Summary
Sesamum spp. (sesame) are known to accumulate a variety of lignans in a lineage‐specific manner. In cultivated sesame (Sesamum indicum), (+)‐sesamin, (+)‐sesamolin and (+)‐sesaminol triglucoside are the three major lignans found richly in the seeds. A recent study demonstrated that SiCYP92B14 is a pivotal enzyme that allocates the substrate (+)‐sesamin to two products, (+)‐sesamolin and (+)‐sesaminol, through multiple reaction schemes including oxidative rearrangement of α‐oxy‐substituted aryl groups (ORA). In contrast, it remains unclear whether (+)‐sesamin in wild sesame undergoes oxidation reactions as in S. indicum and how, if at all, the ratio of the co‐products is tailored at the molecular level. Here, we functionally characterised SrCYP92B14 as a SiCYP92B14 orthologue from a wild sesame, Sesamum radiatum, in which we revealed accumulation of the (+)‐sesaminol derivatives (+)‐sesangolin and its novel structural isomer (+)‐7´‐episesantalin. Intriguingly, SrCYP92B14 predominantly produced (+)‐sesaminol either through ORA or direct oxidation on the aromatic ring, while a relatively low but detectable level of (+)‐sesamolin was produced. Amino acid substitution analysis suggested that residues in the putative distal helix and the neighbouring heme propionate of CYP92B14 affect the ratios of its co‐products. These data collectively show that the bimodal oxidation mechanism of (+)‐sesamin might be widespread across Sesamum spp., and that CYP92B14 is likely to be a key enzyme in shaping the ratio of (+)‐sesaminol‐ and (+)‐sesamolin‐derived lignans from the biochemical and evolutionary perspectives.
Keywords: Sesamum radiatum, Sesamum indicum, lignan, CYP92B14, (+)‐7´‐episesantalin, (+)‐sesangolin, (+)‐sesamin, (+)‐sesamolin, (+)‐sesaminol, oxidative rearrangement of α‐oxy‐substituted aryl groups (ORA)
Significance Statement
In Sesamum indicum (sesame), SiCYP92B14 oxidises (+)‐sesamin, a health‐promoting compound, to either (+)‐sesaminol or (+)‐sesamolin via two distinct reaction mechanisms. The identification and biochemical characterisation of SrCYP92B14 from wild sesame, S. radiatum, revealed that SrCYP92B14 oxidises (+)‐sesamin predominantly to (+)‐sesaminol, which corroborates with the lignan profile of S. radiatum, thus highlighting the relevance of CYP92B14 proteins in shaping the molar ratio of (+)‐sesaminol and (+)‐sesamolin derivatives that accumulate in Sesamum spp.
INTRODUCTION
Lignans are a class of specialised metabolites that are derived from a central precursor, pinoresinol, which is produced by the oxidative coupling of two coniferyl alcohol molecules (Davin et al., 1997; Umezawa, 2003; Suzuki and Umezawa, 2007). Lignans are widespread across the plant kingdom, and occur in the seeds of Sesamum indicum (sesame), Olea europaea (olive), Carthamus tinctorius (safflower), Linum usitatissimum (flaxseed) and various other oil crops (Gunstone, 2011). While lignans have been regarded as potent phytochemicals used in the defense against microorganisms or allelochemicals that inhibit the growth of neighbouring plants (Yamauchi et al., 2015), as yet, the molecular basis for the biological significance of lignans in plants has not been fully elucidated. Moreover, lignans are also known to exhibit various health‐promoting activities (Dar and Arumugam, 2013). For example, (+)‐sesamin, (+)‐sesamolin and (+)‐sesaminol interfere in arachidonic acid biosynthesis by inhibiting delta 5‐desaturase, therefore reducing the formation of pro‐inflammatory mediators (Shimizu et al., 1991). In addition, (+)‐sesamolin, sesamol and (+)‐sesamolinol were observed to inhibit lipid peroxidation in rat liver and kidney (Kang et al., 1998). (+)‐Sesamin, sesamol and other lignans have also been shown to have radical scavenging activity in vitro (Suja et al., 2004). Moreover, (+)‐sesamin has been identified as a phytoestrogen, a precursor of the mammalian lignans enterolactone and enterodiol, and to exhibit protective activity against breast and prostate cancers. It is also known to interfere with the NF‐κB, STAT3, JNK, ERK1/2 and p53 signalling pathways (Majdalawieh et al., 2017).
Major lignans that accumulate in seeds of S. indicum, cultivated sesame, include (+)‐sesamin, (+)‐sesamolin and (+)‐sesaminol triglucosides (Beroza and Kinman, 1955; Fukuda et al., 1988; Kamal‐Eldin and Appelqvist, 1994; Bedigian, 2003, 2010a; Moazzami and Kamal‐Eldin, 2006; Figure S1). The enzymes involved in the biosynthesis of lignans have been well characterised in S. indicum (Figure 1). (+)‐Pinoresinol is subjected to sequential oxygenation reactions to be converted to (+)‐sesamin by the piperitol/sesamin synthase SiCYP81Q1, a cytochrome P450 monooxygenase (CYP) that forms two methylenedioxy bridges (MDBs) (Ono et al., 2006). Likewise, SrCYP81Q2 was identified from a wild sesame, Sesamum radiatum, as an orthologue of SiCYP81Q1 and shown to oxygenate (+)‐pinoresinol in the production of (+)‐sesamin (Ono et al., 2006). However, SaCYP81Q3 from another wild sesame species Sesamum alatum was recently shown to oxygenate (+)‐epipinoresinol and catalyse MDB formation to produce (+)‐pluviatilol, a putative intermediate of (+)‐alatumin and (+)‐2‐episesalatin (Ono et al., 2006, 2018). In S. indicum, (+)‐sesamin is further oxygenated via two distinct mechanisms by another CYP, SiCYP92B14, to co‐generate (+)‐sesamolin and (+)‐sesaminol. (+)‐Sesamolin is generated through a scheme designated as oxidative rearrangement of α‐oxy‐substituted aryl groups (ORA), which involves oxygenation at the C1 position, followed by rearrangement of the aromatic ring system. (+)‐Sesaminol, however, is generated either through ORA or by direct oxidation at the C6 position of the aromatic ring (Figure 1; Murata et al., 2017). Lastly, (+)‐sesaminol is glucosylated sequentially through reactions catalysed by the UDP‐glucosyltransferases (UGT) UGT71A9, UGT94D1, UGT94AA2 and UGT94AG1, thereby accumulating as a water‐soluble sesaminol triglucoside (STG; Noguchi et al., 2008; Ono et al., 2020).
Figure 1.
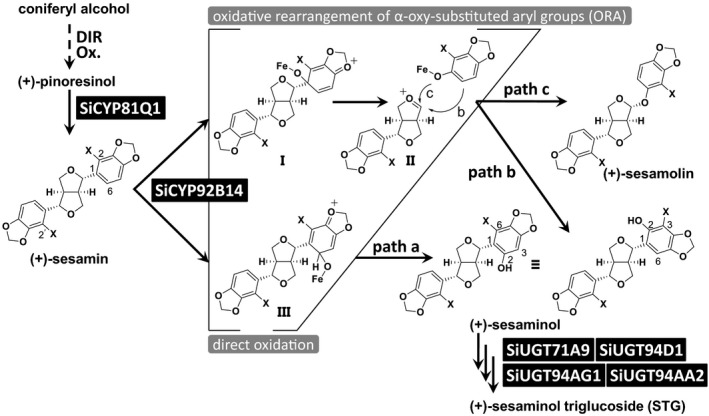
Lignan biosynthetic pathway in Sesamum indicum.
Identified enzymes are indicated in black boxes. A dashed arrow indicates the possible route catalysed by putative dirigent protein (DIR) and oxide (Ox.). SiCYP92B14 is a pivotal enzyme that allocates the substrate (+)‐sesamin to (+)‐sesamolin and (+)‐sesaminol through three separate paths via either direct oxidation (path a) or oxidative rearrangement of α‐oxy‐substituted aryl groups (ORA; paths b and c). I, II and III; intermediate compounds. Atom labels ‘X’ represent either hydrogen ‘H’ or chemically introduced deuterium ‘D’ as identified in Murata et al. (2017).
There are more than 20 wild relatives of S. indicum (Bedigian, 2010b). Previous analyses of lignan profiles demonstrated that wild sesame accumulates lignans with unique structures that are often lineage‐specific (Jones et al., 1962; Fukuda et al., 1988; Kamal‐Eldin and Appelqvist, 1994; Moazzami and Kamal‐Eldin, 2006; Figures 2c and S1). For example, S. alatum seeds accumulate (+)‐alatumin and (+)‐2‐episesalatin, both of which have one instead of two MDBs, as in (+)‐sesamin, whereas (+)‐sesamolin and (+)‐sesaminol are undetectable (Kamal‐Eldin and Yousif, 1992; Kamal‐Eldin et al., 1994; Ono et al., 2018). Moreover, another wild sesame, Sesamum angolense, primarily accumulates (+)‐sesamin and an O‐methylated form of (+)‐sesaminol, (+)‐sesangolin (Jones et al., 1962). These reports indicate that the catalytic properties of lignan biosynthetic enzymes vary widely and contribute to structurally diverse lignan profiles specialised for each sesame species.
Figure 2.
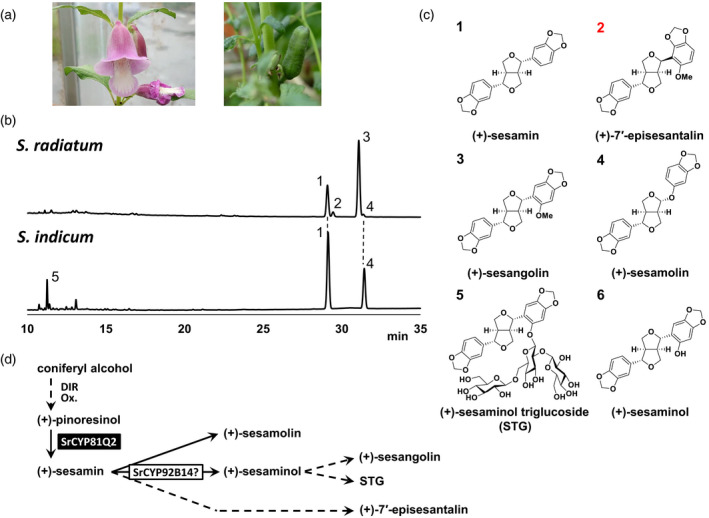
Sesamum radiatum exhibits a unique lignan profile.
(a) Sesamum radiatum flower (left) and capsule (right).
(b) High‐performance liquid chromatography (HPLC) chromatograms of S. radiatum and S. indicum seed extracts. UV absorbance was detected at 280 nm. Numbers next to peaks indicate lignans corresponding to those in (c).
(c) Major lignans that are generated in S. indicum and S. radiatum. Note that (+)‐7´‐episesantalin is a novel lignan identified from S. radiatum in this study (highlighted with a red number).
(d) Possible lignan biosynthetic pathway in S. radiatum. Dashed arrows indicate possible biosynthetic routes of lignans. SrCYP81Q2 responsible for (+)‐sesamin biosynthesis from (+)‐pinoresinol (Ono et al., 2006).
In contrast to the above‐mentioned sesame species, lignan profiles in S. radiatum have been poorly studied; virtually no information has been available with respect to the amount of lignans that accumulate in S. radiatum seeds (Bedigian et al., 1985). Using liquid chromatography‐mass spectrometry (LC‐MS), we revealed that S. radiatum seeds accumulate a considerable level of (+)‐sesangolin and its novel isomer (+)‐7´‐episesantalin, both of which are (+)‐sesaminol derivatives. Moreover, (+)‐sesamin and, to a much lesser extent, (+)‐sesamolin were detected in S. radiatum seeds. This unique lignan profile of S. radiatum prompted us to identify and biochemically characterise a SiCYP92B14 orthologue from S. radiatum (SrCYP92B14), and uncover the molecular basis for the metabolic diversity of lignan profiles in S. indicum and S. radiatum.
RESULTS
Sesamum radiatum seeds accumulate (+)‐sesamin and two (+)‐sesaminol‐derived lignans (+)‐sesangolin and (+)‐7´‐episesantalin
Previous qualitative analysis of lignans by thin‐layer chromatography claimed that S. radiatum (Figure 2a) seeds accumulated (+)‐sesamin, whereas no spots corresponding to (+)‐sesamolin were detected (Kamal‐Eldin et al., 1994). However, the data were unclear, as the results not only showed multiple unassigned bands on the chromatographic assay, but also the author commented later in a book chapter that Sesamum latifolium was misassigned as S. radiatum in the original report (Kamal‐Eldin et al., 1994; Kamal‐Eldin, 2010). Therefore, to seek further confidence in the lignan profile of S. radiatum, we thoroughly investigated the seed contents of this species using LC‐MS. UV spectral analysis of the seed extract detected two major peaks at retention times of 28.9 (peak 1) and 31.0 (peak 3), and two minor peaks at 29.1 (peak 2) and 31.3 (peak 4; Figures 2b and S2). MS and co‐chromatogram analyses with lignan standards revealed that peak 1 and the smaller peak 4 corresponded to (+)‐sesamin and (+)‐sesamolin, respectively, both of which were also generally found in seeds of S. indicum. Peaks 2 and 3 shared the identical fragment ion m/z 367.1163 [M+H‐H2O]+. Unexpectedly, LC‐MS analyses combined with nuclear magnetic resonance (NMR) experiments identified major peak 3 as (+)‐sesangolin, which is previously found in Sesamum angustifolium (Kamal‐Eldin et al., 1994) and S. angolense (Jones et al., 1962). Moreover, peak 2 turned out to be a (+)‐sesangolin isomer, (7α,7´β,8α,8´α)‐6´‐methoxy‐3,4:2´,3´‐bis(methylenedioxy)‐7,9´:7´,9‐diepoxylignan, and we designated this novel lignan as (+)‐7´‐episesantalin (Figure 2c; Table S1).
Identification of a SiCYP92B14 homologous protein in Sesamum radiatum
The finding that S. radiatum seeds accumulate (+)‐sesaminol derivatives (+)‐sesangolin and (+)‐7´‐episesantalin allowed us to speculate that S. radiatum contains an enzyme that is functionally similar to but distinct from SiCYP92B14 (Figure 2d; Murata et al., 2017). Thus, we conducted RNA‐Seq analysis of developing S. radiatum seeds to investigate the presence of expressed sequences with high similarity to that of SiCYP92B14. A BLAST analysis identified a sequence that was 91% identical to that of SiCYP92B14 at the amino acid level, and designated the gene as SrCYP92B14. During seed development, the expression of SrCYP92B14 was upregulated at 35 days after pollination (DAP) and reached its peak at 42 DAP, followed by a rapid decrease at 49 DAP. The expression profile of SrCYP92B14 roughly matched that of the piperitol/sesamin synthase gene SrCYP81Q2 (Ono et al., 2006), which was also expressed in the latter half of the seed development and reached its peak at 42 DAP (Figure 3a). These data suggest that SrCYP81Q2 and SrCYP92B14 are coordinately involved in lignan biosynthesis during seed development in S. radiatum. Moreover, the expression of SrCYP98A20‐like (a potent p‐coumarate 3‐hydroxylase), encoding another P450 monooxygenase with remote functional relevance to lignan biosynthesis, and a cytochrome P450 reductase gene SrCPR1 (that was also identified in this study) were constant throughout seed development. Phylogenetic analysis revealed that SrCYP92B14 exhibited higher sequence similarity to SiCYP92B14 than SIN_1021319, which was the closest homologue of SiCYP92B14 among S. indicum CYP proteins, supporting the view that SrCYP92B14 was a gene product orthologous to SiCYP92B14 (Figure 3b).
Figure 3.
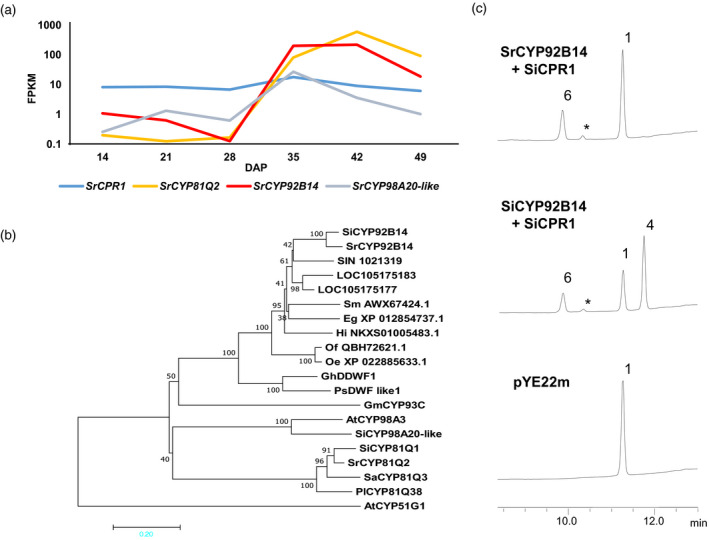
SrCYP92B14 catalyses oxidation of (+)‐sesamin with a catalytic outcome distinct from that of SiCYP92B14.
(a) RNA‐Seq analysis of SrCYP92B14 during Sesamum radiatum seed development. The expression profiles of SrCYP92B14, SrCYP81Q2, SrCPR1 and SrCYP98A20‐like during S. radiatum seed development were analysed by RNA‐Seq. FPKM; fragments per kilobase megareads. DAP, days after pollination.
(b) Phylogenetic analysis of SrCYP92B14 by the neighbour‐joining method using MEGA7. Bootstrap values (1000 replicates) are shown next to the branches. Scale bar represents the rate of substitutions/site.
(c) SrCYP92B14 oxidised (+)‐sesamin primarily to (+)‐sesaminol. Yeast cells expressing SrCYP92B14 together with SiCPR1 were subjected to bioassays using (+)‐sesamin as a substrate. Yeast cells harbouring the empty vector pYE22m were used as negative controls. Bioassay products after 48 h incubation were analysed on high‐performance liquid chromatography (HPLC). Numbers next to peaks indicate lignans corresponding to those in Figure 1c. 1: (+)‐Sesamin; 4: (+)‐sesamolin; 6: (+)‐sesaminol. Asterisks indicate (+)‐2‐episesaminol produced regardless of the expressed CYPs in the bioassays.
Next, we performed biochemical characterisation of SrCYP92B14 using a yeast expression system. The data showed that SrCYP92B14 catalyses the oxidation of (+)‐sesamin to produce (+)‐sesaminol (Figure 3c). Conversely, in clear contrast to SiCYP92B14, SrCYP92B14 barely generated (+)‐sesamolin from (+)‐sesamin. The results were consistent with the finding that S. radiatum seeds predominantly accumulate the (+)‐sesaminol derivative (+)‐sesangolin and, to a lesser extent, (+)‐sesamolin (Figures 2b and S2). An investigation into substrate preference revealed that, as in the case of SiCYP92B14, SrCYP92B14 was not involved in oxidising structural isomers of (+)‐sesamin (Figure S3). Notably, (+)‐2‐episesaminol was generated spontaneously (or non‐enzymatically) from (+)‐sesaminol in our bioassays. Likewise, (+)‐samin and sesamol were also produced from (+)‐sesamolin independently of CYPs in the assay. Taken together, SrCYP92B14 was found to be a unique sesamin monooxygenase, which preferentially generates (+)‐sesaminol as a product.
Comparison of predicted SiCYP92B14 and SrCYP92B14 structures
Because SiCYP92B14 and SrCYP92B14 shared fairly high sequence similarity at the amino acid level, the key amino acids responsible for the difference in the catalytic outcome likely lay within the polymorphic regions. To gain further insight into the structure−activity relationship of CYP92B14 proteins, we used homology modelling to build their structures. As the structures of plant CYPs bound to ligands with molecular sizes similar to (+)‐sesamin have been poorly characterised, we chose human CYP2R1, vitamin D 25‐hydroxylase, as a template (Figure S4). Predicted CYP92B14 model structures and their secondary structure information were presented in Figure 4.
Figure 4.
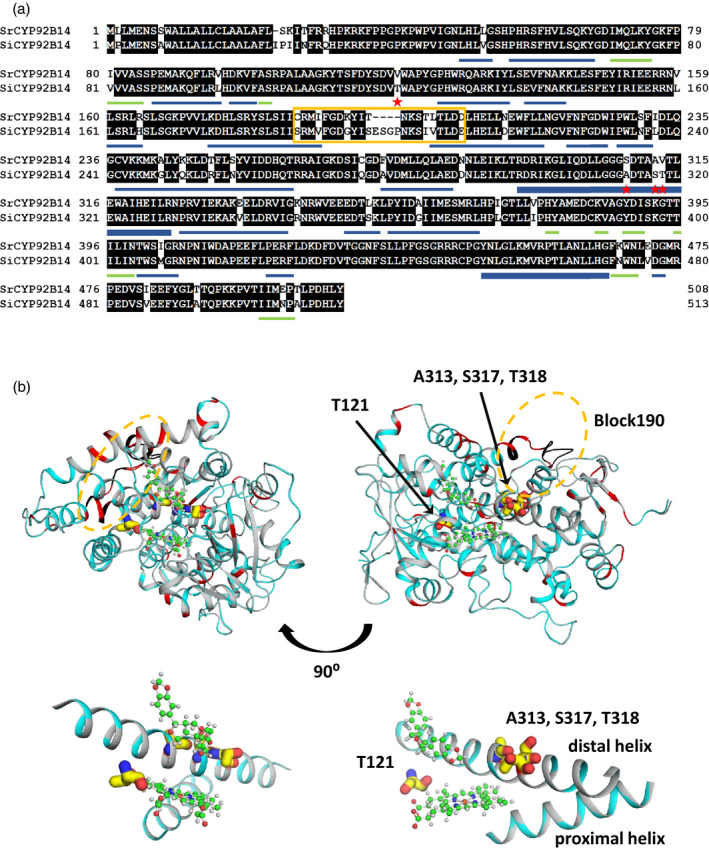
Sequence and structural comparison of SrCYP92B14 and SiCYP92B14.
(a) Sequence comparison between SrCYP92B14 and SiCYP92B14 proteins. Sequences were aligned with GENETYX ver. 12. Red stars and yellow box depict the exchanged residues in SrCYP92B14 and SiCYP92B14 mutant proteins. The secondary structure of CYP92B14 was defined by DSSP programme (Kabsch and Sander, 1983). Helical (α, 310 and PI helices) and β sheet structures are represented by blue and green bars, respectively. The proximal and distal helices are represented by bold blue bars.
(b) Model structures of SrCYP92B14 and SiCYP92B14. The model structures were drawn by PyMOL
ver. 2.4.0a0. Ribbon model structures of SrCYP92B14 and SiCYP92B14 are represented in grey and cyan, respectively. Non‐conserved residues and exchanged regions are coloured in red and black, respectively. Heme and (+)‐sesamin are represented by ball and stick models. The substituted residues that were shown as red stars in (a) are represented by sticks. Partial enlargement view of the heme active site including (+)‐sesamin, proximal and distal helices; the substituted residues are shown at the bottom.
The model structures indicated that amino acid residues polymorphic between SiCYP92B14 and SrCYP92B14 were located mostly at the protein surface. Notably, the loop that connects helices, depicted as a yellow box (Block190), is a region with the lowest similarity between SiCYP92B14 and SrCYP92B14 (Figure 4a). In addition, the model structures revealed that there were additional polymorphic amino acid residues around the heme active site.
In CYP and other heme proteins, the heme active site is sandwiched by two helices, the proximal and the distal helices, and the structural features of these two helices, together with the heme active site and ligand surroundings, generally define the catalytic properties of the enzyme (Schuler and Berenbaum, 2013; Poulos, 2014). Based on these concepts, we selected the following regions and amino acid residues for amino acid substitution experiments, and tested whether any of the residues play crucial roles in the catalytic outcome of CYP92B14 proteins: (i) Block190; (ii) A313, S317 and T318 in SiCYP92B14, corresponding to S308, A312 and V313 in SrCYP92B14, respectively, located in the distal helix (Figure 4b); and (iii) T121 in SiCYP92B14, corresponding to V120 in SrCYP92B14, a residue adjacent to the heme propionate group constituting the active centre of CYP92B14 proteins (Figure 4b).
(+)‐Sesamin oxidation analyses of SiCYP92B14 and SrCYP92B14 mutant proteins
To test the contribution of the polymorphic amino acid residues of CYP92B14 proteins to their catalytic preference, we evaluated the enzymatic properties of mutant CYP92B14 proteins using the ratio of the products: (i) (+)‐sesaminol through direct oxidation at the C6 position (path a); (ii) (+)‐sesaminol through ORA (path b) at the C1 position; and (iii) (+)‐sesamolin through ORA (path c) at the C1 position (Figure 1). In the initial set of experiments, we introduced amino acid substitutions into SiCYP92B14 at the residues mentioned above, and replaced the residues corresponding to those of SrCYP92B14 (SiCYP92B14_T121V, A313S, S317A and T318V). We also constructed a set of SrCYP92B14 mutant plasmid constructs carrying the reciprocal amino acid substitutions replacing the residues corresponding to those of SiCYP92B14 (SrCYP92B14_V120T, S308A, A312S and V313T). Furthermore, we also constructed chimeric proteins with the residues spanning position 190 exchanged between SiCYP92B14 and SrCYP92B14 (SiCYP92B14_Block190 and SrCYP92B14_Block190). These mutant proteins were expressed in yeast cells and subjected to bioassay experiments (Figure S5).
The bioassay experiments demonstrated that the single amino acid substitutions SiCYP92B14_A313S, S317A and T318V and replacements in Block190 resulted in a slight increase in the product ratio of (+)‐sesaminol and (+)‐2‐episesaminol (Sesaminol Total) over the total amount of co‐products (Product Total). Compared with these mutants, the amino acid substitution SiCYP92B14_T121V in the heme active site led to a more apparent increase in the ratio of Sesaminol Total over Product Total (Table S2). These results suggest that T121 plays an important role in the biosynthetic catalysis of (+)‐sesamolin. To better understand the integral effect of amino acid substitutions in the distal helix and heme active sites, we prepared mutant proteins carrying multiple amino acid substitutions, SiCYP92B14_A313S/S317A/T318V and SiCYP92B14_T121V/A313S/S317A/T318V. Considering the subtle effect of the replacement of 11 amino acids in Block190 compared with the single amino acid substitutions in T121, A313, S317 and T318, we excluded Block190 from the list of target sites for the following combined amino acid substitution experiments.
Among other mutant proteins, SiCYP92B14_T121V/A313S/S317A/T318V showed an approximately 3.3‐fold increase in the ratio of Sesaminol Total over Product Total compared with that of wild‐type SiCYP92B14 (SiCYP92B14 WT) (Table 1). The result demonstrates that there are key amino acid changes that are responsible for the product ratio of SiCYP92B14. In contrast, none of the amino acid substitutions to SrCYP92B14 was successful in modifying the product ratio, indicating that a more thorough investigation of amino acid substitutions is required to enhance the sesamolin biosynthetic activity of SrCYP92B14.
Table 1.
Molar ratios of Sesaminol Total to Product Total in (+)‐sesamin oxidation bioassays
| CYP | Sesaminol Total/Product Total |
|---|---|
| SiCYP92B14 WT | 0.16 ± 0.002 |
| SiCYP92B14_Block190 | 0.22 ± 0.016 |
| SiCYP92B14_A313S/S317A/T318V | 0.37 ± 0.003 |
| SiCYP92B14_T121V | 0.30 ± 0.018 |
| SiCYP92B14_T121V/A313S/S317A/T318V | 0.53 ± 0.009 |
Product Total: sum of Sesaminol Total and Sesamolin Total. Sesaminol Total: sum of (+)‐sesaminol and (+)‐2‐episesaminol. Sesamolin Total: sum of (+)‐sesamolin, (+)‐samin and sesamol. Experiments were performed in triplicate (n = 3).
2H‐labelled (+)‐sesamin oxidation in SiCYP92B14 and SrCYP92B14 mutant proteins
We previously showed that SiCYP92B14 produces (+)‐sesamolin through ORA, whereas (+)‐sesaminol is produced via two separate paths, ORA and direct oxidation of the aromatic ring (Murata et al., 2017). Because SrCYP92B14 and its mutant proteins primarily generated (+)‐sesaminol and barely generated (+)‐sesamolin (Tables 1 and S2), we speculated that SrCYP92B14 lacks the ability to catalyse ORA and therefore possibly generates (+)‐sesaminol only by direct oxidation. To test this hypothesis, the reaction mechanism of SrCYP92B14 was analysed in detail using (+)‐2,2′‐2H2‐sesamin as a substrate with the positions of the 2H‐labelling in the enzyme assay products assessed by NMR (Murata et al., 2017). Unexpectedly, the data showed that SrCYP92B14 generated (+)‐sesaminol by both the ORA (path b) and direct oxidation (path a) schemes (Table 2; Figure S6). In the case of SiCYP92B14, (+)‐sesamin was oxidised primarily by ORA (93%), followed by direct oxidation (7%). In contrast, SrCYP92B14 oxidised (+)‐sesamin at the C6 position of the aromatic ring by direct oxidation (71%) and ORA (29%), the latter of which was triggered by the oxidation of the C1 position of the aromatic ring. Amino acid substitutions in SiCYP92B14 in Block190 resulted in only a slight increase in the ratio of Sesaminol Total over Product Total compared with that of SiCYP92B14 WT, which was explained by the minor increase in the ratio of (+)‐sesaminol both by direct oxidation (from 7% to 11%; path a) and ORA (from 11% to 15%; path b) simultaneously. In clear contrast, SiCYP92B14_A313S/S317A/T318V harbouring the amino acid substitutions in the distal helix showed a more significant increase in the ratio of Sesaminol Total over Product Total, expressing a change in the sum of direct oxidation (path a) and ORA (path b; from 18% to 44%). As a result, SiCYP92B14_A313S/S317A/T318V led to a substantial change (from 13% to 50%) in the ratio of (+)‐sesaminol production (path b) over (+)‐sesamolin production (path c) in the context of ORA. SiCYP92B14_T121V/A313S/S317A/T318V thereby produced the highest ratio of Sesaminol Total over Product Total tested in this study, which was facilitated by the synergistic effect of amino acid substitutions in the distal helix (A313, S317 and S317) and T121 (Figure S7).
Table 2.
Molar ratios of (+)‐sesamolin and (+)‐sesaminol produced through paths a, b and c by CYP92B14 proteins using 2H‐labelled (+)‐sesamin as a substrate
| CYP | Ratio (%) | |||
|---|---|---|---|---|
| sesaminol | sesamolin | b/c | ||
| Direct oxidation (a) | ORA (b) | ORA (c) | ||
| SrCYP92B14 WT | 71 | 29 | trace | – |
| SiCYP92B14 WT | 7 | 11 | 82 | 13 |
| SiCYP92B14_Block190 | 11 | 15 | 74 | 20 |
| SiCYP92B14_A313S/S317A/T318V | 16 | 28 | 56 | 50 |
| SiCYP92B14_T121V | 4 | 24 | 72 | 33 |
| SiCYP92B14_T121V/A313S/S317A/T318V | 8 | 41 | 51 | 80 |
| SrCYP92B14_V120T | 87 | 13 | trace | – |
ORA, Oxidative rearrangement of α‐oxy‐substituted aryl groups.
DISCUSSION
Lignans derived from the oxidation of (+)‐sesamin can be categorised into two major groups, sesaminol‐type and sesamolin‐type, based on the positions to which the oxygen molecule is attached. In many cases, (+)‐sesaminol is subjected to further modifications, O‐methylation and O‐glucosylation (Kamal‐Eldin and Appelqvist, 1994; Katsuzaki et al., 1994; Figure S1). However, because the quantitative levels of lignans in various Sesamum spp. have been poorly investigated (Bedigian et al., 1985; Bedigian, 2003; Kamal‐Eldin, 2010), the molar ratio of sesaminol‐type over sesamolin‐type lignans in various Sesamum spp. has not been well understood.
Although a previous study purported to characterise the lignans in S. radiatum (Kamal‐Eldin et al., 1994), the seeds used in that investigation turned out to be those of S. latifolium (Kamal‐Eldin, 2010). Here, we show that S. radiatum seeds predominantly contain (+)‐sesangolin and (+)‐sesamin, with much smaller amounts of (+)‐sesamolin and a (+)‐sesangolin isomer (+)‐7´‐episesantalin. Moreover, we determined that SrCYP92B14 generated far more (+)‐sesaminol than (+)‐sesamolin from the substrate (+)‐sesamin (Figure 3c; Table 2). This enzymatic property of SrCYP92B14 was in clear contrast to that of SiCYP92B14, which preferentially generated (+)‐sesamolin over (+)‐sesaminol from (+)‐sesamin. The differences in the ratio of SiCYP92B14 and SrCYP92B14 co‐products were consistent with the lignan profiles between S. indicum and S. radiatum (Figures 2b and S2). From these data, we concluded that the oxygenation of (+)‐sesamin by CYP92B14 proteins is the pivotal branch point for shaping lignan profiles in S. indicum and S. radiatum. In turn, the data also suggest that SrCYP92B14 is involved in (+)‐sesangolin biosynthesis by primarily producing (+)‐sesaminol in S. radiatum (Figure S8).
In this study, (+)‐7´‐episesantalin was identified from S. radiatum as a novel structural isomer of (+)‐sesangolin. Because the position of a MDB in the aromatic ring attached to the C7´ position of the furofuran ring system is inconsistent to that of (+)‐sesamin, we speculate that (+)‐7´‐episesantalin is another example, in addition to (+)‐sesamolin and (+)‐sesaminol, of oxidation products produced via ORA. Future experiments will reveal whether the biosynthesis of (+)‐7´‐episesantalin requires epimerisation at the C7´ position to occur prior to O‐methylation or involves other mechanisms.
The NMR analysis of the oxygenation products of 2H‐labelled (+)‐sesamin revealed that the proportion of direct carbon oxidation in the aromatic ring of (+)‐sesamin over ORA was much larger when catalysed by SrCYP92B14 compared with SiCYP92B14 (Figure 5). The increased ratio of direct oxidation (path a) likely contributed to the preferred (+)‐sesaminol production by SrCYP92B14 compared with SiCYP92B14 (Table 2). The ratio between (+)‐sesamolin and (+)‐sesaminol through ORA (paths b and c) depends on the ratio between C‐O and C‐C bond formation, respectively, via the common oxonium intermediate II (Figure 1). Therefore, we postulate that the difference in the overall catalytic outcome via ORA and direct oxidation between SiCYP92B14 and SrCYP92B14 lies within various parameters of the active site environment, such as the amino acid residues surrounding heme and the substrate (+)‐sesamin, hydrogen bond network, charge repulsion, and water accessibility. This hypothesis is clearly supported by our data, in which amino acid substitutions in the distal helix located in heme surroundings in SiCYP92B14 to those of SrCYP92B14 led to the increase in the product ratio of (+)‐sesaminol over (+)‐sesamolin through ORA (Table 2). Alternatively, the product ratio of (+)‐sesaminol (7%) through direct oxidation (path a) was decreased in SiCYP92B14_T121V (4%) compared with SiCYP92B14 WT, whereas it was increased in SrCYP92B14_V120T (87%) compared with SrCYP92B14 WT (71%; Tables 1 and 2). These results were unexpected, as the amino acid substitution introduced to SiCYP92B14 was designed to mimic the active site environment of SrCYP92B14, which primarily oxidises (+)‐sesamin by direct oxidation. The results suggest that a hydrophilic group in Thr stabilises hydrogen bond networks in the heme propionate (Guallar and Olsen, 2006) that shapes the active site environment of SiCYP92B14 to favour direct oxidation (path a).
Figure 5.
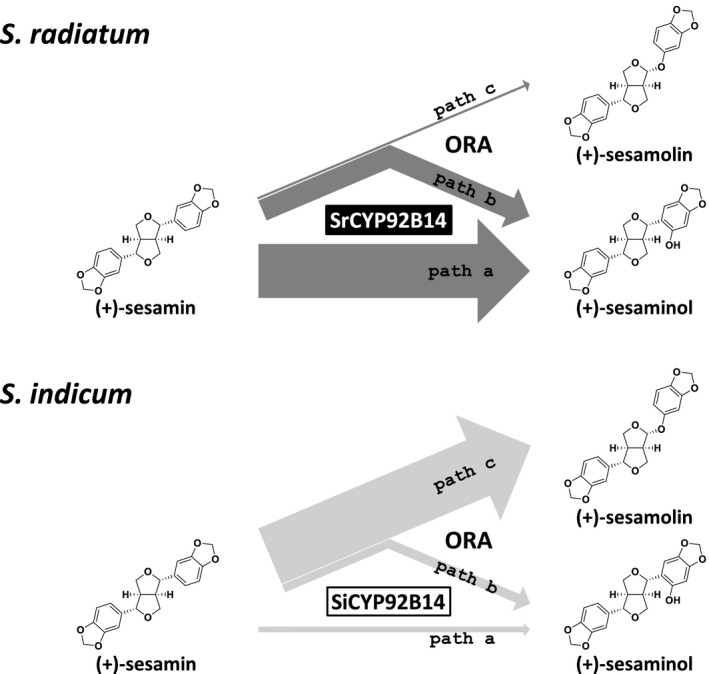
Catalytic outcome of SrCYP92B14 compared with SiCYP92B14.
(+)‐Sesamolin and (+)‐sesaminol are generated by the oxidation of (+)‐sesamin catalysed by CYP92B14 proteins. The catalytic outcomes of SrCYP92B14 and SiCYP92B14 defined in Table 2 are represented by the width of dark grey and light grey arrows that are proportional to the product ratios.
CYPs associated with specialised metabolic processes often catalyse multiple oxygenation reactions, including hydroxylation, epoxidation, ether formation and ring contraction (Mizutani and Sato, 2011; Cochrane and Vederas, 2014). For example, in the case of MycG, which catalyses both hydroxylation and epoxidation reactions in mycinamicin biosynthesis, substrate translocation‐coupled movement of F286 is vital for substrate discrimination (Li et al., 2012). Moreover, catalysis of hydroxylation and ether formation by AurH in aureothin biosynthesis involves an induced‐fit mechanism triggered by substrate binding, and adoption of the steric conformation is necessary for the second oxygenation‐heterocyclisation step (Richter et al., 2012). Thus, changes in steric pressure by amino acid substitution lead to alterations in oxidation positions (Zocher et al., 2011). These data show that the orientation movement of the substrate and/or its neighbouring environment establishes substrate recognition in CYP proteins. These assumptions might also be applicable to CYP92B14 proteins; the amino acid residues that participate in such structural perturbation would contribute to the ratio of co‐products in (+)‐sesamin oxidation. Alternatively, single amino acid substitution in A313 or S317, located in the distal helix of SiCYP92B14, might lead to the movement of the distal helix toward the active site or changes in the stability of the helix. To date, very few crystal structures of plant CYP proteins have been deposited on the public database (Lee et al., 2008; Li et al., 2008; Fujiyama et al., 2019; Gu et al., 2019). Further studies on the crystal structures of CYP92B14 or other plant CYPs will provide insights into the detailed mechanism of substrate recognition and control of catalytic outcome at the atomic level.
We showed that (+)‐sesamin and (+)‐sesangolin are the major lignans accumulated in S. radiatum seeds. (+)‐Sesangolin is present in the seed oil of the selected wild sesames S. angolense and S. angustifolium. It is remarkable that S. angolense and S. angustifolium seeds contain (+)‐sesamin (3.73 and 0.28%), (+)‐sesangolin (3.3 and 2.92%), and a considerable amount of (+)‐sesamolin (2.69 and 0.17%) in their seed oils, respectively (Jones et al., 1962; Kamal‐Eldin and Appelqvist, 1994). As revealed by this study, these lignan profiles of S. angolense and S. angustifolium differ from that of S. radiatum in that S. angolense and S. angustifolium accumulate both (+)‐sesaminol derivatives, and (+)‐sesamolin. These divergent metabolic traits might reflect the widespread nature of CYP92B14 family proteins in Sesamum spp., which may govern the molar ratio of (+)‐sesaminol, (+)‐sesamolin and (+)‐sesamin accumulation by modulating the ratio of ORA/direct oxygenation.
Metabolic traits that are beneficial to humans, such as colour, taste and flavour, have been selected artificially through domestication and breeding processes. The presence of (+)‐sesamolin together with high levels of triacylglycerol might be one of these beneficial traits that has arisen in cultivated sesame plants (Kamal‐Eldin et al., 1992; Pathak et al., 2014). This might relate to the fact that sesamol is a catabolite of (+)‐sesamolin and provides outstanding antioxidative activity to sesame seed oil (Wan et al., 2015), thereby extending the storage period of sesame seeds and their extracted oil products. In this context, ORA through path c to generate (+)‐sesamolin (Figure 1) by SiCYP92B14 might have been improved during the domestication of sesame, because SiCYP92B14_T121V/A313S/S317A/T318V catalysed a substantially low ratio of ORA through path c but an enhanced ratio of ORA through path b (Table 2; Figure S7). These notions are supported by the results that SrCYP92B14 exhibited extremely low activity for generating (+)‐sesamolin, and there existed only a trace amount of (+)‐sesamolin in the seeds of S. radiatum. Therefore, promiscuous regio‐specificity for C1 and C6 in the CYP92B14 protein have been the likely target for the domestication of sesame. The profile for specialised metabolic processes is generally considered to result from the combined substrate specificities of the biosynthetic enzymes involved. However, our results suggest that the metabolic profile can also be influenced by differences in the catalytic outcome from biosynthetic enzymes that share identical substrates.
The diversity of lignan profiles in various Sesamum spp. indicates that there are distinct sets of metabolic enzymes in each Sesamum spp. that are involved in hydroxylation, methylation and glycosylation reactions of lignans. In this study, we found that the catalytic outcome of SrCYP92B14 is biochemically relevant to the lignan profile of S. radiatum (Figures 2b and S2; Table 2). It is plausible that the various UGTs and O‐methyltransferases that are required for the biosynthesis of lignans that have accumulated in respective Sesamum spp. [e.g. STG in S. indicum and (+)‐sesangolin in S. radiatum] might have evolved based on the molecular evolution of the catalytic outcome of CYP92B14 (Figure S8). The RNA‐Seq of S. radiatum would pave the platform to mine other lignan‐modifying enzymes coordinately involved with SrCYP92B14 in specialised lignan metabolism within this species and its phylogenetic relatives.
Altogether, we have provided lines of molecular evidence as to how SrCYP92B14 from a wild sesame S. radiatum oxidises (+)‐sesamin. We also identified amino acid residues that affect the molar ratio of (+)‐sesamolin and (+)‐sesaminol, two major classes of (+)‐sesamin‐derived lignans in Sesamum spp. Therefore, the chemical diversity of lignans across Sesamum spp. and other plants might be explained partly by the catalytic outcome of CYP92B14 proteins. Future identification and biochemical characterisation of CYP92B14 proteins from other wild sesame species, as well as various other lignan‐containing plants, will better highlight the biochemical relevance of CYP92B14 proteins in plant lignan biosynthesis and help to uncover the biochemical origins of ORA reactions catalysed by CYP92B14.
EXPERIMENTAL PROCEDURES
Plant materials
Plants of S. indicum cv. Masekin were cultivated in a field in Osaka. Sesamum radiatum plants were grown in a greenhouse at the University of Toyama.
Chemicals
(+)‐Sesaminol was purchased from Nagara Science (Japan). (+)‐Sesamin, (+)‐sesamolin and (+)‐2,2´‐2H2‐sesamin were prepared according to previously reported methods (Murata et al., 2017). The actual 2H‐labelling yield of the C2 and C2´ protons of (+)‐sesamin was 70%, by comparison with the hydrogen intensity of H‐2,2´and H‐9a,9a´ in a 1H‐NMR experiment (Figure S6).
NMR analyses of stable isotope‐labelled compounds
All NMR experiments were performed on an AVANCE III HD 800 system equipped with a 5‐mm TCI cryogenic probe and Z‐axis gradient (Bruker BioSpin AG, Fällanden, Switzerland) at ambient temperature using 0.4–1 mg samples dissolved in CDCl3. Data analyses were carried out using Bruker TopSpin 3.2 software (Bruker Biospin AG). The acquisition parameters were as follows: number of data points 65K; spectral width 16 025 Hz; relaxation delay 10 sec; number of scans 64; receiver gain 3.2.
Lignan profile analyses
A 50‐mg sample of sesame seeds was frozen in liquid nitrogen and homogenised to a fine powder using a TissueLyser II (Qiagen, Tokyo, Japan). One millilitre of 80% ethanol was added to the homogenised samples and the samples were rotated at room temperature for 1 h. The extracted fraction was filtrated after centrifugation. The filtered fraction was analysed using an ion‐trap time‐of‐flight mass spectrometer (Shimadzu LCMS‐IT‐TOF, Shimadzu Corp., Kyoto, Japan) equipped with a photodiode array detector (Shimadzu, Japan). Each component was separated using a YMC Triart C18 column (TA12S03‐1503WT, 150 mm × 3 mm, 3 µm i.d.) with mobile phases A, 0.1% HCO2H‐H2O; and B, 0.1% HCO2H‐CH3CN in a linear gradient elution (0‐10‐32.5‐33‐40 min and 10‐40‐70‐10‐10% B, respectively) at a flow rate of 0.3 ml min−1.
Homology modelling
The three‐dimensional structural analyses were performed using Discovery Studio 4.0 (BIOVIA, San Diego, CA, USA). The initial structural models of SiCYP92B14 and SrCYP92B14 were constructed using the crystal structure of vitamin D2‐bound form of CYP2R1 as a template (PDB code: 3czh). (+)‐Sesamin‐bound structures of SiCYP92B14 and SrCYP92B14 were created by replacing the substrate from vitamin D2 to (+)‐sesamin. The constructed (+)‐sesamin‐bound structures of SiCYP92B14 and SrCYP92B14 were energy‐minimised and optimised by molecular dynamics simulation (Figure S4).
RNA‐Seq analysis
Total RNA was extracted at six stages during the development of S. radiatum seeds (14, 21, 28, 35, 42 and 49 DAP) using RNeasy Plant Kit (Qiagen). The quality of each RNA sample was evaluated using a BioAnalyzer (Agilent Technologies, Tokyo, Japan) with an RNA6000 Nano Chip. One microgram of total RNA from each sample was used to construct cDNA libraries using a TruSeq Stranded mRNA LT kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. The resulting cDNA library was validated using a BioAnalyzer with a DNA1000 Chip and quantified using a NEBNext Library Quant kit for Illumina (New England Biolabs, Tokyo, Japan). Paired‐end sequencing (2 × 101 cycles) was performed using a HiSeq 1500 sequencing system (Illumina) in Rapid mode. Total reads were extracted using CASAVA v1.8.2 (Illumina), then polymerase chain reaction (PCR) duplicates, adaptor sequences and low‐quality reads were removed from the extracted reads. Briefly, reads were judged as PCR duplicates when: (i) the first 10 bases in multiple reads were identical; and (ii) the reads showed > 90% overall similarity. Base calling from the 5´ to the 3´ end was performed until the frequency of accurately called bases dropped to 0.5. The remaining reads were assembled using Trinity ver. 2.4.0 by normalisation with maximum coverage to 30. For each sample, the fragments per kilobase megareads (FPKM) values were calculated for contigs containing cDNA sequences of SrCYP92B14, SrCYP81Q2 and SrCPR1 using Bowtie.
Molecular cloning of SrCYP92B14, and vector construction of SiCYP92B14 and SrCYP92B14 mutants
cDNAs of SrCYP92B14 and SrCPR1 amplified with cDNAs derived from S. radiatum seeds were used as the PCR template. According to previous methods, amplified fragments of cDNAs from SrCYP92B14 and SrCPR1 were subcloned in the yeast expression vectors, pYE22m and pJHXSBP‐His, respectively (Ono et al, 2006). Individual genes cloned in each vector were verified by DNA sequencing of both strands. Expression vectors for SiCYP92B14 and SrCYP92B14 mutants were generated from the wild‐type by PCR‐based mutagenesis, as described previously (Noguchi et al., 2014; Ono et al., 2018). All primer sets used for PCR are described in Table S3.
Phylogenetic analysis
Phylogenetic analysis of SrCYP92B14 was performed using the neighbour‐joining method with MEGA7 (Kumar et al., 2016).
Bioassay experiments of (+)‐sesamin oxidation in yeast cells
All transformations were carried out using the yeast INVsc strain (Invitrogen, Japan), according to a conventional method. Transformants expressing SiCYP92B14 or SrCYP92B14 and SiCPR1 were grown in Synthetic Defined liquid media at 30°C. A 50‐μl sample of stationary phase culture cells was transferred to 500 μl of fresh medium in 24‐well plates supplemented with 100 µm of (+)‐sesamin. After 48 h of culture, the cultures were mixed with an equal volume of acetonitrile (final 50% acetonitrile) and centrifuged. The filtered supernatant fractions were analysed by high‐performance liquid chromatography (HPLC) equipped with a photodiode array detector (Waters, Milford, MA, USA). Product ratios were calculated from the HPLC spectrum at 283 nm absorption, based on the peak areas of lignan standards.
2H‐labelled (+)‐sesamin oxidation in yeast cells
For the 2H‐labelling studies, 8–10 ml of the overnight cultured yeast cells expressing CYP92B14 and SiCPR1 was inoculated into 150 ml of liquid medium in a 1‐L Erlenmeyer flask. After 2–3 h of inoculation, 2H‐labelled (+)‐sesamin was added to a final concentration of 50 µm. A total of 450 ml culture was further incubated for 3–4 days and extracted twice with an equal volume of ethyl acetate. The organic layer was dehydrated with magnesium sulphate, dried in vacuo to dryness, and reconstituted with 400 µl of 50% acetonitrile. The extract was fractionated by HPLC, and the fractions containing enzyme reaction products were subjected to MS and NMR analyses. The hydrogen intensities of C2, C2´ and C6 protons of (+)‐sesaminol or C2 and C2´ protons of (+)‐sesamolin were measured to determine the product ratios by NMR.
ACCESSION NUMBERS
The nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers LC484020 (SrCYP92B14), LC534417 (SrCYP98A20‐like) and LC484021 (SrCPR1).
RNA‐seq data for Sesamum radiatum seeds were deposited on Sequence Read Archive under the accession numbers SRR11809384‐SRR11809395 (PRJNA633647).
Accession numbers of the genes subjected to phylogenetic analysis are as follows; SiCYP92B14, LC199944; SiCYP98A20‐like, NP_001291339; SiCYP81Q1, BAE48234; SrCYP81Q2, BAE48235; SaCYP81Q3, BAE48236; PlCYP81Q38, BAP46307; GmCYP93C, SBU44858; AtCYP51G1, NP_172633; AtCYP98A3, OAP09214; PsDWF_like1, AAG44132; and GhDDWF1, ABA01477.
Author contributions
EH, JM, EO, MPY and MH designed the research. KH and MPY prepared sesame seed samples. EO and HT cloned SrCYP92B14 and SrCPR1, and constructed expression vectors. JM, EO and AS performed RNA‐Seq analyses. EH, JM and MH performed (+)‐sesamin conversion assays. EH and MH analysed (+)‐sesamin conversion assays. EH performed Western blots. MH conducted chemical synthesis. MH performed NMR and homology modelling analyses. EH, JM, EO, MPY and MH wrote the paper. EH, JM and EO contributed equally to this work.
Conflict of interest
E.O. and H.T. are employees of Suntory Global Innovation Center, Ltd. Suntory Foundation for Life Sciences is a non‐profit organisation. All other authors declare no competing financial interests.
Supporting information
Figure S1. Sesamum spp. accumulate lineage‐specific lignans.
Figure S2. Lignan contents and putative lignan biosynthetic pathways in S. radiatum and S. indicum seeds.
Figure S3. Substrate preference of SrCYP92B14.
Figure S4. Model structure determination of CYP92B14 proteins.
Figure S5. Heterologous expression of CYP92B14 proteins in yeast.
Figure S6. 2H‐labelled (+)‐sesamin oxidation catalysed by SrCYP92B14 and SiCYP92B14.
Figure S7. Residues in heme active site are important for modulating the ratio of path b to path c in CYP92B14 proteins.
Figure S8. CYP92B14 are the key enzymes that shape the molar ratios of (+)‐sesamolin, (+)‐sesaminol and their derivatives.
Table S1. NMR data of (+)‐sesangolin and (+)‐7´‐episesantalin.
Table S2. Molar ratios of Sesaminol Total to Product Total in (+)‐sesamin oxidation bioassays.
Table S3. Primer sets used for PCR.
ACKNOWLEDGEMENTS
The authors are grateful to Yoko Ohta for technical assistance. This work is supported in part by JSPS KAKENHI 19K05857 (to M.H.) and 18K05571 (to M.P.Y.).
Contributor Information
Masayuki P. Yamamoto, Email: mpyama@sci.u-toyama.ac.jp.
Manabu Horikawa, Email: horikawa@sunbor.or.jp.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Bedigian, D. (2003) Evolution of sesame revisited; domestication, diversity and prospects. Gen. Resour. Crop Evol. 50, 779–787. [Google Scholar]
- Bedigian, D. (2010a) Introduction: history of the cultivation and use of sesame In Sesame: The genus Sesamum (Bedigian D., ed). Boca Raton: CRC Press, pp. 1–31. [Google Scholar]
- Bedigian, D. (2010b) Cultivated Sesame and Wild Relatives in the Genus Sesamum L In Sesame: The genus Sesamum (Bedigian D., ed). Boca Raton: CRC Press, pp. 33–77. [Google Scholar]
- Bedigian, D. , Seigler, D.S. and Harlan, J.R. (1985) Sesamin, sesamolin and the origin of sesame. Biochem. System. Ecol. 13, 133–139. [Google Scholar]
- Beroza, M. and Kinman, M.L. (1955) Sesamin, sesamolin, and sesamol content of the oil of sesame seed as affected by strain, location grown, ageing, and frost damage. J. Am. Oil Chem. Soc. 71, 348–350. [Google Scholar]
- Cochrane, R.V. and Vederas, J.C. (2014) Highly selective but multifunctional oxygenases in secondary metabolism. Acc. Chem. Res. 47, 3148–3161. [DOI] [PubMed] [Google Scholar]
- Dar, A.A. and Arumugam, N. (2013) Lignans of sesame: purification methods, biological activities and biosynthesis ‐ A review. Bioorg. Chem. 50, 1–10. [DOI] [PubMed] [Google Scholar]
- Davin, L.B. , Wang, H.B. , Crowell, A.L. , Bedgar, D.L. , Martin, D.M. , Sarkanen, S. and Lewis, N.G. (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science, 275, 362–366. [DOI] [PubMed] [Google Scholar]
- Fujiyama, K. , Hino, T. , Kanadani, M. , Watanabe, B. , Lee, H.J. , Mizutani, M. and Nagano, S. (2019) Structural insights into a key step of brassinosteroid biosynthesis and its inhibition. Nat. Plants, 5, 589–594. [DOI] [PubMed] [Google Scholar]
- Fukuda, Y. , Osawa, T. , Kawagishi, S. and Namiki, M. (1988) Comparison of contents of Sesamolin and Lignan antioxidants in sesame seeds cultivated in Japan. Nippon Shokuhin Kogyo Gakkaishi, 35, 483–486. [Google Scholar]
- Gu, M. , Wang, M. , Guo, J. , Shi, C. , Deng, J. , Huang, L. , Huang, L. and Chang, Z. (2019) Crystal structure of CYP76AH1 in 4‐PI‐bound state from Salvia miltiorrhiza. Biochem. Biophys. Res. Commun. 511, 813–819. [DOI] [PubMed] [Google Scholar]
- Guallar, V. and Olsen, B. (2006) The role of the heme propionates in heme biochemistry. J. Inorg. Biochem. 100, 755–760. [DOI] [PubMed] [Google Scholar]
- Gunstone, F.D. (2011) Vegetable Oils in Food Technology: Composition, Properties and Uses, 2nd edn. (Gustone F.D., ed). Hoboken: Wiley‐Blackwell. [Google Scholar]
- Jones, W.A. , Beroza, M. and Becker, E.D. (1962) Isolation and structure of sesangolin, a constituent of Sesamum angolense (Welw.). J. Org. Chem. 27, 3232–3235. [Google Scholar]
- Kabsch, W. and Sander, C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen‐bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kamal‐Eldin, A. (2010) Chemical studies on the lignans and other minor constituents of sesame seed oil In Sesame: The genus Sesamum (Bedigian D., ed). Boca Raton: CRC Press, pp. 79–92. [Google Scholar]
- Kamal‐Eldin, A. and Appelqvist, L.A. (1994) Variation in the composition of sterols, tocopherols and lignans in seed oils from four sesamum species. J. Am. Oil Chem. Soc. 71, 149–156. [Google Scholar]
- Kamal‐Eldin, A. , Appelqvist, L.Å. and Yousif, G. (1994) Lignan analysis in seed oils from four Sesamum species: Comparison of different chromatographic methods. J. Am. Oil Chem. Soc. 71, 141–147. [Google Scholar]
- Kamal‐Eldin, A. and Yousif, G. (1992) A furofuran lignan from Sesamam alatum . Phytochemistry, 31, 2911–2912. [Google Scholar]
- Kamal‐Eldin, A. , Yousif, G. , Iskander, G.M. and Appelqvist, L.A. (1992) Seed lipids of Sesamum indicum, L. and related wild species in Sudan I: fatty acids and Triacylglycerols. Fat Sci. Technol. 94, 254–259. [Google Scholar]
- Kang, M.H. , Naito, M. , Tsujihara, N. and Osawa, T. (1998) Sesamolin inhibits lipid peroxidation in rat liver and kidney. J. Nutr. 128, 1018–1022. [DOI] [PubMed] [Google Scholar]
- Katsuzaki, H. , Kawakishi, S. and Osawa, T. (1994) Sesaminol glucosides in sesame seeds. Phytochemistry, 35, 773–776. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.S. , Nioche, P. , Hamberg, M. and Raman, C.S. (2008) Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature, 455, 363–368. [DOI] [PubMed] [Google Scholar]
- Li, L. , Chang, Z. , Pan, Z. , Fu, Z.Q. and Wang, X. (2008) Modes of haem binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. USA, 105, 13883–13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Tietz, D.R. , Rutaganira, F.U. , Kells, P.M. , Anzai, Y. , Kato, F. , Pochapsky, T.C. , Sherman, D.H. and Podust, L.M. (2012) Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions. J. Biol. Chem. 287, 37880–37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalawieh, A.F. , Massri, M. and Nasrallah, G.K. (2017) A comprehensive review on the anti‐cancer properties and mechanisms of action of sesamin, a lignan in sesame seeds (Sesamum indicum). Eur. J. Pharmacol. 815, 512–521. [DOI] [PubMed] [Google Scholar]
- Mizutani, M. and Sato, F. (2011) Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 507, 194–203. [DOI] [PubMed] [Google Scholar]
- Moazzami, A.A. and Kamal‐Eldin, A. (2006) Sesame seed is a rich source of dietary lignans (2006). J. Am. Oil Chem. Soc. 83, 719–723. [Google Scholar]
- Murata, J. , Ono, E. , Yoroizuka, S. et al (2017) Oxidative rearrangement of (+)‐sesamin by CYP92B14 co‐generates twin dietary lignans in sesame. Nat. Commun. 8, 2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, A. , Fukui, Y. , Iuchi‐Okada, A. , Kakutani, S. , Satake, H. , Iwashita, T. , Nakao, M. , Umezawa, T. and Ono, E. (2008) Sequential glucosylation of a furofuran lignan, (+)‐sesaminol, by Sesamum indicum UGT71A9 and UGT94D1 glucosyltransferases. Plant J. 54, 415–427. [DOI] [PubMed] [Google Scholar]
- Noguchi, A. , Horikawa, M. , Murata, J. , Tera, M. , Kawai, Y. , Ishiguro, M. , Umezawa, T. , Mizutani, M. and Ono, E. (2014) Mode‐of‐action and evolution of methylenedioxy bridge forming P450s in plant specialized metabolism. Plant Biotechnol. 31, 493–503. [Google Scholar]
- Ono, E. , Murata, J. , Toyonaga, H. , Nakayasu, M. , Mizutani, M. , Yamamoto, M.P. , Umezawa, T. and Horikawa, M. (2018) Formation of a methylenedioxy bridge in (+)‐epipinoresinol by CYP81Q3 corroborates with diastereomeric specialization in sesame lignans. Plant Cell Physiol. 59, 2278–2287. [DOI] [PubMed] [Google Scholar]
- Ono, E. , Nakai, M. , Fukui, Y. et al (2006) Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)‐sesamin. Proc. Natl. Acad. Sci. USA, 103, 10116–10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, E. , Waki, T. , Oikawa, D. et al (2020) Glycoside‐specific glycosyltransferases catalyze regioselective sequential glucosylations for a sesame lignan, sesaminol triglucoside. Plant J. 101, 1221–1233. [DOI] [PubMed] [Google Scholar]
- Pathak, N. , Rai, A.K. , Saha, S. , Walia, S. , Sen, S.K. and Bhat, K.V. (2014) Quantitative dissection of antioxidative bioactive components in cultivated and wild sesame germplasm reveals potentially exploitable wide genetic variability. J. Crop Sci. Biotech. 17, 127–139. [Google Scholar]
- Poulos, T.L. (2014) Heme enzyme structure and function. Chem. Rev. 114, 3919–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M. , Busch, B. , Ishida, K. , Moore, B.S. and Hertweck, C. (2012) Pyran formation by an atypical CYP‐mediated four‐electron oxygenation‐cyclization cascade in an engineered aureothin pathway. ChemBioChem, 13, 2196–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.A. and Berenbaum, M.R. (2013) Structure and function of cytochrome P450s in insect adaptation to natural and synthetic toxins: insights gained from molecular modeling. J. Chem. Ecol. 39, 1232–1245. [DOI] [PubMed] [Google Scholar]
- Shimizu, S. , Akimoto, K. , Shinmen, Y. , Kawashima, H. , Sugano, M. and Yamada, H. (1991) Sesamin is a potent and specific inhibitor of delta 5 desaturase in polyunsaturated fatty acid biosynthesis. Lipids, 26, 512–516. [DOI] [PubMed] [Google Scholar]
- Suja, K.P. , Jayalekshmy, A. and Arumughan, C. (2004) Free radical scavenging behavior of antioxidant compounds of sesame (Sesamum indicum L.) in DPPH˙ system. J. Agric. Food Chem. 52, 912–915. [DOI] [PubMed] [Google Scholar]
- Suzuki, S. and Umezawa, T. (2007) Biosynthesis of lignans and norlignans. J. wood Sci. 53, 273–284. [Google Scholar]
- Umezawa, T. (2003) Diversity in lignan biosynthesis. Phytochem. Rev. 2, 371–390. [Google Scholar]
- Wan, Y. , Li, H. , Fu, G. , Chen, X. , Chen, F. and Xie, M. (2015) The relationship of antioxidant components and antioxidant activity of sesame seed oil. J. Sci. Food Agric. 5, 2571–2578. [DOI] [PubMed] [Google Scholar]
- Yamauchi, S. , Ichikawa, H. , Nishiwaki, H. and Shuto, Y. (2015) Evaluation of plant growth regulatory activity of furofuran lignan bearing a 7,9´:7´,9‐diepoxy structure using optically pure (+)‐ and (‐)‐enantiomers. J. Agric. Food Chem. 63, 5224–5228. [DOI] [PubMed] [Google Scholar]
- Zocher, G. , Richter, M.E. , Mueller, U. and Hertweck, C. (2011) Structural fine‐tuning of a multifunctional cytochrome P450 monooxygenase. J. Am. Chem. Soc. 133, 2292–2302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sesamum spp. accumulate lineage‐specific lignans.
Figure S2. Lignan contents and putative lignan biosynthetic pathways in S. radiatum and S. indicum seeds.
Figure S3. Substrate preference of SrCYP92B14.
Figure S4. Model structure determination of CYP92B14 proteins.
Figure S5. Heterologous expression of CYP92B14 proteins in yeast.
Figure S6. 2H‐labelled (+)‐sesamin oxidation catalysed by SrCYP92B14 and SiCYP92B14.
Figure S7. Residues in heme active site are important for modulating the ratio of path b to path c in CYP92B14 proteins.
Figure S8. CYP92B14 are the key enzymes that shape the molar ratios of (+)‐sesamolin, (+)‐sesaminol and their derivatives.
Table S1. NMR data of (+)‐sesangolin and (+)‐7´‐episesantalin.
Table S2. Molar ratios of Sesaminol Total to Product Total in (+)‐sesamin oxidation bioassays.
Table S3. Primer sets used for PCR.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.


