Abstract
Background
Fortetropin is a proteo-lipid complex made from fertilized egg yolk and, in young men, has been shown to increase lean body mass.
Methods
The purpose of this study was to examine the effects of 21 days of Fortetropin supplementation on the fractional synthetic rate (FSR) of muscle protein in 10 healthy, older men and 10 women (66.4 ± 4.5 y). We used 2H2O labeling to measure FSR of multiple muscle protein ontologies. D3-creatine dilution was used to determine muscle mass at baseline. Subjects ingested 70% 2H2O for 21 day and saliva samples were collected to determine body 2H2O enrichment. A microbiopsy was obtained from the m. vastus lateralis on Day 21. Subjects were randomly assigned to Fortetropin (19.8 g/d) or placebo (cheese powder, 19.8 g/d).
Results
Restricting kinetic data to proteins with ≥2 peptides measured in at least 4 subjects per group resulted in 117 proteins meeting these criteria. The mean FSR for a majority of proteins in several muscle gene ontologies was higher in the Fortetropin group compared to placebo (32/38 myofibril proteins, 33/44 sarcoplasmic proteins, and 12/17 mitochondrial proteins) and this proportion was significantly different between groups using a binomial test and were independent of sex or baseline muscle mass.
Conclusions
The overall magnitude of the difference in muscle protein FSR of Fortetropin from placebo was 18%, with multiple gene ontologies affected. While these results should be confirmed in larger cohorts, they suggest that Fortetropin supplementation is effective for promoting muscle protein synthesis in older people.
Keywords: Aging muscle, Muscle protein synthesis, Sarcopenia
Sarcopenia has been defined as the age-related loss of skeletal muscle mass (1). In older people, low muscle mass is strongly associated with reduced functional capacity and an increased risk of disability (2). Recent studies demonstrate that muscle mass has a powerful effect on risk of disability, fall, and poor functional capacity (2). While basal rates of muscle protein synthesis may not change with age (3), sarcopenia is, at least in part, a result of a reduced rate of protein synthesis after a protein containing meal, referred to as anabolic resistance (4). This age-related reduction in postprandial muscle protein synthesis rate has a number of causes, including decreases in testosterone and growth hormone levels, insulin resistance, reduced levels of physical activity, and more. However, skeletal muscle is also a remarkably plastic tissue with multiple pathways that can stimulate hypertrophy or cause atrophy.
Myostatin is a negative regulator of muscle growth (also referred to as growth differentiation factor-8 or GDF-8), is a member of the transforming growth factor-β superfamily of growth and differentiation factors, and has become an important target for pharmaceutical companies as a way to increase muscle protein synthesis and growth. Anti-myostatin drugs increase muscle size and strength in preclinical studies. Clinical studies with anti-myostatin therapy or activin II receptor blockade in older people have shown significant increases in lean body mass and small increases in functional capacity (5,6). Fortetropin is a proteo-lipid complex made from fertilized egg yolk and Sharp and colleagues (7) demonstrated that Fortetropin provided as a supplement lowered circulating myostatin levels in rodents and in young men in combination with resistance exercise also lowered myostatin, increased lean body mass, and increased mTOR signaling compared to placebo. The functional significance and interpretation of circulating myostatin levels is uncertain, however.
The rate of synthesis of multiple muscle proteins in vivo can be measured using tandem mass spectrometric analysis of labeling patterns after ingestion of relatively small amounts of 2H2O to enrich total body water. Deuterium is incorporated through intermediary metabolic pathways into free amino acids which then enter newly synthesized proteins. In this way, a proteomic approach to changes in the fractional synthetic rate (FSR) of proteins is possible. We previously demonstrated in rats (8) that a selective androgen receptor modulator (a muscle anabolic drug) had a potent dose–responsive effects on the FSR of multiple muscle proteins, particularly in the myofibrillar and glycolytic gene ontologies, and that the short-term increases in FSR were strongly related to longer-term muscle hypertrophy. We have also shown in human subjects, using the heavy water labeling approach with tandem mass spectrometric analyses, that resistance exercise training in obese older men increased FSR of multiple muscle proteins across all ontologies (9) and that a sprint exercise training regimen in young men and women increased FSR of muscle proteins, mostly in the glycolytic and structural protein ontologies (10).
We hypothesized here that compared to controls, daily consumption of Fortetropin supplements would increase the FSR of skeletal muscle proteins.
Methods
This study was approved by the institutional review board of University of Arkansas for Medical Sciences. A total of 20 healthy men and women were recruited, provided informed consent, and were enrolled in the study. Subjects (mean age 66.4 ± 4.5 years) were randomly assigned to a treatment (FO) or control group (CO). Those in the treatment group consumed Fortetropin (19.8 g/d) and the placebo control group consumed cheese powder 19.8 g/d that was matched for macronutrient and energy to Fortetropin (egg yolk) for 21 days. Central randomization was accomplished by the diet staff which prepared the Fortetropin and placebo supplements and placed them in similar looking containers with a specific study number. The study investigators, staff, or research volunteers were not made aware of the study group assignments. No dietary controls were required and subjects were asked to maintain their normal activity patterns during the 21-day treatment period but food intake was not controlled or assessed. Three days before the initiation of FO or CO, each participant ingested a 30 mg capsule of D3-creatine for the measurement of muscle mass. After 3 days, subjects reported to the laboratory and produced a fasting urine sample for later analysis of D3-creatinine enrichment, creatine, and creatinine concentrations (11), which were used for the estimation of baseline muscle mass. On Days 1 and 21, a blood sample was collected for determination of circulating myostatin levels by ELISA (GDF-8/Myostatin Quantikine ELISA Kit, R&D Systems).
On the first four days of treatment, subjects ingested three 50 mL quantities of 70% 2H2O to rapidly (bolus) increase body water enrichment. Thereafter (Days 5–21), subjects consumed 50 mL of 70% 2H2O to maintain a constant body water enrichment. Saliva samples were collected on Days 7, 14, and 21 for determination of 2H2O enrichment (12). A microbiopsy (13) (approximately 10 mg) was collected and immediately frozen for determination of muscle proteome-wide FSR (14).
Muscle proteome dynamics was measured using previously described methods (8,14,15). Briefly, muscle biopsy tissue was suspended in 0.08% SDS at a 10:1 volume:weight ratio, and vortexed at low speed overnight (16 h) to extract cellular proteins. The SDS-soluble proteins were precipitated by overnight incubation at −20°C in ethanol (5:1 ethanol:extraction buffer) followed by centrifugation at 16 000g for 45 min. Pelleted proteins were rinsed twice with 90% ethanol, allowed to air dry, and resuspended in 8 M urea prior to trypsin digestion. Up to 80 µg of SDS-soluble protein sample was denatured using Protease-Max surfactant (0.1%; Promega, Madison, WI) and 4 M urea in 25 mM ammonium bicarbonate (pH 8). Proteins were reduced with TCEP (5 mM) for 20 min at room temperature with vortexing and then incubated with iodoacetamide (10 mM) in the dark for 20 min to chemically modify reduced cysteines. Proteins were then digested with trypsin (Promega) at 37°C overnight using a 1:25 trypsin:protein mass ratio. The following day, formic acid was added to a total concentration of 5%, and samples were centrifuged at 14 000g for 30 min. The supernatant was transferred to a fresh tube, desalted with a C18 spec tip (Varian, Palo Alto, CA), dried via vacuum centrifugation, and resuspended in 0.1% formic acid/3% acetonitrile prior to LC/MS analysis.
Trypsin-digested peptides were analyzed on a 6550 QTOF (quadrupole time-of-flight) mass spectrometer with a 1260 Chip Cube nano ESI source (Agilent Technologies, Santa Clara, CA). Each sample was analyzed once for protein/peptide identification in data-dependent MS/MS mode and once for peptide isotope analysis in MS mode. Acquired MS/MS spectra were extracted and searched using Spectrum Mill Proteomics Workbench software (Agilent Technologies) and mouse protein database (UniProt.org). Search results were validated with a global false discovery rate of 1%. A filtered list of peptides was collapsed into a nonredundant peptide formula database containing peptide elemental composition, mass, and retention time. This was used to extract mass isotope abundances (M0–M3) of each peptide from MS-only acquisition files with Mass Hunter Qualitative Analysis software (Agilent Technologies). An in-house software was used to calculate peptide elemental composition and curve fit parameters for predicting peptide isotope enrichment (EM0) based on precursor body water enrichment (p) and the number (n) of amino acid C-H positions per peptide actively incorporating hydrogen (H) and deuterium (D) from body water.
Subsequent data handling was performed using python-based scripts, with input of average body water enrichment for each participant, to yield fractional synthesis data at the protein level. FSR data were filtered to only include protein measurements with ≥2 peptide isotope measurements per protein measured in at least four subjects per group. Additional details of the FSR calculations and data filtering criteria were as described in detail previously (10,14).
The sample analysis was performed on de-identified samples. Statistical analyses were performed for the different groups of proteins by t-test with Benjamini–Hochberg correction for multiple comparisons, as well as by Binomial test on magnitude of change for gene ontological groups of myofibril, cytoplasmic, and mitochondrial proteins.
Results
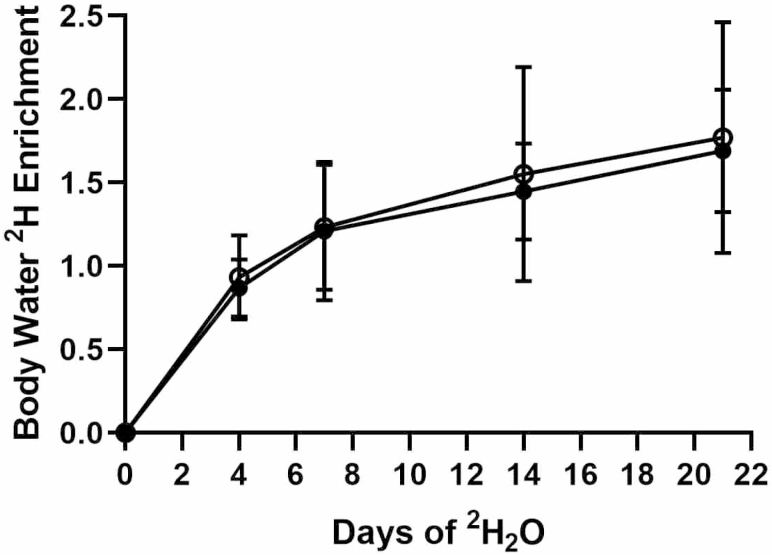
All 20 subjects enrolled completed all aspects of this study. Enrichment of 2H2O increased over 3 weeks to approximately 1.5% of total body water and was not different between the two study groups (Figure 1). No differences in baseline muscle mass between FO (30.81 ± 8.46 kg) and CO (26.06 ± 9.30 kg) were observed, data represent average ± SD.
Figure 1.
Total body 2H2O enrichment during 21 days of daily intake of 70% 2H2O. No differences between groups was observed. Fortetropin: closed circles, Control: open circles.
Muscle protein FSR
MS/MS analysis identified 210 muscle proteins with ≥2 peptides/protein. Using analytic criteria of ≥2 peptides per protein measured in at least 4 subjects per group, the kinetic data comprise 109 muscle proteins. Fractional synthesis for each participant was calculated using the average body water used as the precursor enrichment and FSR (% per week) was calculated as −ln(1−f)/t, where (f) is fractional synthesis and (t) the duration in weeks of label.
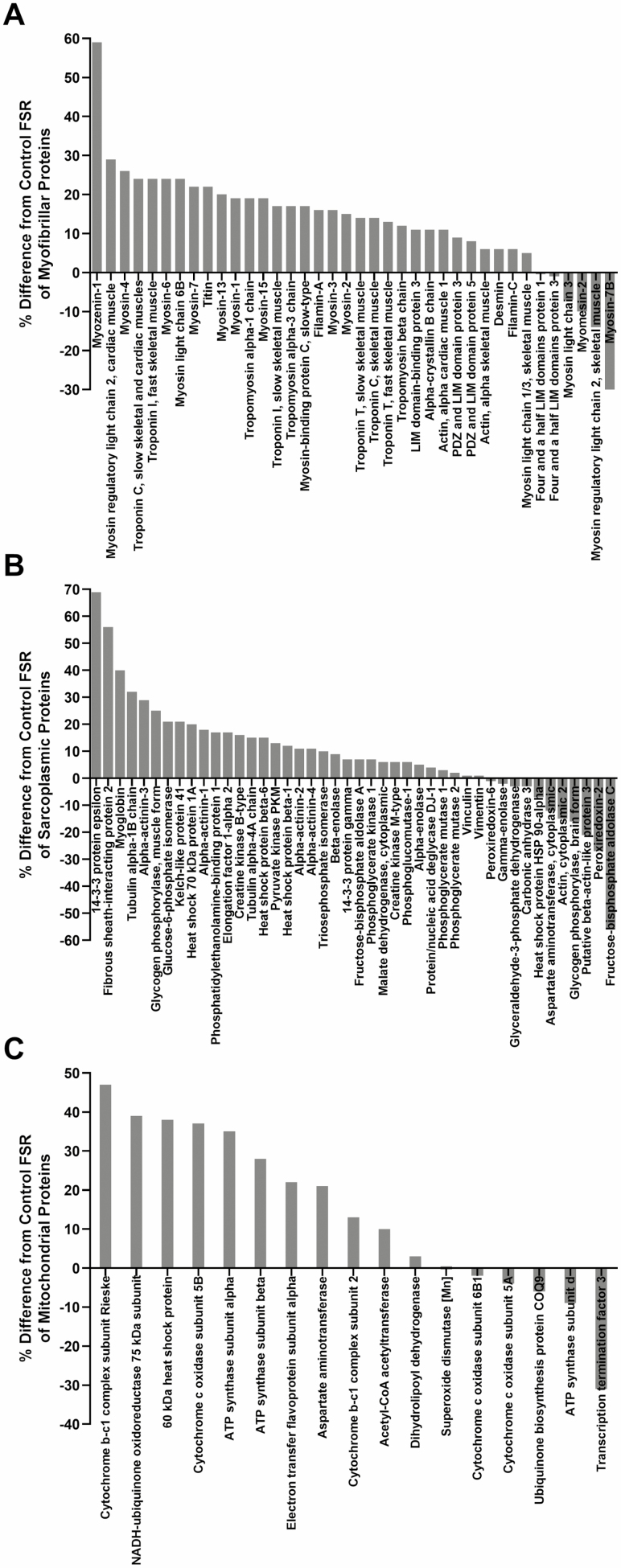
Fractional synthetic rate was measured for all samples, however the number of subjects used for each protein measurement varies based on the analytical criteria of requiring two or more peptides per protein. Each protein must be measured in four or more subjects per group. The FSR of myofibril, sarcoplasmic, and mitochondrial proteins measured in FO and CO groups is given in Table 1 (sample size for each protein is shown). Although there were no significant differences using a Bonferroni correction among individual proteins, an overall 18 ± 13% greater FSR was observed in the FO group for the average of 109 muscle proteins. The average FSR for a majority of proteins in several gene ontologies was higher in the FO group (32/38 myofibril proteins, 33/44 sarcoplasmic proteins, and 12/17 mitochondrial proteins as shown in Figure 2A, B, and C, respectively) and these ontology proportions were each different from control (p < .05 by the binomial test).
Table 1.
Fractional Synthetic Rate (FSR) of Individual Muscle Proteins (±SD) by Gene Ontology for Fortetropin and Control Groups
| Muscle Protein | Fortetropin FSR | Control FSR | % Difference |
|---|---|---|---|
| Myofibrillar | |||
| Myozenin-1 | 23.56 ± 5.35 (6) | 14.85 ± 5.22 (7) | 59 |
| Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | 23.51 ± 6.39 (10) | 18.20 ± 5.34 (10) | 29 |
| Myosin-4 | 9.82 ± 3.51 (10) | 7.79 ± 3.29 (10) | 26 |
| Troponin C, slow skeletal and cardiac muscles | 19.37 ± 4.67 (10) | 15.65 ± 4.53 (10) | 24 |
| Troponin I, fast skeletal muscle | 16.00 ± 6.15 (8) | 12.94 ± 6.94 (8) | 24 |
| Myosin-6 | 7.72 ± 3.89 (9) | 6.24 ± 2.42 (9) | 24 |
| Myosin light chain 6B | 9.77 ± 2.59 (10) | 7.91 ± 2.06 (10) | 24 |
| Myosin-7 | 8.19 ± 2.30 (10) | 6.73 ± 2.46 (10) | 22 |
| Titin | 18.17 ± 5.56 (10) | 14.96 ± 5.05 (10) | 22 |
| Myosin-13 | 7.65 ± 2.02 (10) | 6.36 ± 2.21 (10) | 20 |
| Myosin-1 | 22.76 ± 5.62 (10) | 19.06 ± 5.26 (10) | 19 |
| Tropomyosin α-1 chain | 9.07 ± 2.20 (10) | 7.60 ± 2.22 (10) | 19 |
| Myosin-15 | 11.66 ± 2.69 (10) | 9.82 ± 3.20 (9) | 19 |
| Troponin I, slow skeletal muscle | 8.84 ± 2.34 (10) | 7.53 ± 2.26 (10) | 17 |
| Tropomyosin α-3 chain | 6.19 ± 3.47 (9) | 5.30 ± 1.93 (10) | 17 |
| Myosin-binding protein C, slow-type | 9.73 ± 2.14 (10) | 8.34 ± 2.74 (10) | 17 |
| Filamin-A | 11.23 ± 1.73 (8) | 9.65 ± 1.33 (6) | 16 |
| Myosin-3 | 9.55 ± 1.63 (10) | 8.23 ± 2.23 (10) | 16 |
| Myosin-2 | 9.49 ± 1.82 (10) | 8.25 ± 2.11 (10) | 15 |
| Troponin T, slow skeletal muscle | 24.15 ± 7.06 (7) | 21.12 ± 14.86 (9) | 14 |
| Troponin C, skeletal muscle | 13.73 ± 6.34 (10) | 12.05 ± 9.02 (10) | 14 |
| Troponin T, fast skeletal muscle | 12.42 ± 3.48 (10) | 11.01 ± 4.75 (10) | 13 |
| Tropomyosin-β chain | 4.28 ± 1.07 (10) | 3.82 ± 1.74 (10) | 12 |
| LIM domain-binding protein 3 | 8.67 ± 1.45 (8) | 7.80 ± 1.40 (6) | 11 |
| α-Crystallin B chain | 13.78 ± 5.09 (10) | 12.39 ± 4.96 (10) | 11 |
| Actin, α-cardiac muscle 1 | 24.38 ± 5.28 (10) | 21.94 ± 640 (9) | 11 |
| PDZ and LIM domain protein 3 | 7.05 ± 2.43 (9) | 6.49 ± 2.33 (10) | 9 |
| PDZ and LIM domain protein 5 | 2.68 ± 1.47 (10) | 2.47 ± 1.28 (10) | 8 |
| Actin, α-skeletal muscle | 4.43 ± 1.11 (10) | 4.16 ± 1.52 (10) | 6 |
| Desmin | 32.83 ± 13.77 (10) | 30.87 ± 17.79 (10) | 6 |
| Filamin-C | 18.55 ± 7.65 (9) | 17.50 ± 8.55 (10) | 6 |
| Myosin light chain 1/3, skeletal muscle isoform | 3.99 ± 0.75 (10) | 3.79 ± 0.84 (10) | 5 |
| Four and a half LIM domains protein 1 | 25.30 ± 9.19 (6) | 25.39 ± 11.09 (10) | 0 |
| Four and a half LIM domains protein 3 | 26.19 ± 0.128 (4) | 26.52 ± 2.29 (5) | −1 |
| Myosin light chain 3 | 8.47 ± 2.22 (8) | 9.08 ± 3.66 (8) | −7 |
| Myomesin-2 | 19.33 ± 3.85 (8) | 21.47 ± 7.03 (7) | −10 |
| Myosin regulatory light chain 2, skeletal muscle isoform | 4.92 ± 1.52 (10) | 5.72 ± 3.81 (10) | −14 |
| Myosin-7B | 5.28 ± 2.71 (5) | 7.52 ± 3.91 (5) | −30 |
| Mean magnitude of increase | 17 | ||
| Binomial test two-tailed p-value | p < .0001 | ||
| Sarcoplasmic | |||
| 14-3-3 Protein epsilon | 17.06 ± 7.25 (5) | 10.12 ± 5.34 (6) | 69 |
| Fibrous sheath-interacting protein 2 | 14.32 ± 3.72 (5) | 9.18 ± 4.42 (5) | 56 |
| Myoglobin | 5.42 ± 2.52 (10) | 3.87 ± 1.96 (10) | 40 |
| Tubulin α-1B chain | 25.69 ± 4.45 (4) | 19.44 ± 7.44 (4) | 32 |
| α-Actinin-3 | 5.72 ± 1.49 (10) | 4.44 ± 1.62 (10) | 29 |
| Glycogen phosphorylase, muscle form | 13.96 ± 3.37 (9) | 11.21 ± 3.94 (10) | 25 |
| Glucose-6-phosphate isomerase | 6.37 ± 2.32 (4) | 5.25 ± 2.60 (6) | 21 |
| Kelch-like protein 41 | 20.09 ± 4.27 (4) | 16.60 ± 3.46 (5) | 21 |
| Heat shock 70 kDa protein 1A | 18.68 ± 5.52 (4) | 15.63 ± 2.17 (6) | 20 |
| α-Actinin-1 | 6.21 ± 1.60 (8) | 5.25 ± 1.63 (6) | 18 |
| Phosphatidylethanolamine-binding protein 1 | 11.76 ± 4.59 (6) | 10.04 ± 2.55 (4) | 17 |
| Elongation factor 1-α 2 | 18.51 ± 2.31 (6) | 15.81 ± 4.00 (6) | 17 |
| Creatine kinase B-type | 5.80 ± 1.88 (10) | 4.99 ± 2.95 (10) | 16 |
| Tubulin alpha-4A chain | 24.91 ± 7.07 (4) | 21.62 ± 2.98 (5) | 15 |
| Heat shock protein β-6 | 24.65 ± 13.41 (9) | 21.40 ± 8.32 (10) | 15 |
| Pyruvate kinase PKM | 7.02 ± 2.17 (7) | 6.24 ± 2.10 (8) | 13 |
| Heat shock protein β-1 | 18.53 ± 4.46 (9) | 16.56 ± 5.28 (9) | 12 |
| α-Actinin-2 | 6.29 ± 1.54 (10) | 5.66 ± 1.81 (10) | 11 |
| α-Actinin-4 | 5.69 ± 1.57 (8) | 5.13 ± 1.70 (6) | 11 |
| Triosephosphate isomerase | 7.58 ± 1.61 (9) | 6.92 ± 2.75 (10) | 10 |
| β-Enolase | 8.63 ± 3.77 (10) | 7.88 ± 3.36 (10) | 9 |
| 14-3-3 Protein gamma | 17.12 ± 6.74 (4) | 15.97 ± 3.78 (5) | 7 |
| Fructose-bisphosphate aldolase A | 10.01 ± 2.67 (10) | 9.35 ± 3.60 (10) | 7 |
| Phosphoglycerate kinase 1 | 3.89 ± 2.32 (6) | 3.63 ± 2.14 (7) | 7 |
| Malate dehydrogenase, cytoplasmic | 6.94 ± 2.92 (8) | 6.53 ± 3.60 (10) | 6 |
| Creatine kinase M-type | 6.98 ± 1.73 (10) | 6.57 ± 2.73 (10) | 6 |
| Phosphoglucomutase-1 | 8.42 ± 1.08 (8) | 7.97 ± 2.62 (9) | 6 |
| α-Enolase | 7.08 ± 3.37 (10) | 6.76 ± 3.41 (10) | 5 |
| Protein/nucleic acid deglycase DJ-1 | 8.07 ± 1.68 (5) | 7.72 ± 0.91 (5) | 4 |
| Phosphoglycerate mutase 1 | 4.93 ± 1.21 (9) | 4.78 ± 1.79 (10) | 3 |
| Phosphoglycerate mutase 2 | 4.00 ± 1.80 (10) | 3.93 ± 1.73 (10) | 2 |
| Vinculin | 16.17 ± 3.26 (8) | 15.93 ± 2.34 (6) | 1 |
| Vimentin | 23.34 ± 6.66 (7) | 23.19 ± 8.61 (8) | 1 |
| Peroxiredoxin-6 | 9.49 ± 1.97 (7) | 9.58 ± 5.05 (8) | −1 |
| γ-Enolase | 7.19 ± 2.50 (10) | 7.33 ± 4.23 (10) | −2 |
| Glyceraldehyde-3-phosphate dehydrogenase | 8.34 ± 1.84 (10) | 8.57 ± 7.51 (9) | −3 |
| Carbonic anhydrase 3 | 4.42 ± 1.73 (10) | 4.58 ± 2.01 (10) | −3 |
| Heat shock protein HSP 90-α | 10.34 ± 2.73 (8) | 11.08 ± 3.48 (4) | −7 |
| Aspartate aminotransferase, cytoplasmic | 10.43 ± 3.39 (8) | 11.86 ± 2.37 (10) | −12 |
| Actin, cytoplasmic 2 | 4.48 ± 2.55 (8) | 5.25 ± 3.07 (6) | −15 |
| Glycogen phosphorylase, brain form | 14.39 ± 4.60 (7) | 17.14 ± 6.71 (6) | −16 |
| Putative β-actin-like protein 3 | 3.30 ± 1.47 (8) | 4.20 ± 0.54 (6) | −21 |
| Peroxiredoxin-2 | 6.91 ± 4.17 (5) | 9.42 ± 5.41 (8) | −27 |
| Fructose-bisphosphate aldolase C | 8.40 ± 1.06 (6) | 19.16 ± 17.84 (8) | −56 |
| Mean magnitude of increase | 16 | ||
| Binomial test two-tailed p-value | p < .0005 | ||
| Mitochondrial | |||
| Cytochrome b-c1 complex subunit Rieske, mitochondrial | 14.72 ± 5.43 (6) | 10.02 ± 3.05 (4) | 47 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 79.78 ± 24.34 (4) | 57.49 ± 36.90 (5) | 39 |
| 60 kDa heat shock protein, mitochondrial | 8.47 ± 3.77 (8) | 6.11 ± 1.41 (4) | 38 |
| Cytochrome c oxidase subunit 5B, mitochondrial | 11.93 ± 4.80 (5) | 8.68 ± 5.10 (5) | 37 |
| ATP synthase subunit α, mitochondrial | 12.70 ± 3.80 (5) | 9.38 ± 1.87 (7) | 35 |
| ATP synthase subunit β, mitochondrial | 10.57 ± 3.12 (10) | 8.27 ± 2.59 (10) | 28 |
| Electron transfer flavoprotein subunit α, mitochondrial | 11.77± 5.65 (8) | 9.65 ± 4.06 (8) | 22 |
| Aspartate aminotransferase, mitochondrial | 7.26 ± 2.50 (6) | 5.99 ± 1.38 (5) | 21 |
| Cytochrome b-c1 complex subunit 2, mitochondrial | 10.87 ± 3.93 (4) | 9.65 ± 1.87 (5) | 13 |
| Acetyl-CoA acetyltransferase, mitochondrial | 8.65 ± 2.61 (10) | 7.85 ± 1.01 (10) | 10 |
| Dihydrolipoyl dehydrogenase, mitochondrial | 10.66 ± 3.71 (8) | 10.31 ± 3.87 (9) | 3 |
| Superoxide dismutase [Mn], mitochondrial | 11.89 ± 3.85 (8) | 11.87 ± 4.58 (8) | 0.1 |
| Cytochrome c oxidase subunit 6B1 | 9.76 ± 3.72 (8) | 9.93 ± 5.99 (7) | −2 |
| Cytochrome c oxidase subunit 5A, mitochondrial | 10.70 ± 2.39 (8) | 11.12 ± 3.27 (9) | −4 |
| Ubiquinone biosynthesis protein COQ9, mitochondrial | 6.56 ± 3.16 (4) | 7.02 ± 3.98 (5) | −6 |
| ATP synthase subunit d, mitochondrial | 4.48 ± 1.59 (4) | 4.95 ± 1.89 (7) | −9 |
| Transcription termination factor 3, mitochondrial | 5.62 ± 1.03 (6) | 8.13 ± 5.03 (5) | −31 |
| Mean magnitude of increase | 25 | ||
| Binomial test two-tailed p-value | p < .05 |
% Differences and average % differences by ontology are provided.
Figure 2.
Relative differences (%) in the fractional rate of synthesis for individual muscle proteins by genetic ontology between Fortetropin versus control groups (A) myofibrillar, (B) sarcoplasmic, (C) mitochondrial.
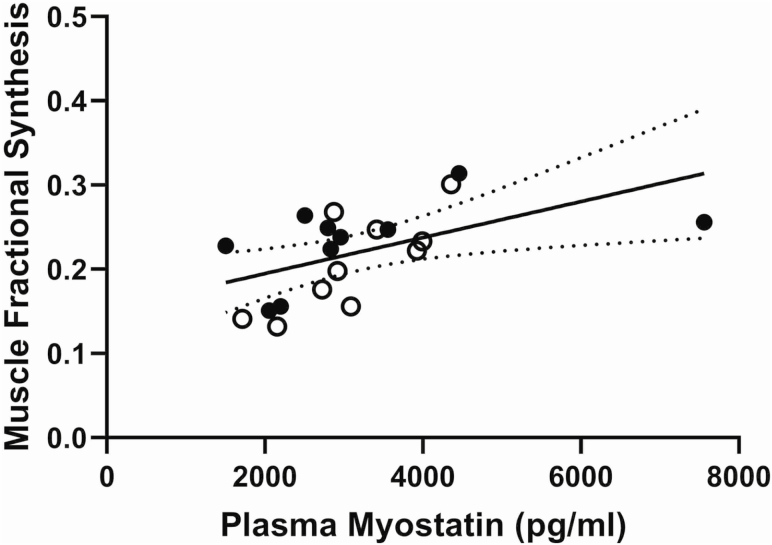
No change in circulating myostatin during the 21-day treatment period and no differences in myostatin concentrations between the two groups were seen. FO: 3.09 ± 1.29 ng/mL, CO: 2.96 ± 0.69 ng/mL at baseline; FO: 3.24 ± 1.72 ng/mL, CO: 3.12 ± 0.83 ng/mL after 21 days of treatment (data represent mean ± SD). A significant positive correlation (r = 0.5327, p < .05) between circulating myostatin concentrations and the average muscle protein fractional synthesis on Day 21 was seen (Figure 3) with a significant correlation of similar strength for Day 1 myostatin levels (r = 0.4688, p < .05).
Figure 3.
The relationship between circulating myostatin levels on Day 21 versus average muscle protein FSR. Fortetropin: closed circles, Control: open circles (r = 0.5327, p < .05).
Discussion
The major finding of this double-blinded study is that, compared to placebo (cheese powder), daily use of the nutritional supplement Fortetropin resulted in a significant increase in the rate of synthesis muscle proteins as a group in healthy older men and women. The increased muscle protein FSR was observed in the major ontologies of muscle proteins—sarcoplasmic, myofibrillar, and mitochondrial proteins. Although significant increases in FSR were not observed for individual muscle proteins, the majority of proteins in all three ontologies had higher mean FSRs in the Fortetropin group (32/38 myofibril proteins, 33/44 sarcoplasmic proteins, and 12/17 mitochondrial proteins), each of which were statistically significant by the binomial test.
This study is consistent with a previous study demonstrating an effect of Fortetropin consumption to increase lean body mass in young subjects. While the mechanism of this increase in muscle protein FSR was not examined, Sharp and colleagues (7) showed that in combination with exercise in rats and resistance trained young men, Fortetropin increased mTOR expression. Fortetropin also resulted in a decrease in circulating myostatin levels. Although we did not observe any effect on circulating myostatin levels, Fortetropin has been shown to decrease circulating myostatin levels (7,16) in healthy subjects. The consensus of published data suggests an age associated decrease in circulating myostatin levels (17,18), although the role of myostatin in aging has not been established and the functional interpretation of circulating myostatin remains uncertain. We found here that in a group of healthy older subjects, circulating myostatin levels were positively associated with muscle protein FSR. A positive correlation is, however, contrary to the hypothesized relationship (myostatin is expected to inhibit muscle protein synthesis). This relationship was similar and significant whether Day 1 or Day 21 levels of myostatin were used. Myostatin has a role in increasing muscle ubiquitination and rate of muscle protein breakdown (19). The relationship between muscle protein synthesis rates and circulating myostatin may be a result of higher breakdown of muscle proteins and the resultant increase in free amino acids which have a stimulatory effect on muscle protein FSR. Our data do not support a direct role of circulating myostatin in the effects of Fortetropin to increase FSR of muscle proteins.
2H2O labeling for determination of protein FSR provides an integrated measurement during the period of labeling that includes both fed and fasted state assessments. In the present study, 2H2O was consumed by subjects for a 21-day period and a broad effect of Fortetropin was observed across the muscle proteome, with a significant increase in the FSR of myofibrillar, mitochondrial, and sarcoplasmic proteins. Using this heavy water labeling-tandem mass spectrometric method in rats, we previously showed that a selective androgen receptor modulator resulted in a substantial and dose–responsive increase in muscle protein FSR, more so in glycolytic and myofibrillar than in mitochondrial protein ontologies, after 10 days that was strongly associated with increased muscle mass after 28 days of use (8). These findings demonstrated that the muscle protein synthetic anabolic response is predictive of the magnitude of muscle hypertrophy. Using heavy water labeling, we have previously demonstrated that resistance exercise increases skeletal muscle protein FSR by about 25% in older men (9), comparable to the 18% increase seen in the present study.
In our study, subjects ingested 2H2O during the entire treatment period of 21-days and as a result, the FSR values represent the average rate of synthesis during that period of time, including both postprandial and postabsorptive conditions. As a result, it is not possible to know if the anabolic effect of Fortetropin occurred in combination with meals or in the postabsorptive condition. Anabolic resistance describes the reduced stimulation of muscle protein synthesis in response to specific amount of protein or essential amino acids in older subjects compared to healthy young people (4) and has been suggested as important in the etiology of sarcopenia. Indeed, levels of protein intake are associated with lean body mass (20) and healthy older people have a higher requirement for dietary protein (21) than is currently recommended for the general population. The subjects in our study were healthy and diet was not controlled during the supplementation period. The macronutrient content of Fortetropin and placebo was the same and the subjects and investigators were blinded as to which study group each participant was part of. The higher rates of muscle protein synthesis suggest that Fortetropin may work to overcome anabolic resistance of aging. Stimulation of muscle protein FSR will result in improvements in muscle mass. Recent studies now demonstrate that muscle mass is strongly associated with health-related outcomes, risk of disability, and mortality in older men (2,22).
In conclusion, in healthy older men and women the daily use of Fortetropin resulted in higher synthesis rates of muscle proteins of multiple ontologies compared to a control group. The effects were independent of sex or initial level of muscle mass. The ability to measure integrated synthesis rates of large numbers of proteins over several weeks’ time by the dynamic proteomics method, using tandem mass spectrometric analyses with long-term heavy water labeling, provided here a sensitive approach for detecting subtle differences in global protein synthesis rates.
These data suggest Fortetropin has an anabolic effect on muscle protein synthesis in older people and warrants testing as a therapy for sarcopenia. The long-term effects of Fortetropin on muscle mass and function in older people is unknown and should be explored.
Acknowledgment
This study was fully supported by a grant from Myos Rens Technology. Myos Rens Technology provided the Fortetropin and Placebo supplements.
Conflict of Interest
Authors declare no conflict of interest. None of the authors served as a consultant or on an advisory board for Myos Rens Technology, Inc.
Author Contributions
William Evans: designed research, conducted research, analyzed data, wrote paper, and had primary responsibility for final content. Mahalakshmi Shankaran: designed research, conducted research, analyzed data, and wrote paper. Edna Nyangau: conducted research and analyzed data. Tyler Field: conducted research and analyzed data. Hussein Mohammed: conducted research and analyzed data. Robert Wolfe: conducted research, analyzed data, and wrote paper. Scott Schutzler: conducted research and analyzed data. Marc Hellerstein: Designed research, conducted research, analyzed data, and wrote paper.
References
- 1. Evans WJ. What is sarcopenia? J Gerontol A Biol Sci Med Sci. 1995;50(Spec No.):5–8. doi: 10.1093/gerona/50a.special_issue.5. [DOI] [PubMed] [Google Scholar]
- 2. Cawthon PM, Orwoll ES, Peters KE, et al. Strong relation between muscle mass determined by D3-creatine dilution, physical performance, and incidence of falls and mobility limitations in a prospective cohort of older men. J Gerontol A Biol Sci Med Sci. 2019;74:844–852. doi: 10.1093/gerona/gly129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Volpi E, Sheffield-Moore M, Rasmussen BB, Wolfe RR. Basal muscle amino acid kinetics and protein synthesis in healthy young and older men. JAMA. 2001;286:1206–1212. doi: 10.1001/jama.286.10.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moore DR, Churchward-Venne TA, Witard O, et al. Protein ingestion to stimulate myofibrillar protein synthesis requires greater relative protein intakes in healthy older versus younger men. J Gerontol A Biol Sci Med Sci. 2015;70:57–62. doi: 10.1093/gerona/glu103 [DOI] [PubMed] [Google Scholar]
- 5. Becker C, Lord SR, Studenski SA, et al. Myostatin antibody (LY2495655) in older weak fallers: a proof-of-concept, randomised, phase 2 trial. Lancet Diabetes Endocrinol. 2015;3:948–957. doi: 10.1016/S2213-8587(15)00298-3 [DOI] [PubMed] [Google Scholar]
- 6. Rooks D, Praestgaard J, Hariry S, et al. Treatment of sarcopenia with bimagrumab: results from a phase ii, randomized, controlled, proof-of-concept study. J Am Geriatr Soc. 2017;65:1988–1995. doi: 10.1111/jgs.14927 [DOI] [PubMed] [Google Scholar]
- 7. Sharp MH, Lowery RP, Mobley CB, et al. The effects of fortetropin supplementation on body composition, strength, and power in humans and mechanism of action in a rodent model. J Am Coll Nutr. 2016;35:679–691. doi: 10.1080/07315724.2016.1142403 [DOI] [PubMed] [Google Scholar]
- 8. Shankaran M, Shearer TW, Stimpson SA, et al. Proteome-wide muscle protein fractional synthesis rates predict muscle mass gain in response to a selective androgen receptor modulator in rats. Am J Physiol Endocrinol Metab. 2016;310:E405–E417. doi: 10.1152/ajpendo.00257.2015 [DOI] [PubMed] [Google Scholar]
- 9. Murphy CH, Shankaran M, Churchward-Venne TA, et al. Effect of resistance training and protein intake pattern on myofibrillar protein synthesis and proteome kinetics in older men in energy restriction. J Physiol. 2018;596:2091–2120. doi: 10.1113/JP275246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shankaran M, King CL, Angel TE, et al. Circulating protein synthesis rates reveal skeletal muscle proteome dynamics. J Clin Invest. 2016;126:288–302. doi: 10.1172/JCI79639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shankaran M, Czerwieniec G, Fessler C, et al. Dilution of oral D3 -creatine to measure creatine pool size and estimate skeletal muscle mass: development of a correction algorithm. J Cachexia Sarcopenia Muscle. 2018;9:540–546. doi: 10.1002/jcsm.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCabe BJ, Bederman IR, Croniger C, Millward C, Norment C, Previs SF. Reproducibility of gas chromatography-mass spectrometry measurements of 2H labeling of water: application for measuring body composition in mice. Anal Biochem. 2006;350:171–176. doi: 10.1016/j.ab.2006.01.020 [DOI] [PubMed] [Google Scholar]
- 13. Hayot M, Michaud A, Koechlin C, et al. Skeletal muscle microbiopsy: a validation study of a minimally invasive technique. Eur Respir J. 2005;25:431–440. doi: 10.1183/09031936.05.00053404 [DOI] [PubMed] [Google Scholar]
- 14. Holmes WE, Angel TE, Li KW, Hellerstein MK. Dynamic proteomics: in vivo proteome-wide measurement of protein kinetics using metabolic labeling. Methods Enzymol. 2015;561:219–276. doi: 10.1016/bs.mie.2015.05.018 [DOI] [PubMed] [Google Scholar]
- 15. Decaris ML, Gatmaitan M, FlorCruz S, et al. Proteomic analysis of altered extracellular matrix turnover in bleomycin-induced pulmonary fibrosis. Mol Cell Proteomics. 2014;13:1741–1752. doi: 10.1074/mcp.M113.037267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Colkler C. Effect on serum myostatin levels of high-grad handled fertile egg yolk powder. J Am Coll Nutr. 2009;28:309. (abstract) [Google Scholar]
- 17. Barrios-Silva LV, Parnell M, Shinwari ZB, et al. Activin subfamily peptides predict chronological age in humans. Physiol Rep. 2018;6:e13823. doi: 10.14814/phy2.13823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schafer MJ, Atkinson EJ, Vanderboom PM, et al. Quantification of GDF11 and myostatin in human aging and cardiovascular disease. Cell Metab. 2016;23:1207–1215. doi: 10.1016/j.cmet.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang DT, Yang YJ, Huang RH, Zhang ZH, Lin X. Myostatin activates the ubiquitin-proteasome and autophagy-lysosome systems contributing to muscle wasting in chronic kidney disease. Oxid Med Cell Longev. 2015;2015:684965. doi: 10.1155/2015/684965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Houston DK, Nicklas BJ, Ding J, et al. ; Health ABC Study Dietary protein intake is associated with lean mass change in older, community-dwelling adults: the Health, Aging, and Body Composition (Health ABC) Study. Am J Clin Nutr. 2008;87:150–155. doi: 10.1093/ajcn/87.1.150 [DOI] [PubMed] [Google Scholar]
- 21. Campbell WW, Trappe TA, Wolfe RR, Evans WJ. The recommended dietary allowance for protein may not be adequate for older people to maintain skeletal muscle. J Gerontol A Biol Sci Med Sci. 2001;56:M373–M380. doi: 10.1093/gerona/56.6.m373 [DOI] [PubMed] [Google Scholar]
- 22. Cawthon PM, Blackwell T, Cummings SR, et al. Muscle mass assessed by D3-Creatine dilution method and incident self-reported disability and mortality in a prospective observational study of community dwelling older men [Published online ahead of print May 22, 2020]. J Gerontol A Biol Sci Med Sci. 2020. doi: 10.1093/gerona/glaa111 [DOI] [PMC free article] [PubMed] [Google Scholar]