Abstract
Telomere length (TL) shortens over time in most human cell types and is a potential biomarker of aging. However, the causal association of TL on physical and cognitive traits that decline with age has not been extensively examined in middle-aged adults. Using a Mendelian randomization (MR) approach, we utilized genetically increased TL (GI-TL) to estimate the impact of TL on aging-related traits among U.K. Biobank (UKB) participants (age 40–69 years). We manually curated 53 aging-related traits from the UKB and restricted to unrelated participants of British ancestry (n = 337,522). We estimated GI-TL as a linear combination of nine TL-associated single nucleotide polymorphisms (SNPs), each weighted by its previously-reported association with leukocyte TL. Regression models were used to assess the associations between GI-TL and each trait. We obtained MR estimates using the two-sample inverse variance weighted (IVW) approach. We identified six age-related traits associated with GI-TL (Bonferroni-corrected threshold p < .001): pulse pressure (PP) (p = 5.2 × 10-14), systolic blood pressure (SBP) (p = 2.9 × 10-15), diastolic blood pressure (DBP) (p = 5.5 × 10-6), hypertension (p = 5.5 × 10-11), forced expiratory volume (FEV1) (p = .0001), and forced vital capacity (FVC) (p = 3.8 × 10-6). Under MR assumptions, one standard deviation increase in TL (~1,200 base pairs) increased PP, SBP, and DBP by 1.5, 2.3, and 0.8 mmHg, respectively, while FEV1 and FVC increased by 34.7 and 52.2 mL, respectively. The observed associations appear unlikely to be due to selection bias based on analyses including inverse probability weights and analyses of simulated data. These findings suggest that longer TL increases pulmonary function and blood pressure traits among middle-aged UKB participants.
Keywords: Telomeres, Mendelian randomization, Polygenic scores, Blood pressure, Pulmonary function
Telomere length (TL) is a proposed biomarker and potential cause of biological aging (1). Telomeres are DNA–protein complexes that protect the ends of chromosomes from degradation and fusion. The DNA component, a 6-nucleotide repeat sequence, shortens with each cell division (2). Thus, TL decreases as human age in most cell types (3). Meta-analyses of observational studies of leukocyte TL suggest that TL is associated with all-cause mortality (4) and risk of age-related chronic diseases including cardiovascular disease (5), type II diabetes (6), Alzheimer’s disease (7), and some cancers (8). These meta-analyses acknowledge that there is substantial heterogeneity across studies due to differences in TL measurement, study design, and adjustment for confounding factors, potentially affecting the validity of these associations.
Epidemiological studies of TL are susceptible to biases caused by confounding factors, such as technical variation in TL measurement, environment, lifestyle, and cell type composition. We can apply Mendelian randomization (MR) as an alternative approach for estimating the association of TL on disease susceptibility and utilize genetic variants (SNPs) associated with leukocyte TL rather than measured TL itself, potentially avoiding biases caused by confounding factors and reverse causation (9). Providing certain assumptions are satisfied, we can use the SNPs associated with TL to estimate the non-confounded causal relationship between TL and a health outcome. These assumptions include (i) the SNPs are associated with TL, (ii) SNPs only affect the outcome via its effect on TL, and (iii) SNPs are not associated with confounders that are associated with both TL and the outcome (10).
SNPs associated with leukocyte TL have been identified in prior meta-analyses of genome-wide association studies (GWAS) (11–13). The ENGAGE consortium identified seven SNPs independently associated with TL, five of which were located in regions containing genes involved in telomere regulation and maintenance (11). Several prior studies have applied MR approaches by utilizing genetic determinants of TL to assess the potential causal contribution of TL to overall and cause-specific mortality (14), the risk of cancer (15), and noncommunicable diseases (16). However, the causal relationship between TL and age-related risk factors, which often increase the risk of age-related chronic disease and early mortality, is not well established.
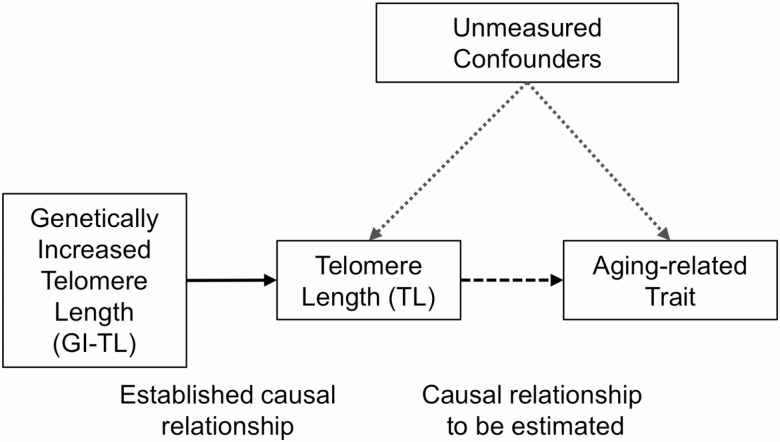
While TL is an important component of biological and cellular aging, we do not know whether variation in TL contributes to systemic aging across many organ systems and the extent to which TL is causally associated with age-related physical and cognitive decline in middle-aged adults. To address this question, we estimated genetically increased TL (GI-TL) and assessed its association with more than 50 aging-related traits in 337,522 unrelated U.K. Biobank (UKB) participants of British ancestry (age 40–69 years). Using a MR approach, we then estimated causal associations between TL and these aging-related traits using SNP summary statistics from the ENGAGE consortium and UKB cohort (Figure 1).
Figure 1.
Causal diagram showing the hypothesized impact of telomere length on aging-related traits in the U.K. Biobank cohort.
Methods
Study Sample
The UKB cohort is a prospective study of ~500,000 middle-aged adults from the United Kingdom aged 40–69 years (recruited from 2006 to 2010). Postal invitations to participate were mailed to more than 9 million people registered with the U.K. National Health Service, who resided within 40 km of one of the 22 assessment centers (17). Of the adults invited, 5.45% responded and participated (18). At baseline, participants completed touchscreen questionnaires, physical exams, and verbal interviews, and a blood sample was obtained. All participants were genotyped at ~800,000 SNPs on either the custom U.K. BiLEVE Axiom Array or U.K. Biobank Axiom Array, with the two arrays sharing ~95% of SNPs. Further information related to array design, genotyping, imputation, and quality control is summarized elsewhere (19). For this analysis, we used version 2 of the imputed genotype data for the full UKB cohort. All nine SNPs included in our analysis were imputed based on the Haplotype Reference Consortium panel; thus, none of the problems reported by U.K. Biobank for the UK10K + 1000 Genomes reference panel affected our analysis. We restricted our analysis to unrelated individuals with both self-reported and genetically predicted British ancestry based on the principal component (PC) analysis of genotypes. Our final sample size was 337,522 participants.
Curation of Age-Related Traits from UKB
We searched the UKB data showcase to manually curate a set of aging-associated traits from the cognitive function, physical measures, and self-reported touchscreen questionnaire variables to capture the following domains of health (at baseline): cardiovascular, pulmonary, anthropometric, physical activity, sensory (hearing and eyesight), oral, musculoskeletal, pain and general health. Among the clinically measured physical measurement variables, we extracted traits related to blood pressure (BP), arterial stiffness, spirometry, anthropometry, bone density, and hand grip strength. We selected cognitive function variables to capture traits related to visual memory, fluid intelligence, and short-term memory, as described in Bakrania and colleagues (20). The remaining aging-related traits were extracted from the self-reported touchscreen questionnaire that UKB participants completed at baseline, and these traits included those related to pain, weight change, activity, fractures, oral health, hearing loss, eyesight, and general health. We extracted a total of 48 aging-related traits.
Derivation of Frailty Index (FI)
FIs have been proposed to capture the variation in health among aging individuals. We applied the algorithm proposed by Williams and colleagues to compute a FI for each participant (21). Briefly, 49 self-reported questionnaire variables related to health, disease, disability, and mental well-being were extracted and coded into binary (0 or 1) or ordinal variables (values ranging from 0 to 1). For each UKB participant, FI was computed from the sum of the observations across these 49 variables and divided by 49 and expressed as a percent. For participants with missing data for one or more variables, values for missing observations were imputed using multiple imputation by chained equations (22).
Estimation of GI-TL
We identified SNPs associated with leukocyte TL from three prior genome-wide meta-analyses (11–13). We selected nine SNPs that were associated with leukocyte TL (p < 5 × 10−8) and not in linkage disequilibrium with each other (see Supplementary Table S1). All seven SNPs that reached genome-wide significance from Codd and colleagues were included (genome-wide meta-analysis of 37,684 individuals) (11). Additionally, the SNP annotating to DCAF4 identified by Mangino and colleagues in a meta-analysis of 20,022 individuals (12) and the SNP annotating to CTC1 identified by Mangino and colleagues in a meta-analysis of 9160 individuals (13) were also included because they reached genome-wide significance (p < 5 × 10−8) and were not in linkage disequilibrium with any of the other SNPs. For each SNP, we extracted the association estimate (ie, beta coefficient and standard error) corresponding to the impact of a one allele increase on leukocyte TL in terms of the standard deviation (SD) (~1,200 base pairs [bp]) from the ENGAGE telomere consortium (11). GI-TL was estimated as a weighted linear combination of the TL-associated SNPs, corresponding to the sum of the weighted values for each SNP and expressed in terms of SD TL. GI-TL was interpreted as the additional TL relative to an arbitrary TL value, corresponding to GI-TL = 0 (Supplementary Figure 1) (23,24). There was a very small proportion of missing data for each of the nine SNPs after imputation (<1% for each SNP), and participants missing one or more genotypes for the TL-associated SNPs were excluded (n = 7624).
Statistical Analysis
Each aging-related trait was expressed as a binary or continuous variable. For individuals taking BP medication at baseline, we added 15 mmHg and 10 mmHg to their baseline systolic (SBP) and diastolic (DBP) BP, respectively (25), and averaged the first and second measurements. For continuous variables, we checked the distributions for outlying or implausible values for SBP, DBP, forced expiratory volume (FEV1), forced vital capacity (FVC), peak expiratory flow (PEF1), and pulse rate, and we removed implausible values for FEV1 (<450 mL, n = 296), FVC (<600 mL, n = 325), and PEF1 (<50 L/min, n = 4644). We computed three additional variables: waist-to-hip ratio from waist and hip measurements, pulse pressure (PP) from the difference between SBP and DBP, and FV-ratio of FEV1 to FVC. Hypertension was defined as at least 140 mmHg SBP or at least 90 mmHg DBP or self-reported BP medication use. We computed genotyping PCs among the selected UKB participants using EIGENSOFT (26–28) implemented in PLINK 1.9.
Logistic or linear multivariable regression models were used to estimate associations between GI-TL and each aging-related trait. We conducted analyses stratified by gender and median age (< or ≥58 years). A Bonferroni correction was applied to account for multiple testing at α = 0.05. We utilized Cox proportional hazards models to estimate the association between GI-TL and mortality. Follow-up began when the participants conducted their baseline assessment and ended at whichever happened first: death, departure from study, or censoring date (November 30, 2015). All analyses included the following covariates: age, sex, and first 10 genotyping PCs.
Mendelian Randomization Analysis
We used a two-sample inverse-weighted variance (IVW) approach to obtain estimates of the impact of TL on aging-related traits, as described previously (9,29). For each SNP included in the GI-TL score (k = 9 SNPs), the beta coefficient (Xk) and its standard error (se) () were obtained from the summary statistics from the GWAS of leukocyte TL from the ENGAGE consortium, and the per allele association estimate (Yk) and its se () with the aging-related trait were obtained from the selected UKB cohort. The IVW MR estimate and its se were computed as follows:
All MR analyses were executed using the MendelianRandomization package in R (30). We compared the MR-estimates from IVW to MR-estimates obtained from the maximum likelihood (9,31) and weighted median (32) approaches to evaluate the consistency of the MR estimates. From MR-Egger regression, we extracted intercept-test for directional pleiotropy to evaluate potential violation of MR assumptions (32,33). To examine horizontal pleiotropic outliers, we applied Mendelian Randomization Pleiotropy RESidual Sum and Ouliter (MR-PRESSO) test (34).
Inverse Probability Weighting (IPW)
Selection can be a form of collider bias in MR studies when confounders are present. Selection into the study based on the outcome or/and the risk factor of interest can bias the MR effect estimate (Supplementary Figure 2), potentially violating the assumption that the instrumental variable and confounder(s) are marginally independent (35,36). UKB participants are more likely to be female, healthier, older, and of higher social and economic status than the target source population (18). We applied an IPW approach to create a pseudo-population that was unaffected by selection on factors like age, sex, smoking status, and BP (37), in order to estimate the average causal effect of TL (on BP and pulmonary traits) in the general population (36). We computed IPWs as followed, using the percent of the source population belonging to a particular group based on a set of characteristics (up to four levels: i, j, k, l) and the percent of UKB participants belonging to that group:
Using Web Table 1 in Fry and colleagues (18), we extracted information on the gender and age group distribution (40–50, 50–60, 60–70 years) of the UKB source population. Because there were no cross-tabulated data available for the UKB source population, we utilized the Health Survey of England (HSE) 2008 and extracted sets of population-weighted cross-tabulated proportions for groups based on sex, age (40–50, 50–60, and 60–70 years), smoking status, and hypertension from the HSE (38). We fit linear models adjusted for age, sex and first 10 genotyping PCs and included IPWs as weights.
Simulated Data sets to Examine the Impact of Selection Bias on TL and SBP
We conducted simulations to examine how selection on TL, SBP (the strongest association identified), and/or confounders influenced the MR estimates under the hypothesis of no causal effect (βx = 0) and of a true effect (based on the observed association) (βx = 0.12, generated from analysis of standard normalized SBP). Using an approach similar to that described by Gkatzionis and Burgess (36), we generated the risk factor, TL, (Xi) as a linear combination of a genetic risk score, a confounder, and an independent error term (see equation 1), where the genetic risk score, confounder, and random error term are independently drawn from a Ν(0,1) distribution. The nonvarying parameters included percent variation explained (PVE) by genetic instrument for TL (), the estimated amount of variation explained by SNPs associated with TL, and PVE in TL by the confounder that we assumed
| (1) |
We generated the outcome, SBP, using a linear combination of the risk factor (Xi), generated in equation 1, confounder, and an independent error term (see equation 2), where the random error term is drawn from Ν(0,1) distribution. The nonvarying parameters included PVE explained by confounders in SBP that we assumed and the causal effect to be estimated (βx = 0 or βx = 0.12).
| (2) |
For each simulated observation, a selection probability was generated and assigned 0 (not selected) or 1 (selected) based on a Bernoulli trial with that selection probability. We examined two functions to generate the selection probability: the risk factor effect on selection (see equations 3 and 4) and the outcome effect on selection (see equations 5 and 6). We varied the following parameters within the selection probability: the effect of risk factor on selection (γx), the effect of outcome on selection (γy), and the confounder effect on selection (γu) (see Supplementary Table S2). We also included the baseline prevalence of selection into the UKB (γ0 = 0.0545).
Risk factor effect on selection:
| (3) |
| (4) |
Outcome effect on selection:
| (5) |
| (6) |
For each simulated data set, we generated a random sample of 10,000,000 observations, similar to the size of the UKB source population. Among the observations with S = 1, we randomly selected 500,000 observations. To obtain the causal effect estimate of the risk factor on the outcome, we conducted linear regressions of the genetic instrument (gene) on the risk factor (X) and the genetic instrument (gene) on the outcome (Y), and then, computed the ratio estimate and its standard error (se). We ran 1000 simulations per scenario and extracted the mean and SD of the ratio estimates, as well as, the Type I error rate (α = 0.05) and empirical power (at 95%). All analyses were performed in R 3.4.3.
Results
We analyzed 53 aging-related traits among the 337,522 UKB participants (48 from UKB and five that we derived [waist-hip ratio, PP, FV-ratio, FI, and hypertension]). The median age of participants included was 58 years (range: 39–72 years), and there were more women (53.7%) than men. Sixteen aging-related traits were associated with GI-TL at a significance threshold of p = .05 (Supplementary Table S3), but only six traits surpassed the Bonferroni-corrected significance threshold (p < .001) (Figure 2). GI-TL was associated with better pulmonary function. FEV1 and FVC increased by 34.7 mL (95% confidence interval [CI] [16.9, 52.6], p = .0001) and 52.2 mL (95% CI [30.1, 74.4], p = 3.8 × 10-6) per one SD increase in TL (corresponding to ~1,200 bp), respectively. The association between GI-TL and FVC was stronger among men (β = 79.4 mL, 95% CI [41.1, 117.6], p = 4.8 × 10-5) compared to women (β = 29.4 mL, 95% CI [4.5, 54.3], p = .02) (interaction p = .02) (Supplementary Tables S4 and S5, and Figure S3). When stratified by median age, GI-TL was positively associated with FEV1 (β = 49.7 mL, 95% CI [25.5, 73.8], p = 5.7 × 10-5) and FVC (β = 69.9 mL, 95% CI [40.1, 99.7], p = 4.3 × 10-6) among individuals aged ≥58 years but FEV1 and FVC were not associated with GI-TL among individuals younger than 58 years (p > .05) (Supplementary Tables S6 and S7, and Figure S4), with interaction p-values of .05 and .07, respectively.
Table 1.
Reported Associations between Genetically Increased Telomere Length (GI-TL) and Aging-Related Traits among U.K. Biobank Participants
| Domain | Aging-Related Trait | N | Mean (SD) | Absent | Present | Direction with Age | β GI-TL (95% CI) | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| General health | Longstanding illness, disability, or infirmity | 329,975 | 222,683 (67.5%) | 107,292 (32.5%) | Positive | 1.04 (0.98, 1.12) | ||
| Other serious medical condition/disability | 331,581 | 263,517 (79.5%) | 68,064 (20.5%) | Positive | 1.05 (0.97, 1.13) | |||
| Fair or Poor Health Rating (good/excellent vs poor/fair) | 336,341 | 253,727 (75.4%) | 82,614 (24.6%) | Positive | 1.07 (1.00, 1.15) * | |||
| Frailty Score (%) | 304,655 | 10.2 (6.8) | Positive | 0.20 (–0.01, 0.41) | ||||
| Anthropometry | Body mass index (kg/m2) (BMI) | 336,041 | 27.4 (4.7) | Positive | –0.08 (–0.22, 0.06) | |||
| Body fat percentage (%) | 331,423 | 31.4 (8.5) | Positive | –0.30 (–0.49, –0.10) ** | ||||
| Trunk fat percentage (%) | 331,314 | 31.1 (8.0) | Positive | –0.33 (–0.55, –0.11) ** | ||||
| Waist-to-hip ratio | 336,265 | 0.9 (0.1) | Positive | 0.00 (0.00, 0.00) | ||||
| Weight gained compared with 1 y ago | 281,473 | 188,487 (67.0%) | 92,986 (33.0%) | Negative | 0.97 (0.90, 1.04) | |||
| Weight lost compared with 1 y ago | 239,130 | 188,487 (78.8%) | 50,643 (21.2%) | Negative | 1.01 (0.92, 1.10) | |||
| Cardiovascular | Central arterial stiffness (ASI) | 109,862 | 9.4 (3.1) | Positive | 0.20 (0.04, 0.35) * | |||
| Diastolic blood pressure (mmHg) | 317,797 | 83.2 (10.8) | Positive | 0.77 (0.44, 1.10) *** | ||||
| Pulse rate (beats per minute) | 317,485 | 69.2 (11.1) | Positive | –0.24 (–0.59, 0.10) | ||||
| Systolic blood pressure (mmHg) | 317,677 | 139.7 (19.6) | Positive | 2.30 (1.73, 2.87) *** | ||||
| Pulse pressure (mmHg) | 317,478 | 56.4 (13.8) | Positive | 1.50 (1.11, 1.89) *** | ||||
| Hypertension | 317,478 | 159,771 (50.3%) | 157,707 (49.7%) | Positive | 1.24 (1.17, 1.33) *** | |||
| Cognitive | Fluid Intelligence (score) | 108,931 | 6.2 (2.1) | Negative | –0.11 (–0.22, 0.00) * | |||
| Poor result pairs matching (≥2 incorrect) | 336,714 | 151,985 (45.1%) | 184,729 (54.9%) | Positive | 1.04 (0.98, 1.10) | |||
| Maximum digits remembered (n) | 34,972 | 6.7 (1.3) | Negative | –0.09 (–0.22, 0.03) | ||||
| Pulmonary | Forced expiration volume (mL) (FEV1) | 307,504 | 2,847.4 (781.3) | Negative | 34.72 (16.87, 52.58) *** | |||
| Forced vital capacity (mL) (FVC) | 307,505 | 3,759.4 (993.0) | Negative | 52.23 (30.07, 74.38) *** | ||||
| FEV1/FVC (%) | 306,699 | 75.9 (6.8) | Negative | –0.06 (–0.27, 0.15) | ||||
| Peak expiratory flow (L/min) (PEF1) | 303,201 | 397.9 (127.5) | Negative | 1.90 (–1.28, 5.09) | ||||
| Shortness of breath walking on level ground | 109,624 | 98,135 (89.5%) | 11,489 (10.5%) | Positive | 0.97 (0.82, 1.15) | |||
| Wheeze or whistling in the chest in last year | 331,569 | 262,933 (79.3%) | 68,636 (20.7%) | Positive | 0.92 (0.85, 0.99) * | |||
| Activity | Basal metabolic rate (kJ) | 331,504 | 6,643.9 (1,363.6) | Negative | 18.21 (–7.14, 43.57) | |||
| Slow walking pace (steady/brisk vs slow) | 335,666 | 310,494 (92.5%) | 25,172 (7.5%) | Positive | 0.99 (0.88, 1.12) | |||
| No exercise past 4 wk | 261,724 | 242,133 (92.5%) | 19,591 (7.5%) | Negative | 1.01 (0.89, 1.15) | |||
| Musculoskeletal | Average hand grip strength (kg) | 335,314 | 30.9 (11.0) | Negative | –0.16 (–0.37, 0.06) | |||
| Bone mineral density (T-score) | 298,810 | –0.4 (1.1) | Negative | –0.00 (–0.04, 0.03) | ||||
| Fracture in past 5 y | 335,906 | 303,682 (90.4%) | 32,224 (9.6%) | Positive | 1.08 (0.97, 1.19) | |||
| Falls in the last year | 336,936 | 270,745 (80.4%) | 66,191 (19.6%) | Positive | 0.97 (0.90, 1.04) | |||
| Sleep | Nap during day (never/rarely vs sometimes/usually) | 337,396 | 190,649 (56.5%) | 146,747 (43.5%) | Positive | 0.95 (0.90, 1.02) | ||
| Sleep duration (hours) | 335,997 | 7.2 (1.1) | Positive | –0.01 (–0.05, 0.02) | ||||
| Trouble falling asleep (never vs sometimes/usually) | 337,288 | 80,513 (23.9%) | 256,775 (76.1%) | Positive | 1.04 (0.97, 1.12) | |||
| Sensory | Cataract | 93,871 | 84,778 (90.3%) | 9,093 (9.7%) | Positive | 0.99 (0.81, 1.21) | ||
| Glaucoma | 89,383 | 84,778 (94.8%) | 4,605 (5.2%) | Positive | 0.98 (0.75, 1.28) | |||
| Macular degeneration | 86,733 | 84,778 (97.7%) | 1,955 (2.3%) | Positive | 1.02 (0.68, 1.53) | |||
| Wears glasses or contacts | 337,242 | 35,941 (10.7%) | 301,301 (89.3%) | Positive | 1.10 (0.99, 1.23) | |||
| Difficulty hearing with background noise | 331,073 | 205,833 (62.2%) | 125,240 (37.8%) | Positive | 0.97 (0.91, 1.03) | |||
| Difficulty hearing | 324,281 | 239,263 (73.8%) | 85,018 (26.2%) | Positive | 0.98 (0.92, 1.06) | |||
| Oral | Bleeding gums | 238,197 | 204,450 (85.8%) | 33,747 (14.2%) | Negative | 1.08 (0.98, 1.20) | ||
| Dentures | 248,424 | 204,450 (82.3%) | 43,974 (17.7%) | Positive | 1.03 (0.93, 1.13) | |||
| Loose teeth | 211,814 | 204,450 (96.5%) | 7,364 (3.5%) | Positive | 0.74 (0.60, 0.92) ** | |||
| Mouth ulcers | 238,899 | 204,450 (85.6%) | 34,449 (14.4%) | Negative | 1.11 (1.00, 1.23) * | |||
| Painful gums | 210,701 | 204,450 (97.0%) | 6,251 (3.0%) | Positive | 0.77 (0.61, 0.96) * | |||
| Toothache | 210,672 | 204,450 (97.0%) | 6,222 (3.0%) | Negative | 1.04 (0.83, 1.31) | |||
| Pain | Back pain | 176,299 | 136,296 (77.3%) | 40,003 (22.7%) | Positive | 1.00 (0.90, 1.10) | ||
| Headache | 203,359 | 136,296 (67.0%) | 67,063 (33.0%) | Negative | 0.97 (0.89, 1.05) | |||
| Hip pain | 145,616 | 136,296 (93.6%) | 9,320 (6.4%) | Positive | 0.91 (0.75, 1.09) | |||
| Knee pain | 156,698 | 136,296 (87.0%) | 20,402 (13.0%) | Positive | 1.04 (0.91, 1.19) | |||
| Neck or shoulder pain | 185,347 | 136,296 (73.5%) | 49,051 (26.5%) | Positive | 1.11 (1.02, 1.22) * | |||
| Other pain | 151,134 | 136,296 (90.2%) | 14,838 (9.8%) | Negative | 0.960.83, 1.12) |
Note: Depending on aging-related trait, linear or logistic models were analyzed, and association estimate (β or odds ratio [OR]), 95% confidence interval (95% CI), and p-value (p) were extracted. All models were adjusted for age, sex, and genotyping PCs (1–10). Reported β and OR corresponds to one standard deviation (SD) increase in GI-TL (one SD in GI-TL is approximately 1,200 base pairs). Three reported significance thresholds corresponded to Bonferroni-corrected significance p < .001 (***), p < .01 (**), and p < .05 (*). GI-TL = genetically increased TL.
Figure 2.
Quantile-quantile plot of p-values for observed associations between aging-related traits and genetically increased TL (GI-TL) among U.K. Biobank participants. Dashed and dotted lines correspond to Bonferroni-corrected significance (p < .001) and nominal significance (p < .05), respectively. Black solid line represents the expected distribution of p-values under the null.
Half of the UKB participants were defined as hypertensive (49.7%), and age was correlated with SBP (Pearson’s r = 0.33), DBP (r = 0.07), and PP (r = 0.42). We observed that GI-TL was associated with increased SBP (β = 2.3 mmHg, 95% CI [1.7, 2.9], p = 2.9 × 10-15), DBP (β = 0.8 mmHg, 95% CI [0.4, 1.1], p = 5.5 × 10-6), and PP (β = 1.5 mmHg, 95% CI [1.1, 1.9], p = 5.2 × 10-14) per one SD increase in TL. GI-TL also increased the risk of hypertension (OR = 1.24, 95% CI [1.17, 1.33], p = 5.5 × 10-11). GI-TL was positively associated with SBP, PP, and DBP among both men and women, and there was not strong evidence that these associations differed by sex (Supplementary Tables S4 and S5, and Figure S3). When we stratified by median age (< or ≥58 years), GI-TL was positively associated with SBP, PP, and DBP within both strata (Supplementary Tables S6 and S7, and Figure S4). When we excluded individuals with self-reported BP medication use (as a sensitivity analysis), the associations persisted between GI-TL and BP traits: SBP (β = 1.8 mmHg, 95% CI [1.2, 2.4], p = 2.6 × 10-10), DBP (β = 0.6 mmHg, 95% CI [0.2, 0.9], p = 5.8 × 10-4), PP (β = 1.2 mmHg, 95% CI [0.8, 1.6], p = 2.2 × 10-9), and hypertension (OR = 1.21, 95% CI [1.13, 1.30], p = 3.4 × 10-8).
We examined GI-TL and mortality among our middle-aged UKB study participants because we observed that GI-TL was not pervasively associated with aging-related traits and a prior population-based study of Danish adults did not observe an association between GI-TL and mortality (14). Before December 2015, 9196 deaths occurred, and GI-TL was not associated with overall mortality, death from ischemic heart disease (n = 1053 deaths), cerebrovascular disease (n = 327), or chronic lower respiratory disease (n = 227) (Supplementary Table S8). GI-TL was associated with decreased risk of mortality from idiopathic pulmonary fibrosis (HR = 0.10, 95% CI [0.02, 0.55], p = .008, n = 106).
In an exploratory sensitivity analysis, we sought to examine whether our results were influenced by the selection bias related to participation in the UKB cohort. To do this, we applied IPWs in linear regression models to examine the association between GI-TL and the six significant aging-related traits (see Methods). When we applied IPWs based on age only (derived from the UKB source population), we observed that the associations were slightly attenuated but still highly significant between GI-TL and SBP (β = 2.2, 95% CI [1.6, 2.7], p = 1.7 × 10−14) and PP (β = 1.3, 95% CI [1.0, 1.7], p = 4.8 × 10−13) (Supplementary Table S9). When we applied IPWs based on age, sex, and hypertension status (derived from the HSE), the association between GI-TL and SBP and PP were attenuated by 13.0% (β = 2.0, 95% CI [1.5, 2.5], p = 1.4 × 10−13) and 16.7% (β = 1.3, 95% CI [0.9, 1.6], p = 4.1 × 10−12), respectively. The associations between FVC and GI-TL were strengthened when IPWs were applied. Overall, the results we observed persisted after the inclusion of IPWs to account for underlying factors affecting selection into and participation in the UKB cohort.
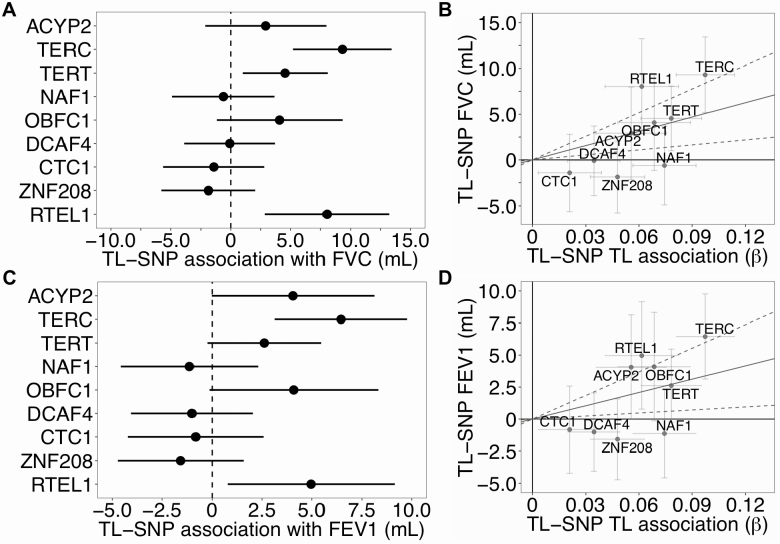
These pulmonary and BP traits were further examined using a two-sample MR analysis. Provided the MR assumptions are satisfied (39), MR-estimates can be interpreted as the estimated causal effect of an one SD increase in TL (~1,200 bp) on each trait (11). Using the IVW MR method, FEV1 and FVC increased by 34.7 mL (95% CI [7.9, 61.5], p = .01) and 52.2 mL (95% CI [18.2, 86.2], p = .003) per one SD increase in TL, respectively (Figure 3 and Supplementary Table S10). For three out of the nine SNPs, the long TL allele was positively associated with FVC: TERC (β = 9.0 mL, 95% CI [5.0, 13.1], p = 1.3 × 10−5), TERT (β = 4.5 mL, 95% CI [1.0, 8.0], p = .01) and RTEL1 (β = 8.0 mL, 95% CI [2.9, 13.2], p = .002). For FEV1, per-allele associations were observed for TERC (β = 6.4 mL, 95% CI [3.2, 9.7], p = 1.2 × 10−4) and RTEL1 (β = 5.0 mL, 95% CI [0.9, 9.1], p = .02) (Supplementary Table S11).
Figure 3.
Long telomere length alleles increased pulmonary function traits among U.K. Biobank participants. Forest plots (right) and scatterplots (left) of associations between telomere length (TL) associated SNPs and pulmonary function traits, FEV1 and FVC, among U.K. Biobank participants. Forest plots show association estimates with long allele (and 95% confidence interval [CI]). Scatterplots present the summary estimate of each SNP with TL (and 95% CI) and individual estimate of each SNP and trait (and 95% CI). Line captures the IVW MR estimate of the causal association of TL on each trait (and 95% CI). FEV1 = Forced expiratory volume; FVC = Forced vital capacity; IVW = Inverse variance weighted; MR = Mendelian randomization.
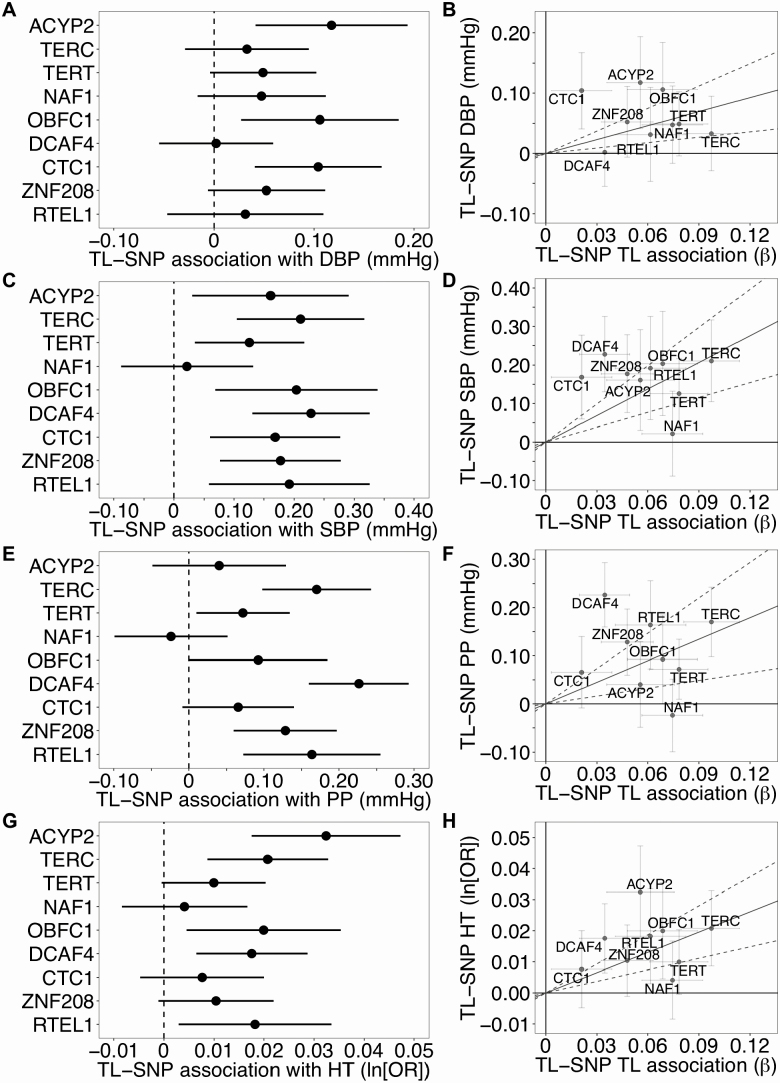
MR analysis indicated that a one SD increase in TL increased SBP, DBP, and PP by 2.3 mmHg (95% CI [1.3, 3.3], p = 8.2 × 10−6), 0.8 mmHg (95% CI [0.3, 1.2], p = .001), and 1.5 mmHg (95% CI [0.5, 2.5], p = .002), respectively (Figure 4 and Supplementary Table S10). One SD increase in TL increased the risk of hypertension (OR = 1.24, 95% CI [1.13, 1.36], p = 5.6 × 10−6). SBP was positively associated with eight of the nine TL-associated SNPs while DBP was positively associated with three of the nine SNPs (p < .05). DBP was not strongly associated with either the TERT or TERC SNPs (p > .05). Six of the nine TL-associated SNPs were positively associated with PP and five were positively associated with hypertension (p < .05) (Supplementary Table S11).
Figure 4.
Long telomere length alleles increased blood pressure traits among U.K. Biobank participants. Forest plots (right) and scatterplots (left) of associations between telomere length (TL) associated SNPs and blood pressure traits, SBP, DBP, PP, and hypertension, among U.K. Biobank participants. Forest plots show association estimates with long allele (and 95% CI). Scatterplots present the summary estimate of each SNP with TL (and 95% CI) and individual estimate of each SNP and trait (and 95% CI). Line captures the IVW MR estimate of the causal association of TL on each trait. DBP = Diastolic blood pressure; IVW = Inverse variance weighted; MR = Mendelian randomization; PP = Pulse pressure; SBP = Systolic blood pressure.
In addition to the IVW MR method, we also obtained MR estimates using several alternative MR approaches (Supplementary Table S10). For both BP and the pulmonary traits, the maximum likelihood approach yielded similar MR-estimates and p-values. Using MR-Egger, the estimated intercept differed from zero for SBP (p = .001), suggesting potential bias in the MR-estimate. However, the MR-estimates obtained from the weighted median approach were similar to IVW for all traits. When we applied the MR-PRESSO test to identify horizontal pleiotropic outliers, SNPs annotating to NAF1 and DCAF4 were identified as outliers for SBP and PP (p < .05), however, the removal of the outliers increased the IVW MR-estimates for both traits.
We utilized simulated data sets to examine different strengths of selection bias on our results in order to support that our observed results are not likely affected by selection. In data sets simulated under the null hypothesis of no causal effect (β = 0), selection on a continuous outcome (such as SBP) did not introduce bias or affect the false-positive rate (Supplementary Table S12). However, strong selection on a risk factor (eg, TL) increased the false-positive rate and biased the MR-estimate away from the null, with the direction of the bias depending on the direction of the confounder effects (Supplementary Table S13). Notably, this selection would have to be extremely strong (eg, yx = −2 or yx = 2) in order to generate the association of the magnitude we observe between TL and SBP (βx = 0.12). Under the alternative hypothesis of a true effect between SBP and TL (βx = 0.12), selection on the outcome attenuated causal effect estimate but did not decrease the empirical power to detect a true association of this magnitude between SBP and TL (Supplementary Table S14). When there was selection on the risk factor (again under the alternative hypothesis with βx = 0.12), two different scenarios were observed (Supplementary Table S15). First, if the confounder effect on the risk factor and the outcome was in the same direction, then selection on the risk factor attenuated the association between SBP and TL and decreased the empirical power when there was also selection on the confounder (yu = −1 or yu = 1). Second, if the confounder effect on the risk factor and outcome were in opposite directions, then strong selection on the risk factor biased the association between SBP and TL upward (similar in nature to the bias observed in Supplementary Table S13).
Discussion
In this work, we examined the association between GI-TL and more than 50 aging-related traits among the middle-aged UKB participants of British ancestry. While most aging-related traits, notably cognitive traits, did not show clear associations with GI-TL, we observed positive associations for GI-TL with pulmonary function and BP traits. MR results suggest that longer TL may be causally associated with increased BP (SBP, DBP, and PP) and pulmonary function (FEV1 and FVC). Additionally, the associations of GI-TL with FEV1 and FVC were stronger among older UKB participants, suggesting longer TL may be a protective factor against less age-related pulmonary decline.
Compared to our results, prior studies have observed similar associations between measured TL and pulmonary function. Among 46,396 Danish individuals, longer measured TL was associated with higher FEV1 and FVC (40). A meta-analysis of 14 European studies identified positive associations between measured TL and FEV1 and FVC (41). While FEV1 and FVC have not been examined in MR studies of TL, a large MR study of TL found evidence for an association between long TL and decreased risk of interstitial lung disease (similar to idiopathic pulmonary fibrosis) (16). Idiopathic pulmonary fibrosis is a hallmark of telomeropathy syndromes, characterized by very short TL due to mutations in telomere maintenance genes, such as TERT and TERC (42,43). The relationship between TL and pulmonary diseases indicates that TL may be a biologically important factor that contributes to the age-related decline of pulmonary function.
While our finding that GI-TL increased BP traits was unexpected, prior studies have observed inconsistent associations between TL and BP traits, with several null results reported (44–49). Measured TL was positively associated with SBP in adult NHANES participants with SBP above 140 mmHg (50), in Costa Rican elderly adults (51), and in Australian adults 40–44 years (52). Measured TL was inversely associated with DBP in NHANES (50) and Costa Rican elderly adults (51). In Danish population-based cohorts, longer measured TL was inversely associated with SBP while more TL shortening alleles were associated with decreased SBP (14). Correspondingly, the percent of individuals with hypertension decreased with increasing TL shortening alleles (53). The opposite associations between GI-TL and measured TL for BP traits suggest that these potential differences may be due to residual confounding (due to biological, technical, and/or environmental effects) and/or biological pleiotropy related to GI-TL. However, the biological mechanism between TL and BP traits remains to be determined, and the positive relationship between GI-TL and BP may reflect the dynamics among TL maintenance, TL function, and cellular senescence in vascular aging.
The UKB sample size is very large, which is critical for precise MR-based estimation. UKB also has a uniform and rigorously collected set of aging-related traits. GI-TL is constructed based on several SNPs that are near genes with clear relevance to TL biology, making GI-TL an instrumental variable with strong biological connection to TL (even though the amount of variation of leukocyte TL explained by TL-associated SNPs is between 2% and 3% (16)). However, it is unknown how well SNP associated with leukocyte TL predict TL in other tissue types, so it is unclear whether these SNPs are strong instrumental variables for TL in other tissue types. A central limitation of MR is that it is not possible to completely rule out the possibility of biological pleiotropy between our TL-associated SNPs and aging-related traits.
Another limitation is the nonrepresentativeness of the UKB sample, potentially reducing the external validity of our results; this limitation we attempted to address using simulated data and IPWs. We utilized IPWs to create pseudo-populations that are unaffected by selection on certain factors and more closely resemble the UKB source population. While all association estimates remained highly significant after the inclusion of IPWs, the association between GI-TL and BP traits slightly decreased and the association between GI-TL and pulmonary traits increased, suggesting that IPWs should be further explored for other characteristics that may influence participation (eg, pulmonary function) and further developed for large cohorts such as UKB to enhance the validity and reliability of MR estimates. From our analysis of simulated data to examine selection as a source of collider bias, we concluded that neither selection on the outcome (eg, SBP) nor on the risk factor (eg, TL) were likely to explain our observed MR association between TL and SBP. While we cannot create IPWs based on TL, selection on TL would have to be very strong to produce the observed association (under the null hypothesis of no effect), which in our view is not a biologically plausible explanation for the observed associations. These results based on simulated data were consistent with what was observed in the simulations reported by Gkatzionis and Burgess (36).
In conclusion, among unrelated middle-aged UKB participants of British ancestry, GI-TL was associated with increased pulmonary function but also with higher BP. These findings provide evidence for a causal association between TL and these aging-related traits. Future longitudinal studies should evaluate age-related temporal changes in pulmonary function and BP traits and how these are related to TL.
Supplementary Material
Acknowledgments
We would like to thank the U.K. Biobank leadership, staff, and participants for their contributions to this research and cohort.
Funding
This work was supported by active and past National Institute of Health grants (R35ES028379, R01ES020506, and U01HG007601 to B.L.P.). Fellowship support for K.D. was provided by the National Institute of Aging Specialized Demography and Economics of Aging Training Program and NIH Research Supplement to Promote Diversity in Health-Related Research (T32AG000243 and R35ES028379-02S1 to K.D.) at the University of Chicago.
Conflict of Interest
None reported.
Author Contributions
All authors (K.D., L.T., and B.L.P) contributed to the design, analysis, writing, and review of this work.
References
- 1. Blackburn EH, Epel ES, Lin J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350:1193–1198. doi:10.1126/science.aab3389 [DOI] [PubMed] [Google Scholar]
- 2. d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi:10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 3. Daniali L, Benetos A, Susser E, et al. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun. 2013;4:1597. doi:10.1038/ncomms2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mons U, Müezzinler A, Schöttker B, et al. Leukocyte telomere length and all-cause, cardiovascular disease, and cancer mortality: results from individual-participant-data meta-analysis of 2 large prospective cohort studies. Am J Epidemiol. 2017;185:1317–1326. doi:10.1093/aje/kww210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haycock PC, Heydon EE, Kaptoge S, Butterworth AS, Thompson A, Willeit P. Leucocyte telomere length and risk of cardiovascular disease: systematic review and meta-analysis. BMJ. 2014;349:g4227. doi:10.1136/bmj.g4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Willeit P, Raschenberger J, Heydon EE, et al. Leucocyte telomere length and risk of type 2 diabetes mellitus: new prospective cohort study and literature-based meta-analysis. PLoS One. 2014;9:e112483. doi:10.1371/journal.pone.0112483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forero DA, González-Giraldo Y, López-Quintero C, Castro-Vega LJ, Barreto GE, Perry G. Meta-analysis of telomere length in Alzheimer’s disease. J Gerontol A Biol Sci Med Sci. 2016;71:1069–1073. doi:10.1093/gerona/glw053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang X, Zhao Q, Zhu W, et al. The association of telomere length in peripheral blood cells with cancer risk: a systematic review and meta-analysis of prospective studies. Cancer Epidemiol Biomarkers Prev. 2017;26:1381–1390. doi:10.1158/1055-9965.EPI-16-0968 [DOI] [PubMed] [Google Scholar]
- 9. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–665. doi:10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boef AG, Dekkers OM, le Cessie S. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol. 2015;44:496–511. doi:10.1093/ije/dyv071 [DOI] [PubMed] [Google Scholar]
- 11. Codd V, Nelson CP, Albrecht E, et al. ; CARDIoGRAM consortium. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–427, 427e421–422. doi:10.1038/ng.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mangino M, Christiansen L, Stone R, et al. DCAF4, a novel gene associated with leucocyte telomere length. J Med Genet. 2015;52:157–162. doi:10.1136/jmedgenet-2014-102681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mangino M, Hwang SJ, Spector TD, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet. 2012;21:5385–5394. doi:10.1093/hmg/dds382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rode L, Nordestgaard BG, Bojesen SE. Peripheral blood leukocyte telomere length and mortality among 64,637 individuals from the general population. J Natl Cancer Inst. 2015;107:djv074. doi:10.1093/jnci/djv074 [DOI] [PubMed] [Google Scholar]
- 15. Rode L, Nordestgaard BG, Bojesen SE. Long telomeres and cancer risk among 95 568 individuals from the general population. Int J Epidemiol. 2016;45:1634–1643. doi:10.1093/ije/dyw179 [DOI] [PubMed] [Google Scholar]
- 16. Haycock PC, Burgess S, Nounu A, et al. ; Telomeres Mendelian Randomization Collaboration Association between telomere length and risk of cancer and non-neoplastic diseases: a Mendelian randomization study. JAMA Oncol. 2017;3:636–651. doi:10.1001/jamaoncol.2016.5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi:10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi:10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi:10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bakrania K, Edwardson CL, Khunti K, Bandelow S, Davies MJ, Yates T. Associations between sedentary behaviors and cognitive function: cross-sectional and prospective findings from the UK Biobank. Am J Epidemiol. 2018;187:441–454. doi:10.1093/aje/kwx273 [DOI] [PubMed] [Google Scholar]
- 21. Williams DM, Jylhävä J, Pedersen NL, Hägg S. A frailty index for UK Biobank participants. J Gerontol A Biol Sci Med Sci. 2019;74:582–587. doi:10.1093/gerona/gly094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med. 2011;30:377–399. doi:10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- 23. Walsh KM, Codd V, Rice T, et al. ; ENGAGE Consortium Telomere Group. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6:42468–42477. doi:10.18632/oncotarget.6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamad R, Walter S, Rehkopf DH. Telomere length and health outcomes: a two-sample genetic instrumental variables analysis. Exp Gerontol. 2016;82:88–94. doi:10.1016/j.exger.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warren HR, Evangelou E, Cabrera CP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi:10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi:10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 27. Galinsky KJ, Loh PR, Mallick S, Patterson NJ, Price AL. Population structure of UK Biobank and Ancient Eurasians reveals adaptation at genes influencing blood pressure. Am J Hum Genet. 2016;99:1130–1139. doi:10.1016/j.ajhg.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galinsky KJ, Bhatia G, Loh PR, et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98:456–472. doi:10.1016/j.ajhg.2015.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang C, Doherty JA, Burgess S, et al. ; GECCO and GAME-ON Network: CORECT, DRIVE, ELLIPSE, FOCI, and TRICL Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum Mol Genet. 2015;24:5356–5366. doi:10.1093/hmg/ddv252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. doi:10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson JR, Minelli C, Abrams KR, Tobin MD, Riley RD. Meta-analysis of genetic studies using Mendelian randomization—a multivariate approach. Stat Med. 2005;24:2241–2254. doi:10.1002/sim.2100 [DOI] [PubMed] [Google Scholar]
- 32. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi:10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–389. doi:10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi:10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol. 2018;47:226–235. doi:10.1093/ije/dyx206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gkatzionis A, Burgess S. Contextualizing selection bias in Mendelian randomization: how bad is it likely to be? Int J Epidemiol. 2018;48:691–701. doi:10.1093/ije/dyy202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nohr EA, Liew Z. How to investigate and adjust for selection bias in cohort studies. Acta Obstet Gynecol Scand. 2018;97:407–416. doi:10.1111/aogs.13319 [DOI] [PubMed] [Google Scholar]
- 38. National Centre for Social Research UCL, Department of Epidemiology and Public Health. Health Survey for England, 2008. 4th edn.Essex, UK: UK Data Service; 2013. [Google Scholar]
- 39. Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16:309–330. doi:10.1177/0962280206077743 [DOI] [PubMed] [Google Scholar]
- 40. Rode L, Bojesen SE, Weischer M, Vestbo J, Nordestgaard BG. Short telomere length, lung function and chronic obstructive pulmonary disease in 46,396 individuals. Thorax. 2013;68:429–435. doi:10.1136/thoraxjnl-2012-202544 [DOI] [PubMed] [Google Scholar]
- 41. Albrecht E, Sillanpää E, Karrasch S, et al. Telomere length in circulating leukocytes is associated with lung function and disease. Eur Respir J. 2014;43:983–992. doi:10.1183/09031936.00046213 [DOI] [PubMed] [Google Scholar]
- 42. Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi:10.1038/nrg3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Armanios M. Telomeres and age-related disease: how telomere biology informs clinical paradigms. J Clin Invest. 2013;123:996–1002. doi:10.1172/JCI66370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bekaert S, De Meyer T, Rietzschel ER, et al. ; Asklepios investigators. Telomere length and cardiovascular risk factors in a middle-aged population free of overt cardiovascular disease. Aging Cell. 2007;6:639–647. doi:10.1111/j.1474-9726.2007.00321.x [DOI] [PubMed] [Google Scholar]
- 45. Brouilette SW, Moore JS, McMahon AD, et al. ; West of Scotland Coronary Prevention Study Group Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–114. doi:10.1016/S0140-6736(07)60071-3 [DOI] [PubMed] [Google Scholar]
- 46. Cheng YY, Kao TW, Chang YW, et al. Examining the gender difference in the association between metabolic syndrome and the mean leukocyte telomere length. PLoS One. 2017;12:e0180687. doi:10.1371/journal.pone.0180687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fitzpatrick AL, Kronmal RA, Gardner JP, et al. Leukocyte telomere length and cardiovascular disease in the cardiovascular health study. Am J Epidemiol. 2007;165:14–21. doi:10.1093/aje/kwj346 [DOI] [PubMed] [Google Scholar]
- 48. Denil SL, Rietzschel ER, De Buyzere ML, et al. ; Asklepios Investigators. On cross-sectional associations of leukocyte telomere length with cardiac systolic, diastolic and vascular function: the Asklepios study. PLoS One. 2014;9:e115071. doi:10.1371/journal.pone.0115071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Révész D, Milaneschi Y, Verhoeven JE, Lin J, Penninx BW. Longitudinal associations between metabolic syndrome components and telomere shortening. J Clin Endocrinol Metab. 2015;100:3050–3059. doi:10.1210/JC.2015-1995 [DOI] [PubMed] [Google Scholar]
- 50. Rehkopf DH, Needham BL, Lin J, et al. Leukocyte telomere length in relation to 17 biomarkers of cardiovascular disease risk: a cross-sectional study of US adults. PLoS Med. 2016;13:e1002188. doi:10.1371/journal.pmed.1002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rehkopf DH, Dow WH, Rosero-Bixby L, Lin J, Epel ES, Blackburn EH. Longer leukocyte telomere length in Costa Rica’s Nicoya Peninsula: a population-based study. Exp Gerontol. 2013;48:1266–1273. doi:10.1016/j.exger.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mather KA, Jorm AF, Milburn PJ, Tan X, Easteal S, Christensen H. No associations between telomere length and age-sensitive indicators of physical function in mid and later life. J Gerontol A Biol Sci Med Sci. 2010;65:792–799. doi:10.1093/gerona/glq050 [DOI] [PubMed] [Google Scholar]
- 53. Scheller Madrid A, Rode L, Nordestgaard BG, Bojesen SE. Short telomere length and ischemic heart disease: observational and genetic studies in 290 022 individuals. Clin Chem. 2016;62:1140–1149. doi:10.1373/clinchem.2016.258566 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.