., Filimonov D.A., Ihlenfeldt W.-D., Gloriozova T.A., Lagunin A.A., Borodina Y.V., Stepanchikova A.V., Nicklaus M.C. PASS biological activity spectrum predictions in the enhanced open NCI database browser. J Chem Inf Comput Sci. 2003;43:228–236. doi: 10.1021/ci020048r. [DOI] [PubMed] [Google Scholar]
- 41.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS‐CoV‐2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 44.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV-2. Nat Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nouchikian L., Lento C., Donovan K., Dobson R., Wilson D.J. Comparing the conformational stability of pyruvate kinase in the gas phase and in solution. J Am Soc Mass Spectrom. 2020;31:685–692. doi: 10.1021/jasms.9b00130. [DOI] [PubMed] [Google Scholar]
- 51.Kufareva I., Abagyan R. Methods of protein structure comparison. In: Orry A.J.W., Abagyan R., editors. Homol. Model. Humana Press; Totowa, NJ: 2011. pp. 231–257. [DOI] [Google Scholar]
- 52.Zhang Z., Shi Y., Liu H. Molecular dynamics simulations of peptides and proteins with amplified collective motions. Biophys J. 2003;84:3583–3593. doi: 10.1016/S0006-3495(03)75090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomes D.E.B., Lins R.D., Pascutti P.G., Lei C., Soares T.A. The role of nonbonded interactions in the conformational dynamics of organophosphorous hydrolase adsorbed onto functionalized mesoporous silica surfaces. J Phys Chem B. 2010;114:531–540. doi: 10.1021/jp9083635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petukhov M., Rychkov G., Firsov L., Serrano L. H-bonding in protein hydration revisited. Protein Sci. 2004;13:2120–2129. doi: 10.1110/ps.04748404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheurer M., Rodenkirch P., Siggel M., Bernardi R.C., Schulten K., Tajkhorshid E., Rudack T. PyContact: rapid, customizable, and visual analysis of noncovalent interactions in MD simulations. Biophys J. 2018;114:577–583. doi: 10.1016/j.bpj.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torshin I.Y., Weber I.T., Harrison R.W. Geometric criteria of hydrogen bonds in proteins and identification of ‘bifurcated’ hydrogen bonds, Protein Eng. Des. Sel. 2002;15:359–363. doi: 10.1093/protein/15.5.359. [DOI] [PubMed] [Google Scholar]
- 57.Suess C., Hausmann R. Gross and histopathological pulmonary findings in a COVID-19 associated death during self-isolation. Int J Leg Med. 2020;134:1285–1290. doi: 10.1007/s00414-020-02319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Opitz B., van Laak V., Eitel J., Suttorp N. Innate immune recognition in infectious and noninfectious diseases of the lung. Am J Respir Crit Care Med. 2010;181:1294–1309. doi: 10.1164/rccm.200909-1427SO. [DOI] [PubMed] [Google Scholar]
- 59.National Research Council (US) Committee on Research Opportunities in Biology . vol. 7. National Academies Press (US); Washington (DC): 1989. https://www.ncbi.nlm.nih.gov/books/NBK217803/ (Opportunities in biology). The Immune System and Infectious Diseases., (1989) [Google Scholar]
- 60.Khanal P., Duyu T., Patil B.M., Dey Y.N., Pasha I., Wanjari M., Gurav S.S., Maity A. Network pharmacology of AYUSH recommended immune-boosting medicinal plants against COVID-19. J Ayurveda Integr Med. 2020 doi: 10.1016/j.jaim.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geier M.R., Geier D.A. Respiratory conditions in coronavirus disease 2019 (COVID-19): important considerations regarding novel treatment strategies to reduce mortality. Med Hypotheses. 2020;140:109760. doi: 10.1016/j.mehy.2020.109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keen P. Effect of binding to plasma proteins on the distribution, activity and elimination of drugs. In: Brodie B.B., Gillette J.R., Ackerman H.S., editors. Concepts biochem. Pharmacol. Springer Berlin Heidelberg; Berlin, Heidelberg: 1971. pp. 213–233.http://link.springer.com/10.1007/978-3-642-65052-9_10 [Google Scholar]
- 63.Urbanelli L., Buratta S., Tancini B., Sagini K., Delo F., Porcellati S., Emiliani C. The role of extracellular vesicles in viral infection and transmission. Vaccines. 2019;7:102. doi: 10.3390/vaccines7030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muñoz-Fontela C., Mandinova A., Aaronson S.A., Lee S.W. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol. 2016;16:741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hedrick S.M., Michelini R.H., Doedens A.L., Goldrath A.W., Stone E.L. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12:649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arthur J.S.C., Ley S.C. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 67.Suryawanshi A., Tadagavadi R.K., Swafford D., Manicassamy S. Modulation of inflammatory responses by Wnt/β-Catenin signaling in dendritic cells: a novel immunotherapy target for autoimmunity and cancer. Front Immunol. 2016;7 doi: 10.3389/fimmu.2016.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson D.S., Chen Y.H. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol. 2012;12:458–463. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon F. Immunomodulatory cytokines: directing and controlling immune activation. Arthritis Res Ther. 2011;13 doi: 10.1186/ar3418. [DOI] [Google Scholar]
- 70.Muller W.A. Getting leukocytes to the site of inflammation. Vet Pathol. 2013;50:7–22. doi: 10.1177/0300985812469883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muller W.A. Mechanisms of leukocyte transendothelial migration. Annu Rev Pathol. 2011;6:323–344. doi: 10.1146/annurev-pathol-011110-130224. https://www.genome.jp/kegg/ Websites: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.