Abstract
The anaerobe Clostridium acetobutylicum belongs to the most important industrially used bacteria. Whereas genome mining points to a high potential for secondary metabolism in C. acetobutylicum, the functions of most biosynthetic gene clusters are cryptic. We report that the addition of supra‐physiological concentrations of cysteine triggered the formation of a novel natural product, clostrisulfone (1). Its structure was fully elucidated by NMR, MS and the chemical synthesis of a reference compound. Clostrisulfone is the first reported natural product with a diphenylsulfone scaffold. A biomimetic synthesis suggests that pentamethylchromanol‐derived radicals capture sulfur dioxide to form 1. In a cell‐based assay using murine macrophages a biphasic and dose‐dependent regulation of the LPS‐induced release of nitric oxide was observed in the presence of 1.
Keywords: clostridia, diphenylsulfone, Griess assay, macrophages, tocopherol
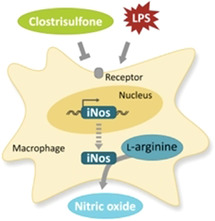
Induction of the industrial anaerobe Clostridium acetobutylicum with cysteine led to the discovery of an unprecedented diarylsulfone natural product named clostrisulfone that likely results from sulfur dioxide capture by chromane‐derived radicals. Its structure was elucidated by NMR and confirmed by synthesis. The tocopherol‐related molecule exerts immunomodulatory activities (see figure).
Anaerobic bacteria, supposedly the oldest creatures on earth, are ubiquitously distributed in oxygen‐free niches such as soils, sediments, and intestines of higher organisms. Among the best‐studied anaerobic bacteria are clostridia, which play major roles in human and animal health, ecology, remediation, and industry. [1] Because of their specialized anoxic catabolism, they are industrially used as solvent producers. An important example is Clostridium acetobutylicum, which has played a major role in the ABE (acetone, butanol, ethanol) Weizmann fermentation process for more than a century. [2] Until recently, however, clostridia—and anaerobes in general—have been neglected as a source of non‐protein natural products. [3] With the advent of massive genome sequencing and bioinformatics, it has become apparent that certain anaerobes have a rich and diverse biosynthetic potential. [4] However, in most cases the gene clusters are silent and need to be activated by means of particular stimuli. Several genetic and chemical approaches to triggering biosynthesis [5] have been applied to clostridia,[ 3a , 6 ] A range of antibiotics, such as closthioamides,[ 3a , 7 ] clostrubins, [8] clostrindoline, [9] antibacterial acyloins, [10] and a new lipopeptide [11] have been isolated from various clostridia. Mining the genome sequence of C. acetobutylicum [12] pointed to a high biosynthetic potential. [4] Apart from a small polyketide that has been implicated in the regulation of cellular differentiation or C. acetobutylicum, [13] so far no secondary metabolites have been reported for these important industrial bacteria. Here, we report the discovery of an unusual, diarylsulfone metabolite from a C. acetobutylicum strain disturbed in sulfur metabolism, verify its unusual structure by synthesis, and evaluate its bioactivities.
To trigger the biosynthesis of cryptic natural products in C. acetobutylicum (DSM 792/ATCC 824), we compared the metabolic profiles of the wild type grown in standard media with those of cultures supplemented with potential elicitors. Generally, no secondary metabolites could be detected in the extracts of standard media. However, when we challenged C. acetobutylicum with supra‐physiological concentrations of l‐cysteine (1 mm) in the culture broth, the metabolic profile changed markedly in the highly unpolar region (Figure 1). We detected a new compound (1) with the sum formula C28H39O4S, deduced from its exact mass (m/z 471.2559 [M+H]+) obtained by HR‐ESI‐MS measurements (Figure S1). To obtain amounts of 1 that would allow for a full structural characterization, we subjected the ethyl acetate extract of 0.5 L culture to reversed‐phase HPLC to yield 0.2 mg.
Figure 1.
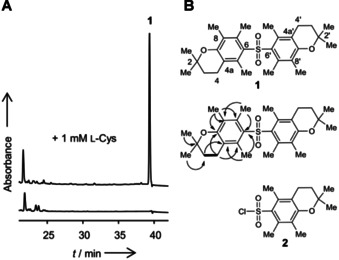
Detection and structural elucidation of clostrisulfone. A) HPLC profiles (UV absorbance at 260 nm) of extracts from C. acetobutylicum cultures with and without cysteine (Cys) supplementation. B) Structure of clostrisulfone (1) and selected 1H‐1H COSY (bold line) and HMBC (arrow) correlations. 1 possesses the same substructure as Pmc chloride (2).
The 1H and 13C NMR spectra of 1 show an unexpectedly low number of signals, which is indicative for a highly symmetric structure (Figures S2 and S3). The DEPT135 and HSQC spectral analysis indicate that 1 possesses five methyl groups (Figures S4 and S6). By the HMBC correlations from 2,2‐Me protons (δH 1.27) to C‐2 (δ 74.0)/C‐3 (δ 31.8) and from methylene protons H‐3 (δ 1.78) to 2,2‐Me carbons (δ 26.3)/C‐3, the gem‐dimethyl moiety is connected to the oxy‐quaternary carbon C‐2 next to C‐3, which is adjacent to a methylene C‐4 (δ 20.4) according to a 1H‐1H COSY coupling signal between H3 and H4 (δ 2.55) (Figures S5 and S7). The HMBC correlations of three aromatic methyl protons and quaternary carbons, 5‐Me (δ H 2.21)/C‐4a (δ 118.5) and C‐6 (δ 134.6), 7‐Me (δ H 2.16)/C‐6 and C‐8 (δ 123.5), and 8‐Me (δ H 2.00)/C‐7 (δ C 134.17), C‐8, C‐8a (δ 153.5) revealed the presence of a fully substituted phenol ring. Finally, the HMBC correlations from H‐4 to C‐4a, C‐5 (δ 134.23), and C‐8a suggested that 1 harbors two 2,2,5,7,8‐pentamethyl‐6‐chromane moieties (Figure 1). Surprisingly, this substructure is the same as the Pmc (2,2,5,7,8‐pentamethylchromane‐6‐sulfonyl) protection group used for the arginine side chain. [14] Since 1 could only be isolated in minute amounts, the presence of the sulfone bridge was initially deduced from HRMS data and the similarity of 13C NMR data of 1 and Pmc chloride (2) (Table S1).
To unequivocally confirm the unusual structure of 1 and to obtain sufficient amounts for bioactivity assays we aimed to synthesize the diarylsulfone. Therefore, we devised a short synthetic route involving a Friedel–Crafts‐type aromatic sulfonylation of chromane 3 with sulfonyl chloride 4, a reagent commonly used for arginine side chain protection. [14] 2,2,5,7,8‐Pentamethyl‐6‐chromane 3 was prepared from 2,3,5‐trimethylphenol and isoprene (Figures S8 and S9). The SnCl4‐mediated coupling of 3 with 4 provided the target molecule (1) in good yield (70 %) (Figures S10 and S11). The proposed structure of clostrisulfone was verified by comparison of physicochemical data of the synthetic compound with the natural product. Owing to its biological origin and key structural feature we named this new compound clostrisulfone (1; Scheme 1).
Scheme 1.
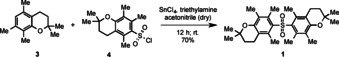
Chemical synthesis of clostrisulfone (1).
The symmetrical diarylsulfone structure of clostrisulfone (1) is highly unusual. Besides sulfadixiamycins B and C from bacterial mangrove endophytes, [15] 1 represents only the second naturally occurring diarylsulfone scaffold, and it is the first reported natural product with a diphenylsulfone substructure. Furthermore, the two chromane substructures of 1 are strikingly similar to 2,2,5,7,8‐pentamethyl‐6‐chromanol, which plays a major role as a synthetic tocopherol (vitamin E) analogue. [16]
It is well conceivable that 1 results from the fusion of sulfur dioxide with two chromane or chromanol building blocks. As genome analyses of C. acetobutylicum did not reveal any obvious gene candidates that could code for the biosynthesis of tocopherol‐like compounds, the pathway to 1 likely differs from known ones. Genome mining did not point to a known biochemical basis for C−S bond formation, either. [17] However, a plausible biosynthetic model could involve sulfur dioxide capture as in sulfadixiamicin biosynthesis, [15] where a flavoprotein has been implicated in xiamycin dimerization and radical‐mediated SO2 incorporation, likely via a semiquinone radical. [18]
Clostridia are sensitive towards SO2, which is an approved additive to preserve a range of foods, such as canned meat. [19] Thus, we reasoned that 1 could result from a sulfur dioxide detoxification pathway involving chromane or chromanol radicals that capture SO2. Such a scenario appears plausible since high concentrations of cysteine, which induce clostrisulfone formation in C. acetobutylicum, could perturb the anaerobes’ sulfur metabolism and could lead to sulfite formation. While still relatively little is known about sulfur metabolism in clostridia, the cysteine addition alters the thiol landscape and thus redox potentials, which may result, among others, in impaired sulfite respiration in clostridia. [20]
To evaluate the potentially increased susceptibility of C. acetobutylicum towards sulfur dioxide we supplemented cultures with either chromane 3 or chromanol 5 and different sulfur sources, gaseous sulfur dioxide, Na2SO3, and 1,4‐diazabicyclo[2.2.2]octane bis(sulfur dioxide) adduct (DABSO), and monitored bacterial growth. Yet, these experiments were not conclusive since chromane 3 and chromanol 5 alone show adverse effects on the growth of the bacteria. Furthermore, the formation of 1 could not be induced with any of the SO2 sources (Tables S2 and S3, Figures S12–14).
Since the in vivo experiments did not provide any insight into the potential biogenesis of 1, we performed various qualitative synthetic experiments and evaluated a potential biomimetic route to 1. First, we screened conditions for the reaction between pentamethylchromane 3 and gaseous sulfur dioxide, sodium sulfite or SO2 released from DABSO. Neither electrochemical oxidations, nor oxidative reactions (with zinc powder, iron(III) chloride, zinc‐copper couple) and radical reaction conditions with different radical starters like azobisisobutyronitrile (AIBN) or dibenzoyl peroxide (DBP) in different solvents (acetonitrile, benzene or carbon tetrachloride) led to detectable amounts of 1 (Figures 2 A and 2B). In contrast, when using chromanol 5 as starting material in the presence of 5 mol‐% of a radical starter like AIBN or DBP at 50 °C, the diarylsulfone was detectable in trace amounts by HRMS‐HPLC. While this approach is potentially biomimetic, alternative avenues in the biosynthesis of 1 cannot be ruled out.
Figure 2.
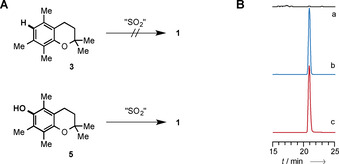
Testing potential biomimetic routes to 1 involving sulfur dioxide capture. A) Radical starters such as azobisisobutyronitrile (AIBN) or dibenzoyl peroxide (DBP) in the presence of Na2SO3 can promote the formation of trace amounts of 1 from chromanol 5, whereas chromane 3 does not show any conversion, even when using other sulfur sources like gaseous sulfur dioxide or DABSO. B) Extracted ion chromatograms (HPLC‐HRMS); a) start of reaction; b) after 24 hours; c) synthetic reference of 1.
As diarylsulfones such as dapsone, bis‐2‐nitrophenylsulfone [21] and diphenylsulfone [22] exhibit antibacterial, antiviral, and cytotoxic activities, we tested 1 in a panel of bioassays. Clostrisulfone did not show any activity against Gram‐positive or Gram‐negative bacteria, mycobacteria, and HIV; it did not show any effect in antifungal, antiviral, cytotoxicity and antiproliferative bioassays, either. Yet, the structural similarity of 1 to tocopherol analogues pointed to a possible anti‐inflammatory activity.
To monitor its pro‐ or anti‐inflammatory potential, we subjected 1 to a cell‐based assay using murine macrophages. Specifically, we pre‐treated the macrophages with increasing concentrations of 1 (0 to 10 μg mL−1) prior to co‐incubation with lipopolysaccharide (LPS, 100 ng mL−1), which induces an inflammatory response and oxidative burst. [23] At the end of the incubation time (24 h) nitric oxide release from the cells was detected by reaction with the Griess reagent (Figure 3). [23] A reduced LPS‐induced NO release would be indicative of an anti‐inflammatory capacity of 1; for a pro‐inflammatory response, contradictory results would be expected.
Figure 3.
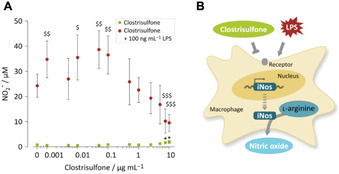
Biphasic and dose‐dependent regulation of LPS‐induced NO release by clostrisulfone in murine macrophages. A) Murine macrophages incubated with increasing concentrations of 1 (0–10 μg mL−1) in the absence (black squares) or presence (grey circles) of LPS (100 ng mL−1). Griess reagent was used to measure NO released by cells. B) Murine macrophages release NO after LPS stimulus via inducible nitric oxide synthase (iNos), which breaks down l‐arginine to generate NO. Interference of 1 with this process may occur at several levels, for example, transcriptional, translational or on functional level by blockage of enzyme activity.
In the inactivated state (incubation in the absence of LPS), only the highest concentrations tested (7.5 and 10 μg mL−1) slightly induce the release of nitric oxide (0.8±0.5 μm nitric oxide in controls vs. 1.9±0.6 μm nitric oxide for 10 μg mL−1 clostrisulfone). All samples treated with LPS released significantly (p<0.05) more nitric oxide compared to LPS‐free controls. While low concentrations of 1 (0.001–0.1 μg mL−1) significantly (p<0.05 to p<0.01) enhance the LPS‐induced NO release compared to controls treated only with LPS (24.3±4.6 μm NO in control vs. 38.7±7.6 μm NO for 0.05 μg mL−1 1), higher concentrations of 1 (7.5–10 μg mL−1) significantly (p<0.001) reduce the LPS‐induced NO release of (9.5±3.4 μm NO for 10 μg mL−1 1). As no cytotoxicity was observed as assessed by MTT assay, the reduction in NO release was not due to reduced cell viability. In conclusion, we observed a biphasic and dose‐dependent regulation of the LPS‐induced release of nitric oxide in murine macrophages in the presence of 1.
In conclusion, by disturbing the thiol landscape in the culture of C. acetobutylicum we have discovered an unprecedented diarylsulfone natural product. The discovery of clostrisulfone is remarkable since this anaerobe has been playing a major role in industrial processes, while the compound has remained hidden. The structure of the highly substituted diarylsulfone is unprecedented among natural products. Its symmetric architecture and a biomimetic synthesis point to a biogenetic origin involving sulfur dioxide capture. The tocopherol‐like substructures may account for the dose‐dependent regulation of the LPS‐induced release of nitric oxide at higher concentrations of 1 and thus indicates immunomodulatory potential. These findings underscore the hidden biosynthetic potential of clostridia and may encourage similar approaches to trigger natural product formation in anaerobes for drug discovery.
Experimental Section
See the Supporting Information.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank A. Perner and H. Heinecke for MS and NMR measurements. Financial support by the BMBF (InfectControl) and the DFG (Leibniz Award) is gratefully acknowledged. Open access funding enabled and organized by Projekt DEAL.
T. Neuwirth, A.-C. Letzel, C. Tank, K. Ishida, M. Cyrulies, L. Schmölz, S. Lorkowski, C. Hertweck, Chem. Eur. J. 2020, 26, 15855.
References
- 1. Bahl H., Dürre P., Clostridia: Biotechnology & Medical Applications, Wiley-VCH, Weinheim, 2001. [Google Scholar]
- 2. Jones D. T., Woods D. R., Microbiol. Rev. 1986, 50, 484–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Lincke T., Behnken S., Ishida K., Roth M., Hertweck C., Angew. Chem. Int. Ed. 2010, 49, 2011–2013; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 2055–2057; [Google Scholar]
- 3b. Behnken S., Hertweck C., Appl. Microbiol. Biotechnol. 2012, 96, 61–67; [DOI] [PubMed] [Google Scholar]
- 3c. Pidot S. J., Coyne S., Kloss F., Hertweck C., Int. J. Med. Microbiol. 2014, 304, 14–22; [DOI] [PubMed] [Google Scholar]
- 3d. Li J. S., Barber C. C., Zhang W., J. Ind. Microbiol. Biotechnol. 2019, 46, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Letzel A. C., Pidot S. J., Hertweck C., Nat. Prod. Rep. 2013, 30, 392–428; [DOI] [PubMed] [Google Scholar]
- 4b. Letzel A. C., Pidot S. J., Hertweck C., BMC Genomics 2014, 15, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a. Scherlach K., Hertweck C., Org. Biomol. Chem. 2009, 7, 1753–1760; [DOI] [PubMed] [Google Scholar]
- 5b. Behnken S., Hertweck C., PLoS ONE 2012, 7, e29609; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5c. Baral B., Akhgari A., Metsä-Ketelä M., Synth. Syst. Biotechnol. 2018, 3, 163–178; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5d. Zhang X., Hindra, Elliot M. A., Curr. Opin. Microbiol. 2019, 51, 9–15. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Behnken S., Lincke T., Kloss F., Ishida K., Hertweck C., Angew. Chem. Int. Ed. 2012, 51, 2425–2428; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 2475–2478; [Google Scholar]
- 6b. Dunbar K. L., Büttner H., Molloy E. M., Dell M., Kumpfmüller J., Hertweck C., Angew. Chem. Int. Ed. 2018, 57, 14080–14084; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 14276–14280. [Google Scholar]
- 7.
- 7a. Kloss F., Chiriac A. I., Hertweck C., Chem. Eur. J. 2014, 20, 15451–15458; [DOI] [PubMed] [Google Scholar]
- 7b. Chiriac A. I., Kloss F., Krämer J., Vuong C., Hertweck C., Sahl H. G., J. Antibicrob. Chemother. 2015, 70, 2576–2588. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Pidot S., Ishida K., Cyrulies M., Hertweck C., Angew. Chem. Int. Ed. 2014, 53, 7856–7859; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 7990–7993; [Google Scholar]
- 8b. Shabuer G., Ishida K., Pidot S. J., Roth M., Dahse H. M., Hertweck C., Science 2015, 350, 670–674. [DOI] [PubMed] [Google Scholar]
- 9. Schieferdecker S., Shabuer G., Knuepfer U., Hertweck C., Org. Biomol. Chem. 2019, 17, 6119–6121. [DOI] [PubMed] [Google Scholar]
- 10. Schieferdecker S., Shabuer G., Letzel A. C., Urbansky B., Ishida-Ito M., Ishida K., Cyrulies M., Dahse H. M., Pidot S., Hertweck C., ACS Chem. Biol. 2019, 14, 1490–1497. [DOI] [PubMed] [Google Scholar]
- 11. Li J. S., Barber C. C., Herman N. A., Cai W., Zafrir E., Du Y., Zhu X., Skyrud W., Zhang W., J. Ind. Microbiol. Biotechnol. 2020, 47, 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nölling J., Breton G., Omelchenko M. V., Makarova K. S., Zeng Q., Gibson R., Lee H. M., Dubois J., Qiu D., Hitti J., Wolf Y. I., Tatusov R. L., Sabathe F., Doucette-Stamm L., Soucaille P., Daly M. J., Bennett G. N., Koonin E. V., Smith D. R., J. Bacteriol. 2001, 183, 4823–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herman N. A., Kim S. J., Li J. S., Cai W., Koshino H., Zhang W., Nat. Commun. 2017, 8, 1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramage R., Green J., Blake A. J., Tetrahedron 1991, 47, 6353–6370. [Google Scholar]
- 15. Baunach M., Ding L., Willing K., Hertweck C., Angew. Chem. Int. Ed. 2015, 54, 13279–13283; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 13477–13481. [Google Scholar]
- 16. Niki E., Kawakami A., Saito M., Yamamoto Y., Tsuchiya J., Kamiya Y., J. Biol. Chem. 1985, 260, 2191–2196. [PubMed] [Google Scholar]
- 17. Dunbar K. L., Scharf D. H., Litomska A., Hertweck C., Chem. Rev. 2017, 117, 5521–5577. [DOI] [PubMed] [Google Scholar]
- 18.
- 18a. Zhang Q., Li H., Yu L., Sun Y., Zhu Y., Zhu H., Zhang L., Li S. M., Shen Y., Tian C., Li A., Liu H. W., Zhang C., Chem. Sci. 2017, 8, 5067–5077; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18b. Baunach M., Ding L., Bruhn T., Bringmann G., Hertweck C., Angew. Chem. Int. Ed. 2013, 52, 9040–9043; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 9210–9213. [Google Scholar]
- 19. Tompkin R. B., Christiansen L. N., Spaparis A. B., Appl. Environ. Microbiol. 1980, 39, 1096–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.
- 20a. André G., Haudecoeur E., Monot M., Ohtani K., Shimizu T., Dupuy B., BMC Microbiol. 2010, 10, 234; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20b. Dubois T., Dancer-Thibonnier M., Monot M., Hamiot A., Bouillaut L., Soutourina O., Martin-Verstraete I., Dupuy B., Infect. Immun. 2016, 84, 2389–2405; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20c. Gu H., Shi K., Liao Z., Qi H., Chen S., Wang H., Li S., Ma Y., Wang J., Microbiol. Res. 2018, 215, 114–125. [DOI] [PubMed] [Google Scholar]
- 21. McMahon J. B., Gulakowski R. J., Weislow O. S., Schultz R. J., Narayanan V. L., Clanton D. J., Pedemonte R., Wassmundt F. W., Buckheit R. W. J., Decker W. D., White E. L., Bader J. P., Boyd M. R., Antimicrob. Agents Chemother. 1993, 37, 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J. J., Duh C. Y., Chen I. S., Planta Med. 2005, 71, 370–372. [DOI] [PubMed] [Google Scholar]
- 23. Schmölz L., Wallert M., Lorkowski S., J. Immunol. Methods 2017, 449, 68–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


