Abstract
Long noncoding RNAs (lncRNAs) have emerged as key regulators in a variety of cellular processes that influence disease states. In particular, many lncRNAs are genetically or epigenetically deregulated in cancer. However, whether lncRNA alterations are passengers acquired during cancer progression or can act as tumorigenic drivers is a topic of ongoing investigation. In this review, we examine the current methodologies underlying the identification of cancer‐associated lncRNAs and highlight important considerations for evaluating their biological significance as cancer drivers.
Keywords: cancer, function, GEMMs, identification, lncRNA
Abbreviations
- AFAP1
actin filament‐associated protein 1
- AFAP1‐AS1
AFAP1 antisense RNA 1
- ANRIL
antisense noncoding RNA in the INK4 locus
- APC
adenomatous polyposis coli protein
- AR
androgen receptor
- ARLNC1
AR‐regulated long noncoding RNA 1
- ASO
antisense oligonucleotide
- BANCR
BRAF‐activated non‐protein coding RNA
- BC
breast cancer
- BCAR4
breast cancer anti‐estrogen resistance 4
- CARLo‐5
cancer‐associated region long noncoding RNA 5
- CASC14
cancer susceptibility candidate‐14
- CCAT1
colon cancer‐associated transcript 1
- CCAT2
colon cancer‐associated transcript 2
- CDKN2A
cyclin‐dependent kinase inhibitor 2A
- CDKN2B
cyclin‐dependent kinase inhibitor 2B
- CNV
copy number variation
- CRISPR
clustered regulatory interspaced short palindromic repeats
- CRISPRa
CRISPR activation
- CRISPRi
CRISPR inhibition
- CRNDE
colorectal neoplasia differentially expressed
- CTLs
cytotoxic T cells
- DANCR
differentiation antagonizing non‐protein coding RNA
- DINO
damage‐induced noncoding
- DMBA
7,12‐Dimethylbenz[a]anthracene
- EPIC1
epigenetically induced lncRNA 1
- ER
estrogen receptor
- ERAR
ER agitation‐related
- FAL1
focally amplified lncRNA 1
- FALEC
focally amplified long noncoding RNA in epithelial cancer
- gadd7
growth‐arrested DNA damage‐inducible gene 7
- GAPLINC
gastric adenocarcinoma predictive long intergenic noncoding RNA
- GAS5
growth arrest‐specific 5
- GATA6
GATA‐binding protein 6
- GEMM
genetically engineered mouse model
- GOF
gain‐of‐function
- gRNA
guide RNA
- GWAS
genome‐wide associated study
- HOTAIR
HOX transcript antisense RNA
- HOTTIP
HOXA distal transcript antisense RNA
- HPV
human papillomavirus
- HULC
highly upregulated in liver cancer
- IκB
inhibitor of kappa B
- LAST
lncRNA‐assisted stabilization of transcripts
- LED
lncRNA activator of enhancer domains
- LINC‐PINT
long intergenic non‐protein coding RNA, p53‐induced transcript
- LNA
locked nucleic acid
- lncGATA6
lncRNA GATA6
- lncPRESS1
lncRNA p53‐regulated and ESC‐associated 1
- LncRNA
long noncoding RNA
- LOF
loss‐of‐function
- LOH
loss of heterozygosity
- LSAMP
limbic system‐associated membrane protein
- LUAD
lung adenocarcinoma
- MALAT1
metastasis‐associated lung adenocarcinoma transcript 1
- MAPK
mitogen‐activated protein kinase
- MaTARs
mammary tumor‐associated RNAs
- MCL1
myeloid cell leukemia sequence 1
- MEFs
mouse embryonic fibroblasts
- MEG3
maternally expressed 3
- MITF
microphthalmia‐associated transcription factor
- MMTV‐PyMT
mouse mammary tumor virus‐polyomavirus middle T antigen
- MYCLos
MYC‐regulated lncRNAs
- NBAT‐1
neuroblastoma‐associated transcript‐1
- ncRNA
noncoding RNA
- NEAT1
nuclear enriched abundant transcript 1
- NF‐κB
nuclear factor‐kappa B
- NKILA
NF‐κB interacting long noncoding RNA
- OIS1
oncogene‐induced senescence 1
- ORF
open reading frame
- Orilnc1
oncogenic RAS‐induced lncRNA 1
- p53BERs
p53‐bound enhancer regions
- p53RE
p53 response element
- PANDA
p21‐associated ncRNA DNA damage activated
- PANDAR
promoter of CDKN1A antisense DNA damage activated RNA
- PanINs
pancreatic intraepithelial neoplasias
- PAS
polyadenylation signal
- PCa
prostate cancer
- PCAT1
prostate cancer‐associated transcript 1
- PCAT19
prostate cancer‐associated transcript 19
- PCATs
prostate cancer‐associated ncRNA transcripts
- PCGEM1
prostate cancer gene expression marker 1
- PDAC
pancreatic ductal adenocarcinoma
- PDX
patient‐derived xenograft
- PINCR
p53‐induced noncoding RNA
- PRAL
p53 regulation‐associated lncRNA
- PR‐lncRNAs
p53‐regulated lncRNAs
- PRNCR1
prostate cancer‐associated noncoding RNA 1
- PTENP1
phosphatase and tensin homolog pseudogene 1
- PURPL
p53 upregulated regulator of p53 levels
- PVT1
plasmacytoma variant translocation 1
- RNAi
RNA interference
- SALNR
senescence‐associated lncRNA
- SAMMSON
survival‐associated mitochondrial melanoma‐specific oncogenic noncoding RNA
- SCC
squamous cell carcinoma
- SCNV
somatic copy number variation
- SNHG15
small nucleolar RNA host gene 15
- SNP
single nucleotide polymorphism
- SPRY4
sprouty RTK signaling antagonist 4
- SPRY4‐IT1
SPRY4 intronic transcript 1
- TCGA
The Cancer Genome Atlas
- THOR
testis‐associated highly conserved oncogenic long noncoding RNA
- TPA
12‐O‐tetradecanoylphorbol‐13‐acetate
- XIST
X‐inactive specific transcript
1. INTRODUCTION
Cancer is a disease of aberrant cell growth arising from a complex genetic landscape of inherited and sporadic mutations and environmental factors. Historically, cancer research has prioritized examining alterations to protein‐coding genes in molecular pathways influencing the hallmarks of cancer. 1 , 2 While these analyses have provided extensive insights into key players in tumorigenesis, protein‐coding sequences account for only 2% of the genome. 3 Both the pervasive transcription of the human genome 4 and the presence of cancer‐associated mutations in noncoding regions 5 have suggested a potential wealth of unexplored cancer targets. Notably, the heterogeneous class of long noncoding RNAs (lncRNAs) occupies a significant space within the noncoding transcriptome, with recent estimates suggesting the existence of over 100, 000 human lncRNA transcripts. 6 , 7 , 8 , 9
LncRNAs are operationally defined as RNA molecules exceeding 200 nucleotides in length that lack protein‐coding potential. 10 , 11 Able to dynamically fold into intricate secondary structures 12 to interact with DNA, proteins, and other RNAs, lncRNAs are diverse in their structure, localization, and pattern of expression, enabling them to regulate the flow of cellular information at many levels. 13 Frequently the targets of transcriptional programs, lncRNAs influence many fundamental cellular processes including cell division, genome maintenance, and pluripotency. 14 , 15 , 16
As lncRNAs are expressed with exquisite cell‐type and disease‐state specificity, they are ideally positioned to act as biomarkers for a number of pathologies, including different cancers. 17 , 18 , 19 Identifying lncRNA expression changes, or their association with recurrent copy number variations (CNVs) or cancer susceptibility single nucleotide polymorphisms (SNPs) have the potential to become useful tools in cancer diagnosis and treatment planning. Beyond their diagnostic and prognostic utility, over the past decade, individual lncRNAs have been mechanistically and functionally dissected, revealing critical roles in cancer‐related pathways at the cellular and organismal levels. These studies have pointed to lncRNAs as operators within proto‐oncogenic and tumor suppressive networks, suggesting that lncRNAs themselves may play active roles in promoting or limiting tumor development. 20 , 21 , 22
Despite growing data supporting the involvement of lncRNAs in tumorigenesis, it is often difficult to surmise whether changes in individual lncRNAs are bona fide drivers of human cancer development and whether targeting altered lncRNAs in patients would be expected to produce therapeutic benefit. Here, we present an overview of how functional lncRNAs in cancer are identified. We highlight promising therapeutic targets based on patient data and on experimental evidence from in vitro and in vivo cancer models. We also discuss important discrepancies to suggest a best‐practice roadmap for further characterization of the roles of lncRNAs in cancer.
2. IDENTIFICATION OF CANCER‐ASSOCIATED LNCRNAS
2.1. Mining global human cancer genomic and transcriptomic data
Integrating genomic and transcriptomic data from diverse human cancers has provided a starting point for the identification of lncRNAs with functional roles in cancer. In particular, recurrent genetic alterations have implicated many genes involved in oncogenesis, and the capacity to identify such genes has expanded in the last several years due to rapid advances in sequencing technologies. These studies have uncovered that many recurrent somatic copy number variations (SCNVs) map to noncoding regions. 23 Notably, analysis of 5000 human tumor samples across 13 cancer types from The Cancer Genome Atlas (TCGA) revealed that, on average, as many as one quarter of all lncRNAs manifest frequent cancer‐related copy number gains or losses. 19 A more recent study probed the copy number of over 10,000 lncRNAs in 80 cancer cell lines across 11 cancer types, identifying 136 lncRNAs involved in focal SCNVs. 24 Importantly, 76 of these lncRNAs lacked copy number changes in flanking protein‐coding genes, suggesting potential lncRNA‐driven genomic alterations in cancer. Cancer risk SNPs in noncoding loci can also point to a potential role for specific lncRNAs in tumorigenesis. One study identified nearly 4000 lncRNAs overlapping disease‐associated SNPs, while another estimated that roughly 12% of all cancer‐associated SNPs mapped within 5 Kb of lncRNA loci (compared to 55% mapping near protein‐coding genes). 18 , 19
Apart from harboring genomic alterations, lncRNAs have also been found to exhibit differential expression patterns in tumor samples compared to normal tissues. A comprehensive meta‐analysis of over 7000 gene expression datasets, including a range of normal and cancer samples, identified as many as 60,000 lncRNAs with altered expression. 18 Notably, many previously unannotated lncRNAs were found in disease‐associated regions and the expression of roughly 8000 lncRNAs clustered with specific cancer or cell lineages, suggesting the potential for lncRNAs to execute cancer‐specific functions. 18 Along similar lines, an analysis of seven cancer types revealed that, on average, 26% of expressed lncRNAs were significantly deregulated in at least one cancer type (15% upregulated and 11% downregulated) with 60% of these altered lncRNAs demonstrating cancer specificity. 19 In addition, a recent study of lncRNA‐associated epigenetic alterations across 20 different cancers identified over 2000 lncRNAs either epigenetically activated or silenced in at least one cancer type. 25 Altogether, these studies led to the consensus that, as a class, lncRNAs are subject to frequent genetic and epigenetic alterations in cancer.
2.2. LncRNA loci with recurrent SCNVs in cancer
In addition to global patterns of lncRNA deregulation in cancer, several individual lncRNAs have been identified based on frequent large‐scale genomic alterations. One of the first cancer‐associated lncRNAs was identified in murine lymphomas due to the frequent translocations and viral insertions involving the as yet uncharacterized Pvt1 (Plasmacytoma Variant Translocation 1) lncRNA, 26 , 27 located approximately 72 Kb downstream of the MYC (Myelocytomatosis) proto‐oncogene. Later studies extended these results to human cancer and demonstrated a correlation between PVT1 genomic amplification and poor prognosis in acute myeloid leukemia and in breast and ovarian cancers, among others (reviewed in 28 ). Significantly, PVT1 amplification is observed frequently in a range of cancer types including in 33% of ovarian cancers, 20% of esophageal cancers, 13% of invasive breast carcinomas, and 7% of lung adenocarcinomas based on TCGA data. 29 Moreover, PVT1 alterations are associated with a significant reduction in overall and disease‐free survival. 29 , 30 , 31
Another prominent example of a lncRNA initially characterized by genomic alterations is FAL1 (Focally Amplified LncRNA 1, also known as FALEC) located on chromosome 1q21. 32 FAL1 copy number gains have been observed across many cancer types, including in approximately 10% of liver cancers, invasive breast carcinomas, and lung adenocarcinomas according to TCGA data. 29 FAL1 amplification and overexpression are associated with late stage tumors and with decreased survival of patients with ovarian cancer. 29 , 32 Similarly, the lncRNA SAMMSON (Survival‐Associated Mitochondrial Melanoma‐Specific Oncogenic Noncoding RNA) was identified in a region of focal amplification on chromosome 3p13‐14 in 10% of melanomas. 33 High SAMMSON copy number and expression levels are correlated with a reduction in disease‐free survival of melanoma patients and associated with resistance to MAPK (Mitogen Activated Protein Kinase) inhibitors. 29 , 30 , 31 , 33
The locus of the lncRNA LOC285194 on chromosome 3q, moreover, is subject to recurrent monoallelic deletions in as many as 80% of osteosarcomas, often followed by loss of heterozygosity (LOH). 34 Loss of LOC285194 is associated with decreased survival in osteosarcoma patients. 34 The focal deletion of PRAL (p53 Regulation‐Associated LncRNA) on chromosome 17p in hepatocellular carcinoma has also been associated with reduced survival. 35 Similarly, recurrent loss of the 9p21 locus, where the lncRNA ANRIL (Antisense Noncoding RNA in the INK4 Locus) resides, is observed in over 50% of glioblastomas, more than 40% of mesotheliomas, and roughly 30% of bladder cancers. 29 Interestingly, a 403 Kb germline deletion encompassing the ANRIL locus is associated with a strong hereditary predisposition to melanoma development. 36
Many regions of recurrent SCNVs, however, harbor both lncRNAs and protein‐coding genes. Therefore, determining the specific contribution of the lncRNA has been challenging. For example, the presence of multiple overlapping transcripts in the ANRIL locus, including the p15INK4B (CDKN2B), p16INK4A (CDKN2A), and p19ARF tumor suppressors, has confounded the role of ANRIL. 16 Analogously, PVT1 is co‐amplified with MYC and the PVT1 gene body contains DNA regulatory elements, which promote MYC expression. 37 Likewise, SAMMSON is expressed near MITF (Microphthalmia‐Associated Transcription Factor), a key factor in melanocyte differentiation, whereas the commonly amplified genomic region in which FAL1 resides contains the proto‐oncogene MCL1 (Myeloid Cell Leukemia Sequence 1). Finally, the LOC285194‐associated region of deletion also harbors the tumor suppressor LSAMP (Limbic System‐Associated Membrane Protein). Given the complex chromatin architecture and transcriptional profiles in these loci, further studies are needed to deconvolve the specific roles of the lncRNAs and to determine whether lncRNAs act in cooperation with or independently of their neighboring protein‐coding genes.
2.3. LncRNA loci with cancer‐associated SNPs
The link between inherited germline variants in lncRNA loci and cancer predisposition or prognosis has been probed extensively in large‐scale genome‐wide associated studies (GWAS). These studies have identified a plethora of lncRNA‐linked SNPs associated with altered cancer risk or patient prognosis.
As an example, the 2 Mb region mapping to 8q24 has emerged as a major hotspot for over a 100 SNPs strongly associated with multiple diseases, including cancers of the breast, colon, ovaries, prostate, and bladder. 38 , 39 , 40 , 41 Many of these SNPs are significantly correlated with cancer development and highly predictive of poor patient outcome. 42 , 43 , 44 , 45 , 46 While MYC is the dominant oncogene in the locus, many of the cancer risk SNPs are linked to the expression of lncRNAs in the surrounding region, including PVT1, 47 CCAT1 (Colon Cancer‐Associated Transcript 1, also known as CARLo‐5), 48 CCAT2 (Colon Cancer‐Associated Transcript 2), 49 PCAT1 (Prostate Cancer‐Associated Transcript 1), 50 PCAT19 (Prostate Cancer‐Associated Transcript 19), 51 and PRNCR1 (Prostate Cancer‐Associated Noncoding RNA 1). 52 The ANRIL locus is another example of a hotspot harboring more than 10 cancer risk SNPs, some of which are correlated with ANRIL expression. 53 , 54 Other lncRNAs linked to cancer SNPs include HOTAIR (HOX Transcript Antisense RNA), 55 HOTTIP (HOXA Distal Transcript Antisense RNA), 52 MALAT1 (Metastasis‐Associated Lung Adenocarcinoma Transcript 1), 52 HULC (Highly Upregulated in Liver Cancer), 52 MEG3 (Maternally Expressed 3), 56 H19, 57 GAS5 (Growth Arrest‐Specific 5), 56 and PTENP1 (Phosphatase And Tensin Homolog Pseudogene 1). 58
Mechanistic investigations of SNPs associated with lncRNAs have suggested that the risk variants may, in some cases, affect regulatory DNA sequences, thereby resulting in altered lncRNA expression. For example, the PCAT1‐linked risk variant rs7463708 was found to increase the activity of a distal enhancer, resulting in increased PCAT1 expression, 50 whereas the PCAT19‐linked SNP rs11672691 was proposed to perturb transcription factor binding sites, resulting in the increased expression of a pro‐metastatic PCAT19 isoform. 51 , 59 Finally, a high‐risk neuroblastoma‐associated SNP rs693940 on chromosome 6p22 was found to contribute to differential CpG methylation and decreased expression of NBAT‐1 (Neuroblastoma‐Associated Transcript‐1, also known as CASC14), a lncRNA with tumor suppressor properties. 60 Apart from these intriguing examples, however, the majority of lncRNA‐associated SNPs lack experimental support that would robustly link the cancer susceptibility variants with deregulation of lncRNA levels or function, and have thus had limited impact on the identification and characterization of functional lncRNAs in cancer.
2.4. LncRNAs differentially expressed in cancer
Global gene expression analyses of normal and cancer samples have also led to the identification of numerous differentially expressed lncRNAs hypothesized to contribute to disease development. Some of the initial analyses revealed frequent upregulation of lncRNAs, such as the imprinted lncRNA H19 in Wilms’ tumors and lung cancer, 61 , 62 , 63 the prostate cancer‐specific lncRNA PCGEM1 (Prostate Cancer Gene Expression Marker 1), 64 the lung metastasis‐promoting lncRNA MALAT1, 65 and the hepatocellular carcinoma overexpressed lncRNA HULC. 66
The differential expression of some of these lncRNAs has been associated with clinical outcomes. For example, altered H19 expression correlates with poor clinical outcomes across various cancer types including breast cancer, non‐small cell lung cancer, and acute myeloid leukemia. 67 , 68 , 69 Moreover, increased expression of PCGEM1 in normal prostate tissue is a prostate cancer risk factor. 64 , 70 At the same time, a large body of literature has cemented the strong correlation between high MALAT1 expression levels and poor patient prognosis across over 20 cancer types. 71 , 72 Finally, high expression of HULC is associated with poor overall survival and distant metastases. 73
Notably, integrated analysis of gene expression and methylation datasets has also led to the identification of differentially expressed lncRNAs arising from cancer‐associated epigenetic changes, including AFAP1‐AS1 (AFAP1 Antisense RNA 1), and EPIC1 (Epigenetically Induced LncRNA1), both identified as hypomethylated and overexpressed in Barrett's esophagus and esophageal adenocarcinoma, and breast cancer, respectively. 25 , 74
Gene expression profiling in cohorts of cancer patients have further fueled the discovery of lncRNAs associated with specific cancer types. Transcriptome sequencing across a cohort of prostate cancer patients identified PCAT1 among 121 unannotated prostate cancer‐associated ncRNA (noncoding RNA) transcripts (PCATs). 75 Similarly, comprehensive lncRNA profiling in colorectal carcinoma led to the identification of CCAT1, 76 , 77 CCAT2, 49 and other CCAT family members, 78 whereas the lncRNA GAPLINC (Gastric Adenocarcinoma Predictive Long Intergenic Noncoding RNA) stood out as aberrantly overexpressed in gastric tumors. 79 A different set of analyses led to the identification of stage‐specific lncRNAs, such as the lncRNA CRNDE (Colorectal Neoplasia Differentially Expressed), 80 a marker of early stages of colorectal cancer development, although the protein‐coding capacity of CRNDE remains an open question. 81 Transcriptome profiling of breast cancer subtypes, moreover, highlighted the sets of lncRNAs which are either differentially expressed in tumor samples compared to normal tissues or uniquely enriched in specific stages or subtypes of breast cancer. Examples include MALAT1, 82 , 83 HOTAIR, 84 and BCAR4 (Breast Cancer Anti‐Estrogen Resistance 4). 85 , 86 In parallel, mouse models of cancer were recently employed for the identification of 30 murine MaTARs (Mammary Tumor‐Associated RNAs), many of which were found to have human counterparts (hMaTARs) with potential clinical significance determined based on differential expression and correlation with cancer subtype and/or hormone receptor status. 87 Interestingly, many of these examples of cancer‐specific lncRNAs were later found to show differential expression across multiple cancer types, hinting at universal roles in cancer pathogenesis.
2.5. LncRNAs in cancer pathways
In addition to profiling tumor samples, many researchers have undertaken diverse functional approaches to identify novel lncRNAs, including dissecting tumor suppressive and pro‐oncogenic transcriptional networks, analyzing various cancer‐related cellular states and processes, and performing genome‐wide functional screens.
Analysis of the p53 (also known as Trp53) transcriptional network, in particular, has revealed a wealth of lncRNAs with potential tumor suppressor functions. By comparing gene expression profiles and p53‐binding patterns in the absence and in the presence of genotoxic or oncogenic stress, known to activate the p53 pathway, as well as in p53‐proficient and ‐deficient cells, researchers have identified multiple direct lncRNA targets of p53. These included lincRNA‐p21; 88 PANDAR (Promoter Of CDKN1A Antisense DNA Damage‐Activated RNA, also known as PANDA); 89 p53BERs (p53‐Bound Enhancer Regions); 90 Linc‐ Pint (Long Intergenic Non‐Protein Coding RNA, P53‐Induced Transcript); 91 LED (LncRNA Activator of Enhancer Domains); 92 PR‐lncRNAs (P53‐Regulated lncRNAs); 93 , 94 DINO (Damage‐Induced Noncoding); 95 lncPRESS1 (LncRNA P53‐Regulated And ESC‐Associated 1); 96 NEAT1 (Nuclear Enriched Abundant Transcript 1); 97 , 98 , 99 PURPL (P53 Upregulated Regulator Of P53 Levels); 100 PINCR (P53‐Induced Noncoding RNA); 101 GUARDIN; 102 and an isoform of Pvt1, Pvt1b. 103 Functional characterizations have suggested that many of these lncRNAs contribute to p53 tumor suppressor activities.
Other lncRNAs have been identified downstream of oncogenic signaling networks, giving insight into their potential functions. For example, Orilnc1 (Oncogenic RAS‐induced lncRNA 1) was identified as a target of oncogenic RAS signaling with a proposed role in promoting cell growth. 104 LncRNA‐OIS1 (Oncogene‐Induced Senescence 1) was found to modulate senescence induced by activation of oncogenic RAS, 105 whereas BANCR (BRAF‐Activated Non‐Protein Coding RNA) was identified as a transcript induced upon the expression of oncogenic BRAFV600E. 106 Analogously, investigation of estrogen receptor (ER) signaling targets identified 33 ER agitation‐related (ERAR) lncRNAs and suggested potential roles in ER‐positive breast cancer. 107 A similar study was performed to examine lncRNAs regulated by androgen receptor (AR) signaling, which identified ARLNC1 (AR‐Regulated Long Noncoding RNA 1) as both a downstream target and upstream effector of AR signaling during prostate cancer progression. 108 MYC‐regulated lncRNAs have also been identified, including a set of MYCLos (MYC‐regulated LncRNAs), 78 LAST (LncRNA‐Assisted Stabilization of Transcripts), 109 DANCR (Differentiation Antagonizing Non‐Protein Coding RNA), 110 and SNHG15 (Small Nucleolar RNA Host Gene 15). 111
Alterations of cancer hallmarks that enable tumorigenesis have also been linked to the functions of specific lncRNAs (reviewed in 112 ). Examples include lncRNA gadd7 (growth‐arrested DNA damage‐inducible gene 7) with a proposed role in suppressing cell cycle progression, 113 SPRY4‐IT1 (SPRY4 Intronic Transcript 1) with a proposed role in inhibiting apoptosis in melanoma, 114 and SALNR (Senescence‐Associated lncRNA), proposed to regulate senescence. 115
Finally, genome‐wide functional screens for lncRNAs involved in promoting or inhibiting specific cellular outcomes important in cancer have aimed to identify candidates for further study. A CRISPR/Cas9‐based genome editing approach used a paired guide RNA (gRNA) strategy to target for deletion a set of 700 human lncRNAs, identifying 51 lncRNAs able to regulate cancer cell growth. 116 Alternatively, CRISPRi (CRISPR inactivation) and CRISPRa (CRISPR activation) screens, involving a nuclease‐dead Cas9 to tether transcriptional repressors or activators to lncRNA loci have provided effective epigenetic loss‐of‐function and gain‐of‐function approaches to query on a genome‐wide level the role of lncRNAs in processes such as cellular proliferation or therapeutic resistance. 117 , 118 , 119 , 120
3. FUNCTIONAL CHARACTERIZATION OF LNCRNAS IN CANCER
3.1. Common approaches and limitations
For the 100 or so lncRNAs identified in the approaches described above, the pressing question has become how to accurately distinguish functional lncRNAs from lncRNAs that are subject to passenger genetic and epigenetic alterations in cancer. RNA interference (RNAi)‐mediated downregulation of lncRNAs has been a common approach for functional characterization. In parallel, antisense oligonucleotides (ASOs) have provided a convenient and efficient loss‐of‐function alternative. While RNAi is most effective for lncRNAs exported to the cytoplasm, ASOs lend broader efficacy by triggering RNase H‐mediated co‐transcriptional RNA cleavage and degradation, in some cases accompanied by transcriptional repression. 121 , 122 Frequently, RNAi and ASO approaches have been performed in parallel with exogenous lncRNA overexpression. Regrettably, few studies have complemented RNAi or ASO loss‐of‐function experiments with knockdown‐resistant lncRNA rescue mutants, missing an important opportunity to both demonstrate specificity and establish a system to investigate the sequence basis for lncRNA function. CRISPR‐based epigenetic inhibition (CRISPRi) and activation (CRISPRa) have also been employed as successful loss‐of‐function and gain‐of‐function approaches, respectively.
Genetically engineered mouse models (GEMMs) of lncRNAs and CRISPR‐based editing of lncRNA loci in cell lines have also brought important insights. In contrast to protein‐coding genes, where genetic approaches aim to perturb the open reading frame (ORF) and, therefore, the functional output of the transcript, methods to target lncRNAs have been, by necessity, more diverse and creative (reviewed in 123 ). Some loss‐of‐function studies have undertaken deletion of the entire gene body, the promoter region, or narrower functional regions, while others have employed introduction of a premature polyadenylation signal (PAS) or polyadenylation cassette (STOP) to terminate transcription. Conversely, gain‐of‐function studies in animal models have involved the introduction of a transgenic lncRNA sequence or amplification of an entire lncRNA locus.
Strikingly, for many lncRNAs, observed phenotypes have varied with the use of alternative approaches. For example, initial RNAi knockdown of the p53‐regulated lncRNA, lincRNA‐p21, suggested that it acts globally to modulate the expression of multiple p53 target genes, whereas subsequent genetic deletion of its promoter in the mouse revealed a more restricted role in promoting the expression of the neighboring p21/CDKN1A gene. 88 , 124 Further investigation involving a locus deletion genetic approach, however, raised doubts about whether the lncRNA plays any functional role at all. 125 The metastasis‐promoting lncRNA HOTAIR has provided additional examples of the complexity in developing lncRNA models. While ectopic expression of HOTAIR in breast cancer cellsinduced global gene expression changes and increased metastases in a xenograft mouse model, supporting an oncogenic function, 84 loss‐of‐function models, including RNAi‐mediated knockdown, a 4 Kb gene body deletion, and a 140 Kb locus deletion have led to significant discrepancies. 126 , 127 , 128 , 129 The differences between alternative models have highlighted the need to use multiple independent and complementary approaches to investigate the functional roles of lncRNAs in cancer biology.
3.2. Multi‐pronged approaches to lncRNA characterization
In this section, we focus on a small set of lncRNAs for which work from multiple groups or involving an array of in vitro and in vivo approaches has revealed exciting functional insights and provided starting points for further exploration of their contributions to tumor development.
3.2.1. MALAT1
MALAT1 remains one of the most studied cancer‐associated lncRNAs, with proposed roles in influencing nuclear speckles, 130 pre‐mRNA splicing, 131 and epigenetically regulating gene transcription. 132 While initial studies pointed to a pro‐metastatic function, 65 further characterization resulted in discrepancies (Figure 1). Three different loss‐of‐function GEMMs, including an insertion of a LacZ reporter and polyadenylation cassette 69 nucleotides downstream of the Malat1 transcription start site, a 3 Kb deletion of the 5’ end and promoter region of Malat1, and a conditional deletion of 7 Kb encompassing the entire Malat1 gene body, revealed that Malat1 is dispensable for organismal development and viability. 133 , 134 , 135 Strikingly, none of the mouse models showed effects on global gene expression, nuclear speckle formation, or alternative pre‐mRNA splicing. This opposed previous findings using RNAi to downregulate MALAT1 levels in cancer cell lines in vitro, 131 , 132 perhaps suggesting a cancer‐specific function. Furthermore, different in vivo models have yielded conflicting results about the function of MALAT1 in cancer. On the one hand, crossing the promoter deletion model 135 to the MMTV‐PyMT (mouse mammary tumor virus‐polyomavirus middle T antigen) mouse model of breast cancer resulted in reduced metastases to the lung, without affecting primary tumor burden, an effect largely recapitulated by ASO‐depletion of Malat1 in vivo. 82 This pro‐metastatic function was also observed in a mouse xenograft model of lung cancer where MALAT1 knockout human lung tumor cells formed fewer tumor nodules. 136 In this model, targeting MALAT1 with ASOs after tumor implantation prevented metastasis formation, pointing to MALAT1 as a viable therapeutic target. 136 On the other hand, crossing the Malat1 premature termination model 134 to the MMTV‐PyMT breast cancer model led to a significant increase in the number and area of metastatic nodules in the lungs. 137 This surprising tumor suppressive effect could be rescued with a Malat1 transgene expressed from the Rosa26 locus. 137 A similar effect was observed in vitro in human breast cancer cells, with the expression of MALAT1 from an exogenous construct rescuing the increased metastatic ability conferred by MALAT1 knockout in clonal cell populations. 137 The debate surrounding the precise contribution of MALAT1 to cancer development is ongoing. It is unclear whether the phenotypic differences arising from MALAT1 loss might be due to differences in experimental setup, such as mouse strain or knockout approach, or reflect the complex biology of MALAT1. Altogether, investigations of MALAT1 using in vitro and in vivo approaches have highlighted the biological and technical complexities associated with studying the functional roles of lncRNAs in cancer. 138 , 139
FIGURE 1.
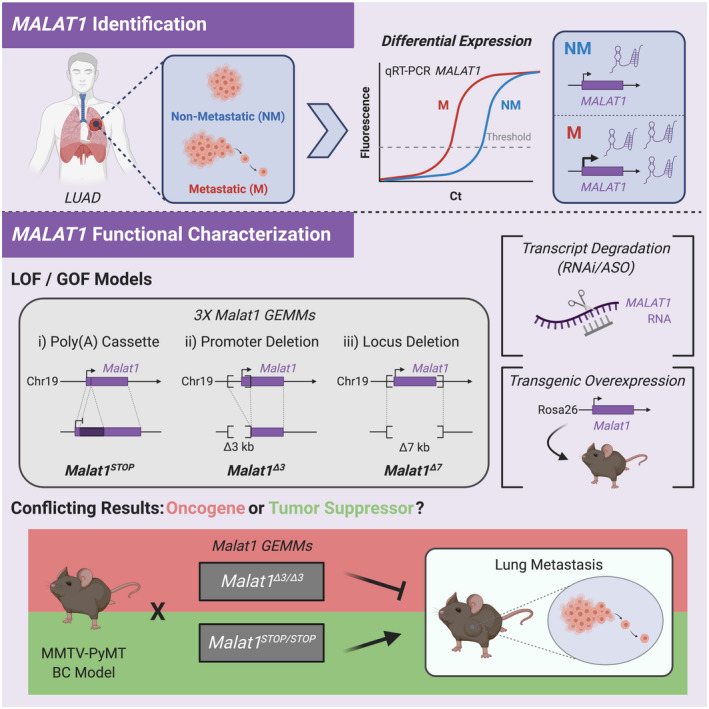
Identification and functional characterization of MALAT1. MALAT1 was identified as upregulated in metastatic (M) LUAD (lung adenocarcinoma) compared to nonmetastatic (NM) tissue. Functional characterization of MALAT1 has utilized various loss‐of‐function (LOF) and gain‐of‐function (GOF) models including polyadenylation cassette insertion (Malat1STOP, 134 ), promoter deletion (Malat1Δ3, 135 ), and locus deletion (Malat1Δ7, 133 ) genetically engineered mouse models (GEMMs), as well as transcript degradation with RNAi and ASO, and transgenic overexpression. Crossing Malat1Δ3 or Malat1STOP GEMMs to the MMTV‐PyMT BC (breast cancer) mouse model has resulted in either oncogenic (red box, 82 ) or tumor suppressor (green box, 137 ) models for Malat1 function, due to observed decreases and increases in lung metastases, respectively.
3.2.2. NEAT1
Similarly to MALAT1, several studies have examined the role of NEAT1 during cancer development, leading to opposing views (Figure 2). Initial studies suggested that NEAT1 levels were elevated in a variety of human cancers relative to normal tissues and correlated with worse prognosis, suggesting a pro‐oncogenic role for NEAT1 ( 140 and reviewed in 141 ). This conclusion was supported by a study of Neat1 knockout mice subjected to chemical induction of skin squamous cell carcinoma with the carcinogen DMBA and the pro‐inflammatory agent TPA. 97 , 142 While Neat1‐deficient animals displayed no obvious phenotypes in the absence of stress, 143 loss of Neat1 conferred resistance to chemically induced squamous cell carcinoma. 97 Interestingly, studies have also suggested that NEAT1 may be a target of the p53 pathway and, therefore, may have tumor suppressive activities in some contexts. 98 , 144 Indeed, tumor suppressive functions of Neat1 were unveiled in primary mouse embryonic fibroblasts (MEFs), where Neat1 knockout led to increased colony formation in an E1A; HrasG12V transformation experiment, as well as in an autochthonous mouse model of pancreatic cancer, where Neat1 deficiency increased the occurrence of premalignant lesions, known as pancreatic intraepithelial neoplasias (PanINs). 99 Interestingly, Malat1 and Neat1 are neighboring genes and studies have suggested that genomic deletion of either lncRNA may impact the epigenetic organization and transcriptional profiles of the entire locus, raising questions about the specificity of each approach. 134
FIGURE 2.

Identification and functional characterization of NEAT1. NEAT1 was initially identified as being upregulated in prostate cancer (PCa) compared to normal (N) tissue, suggesting a potential oncogenic function (top, red box). Later, it was also identified as a p53 target with p53 binding to a conserved p53 Response Element (p53RE) in the NEAT1 promoter, as well as a paraspeckle component induced by cellular stress, suggesting a potential tumor suppressor function (top, green box). Functional characterization of NEAT1 has utilized various loss‐of‐function (LOF) and gain‐of‐function (GOF) models including a polyadenylation cassette insertion genetically engineered mouse model (GEMM), 143 transcript degradation with RNAi or ASO, and exogenous overexpression. The Neat1STOP GEMM has been shown to either decrease 97 or increase 99 tumor growth following chemical induction of SCC (squamous cell carcinoma) or when crossed to a PDAC (pancreatic ductal adenocarcinoma) GEMM, respectively, suggesting either oncogenic (bottom, red box) or tumor suppressor (bottom, green box) models for Neat1 function in cancer
3.2.3. PVT1
As one of the lncRNAs strongly associated with advanced disease and poor patient prognosis, PVT1 has been the subject of extensive investigation (Figure 3). In keeping with the finding that PVT1 is frequently co‐amplified with the MYC proto‐oncogene, Myc‐Pvt1 co‐amplification in a mouse model of breast cancer was found to be more tumorigenic than Myc amplification alone. 145 This study suggested that PVT1 acts in trans to promote MYC protein stability, based on evidence that a 300 Kb genomic deletion of the PVT1 locus in a human colorectal carcinoma cell line resulted in decreased MYC protein levels. 145 However, later studies found evidence for MYC enhancers within the region of deletion, raising questions about the role of the PVT1 locus and its associated RNA in MYC regulation. 37 Subsequent studies confirmed the presence of DNA regulatory elements in the locus but challenged the understanding of PVT1 as a strictly pro‐oncogenic lncRNA. 146 , 147 On the one hand, deletion of a ~600 bp region containing a p53‐binding site and mapping to the 5’ end of PVT1 led to defects in p53‐mediated MYC repression, although the contribution of PVT1 to the p53 response was unclear. 147 On the other hand, CRISPRi‐based inhibition of PVT1 in breast cancer cell lines revealed a role for the PVT1 promoter as a DNA tumor suppressor boundary element that limits MYC promoter accessibility to enhancers within the PVT1 gene body, resulting in restricted MYC expression. 146 In this setting, the PVT1 RNA appeared to be dispensable. 146 In contrast, our group identified a stress‐induced, p53‐dependent isoform of Pvt1, Pvt1b, which is both necessary and sufficient to repress Myc transcription. 103 These findings were recapitulated in vitro using a genetic loss‐of‐function approach to mutate the p53‐binding site required for Pvt1b expression. 103 Importantly, mutagenesis of the Pvt1‐associated p53‐binding site at the time of tumor initiation in an autochthonous mouse model of lung cancer led to larger tumors and indicated a key role for Pvt1b in restraining tumor growth downstream of p53. 103 In the future, it would be interesting to deconvolve the oncogenic and tumor suppressive elements in the PVT1 locus and to differentiate between DNA elements and RNA isoforms with potentially distinct functions.
FIGURE 3.

Identification and functional characterization of PVT1. PVT1 was identified in murine lymphomas following the observation of translocations, viral insertions, and amplifications involving the Pvt1 locus. Functional characterization of PVT1 has utilized various loss‐of‐function (LOF) and gain‐of‐function (GOF) models including amplification genetically engineered mouse models (GEMMs) (Myc/Pvt1AMP, MycAMP, 145 ), locus deletion (PVT1Δ), tumor‐specific mutagenesis of the Pvt1‐associated p53 Response Element (p53RE) (Δp53RE, 103 ), transcript degradation with RNAi and ASO, and CRISPR‐mediated epigenetic activation and inhibition (CRISPRa/i). The increased tumor growth observed in a Myc/Pvt1 co‐amplification GEMM (Myc/Pvt1AMP) compared to Myc amplification alone (MycAMP) when crossed to the MMTV‐Neu BC (breast cancer) GEMM suggests an oncogenic function for Pvt1 (red box, 145 ). However, the increased tumor growth in Pvt1‐associated p53RE mutagenized lung tumors following Cre‐mediated tumor initiation in a Kras‐driven lung adenocarcinoma (LUAD) GEMM suggests a tumor suppressor function (green box, 103 )
3.2.4. XIST
With a critical role in X‐chromosome inactivation and dosage compensation that has been investigated for decades (reviewed in 148 , 149 ), the potential role of XIST (X‐Inactive Specific Transcript) in tumorigenesis has intrigued researchers. Historically, it has been observed that altered chromosome copy numbers and inappropriate dosage compensation are frequently associated with human cancer. Notably, men with Klinefelter syndrome characterized by an extra X‐chromosome have an increased risk of many malignancies including breast cancer and non‐Hodgkin lymphoma, 150 and loss of X‐chromosome inactivation has been observed in breast cancer cell lines 151 and testicular germ cell tumors. 152 In support of these correlative observations, a conditional Xist deletion model in mouse blood cell lineages led to aggressive myeloproliferative neoplasm and myelodysplastic syndrome with complete penetrance, likely as the result of widespread gene expression changes. 153 The tumor suppressive role of XIST was recapitulated in RNAi and overexpression studies in breast cancer cell lines as well as by crossing the Xist knockout to the MMTV‐PyMT mouse model of breast cancer. 154 Further studies should determine the prevalence of XIST and X‐inactivation perturbations in human cancer and investigate the possibility of targeting this pathway as a therapeutic strategy.
3.2.5. ANRIL
High ANRIL expression in tumor tissues has been linked to aggressive pathological features and poor overall survival (reviewed in 155 ). In initial studies, targeted deletion of a 70 Kb region in the Anril locus, which harbors multiple cancer and coronary artery disease‐associated SNPs, led to viable progeny but showed increased mortality during development and as adults. 156 Primary cultures of smooth muscle cells, isolated from mutant mice, exhibited excessive proliferation and diminished senescence, cellular phenotypes consistent both with accelerated coronary disease pathogenesis and increased cancer risk. Mechanistic investigation revealed that the effects were mediated in cis through the reduced expression of Cdkn2a and Cdkn2b and led to the conclusion that the risk region contained key regulatory elements. Subsequent investigation using exogenous overexpression of ANRIL in primary human fibroblasts suggested that the lncRNA may be responsible for CDKN2A/2B repression through the locus‐specific recruitment of the repressive PRC1 complex. 16 Unfortunately, little progress has been made over the past decade in determining whether ANRIL transcription or transcript accumulation is required for its cis‐regulatory function, in part due to the limited conservation of ANRIL sequence and exonic structure between human and mouse.
3.3. Promising lncRNA candidates warranting further investigation
In this section we examine exciting, albeit limited, initial studies of lncRNAs with putative cancer functions, the validation of which could benefit from the development of alternative approaches and further characterization.
3.3.1. SAMMSON
To investigate the role of SAMMSON as a lineage addiction oncogene in melanoma, researchers employed ASO‐mediated knockdown and exogenous overexpression as loss‐of‐function and gain‐of‐function tools. 33 They observed that SAMMSON amplification and increased expression led to altered mitochondrial metabolism and homeostasis. In turn, this caused increased melanoma cell viability and clonogenic potential and resulted in sensitization of melanoma cells to MAPK targeting therapeutics in vitro and in patient‐derived xenograft (PDX) models in vivo. Further mechanistic studies clarified the role of SAMMSON in balancing mitochondrial translation rates. 157 The generation of genetic models of SAMMSON may reveal further insights into its role in melanoma development.
3.3.2. NKILA
NKILA (NF‐κB Interacting Long Noncoding RNA) was identified as both a target and negative modulator of the NF‐κB signaling pathway, with low NKILA levels observed in metastatic breast cancer cell lines and correlated with decreased disease‐free survival in a cohort of breast cancer patients. 158 Mechanistically, a series of deletion mutants demonstrated that NKILA interacts directly and stably with the NF‐κB:IκB complex in the cytoplasm to prevent IκB phosphorylation and suppress activation of the NF‐κB pathway, suggesting a tumor suppressive role for NKILA in limiting inflammatory processes in cancer. 158 A different study from the same group showed that RNAi downregulation of NKILA in cytotoxic T cells (CTLs) led to increased tumor infiltration and reduced tumor volume in a breast cancer PDX mouse model, implicating NKILA as a potential target in the field of cancer immunotherapy. 159
3.3.3. LncGata6
LncGata6 (LncRNA GATA6) was identified as a divergent transcript expressed from the promoter of Gata6, which is specifically enriched in a subset of intestinal stem cells. 160 Deletion of exons 2‐4 of lncGata6 in the mouse did not affect Gata6 levels but resulted in decreased intestinal regeneration due to decreased proliferative capacity of intestinal stem cells. 160 Consistent with the key role of stem cells in intestinal tumorigenesis, genetic and ASO‐mediated depletion of lncGata6/lncGATA6 were found to impair tumor growth in the APCmin mouse model of intestinal adenoma and in a PDX model. 160 Future studies should focus on elucidating the mechanism by which lncGATA6 is upregulated in colorectal cancer and on determining the extent to which it contributes to aberrant Wnt signaling, a known colorectal cancer driver.
3.3.4. DINO
The p53 target lncRNA DINO binds to and stabilizes p53 in a positive feedback loop, enhancing the activation of p53 target genes. 95 Importantly, RNAi knockdown of DINO in human fibroblasts and a deletion of the Dino promoter in MEFs led to impaired cell cycle arrest following genotoxic stress. 95 Interestingly, ectopic expression of DINO in HPV‐positive cervical cancer cells, which suppress p53 stabilization and express DINO at low levels, led to reactivation of dormant p53, resulting in sensitization of the cancer cells to chemotherapeutic agents and vulnerability to metabolic stress. 161 To date, however, evidence that DINO acts as a tumor suppressor in human cancer is limited.
3.3.5. LINC‐PINT
Like DINO, linc‐Pint was also identified as a p53 target. 91 A knockout mouse generated by replacing the linc‐Pint locus with a LacZ reporter cassette yielded smaller pups, suggesting a role for linc‐Pint in early development. 162 Characterization of LINC‐PINT function in cancer suggested a role in limiting cell invasion, with LINC‐PINT overexpression leading to decreased liver metastases in a mouse model. 163 In a transwell migration and invasion assay, invasiveness increased following treatment with LINC‐PINT‐targeting ASOs or following CRISPR‐mediated deletion of a highly conserved LINC‐PINT sequence element. 163 Analysis of the previously generated linc‐Pint knockout mouse 162 in a cancer background could help to support these results. However, the potential role of the LINC‐PINT RNA may be confounded by the identification of a peptide with a function in suppressing cell proliferation encoded by a circular form of LINC‐PINT. 164
3.3.6. THOR
While examples of alternative organismal models for lncRNA function in cancer are limited, in part due to low evolutionary conservation of lncRNAs, investigation of the highly conserved lncRNA THOR (Testis‐Associated Highly Conserved Oncogenic Long Noncoding RNA) in human and zebrafish cancer models has implicated this lncRNA in promoting melanoma development (Figure 4). 165 THOR expression is normally restricted to the testis, but has been found aberrantly overexpressed in multiple cancer types, including lung adenocarcinoma, lung squamous carcinoma, and melanoma. 165 Knockdown of THOR via RNAi and ASOs in lung adenocarcinoma and melanoma cell lines led to decreased proliferation and reduced colony formation. 165 These findings were corroborated in two independently derived lung adenocarcinoma cell lines harboring approximately 3 Kb CRISPR‐mediated deletions within the THOR gene body. Conversely, THOR overexpression gave the opposite phenotype, leading to increased proliferative capacity and anchorage‐independent growth. Importantly, ectopic expression of human THOR in zebrafish cooperated with oncogenic NRAS and p53 loss to promote melanoma development, whereas knockout of THOR in zebrafish embryos delayed mutant NRAS‐induced melanoma formation. 165 Further studies may reveal the potential of using THOR expression as a biomarker or targeting THOR as a therapeutic strategy.
FIGURE 4.

Identification and functional characterization of THOR THOR was identified as a testis‐specific ultra‐conserved lncRNA aberrantly expressed in cancer tissues. 165 Hosono and colleagues generated several in vitro and in vivo loss‐of‐function (LOF) and gain‐of‐function (GOF) models to functionally characterize THOR. LOF models included transcript degradation with RNAi and ASO, and THOR partial locus deletion (THOR−/−) in both human cells injected in severe combined immunodeficiency disease (SCID) mice and in a genetically engineered zebrafish model (THOR−/−) embryonically injected with NRAS to induce melanoma. GOF models included THOR overexpression (OE) in vitro and OE of human THOR (hTHOR) in p53‐deficient zebrafish (p53−/−) embryonically injected with NRAS to induce melanoma. Overexpression of THOR plays an oncogenic role (red box) in cancer by binding to IGF2BP1 and increasing the stability of its mRNA targets to promote cancer progression
4. FUTURE PERSPECTIVES
Identification of lncRNAs that are genetically or epigenetically perturbed in cancer has risen sharply over the past decade. The precipitous increase in the number of cancer‐associated lncRNAs has been accompanied by a growing excitement that many lncRNAs may act as novel drivers of cancer development. Yet, lagging understanding of how lncRNAs function in physiologic and pathologic contexts has limited our insights into the roles of lncRNAs in tumorigenesis. The current literature points to many lncRNAs acting as both oncogenes and tumor suppressors. While these seemingly contradictory findings may stem from differences in experimental models, they may also be reflective of complex and context‐dependent lncRNA biology, analogous to the dual oncogenic and tumor suppressor roles played by cancer‐associated protein‐coding genes. 166 Future studies should prioritize the identification and validation of true dual functions from technical inconsistencies.
LncRNAs make attractive drug targets, particularly in diseases where protein candidates are not amenable to pharmacological inhibition. 167 Both siRNA‐ and ASO‐mediated lncRNA degradation as well as locked nucleic acid (LNA)‐mediated interference with lncRNA function have emerged as clinic‐ready approaches. 168 , 169 The successful deployment of these approaches in cancer, however, is predicated upon robust functional characterization. In the future, it would be essential to develop in vitro and in vivo models that closely recapitulate the recurrent genetic or epigenetic changes of lncRNAs observed in human cancer. In parallel, experiments that uncover the functional elements of perturbed lncRNA loci will inform whether motives or structural features of the lncRNA molecules, the act of their transcription, or underlying DNA elements mediate their roles in disease development. These questions will be best answered through the integration of diverse and complementary approaches and by corroboration from multiple independent studies.
5. COMPETING INTEREST
The authors declare no competing financial interest.
AUTHOR CONTRIBUTIONS
C. Olivero and N. Dimitrova wrote and edited the manuscript. C. Olivero created the figures.
ACKNOWLEDGMENTS
The authors are grateful to Lauren Winkler, Elena Martinez, and Giuseppe Militello for insightful comments. ND and CO are supported by grants from the Pew‐Stewart Foundation for Cancer Research and from the National Cancer Institute (R37 CA230580). Figures were created with BioRender.com.
Olivero C, Dimitrova N. Identification and characterization of functional long noncoding RNAs in cancer. The FASEB Journal. 2020;34:15630–15646. 10.1096/fj.202001951R
REFERENCES
- 1. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57‐70. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646‐674. [DOI] [PubMed] [Google Scholar]
- 3. International Human Genome Sequencing Consortium . Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931‐945. [DOI] [PubMed] [Google Scholar]
- 4. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post‐GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242‐2246. [DOI] [PubMed] [Google Scholar]
- 7. Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559‐1563. [DOI] [PubMed] [Google Scholar]
- 8. The ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484‐1488. [DOI] [PubMed] [Google Scholar]
- 10. Mercer TR, Dinger ME, Mattick JS. Long non‐coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155‐159. [DOI] [PubMed] [Google Scholar]
- 11. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK. Revealing lncRNA structures and interactions by sequencing‐based approaches. Trends Biochem Sci. 2019;44:33‐52. [DOI] [PubMed] [Google Scholar]
- 13. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loewer S, Cabili MN, Guttman M, et al. Large intergenic non‐coding RNA‐RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yap KL, Li S, Munoz‐Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775‐1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan X, Hu Z, Feng Y, et al. Comprehensive genomic characterization of long non‐coding RNAs across human cancers. Cancer Cell. 2015;28:529‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253‐1261. [DOI] [PubMed] [Google Scholar]
- 21. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354‐361.21550244 [Google Scholar]
- 23. Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy‐number alteration across human cancers. Nature. 2010;463:899‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volders PJ, Lefever S, Baute S, et al. Targeted genomic screen reveals focal long non‐coding RNA copy number alterations in cancer cell lines. Noncoding RNA. 2018;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z, Yang B, Zhang M, et al. LncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell‐cycle progression in cancer. Cancer Cell. 2018;33:706‐720.e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cory S, Graham M, Webb E, Corcoran L, Adams JM. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c‐myc oncogene. EMBO J. 1985;4:675‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graham M, Adams JM, Cory S. Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. Nature. 1985;314:740‐743. [DOI] [PubMed] [Google Scholar]
- 28. Colombo T, Farina L, Macino G, Paci P. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoadley KA, Yau C, Hinoue T, et al. Cell‐of‐origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291‐304.e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu X, Feng Y, Zhang D, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non‐coding RNA SAMMSON. Nature. 2016;531:518‐522. [DOI] [PubMed] [Google Scholar]
- 34. Pasic I, Shlien A, Durbin AD, et al. Recurrent focal copy‐number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160‐171. [DOI] [PubMed] [Google Scholar]
- 35. Zhou CC, Yang F, Yuan SX, et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016;63:850‐863. [DOI] [PubMed] [Google Scholar]
- 36. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ‐line deletion, including the entire INK4/ARF locus, in a melanoma‐neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963‐3969. [DOI] [PubMed] [Google Scholar]
- 37. Fulco CP, Munschauer M, Anyoha R, et al. Systematic mapping of functional enhancer‐promoter connections with CRISPR interference. Science. 2016;354:769‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Easton DF, Eeles RA. Genome‐wide association studies in cancer. Hum Mol Genet. 2008;17:R109‐R115. [DOI] [PubMed] [Google Scholar]
- 39. Ghoussaini M, Song H, Koessler T, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962‐966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grisanzio C, Freedman ML. Chromosome 8q24‐associated cancers and MYC. Genes Cancer. 2010;1:555‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. The 8q24 gene desert: an oasis of non‐coding transcriptional activity. Front Genet. 2012;3:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bertucci F, Lagarde A, Ferrari A, et al. 8q24 cancer risk allele associated with major metastatic risk in inflammatory breast cancer. PLoS One. 2012;7:e37943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia‐Closas M, Hall P, Nevanlinna H, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yeager M, Orr N, Hayes RB, et al. Genome‐wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645‐649. [DOI] [PubMed] [Google Scholar]
- 46. Zhang X, Chen Q, He C, et al. Polymorphisms on 8q24 are associated with lung cancer risk and survival in Han Chinese. PLoS One. 2012;7:e41930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meyer KB, Maia AT, O'Reilly M, et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7:e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao X, Wei X, Zhao L, et al. The rs6983267 SNP and long non‐coding RNA CARLo‐5 are associated with endometrial carcinoma. Environ Mol Mutagen. 2016;57:508‐515. [DOI] [PubMed] [Google Scholar]
- 49. Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446‐1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo H, Ahmed M, Zhang F, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet. 2016;48:1142‐1150. [DOI] [PubMed] [Google Scholar]
- 51. Hua JT, Ahmed M, Guo H, et al. Risk SNP‐mediated promoter‐enhancer switching drives prostate cancer through lncRNA PCAT19. Cell. 2018;174:564‐575.e18. [DOI] [PubMed] [Google Scholar]
- 52. Huang X, Zhang W, Shao Z. Association between long non‐coding RNA polymorphisms and cancer risk: a meta‐analysis. Biosci Rep. 2018;38:BSR20180365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6:e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri‐Fard S. ANRIL genetic variants in iranian breast cancer patients. Cell J. 2017;19:72‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Botti G, Collina F, Scognamiglio G, et al. LncRNA HOTAIR polymorphisms association with cancer susceptibility in different tumor types. Curr Drug Targets. 2018;19:1220‐1226. [DOI] [PubMed] [Google Scholar]
- 56. Dong X, Gao W, Lv X, et al. Association between lncRNA GAS5, MEG3, and PCAT‐1 polymorphisms and cancer risk: a meta‐analysis. Dis Markers. 2020;2020:6723487. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Hashemi M, Moazeni‐Roodi A, Sarabandi S, Karami S, Ghavami S. Association between genetic polymorphisms of long noncoding RNA H19 and cancer risk: a meta‐analysis. J Genet. 2019;98:81. [PubMed] [Google Scholar]
- 58. Ge Y, He Y, Jiang M, et al. Polymorphisms in lncRNA PTENP1 and the risk of gastric cancer in a chinese population. Dis Markers. 2017;2017:6807452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gao P, Xia J‐H, Sipeky C, et al. Biology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locus. Cell. 2018;174:576‐589.e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pandey GK, Mitra S, Subhash S, et al. The risk‐associated long noncoding RNA NBAT‐1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722‐737. [DOI] [PubMed] [Google Scholar]
- 61. Hibi K, Nakamura H, Hirai A, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480‐482. [PubMed] [Google Scholar]
- 62. Kondo M, Suzuki H, Ueda R, et al. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193‐1198. [PubMed] [Google Scholar]
- 63. Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747‐749. [DOI] [PubMed] [Google Scholar]
- 64. Srikantan V, Zou Z, Petrovics G, et al. PCGEM1, a prostate‐specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97:12216‐12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ji P, Diederichs S, Wang W, et al. MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene. 2003;22:8031‐8041. [DOI] [PubMed] [Google Scholar]
- 66. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up‐regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330‐342. [DOI] [PubMed] [Google Scholar]
- 67. Shima H, Kida K, Adachi S, et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res Treat. 2018;170:507‐516. [DOI] [PubMed] [Google Scholar]
- 68. Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou Y, Sheng B, Xia Q, Guan X, Zhang Y. Association of long non‐coding RNA H19 and microRNA‐21 expression with the biological features and prognosis of non‐small cell lung cancer. Cancer Gene Ther. 2017;24:317‐324. [DOI] [PubMed] [Google Scholar]
- 70. Petrovics G, Zhang W, Makarem M, et al. Elevated expression of PCGEM1, a prostate‐specific gene with cell growth‐promoting function, is associated with high‐risk prostate cancer patients. Oncogene. 2004;23:605‐611. [DOI] [PubMed] [Google Scholar]
- 71. Amodio N, Raimondi L, Juli G, et al. MALAT1: a druggable long non‐coding RNA for targeted anti‐cancer approaches. J Hematol Oncol. 2018;11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang J, Zhang B, Wang T, Wang H. LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta‐analysis. Int J Clin Exp Med. 2015;8:5499‐5505. [PMC free article] [PubMed] [Google Scholar]
- 73. Chen X, Lun L, Hou H, Tian R, Zhang H, Zhang Y. The value of lncRNA HULC as a prognostic factor for survival of cancer outcome: a meta‐analysis. Cell Physiol Biochem. 2017;41:1424‐1434. [DOI] [PubMed] [Google Scholar]
- 74. Wu W, Bhagat TD, Yang X, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1‐AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956‐966.e954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT‐1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742‐749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim T, Cui R, Jeon YJ, et al. Long‐range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo‐5. Proc Natl Acad Sci U S A. 2014;111:4173‐4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nissan A, Stojadinovic A, Mitrani‐Rosenbaum S, et al. Colon cancer associated transcript‐1: a novel RNA expressed in malignant and pre‐malignant human tissues. Int J Cancer. 2012;130:1598‐1606. [DOI] [PubMed] [Google Scholar]
- 78. Kim T, Jeon YJ, Cui R, et al. Role of MYC‐regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl Cancer Inst. 2015;107:dju505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44‐dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890‐6902. [DOI] [PubMed] [Google Scholar]
- 80. Graham LD, Pedersen SK, Brown GS, et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Szafron LM, Balcerak A, Grzybowska EA, et al. The novel gene CRNDE encodes a nuclear peptide (CRNDEP) which is overexpressed in highly proliferating tissues. PLoS ONE. 2015;10:e0127475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non‐coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7:40418‐40436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Meijer D, van Agthoven T, Bosma PT, Nooter K, Dorssers LC. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res. 2006;4:379‐386. [DOI] [PubMed] [Google Scholar]
- 86. Xing Z, Lin A, Li C, et al. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110‐1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Diermeier SD, Chang KC, Freier SM, et al. Mammary tumor‐associated RNAs impact tumor cell proliferation, invasion, and migration. Cell Rep. 2016;17:261‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell‐cycle promoters. Nat Genet. 2011;43:621‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Melo CA, Drost J, Wijchers PJ, et al. eRNAs are required for p53‐dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524‐535. [DOI] [PubMed] [Google Scholar]
- 91. Marin‐Bejar O, Marchese FP, Athie A, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Leveille N, Melo CA, Rooijers K, et al. Genome‐wide profiling of p53‐regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sanchez Y, Segura V, Marin‐Bejar O, et al. Genome‐wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Younger ST, Kenzelmann‐Broz D, Jung H, Attardi LD, Rinn JL. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Res. 2015;43:4447‐4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schmitt AM, Garcia JT, Hung T, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48:1370‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jain AK, Xi Y, McCarthy R, et al. LncPRESS1 is a p53‐regulated LncRNA that safeguards pluripotency by disrupting SIRT6‐mediated de‐acetylation of histone H3K56. Mol Cell. 2016;64:967‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA‐containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861‐868. [DOI] [PubMed] [Google Scholar]
- 98. Blume CJ, Hotz‐Wagenblatt A, Hullein J, et al. p53‐dependent non‐coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29:2015‐2023. [DOI] [PubMed] [Google Scholar]
- 99. Mello SS, Sinow C, Raj N, et al. Neat1 is a p53‐inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095‐1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li XL, Subramanian M, Jones MF, et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 2017;20:2408‐2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chaudhary R, Gryder B, Woods WS, et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. eLife. 2017;6:e23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hu WL, Jin L, Xu A, et al. GUARDIN is a p53‐responsive long non‐coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492‐502. [DOI] [PubMed] [Google Scholar]
- 103. Olivero CE, Martinez‐Terroba E, Zimmer J, et al. p53 activates the long noncoding RNA Pvt1b to inhibit Myc and suppress tumorigenesis. Mol Cell. 2020;77:761‐774.e768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang D, Zhang G, Hu X, et al. Oncogenic RAS regulates long noncoding RNA Orilnc1 in human cancer. Cancer Res. 2017;77:3745‐3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li L, van Breugel PC, Loayza‐Puch F, et al. LncRNA‐OIS1 regulates DPP4 activation to modulate senescence induced by RAS. Nucleic Acids Res. 2018;46:4213‐4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Flockhart RJ, Webster DE, Qu K, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wu L, Xu Q, Zhang H, et al. A new avenue for obtaining insight into the functional characteristics of long noncoding RNAs associated with estrogen receptor signaling. Sci Rep. 2016;6:31716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhang Y, Pitchiaya S, Cieslik M, et al. Analysis of the androgen receptor‐regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet. 2018;50:814‐824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cao L, Zhang P, Li J, Wu M. LAST, a c‐Myc‐inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. eLife. 2017;6:e30433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lu Y, Hu Z, Mangala LS, et al. MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Res. 2018;78:64‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jiang H, Li T, Qu Y, et al. Long non‐coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018;425:78‐87. [DOI] [PubMed] [Google Scholar]
- 112. Gutschner T, Diederichs S. The hallmarks of cancer: a long non‐coding RNA point of view. RNA Biol. 2012;9:703‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non‐coding RNA gadd7 interacts with TDP‐43 and regulates Cdk6 mRNA decay. EMBO J. 2012;31:4415‐4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Khaitan D, Dinger ME, Mazar J, et al. The melanoma‐upregulated long noncoding RNA SPRY4‐IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852‐3862. [DOI] [PubMed] [Google Scholar]
- 115. Wu CL, Wang Y, Jin B, Chen H, Xie BS, Mao ZB. Senescence‐associated long non‐coding RNA (SALNR) delays oncogene‐induced senescence through NF90 regulation. J Biol Chem. 2015;290:30175‐30192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhu S, Li W, Liu J, et al. Genome‐scale deletion screening of human long non‐coding RNAs using a paired‐guide RNA CRISPR‐Cas9 library. Nat Biotechnol. 2016;34:1279‐1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bester AC, Lee JD, Chavez A, et al. An integrated genome‐wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649‐664.e620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Joung J, Engreitz JM, Konermann S, et al. Genome‐scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Liu SJ, Horlbeck MA, Cho SW, et al. CRISPRi‐based genome‐scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:eaah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Liu SJ, Malatesta M, Lien BV, et al. CRISPRi‐based radiation modifier screen identifies long non‐coding RNA therapeutic targets in glioma. Genome Biol. 2020;21:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lai F, Damle SS, Ling KK, Rigo F. Directed RNase H cleavage of nascent transcripts causes transcription termination. Mol Cell. 2020;77:1032‐1043.e1034. [DOI] [PubMed] [Google Scholar]
- 122. Lee JS, Mendell JT. Antisense‐mediated transcript knockdown triggers premature transcription termination. Mol Cell. 2020;77:1044‐1054.e1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bassett AR, Akhtar A, Barlow DP, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA‐p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Groff AF, Sanchez‐Gomez DB, Soruco MML, et al. In vivo characterization of Linc‐p21 reveals functional cis‐regulatory DNA elements. Cell Rep. 2016;16:2178‐2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Amandio AR, Necsulea A, Joye E, Mascrez B, Duboule D. Hotair is dispensible for mouse development. PLoS Genet. 2016;12:e1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li L, Liu B, Wapinski OL, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Schorderet P, Duboule D. Structural and functional differences in the long non‐coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Tripathi V, Ellis JD, Shen Z, et al. The nuclear‐retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925‐938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. West JA, Davis CP, Sunwoo H, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Eissmann M, Gutschner T, Hammerle M, et al. Loss of the abundant nuclear non‐coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487‐1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis‐regulatory role in the adult. Cell Rep. 2012;2:111‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705‐1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Arun G, Spector DL. MALAT1 long non‐coding RNA and breast cancer. RNA Biol. 2019;16:860‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Sun Y, Ma L. New insights into long non‐coding RNA MALAT1 in cancer and metastasis. Cancers (Basel). 2019;11:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha‐regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yang C, Li Z, Li Y, et al. Long non‐coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: a systematic review and meta‐analysis. Oncotarget. 2017;8:2672‐2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Nassar D, Latil M, Boeckx B, Lambrechts D, Blanpain C. Genomic landscape of carcinogen‐induced and genetically induced mouse skin squamous cell carcinoma. Nat Med. 2015;21:946‐954. [DOI] [PubMed] [Google Scholar]
- 143. Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation‐specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Idogawa M, Ohashi T, Sasaki Y, Nakase H, Tokino T. Long non‐coding RNA NEAT1 is a transcriptional target of p53 and modulates p53‐induced transactivation and tumor‐suppressor function. Int J Cancer. 2017;140:2785‐2791. [DOI] [PubMed] [Google Scholar]
- 145. Tseng YY, Moriarity BS, Gong W, et al. PVT1 dependence in cancer with MYC copy‐number increase. Nature. 2014;512:82‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Cho SW, Xu J, Sun R, et al. Promoter of lncRNA gene PVT1 Is a tumor‐suppressor DNA boundary element. Cell. 2018;173:1398‐1412.e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Porter JR, Fisher BE, Baranello L, et al. Global inhibition with specific activation: how p53 and MYC redistribute the transcriptome in the DNA double‐strand break response. Mol Cell. 2017;67:1013‐1025.e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Brockdorff N, Bowness JS, Wei G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020;34:733‐744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sahakyan A, Yang Y, Plath K. The role of Xist in X‐chromosome dosage compensation. Trends Cell Biol. 2018;28:999‐1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Swerdlow AJ, Schoemaker MJ, Higgins CD, Wright AF, Jacobs PA, UK Clinical Cytogenetics Group . Cancer incidence and mortality in men with Klinefelter syndrome: a cohort study. J Natl Cancer Inst. 2005;97:1204‐1210. [DOI] [PubMed] [Google Scholar]
- 151. Sirchia SM, Ramoscelli L, Grati FR, et al. Loss of the inactive X chromosome and replication of the active X in BRCA1‐defective and wild‐type breast cancer cells. Cancer Res. 2005;65:2139‐2146. [DOI] [PubMed] [Google Scholar]
- 152. Kawakami T, Okamoto K, Sugihara H, et al. The roles of supernumerical X chromosomes and XIST expression in testicular germ cell tumors. J Urol. 2003;169:1546‐1552. [DOI] [PubMed] [Google Scholar]
- 153. Yildirim E, Kirby JE, Brown DE, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727‐742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Xing F, Liu Y, Wu SY, et al. Loss of XIST in breast cancer activates MSN‐c‐met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 2018;78:4316‐4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Kong Y, Hsieh CH, Alonso LC. ANRIL: a lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front Endocrinol (Lausanne). 2018;9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Visel A, Zhu Y, May D, et al. Targeted deletion of the 9p21 non‐coding coronary artery disease risk interval in mice. Nature. 2010;464:409‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vendramin R, Verheyden Y, Ishikawa H, et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat Struct Mol Biol. 2018;25:1035‐1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Liu B, Sun L, Liu Q, et al. A cytoplasmic NF‐kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370‐381. [DOI] [PubMed] [Google Scholar]
- 159. Huang D, Chen J, Yang L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation‐induced cell death. Nat Immunol. 2018;19:1112‐1125. [DOI] [PubMed] [Google Scholar]
- 160. Zhu P, Wu J, Wang Y, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;20:1134‐1144. [DOI] [PubMed] [Google Scholar]
- 161. Sharma S, Munger K. Expression of the long noncoding RNA DINO in human papillomavirus‐positive cervical cancer cells reactivates the dormant TP53 tumor suppressor through ATM/CHK2 signaling. mBio. 2020;11e01190‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sauvageau M, Goff LA, Lodato S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01190‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Marin‐Bejar O, Mas AM, Gonzalez J, et al. The human lncRNA LINC‐PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017;18:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC‐PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Hosono Y, Niknafs YS, Prensner JR, et al. Oncogenic role of THOR, a conserved cancer/testis long non‐coding RNA. Cell. 2017;171:1559‐1572.e1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Shen L, Shi Q, Wang W. Double agents: genes with both oncogenic and tumor‐suppressor functions. Oncogenesis. 2018;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the 'undruggable' cancer targets. Nat Rev Cancer. 2017;17:502‐508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non‐coding RNAs in cancer. Trends Mol Med. 2018;24:257‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lieberman J. Tapping the RNA world for therapeutics. Nat Struct Mol Biol. 2018;25:357‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]


