Abstract
Gaps in the translation of research findings to clinical management have been recognized for decades. They exist for the diagnosis as well as the management of cancer. The international standards for cancer diagnosis are contained within the World Health Organization (WHO) Classification of Tumours, published by the International Agency for Research on Cancer (IARC) and known worldwide as the WHO Blue Books. In addition to their relevance to individual patients, these volumes provide a valuable contribution to cancer research and surveillance, fulfilling an important role in scientific evidence synthesis and international standard setting. However, the multidimensional nature of cancer classification, the way in which the WHO Classification of Tumours is constructed, and the scientific information overload in the field pose important challenges for the translation of research findings to tumour classification and hence cancer diagnosis. To help address these challenges, we have established the International Collaboration for Cancer Classification and Research (IC3R) to provide a forum for the coordination of efforts in evidence generation, standard setting and best practice recommendations in the field of tumour classification. The first IC3R meeting, held in Lyon, France, in February 2019, gathered representatives of major institutions involved in tumour classification and related fields to identify and discuss translational challenges in data comparability, standard setting, quality management, evidence evaluation and copyright, as well as to develop a collaborative plan for addressing these challenges.
Keywords: cancer research, evidence‐based pathology, International Agency for Research on Cancer (IARC), international standards, WHO Tumour Classification
Short abstract
What's new?
The World Health Organization Classification of Tumours has been informing cancer research and clinical practice by providing an international consensus on the tumour criteria for cancer diagnosis. In a new initiative coordinated by the International Agency for Research on Cancer, major institutions are now joining forces to address the remaining challenges in translational research and facilitate the application of research results to clinical practice. The newly‐established International Collaboration for Cancer Classification and Research (IC3R) aims to provide a forum for the coordination of efforts in generating evidence, setting standards, and providing best practice recommendations for tumor classification and cancer research.
List of Abbreviations
- AI
artificial intelligence
- AJCC
American Joint Committee on Cancer
- ctDNA
circulating tumour DNA
- ENCR
European Network of Cancer Registries
- EQA
external quality assessment
- EQUATOR
Enhancing the Quality and Transparency of Health Research
- FIGO
International Federation of Gynaecology and Obstetrics
- GA4GH
Global Alliance for Genomics and Health
- GenQA
Genomics Quality Assessment
- ClinGen
Clinical Genome Resource Somatic Working Group
- GNU
GNU's Not Unix
- GPU
graphics processing unit
- HGVS
Human Genome Variation Society
- IARC
International Agency for Research on Cancer
- ICCR
International Collaboration on Cancer Reporting
- IC3R
The International Collaboration for Cancer Classification and Research
- ICD‐O
International Classification of Diseases for Oncology
- ICD‐10/11
International Classification of Diseases
- IQN Path
Quality Network for Pathology
- ISO
International Organization for Standardization
- MVLD
minimum variant‐level data
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta‐Analyses
- PT
proficiency testing
- R&D
research and development
- SEER
Surveillance, Epidemiology, and End Results
- SNOMED CT
Systematized Nomenclature of Medicine Clinical Terms
- STARD
Standards for Reporting Diagnostic Accuracy Studies
- TNM
Tumour, node, metastasis
- TRIPOD
Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis
- UICC
Union for International Cancer Control
- VCF
variant call format
- WHO
World Health Organization
1. BACKGROUND
Cancer research aims to help people who are at risk of (or who already have) the various forms of cancer, in terms of risk assessment, prevention, early detection, diagnosis and treatment. Most countries have clear pathways for the translation of results of clinical trials of interventions into practice through drug approval and clinical guidelines, and also regulatory control over the use of medical devices. 1 However, diagnostic interpretation, early detection procedures and screening methodologies are often not subjected to the same level of scrutiny.
The primary route for the incorporation of noncommercial knowledge into patient management is through the development of international standards and clinical guidance. The World Health Organization (WHO) Classification of Tumours fulfils an important role for cancer‐related research and practice by providing an international consensus on classification and diagnostic criteria of tumours. The classification comprises a unique synthesis of scientific evidence (both written and illustrated) that underpins the diagnosis of individual tumour types. It is published as a series of books, known to pathologists as the WHO Blue Books, and is now also available as a website (see https://tumourclassification.iarc.who.int). The evidence base for each tumour type comprises peer‐reviewed publications in multiple fields, and although the primary diagnostic method is still histopathology, other disciplines have an increasingly large role to play. The vast number of publications that inform the classification now present a significant problem of information overload, but also an enormous opportunity, as our understanding of cancer continues to progress. It is also true that not all of the information published warrants inclusion in the WHO classification or in clinical guidance—it is of variable quality and relevance, and sifting through this information is now a major task for the biomedical community. Systematic reviews, which have been the bedrock of evidence‐based medicine during the last decades, have proven to provide reliable evidence synthesis and to allow the evaluation of available information (eg, the application of such methods, in addition to the attachment to existing guidelines as the EQUATOR (Enhancing the Quality and Transparency of Health Research) Network guidance (see https://www.equator-network.org/) would certainly help to get results into practice in the field of cancer research and tumour classification. But focus on translational research is also needed to bridge the gap between basic and clinical investigators, and facilitate the application of research results to practice. 1 , 2 , 3 The task is still enormous and needs individuals, organizations and networks committed to bridge the gap.
The International Collaboration for Cancer Classification and Research (IC3R) is intended to provide a forum for the coordination of both the generation and evaluation of evidence for tumour classification. IC3R will provide the framework (see Figure 1) for international collaborative projects committed to set standards, raise quality management and promote evidence‐based practice. Its members will be institutions (not individuals), who will designate representatives to a steering group that will organize meetings and coordinate projects under the auspices of IC3R in order to further promote normalization and comparability in cancer research and provide the necessary evidence to underpin the WHO Classification of Tumours.
FIGURE 1.

IC3R framework [Color figure can be viewed at wileyonlinelibrary.com]
1.1. Vision and aims
IC3R will promote collaboration between organizations to ensure and improve the provision of high‐quality evidence and evidence synthesis, permitting rapid translation of cancer research into clinical diagnostic practice for patients.
IC3R will be tasked with the following:
Harmonizing cancer‐related data
Setting standards for analytical procedures
Identifying and informing the community of critical gaps (eg, nonuniform annotations and classifications and gaps in bioinformatics, computational pathology, clinical chemistry and other areas of research)
Managing quality and establishing best practice
Coordination of developments in artificial intelligence (AI)
Updating the evidence and generating evidence synthesis
Addressing copyright considerations
IC3R will act in close collaboration with other stakeholders in the field. The International Agency for Research on Cancer (IARC) will provide the secretariat and may assist the coordination of international datasets as required, as well as encouraging the generation of standards and procedures for data management that will benefit all parties involved.
The activities of IC3R currently fall into six main categories (although other activities could be added if members identify the need): harmonizing data, identifying gaps, setting standards, improving quality and establishing best practices, updating the evidence, and addressing copyright considerations. In our study, we provide a short description of these categories, the challenges and potential solutions IC3R will focus on.
2. HARMONIZING CANCER‐RELATED DATA
Tumour classification relies on various types of data related to the patient, clinical context, biological specimens and analytical measurements. Harmonizing the structure, collection and registration of data across cases within an institution is essential to ensure comparability and reproducibility, but standardization and harmonization across institutions is even more crucial, to improve the basis of tumour classification and inform cancer care worldwide.
The need for data standardization and harmonization is present at all stages of the tumour classification process. For example, cancer registration standards for data collection are needed to provide alignment for the determination of the date of diagnosis between groups such as the Surveillance, Epidemiology and End Results (SEER) program, the International Association of Cancer Registries (IACR) and the European Network of Cancer Registries (ENCR). Consent on criteria for the definition of case entities is important, as for example in patients with multiple primary tumours. Differences between the TNM (tumour, node, metastasis) staging systems of the American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC), and between TNM and International Federation of Gynaecology and Obstetrics (FIGO) staging are increasingly addressed collectively by these organizations and benefit both patients and research. Standardization is also needed in the pipelines used for the identification of relevant genomic characteristics and the reporting of molecular biomarker assays, where insufficient laboratory certification, absence of standards and lack of confirmatory large‐scale studies for the validation of biomarkers often limits the scope of the results. Some current initiatives aim to address these issues, but do not cover all eventualities, for instance, the International Collaboration on Cancer Reporting (ICCR), a collaborative initiative that involves key international cancer‐ and pathology‐related organizations. ICCR produces internationally standardized and evidence‐based datasets for the pathology reporting of cancer using defined terminology and normalized pathology information on tumour classification and staging, as well as on prognostic and predictive factors (see http://www.iccr-cancer.org/). These datasets are produced in a variety of formats and languages to maximize their worldwide usage.
Large‐scale molecular profiling of tumour DNA, RNA, and epigenetic features has generated informative data for tumour classification. These data are available through various online databases (see Table 1), which have each implemented their own data harmonization procedures. 4 , 5 , 6 Also, for the reporting of genetic variations well‐accepted standards exist, the Variant Call Format (VCF) and the Human Genome Variation Society (HGVS) nomenclature, which have been adopted in a growing number of resources. However, the use of variations of these formats impedes the achievement of clear equivalence between data sources and engenders difficulty in the comparability of research results.
TABLE 1.
Online databases for molecular profiling of tumour DNA, RNA and epigenetic features
| Name | Available at |
|---|---|
| The Catalogue of Somatic Mutations in Cancer (COSMIC) | https://cancer.sanger.ac.uk/cosmic |
| The National Cancer Institute's Genomic Data Commons (NCI GDC) | https://gdc.cancer.gov/ |
| The International Cancer Genome Consortium (ICGC) | https://icgc.org/ |
| The cBioPortal for Cancer Genomics | https://www.cbioportal.org/ |
| The National Center for Biotechnology Information's Gene Expression Omnibus (NCBI GEO) | https://www.ncbi.nlm.nih.gov/geo/ |
| The European Bioinformatics Institute's ArrayExpress Archive of Functional Genomics Data (EBI ArrayExpress) | https://www.ebi.ac.uk/arrayexpress/ |
Many current genomic resources suffer from a lack of harmonization of molecular data, clinical information and biological specimen features. Few of them use recognized international classifications such as the International Classification of Diseases for Oncology (ICD‐O), International Classification of Diseases (ICD‐10/11) or Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT). Many have difficulties due to the diversity of bioinformatics pipelines used for the processing of genomic data, which may create batch effects when aggregating data from different databases to conduct large‐scale studies. Here too, some initiatives to address these issues are ongoing; an example is the proposed guidelines for the harmonization of variant calling across human genetics projects. 7 , 8 The Global Alliance for Genomics and Health (GA4GH) was set up to create frameworks and standards to enable the responsible, voluntary and secure sharing of genomic and health‐related data, and the Clinical Genome Resource (ClinGen) Somatic Working Group has developed a minimum variant‐level data (MVLD) methodology for promoting the harmonization of cancer variant representation and efficient distribution of this information through ClinVar. 9 Beyond standards of nomenclature, the interpretation of the significance of the specific variants in terms of their clinical impact is evolving and improving, but many variants still fall into the challenging category of “variants of unknown significance”. The development of a standardized terminology and dataset for interoperability would accelerate the challenging process of determining the clinical significance of specific gene variants.
These initiatives pave the way for improving database interoperability and facilitating the aggregation of datasets, but important challenges remain in the harmonization of individual data and production of datasets usable for evidence‐based medicine. Data quality metrics and assessments should also be standardized, and internationally approved guidelines should be developed with recommendations for minimum data sets, content and validation, as well as reporting recommendations defining the minimum annotations needed, and quality standards for cancer research databases and tumour classification.
3. IDENTIFYING GAPS
The WHO Classification of Tumours is a taxonomy. It groups tumours into diagnostic entities (tumour types) based on their shared characteristics, just as Linnaeus (1758) identified species based on shared characteristics. There is still a reliance on histology and increasingly on molecular pathology, but radiology, epidemiology and other disciplines contribute to the classification of many tumour types. The classification defines a tumour type as an entity in which multiple parameters (eg, radiological appearance, localization, histopathology, and molecular profile) differ from those of other types, whereas a subtype may not vary by more than one or two parameters, and usually without sufficient evidence that these differences would currently affect the clinical management of the patient.
The WHO Classification of Tumours also identifies tumour types or subtypes where there is currently insufficient information about some characteristics (parameters), but although this lack of information may be indicated in the WHO Blue Books, there is no organized database that systematizes the identified research needs in a way that could drive research to bridge these gaps. As part of IC3R, IARC proposes to maintain an organized record of the current research needs, with contributions from other organizations.
As an example of an as‐yet unmet research need, one of the challenges of personalized medicine is to identify the specific molecular anomalies that define tumour types and subtypes. The WHO Blue Books focus on incorporating the most frequent anomalies, but these studies also provide a steady flow of newly described rarer molecular features that identify tumour types that are not necessarily published, due to insufficient information and validation of these characteristics and their relevance. Similarly, as technologies evolve and the research community applies them in innovative ways, the information about the molecular features used in tumour classification becomes increasingly diverse and complex. For these reasons, evaluations of the existing evidence for mutations and their relationships to clinical features such as diagnosis and prognosis are required for each tumour type and recommendations to guide future research in the production of missing evidence need to be formulated. This information must be reliable and reproducible to permit informed, evidence‐based decisions.
The effort to identify relevant anomalies cannot be fuelled solely by case reports since they have a high risk of bias in their observations due to limitations of the study design. Publication policies often lead editors to accept the first report of a new anomaly, but further identical reports are viewed as less interesting in terms of novelty and the submitted paper can be rejected (an example of publication bias). The establishment of open and interactive database(s) that describe all relevant features and their clinical or pathological relevance is to be encouraged, though a multiplicity of such databases can result in a further need for harmonization (eg, Decipher; https://decipher.sanger.ac.uk). IC3R community could stimulate the development of a forum through which additional knowledge gaps could also be communicated to these databases, which would facilitate the evaluation of the available evidence for the associations of these features with relevant clinical outcomes. The resulting technical developments and research could subsequently be used to inform the WHO Classification of Tumours. Similarly, this community may also provide the potential to build further consortia to provide the research community with the biological and technical infrastructure necessary to validate and replicate key insights, as well as to communicate cutting‐edge developments (eg, future AI‐based analysis or other integrated analysis approaches). Such a database, and the related community, would be a valuable addition to facilitating an appropriate translation of robust findings from the research community into practice.
4. SETTING STANDARDS
The rapid, global development of the healthcare industry and related research has meant that increasingly greater importance is being placed on standardization activities that facilitate compatibility. Standards for technology ensure that any expected component or process will operate safely and predictably, independently of the manufacturer and/or operator. The same considerations apply to the standardization of processes and services. Service standardization in health care is an important adjunct to promoting the provision of high‐quality, specialized expertise and services globally. 10 Systems in several countries provide standards accreditation through national structures and maintain online platforms that monitor quality indicators for nationwide analysis.
In terms of current research in cancer, conclusions related to tumour aetiology often follow the law of large numbers, where increasingly larger sample sizes are used for analysis. Standards are essential to support such scalability in medical research. However, the level of standards implementation is not equally supported or reported across different geographical regions; as a result, the promotion, endorsement and development of such standards are often delegated to national and/or international professional associations 11 that face limitations implementing and monitoring these standards. Stronger international commitment is required to promote the development and subsequent uniform implementation of standards in cancer research.
The Standards for Reporting Diagnostic Accuracy Studies (STARD) 12 constitute a good example of such an initiative, which was led by the professional community, and is now part of the EQUATOR network library of standards for healthcare reporting (see https://www.equator-network.org) mandated by many biomedical journals. The STARD guidelines were first published in 2003 13 and were updated in 2015, 12 , 14 primarily to assist in the completeness and transparency of reporting diagnostic accuracy studies. And a similar process was used to develop the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) recommendations. 15 , 16 Such initiatives are essential parts for an evidence‐based approach that will be used for the development of future standards.
In addition, over the past two decades, the International Organization for Standardization (ISO) has increasingly developed standards for the healthcare sector. At the beginning, these standards were focused primarily on the manufacture of healthcare‐related equipment and medical agents for clinical purposes, later on the healthcare processes themselves and subsequently on the use of reagents and processes for healthcare research, mirroring equivalent industrial standards. In addition, previous ISO norms have been updated to include healthcare data handling and storage. There are now efforts in progress to develop and implement ISO standards for research laboratories, based on the experiences accrued during the creation of the equivalent industrial process standards (see Figure 2).
FIGURE 2.
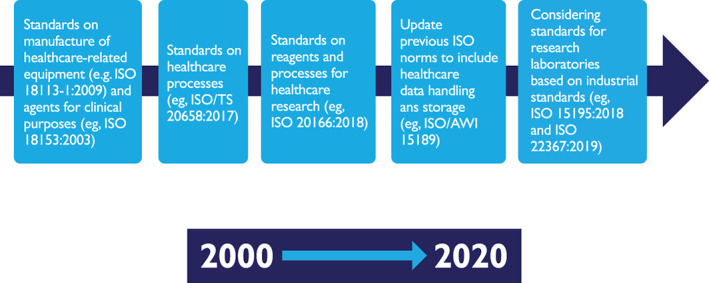
Evolution of the development of ISO standards for the healthcare sector [Color figure can be viewed at wileyonlinelibrary.com]
5. MANAGING QUALITY AND ESTABLISHING BEST PRACTICE
5.1. The need for quality improvement and management
Quality management systems are designed to plan, control and improve the elements of a process that impact on the achievement of the desired results, which includes the safety, effectiveness and accuracy of testing and has a fundamental role in diagnostic science. These systems permit cross comparison between diagnostic services. All cancer research laboratories, including pathology, genetics and other diagnostic services should implement minimum standards and follow the continuous model required by these systems. 17 They also allow the continuous monitoring of performance indicators and the reporting of poor performance in cases where the required standards have not been reached. This gives laboratories the opportunity to investigate the causes and determinants, for instance performing a root cause analysis that would permit the planning of specific actions to rectify the problem and promote improvements. This type of process management ensures reliable test results and that accurate interpretations are provided to patients via the referring clinician. Many processes are performed in diagnostic services to guarantee the accuracy, reliability and traceability of results (eg, it is expected that the correct international nomenclature is used for an external quality assessment (EQA) in a genetics/molecular laboratory 18 ), and they often apply programs as EQA or proficiency testing (PT). It is also important to note that the application of an ISO‐accredited EQA provides evidence of the standard of testing and reporting (including accuracy, reliability and appropriateness) for diagnostic services, as well as benchmarks, and should be undertaken at least annually for every aspect of the service.
5.2. Methods of quality improvement
Quality management systems provide methods for centres to assess their clinical and analytical performance, as well as their interpretation of molecular pathology/genetic testing based on international standards and compared to that of other laboratories. Additionally, EQA programmes offered by external providers facilitate method validation, comparing of results among laboratories, problem identification and compliance with standards and requirements, as well as increases credibility.
The external audit of the diagnostic services is an essential part of any type of quality management programme and is usually undertaken by an EQA or PT provider and leads to the certification of procedures or accreditation of the services based on international ISO standards. 19 These programs are excellent tools to keep procedures and all related variables (staff, equipment and method) well controlled, ensuring that the quality of the service meets minimum professional standards and continuous improvement principles are followed. EQA providers need to be autonomous from professional and national bodies to provide independent external verification of service quality, in addition to a performance guarantee to the laboratory, the host institution and service users. 4 , 18 , 20 , 21 Accreditation bodies use results in EQA performance as an indicator for the quality of a laboratory's routines, and to achieve and maintain certification for a genetics/molecular pathology testing procedure the appropriate EQA(s) accreditation is required. Currently most laboratories use ISO 15189, 4 , 21 , 22 but more appropriate standards for research centres and other histopathology procedures need to be developed. Initiatives such as IC3R can facilitate the collaborative effort needed to produce more specific quality certifications and accreditation in a future.
EQAs are designed to check and occasionally challenge laboratory testing for particular disorders or gene–target combinations for diseases. They cover all aspects of the diagnostic process of the clinical samples, that is, the preanalytical, analytical and postanalytical phases, including genetic counselling. EQAs measure quantitative and/or qualitative indicators depending on the nature of the evaluated component. In molecular pathology, most indicators are qualitative (ie, assessing the presence or absence of a variant), but assessment of molecular changes in blood and other body fluids (liquid biopsy) can be applied in a qualitative and quantitative way, indicating the presence or the load of tumour mutations in cell‐free, circulating tumour DNA (ctDNA) as well as their levels. Analytical, preanalytical and clinical validation of the technologies and methods is therefore similar to other standardized validation procedures in laboratory medicine. 23 Because quantitative liquid biopsy approaches are used not only for diagnosis and staging but also for monitoring patients during therapy and disease course, EQAs are essential to externally verify accuracy, sensitivity and method dependency. Thereby, the formulation of ctDNA fragments (eg, size and association with histone proteins) should be comparable to that in endogenous conditions.
EQA providers have the ability to compare participating laboratories, thus benchmarking the best practices for services. These comparisons between laboratories are incredible helpful in refining laboratory standards and essential to establish best practice guidelines. This is feasible for research laboratories, and some already participate in clinical EQA schemes. To promote this practice at worldwide level, the cancer research and tumour classification community will enable to enhance collaborative projects and improve comparability of results in the field. IC3R constitutes an opportunity to appraise EQA programs in cancer research and promote international comparable evaluations.
5.3. Benefits of external quality assurance schemes
Among their many benefits (see Table 2), EQA programmes improve and validate the overall quality of a service. In the case of a diagnostic, genomic or genetic service, this includes all procedures from sample receipt to genetic referral, including necessary education and training. EQA allows the identification of variations in results and reporting. It encourages a closer interaction between laboratories and clinicians, and often underpins the development of guidelines to promote consistency in the procedures and ensure high‐quality testing for the benefit of patients.
TABLE 2.
Benefits of applying an external quality assessment
|
Several international entities offer EQA programs in the field of molecular pathology, and most are ISO 17043 accredited. Laboratories clearly benefit from selecting an ISO‐accredited EQA provider that covers the procedures of their diagnostic service, but there is not always one available. Pilot EQAs are not eligible for accreditation since they have no performance monitoring, but after the first year of participation, it should be mandatory to include the EQAs in the Quality Management System for both research facilities and diagnostic services in cancer. Genomics Quality Assessment (GenQA) and other EQA providers have proven that such annual participation improves the quality of the diagnostic service. 20 For instance, the recent EQA assessment of DNA extraction from formalin‐fixed and paraffin‐embedded samples by Professor Z. Deans from GenQA showed an improvement in DNA quality and quantity over time. 24
A continuous effort is made by various EQA providers to standardize the procedures and the scoring system of the EQA schemes in molecular pathology 21 so that a large number of diagnostic services can participate. To assist diagnostic and research centres, GenQA provides teaching and education through an online training and competency tool, G‐TACT, for various aspects of a cancer genomics service. 25 To ensure consistency among EQA providers and to develop new operating models, the most relevant EQA providers recently founded the International Quality Network for Pathology (IQN Path). IQNPath has the main purpose of improving the quality of biomarker tests, in particular with regard to molecular pathology.
Although current EQA schemes clearly demonstrate the challenges faced in cancer diagnostic testing and interpretation, as well as the need for better training, competency assessment and standardization worldwide, the results from assessments demonstrate the value of consistent data, sharing of results and collaboration in improving diagnostic practice.
5.4. Quality improvement and research
Improving quality in cancer‐related research is of relevance for tumour classification and diagnostic practice in general. 26 Applying lessons learned from quality management in laboratory medicine could drive improvement in the quality of cancer research and stimulate better translation of research findings into clinical practice. However, this is not always easy. A recent systematic review of the use of circulating tumour DNA (ctDNA) for early cancer detection reached the conclusion that several preanalytical, analytical and postanalytical considerations need to be addressed before biomarkers enter clinical practice. 27 The same issue is shown by data from EQAs, suggesting that the quality of cfDNA testing needs to be significantly improved. 28
The practice of quality management in cancer research laboratories could help to solve such problems. The adoption of ISO standards for cancer research laboratories, perhaps adapted from ISO 15189 for clinical laboratories, could provide a quality management system; training; an environmental policy; guidance for equipment, information systems and materials, as well as defined preanalytical, analytical and postanalytical processes (standard operating procedures); evaluation methods (including publication); and quality assurance. Good scientific practice already mandates much of this and many important research institutions have defined guidelines for effective validation of some procedures as next‐generation sequencing methods and monitoring of analytical procedures. 29 , 30 , 31 Additionally, the European Molecular Genetics Quality Network and GenQA have launched a pilot EQA methodology for next‐generation sequencing in Europe. 26 , 29 , 32 , 33 , 34 Thus, some research laboratories already participate in EQA schemes driven by the above initiatives, but consolidation of this practice is necessary to promote wider, international standardization.
In addition, a more rigorous attitude in reporting of results and scientific publication is desirable. For example, differences in sample collection, extraction, storage and processing (physical and computational) can have a strong effect on downstream analyses and decisions. Reporting of raw data in open‐source databases is a desirable standard that facilitates pooled analyses and accelerates knowledge generation. Many scientific journals have already adopted the recommendations of the EQUATOR network for scientific publications, 35 and IC3Rs will endorse these journals while at the time as it promotes the application of standards in the cancer research community.
6. KEEPING ABREAST OF DEVELOPMENTS IN AI
In recent years, there has been a surge of interest and some significant developments in the emerging area of computational pathology, a rapidly developing discipline in its own right, concerned mainly with the computerized analysis of digitized images of histology slides. This surge of interest has been spurred by the increased use of digital slide scanners in diagnosis 36 , 37 and significant advances in AI algorithms, in particular deep learning algorithms, 38 , 39 , 40 , 41 matched by the availability of powerful graphics processing units (GPUs; originally developed for the gaming industry) and storage hardware at a fraction of the cost compared with that only a few years ago. In combination, computational pathology has been dubbed “the third revolution in pathology”, after immunohistochemistry and next‐generation sequencing 42 and has to be considered in IC3R's strategy.
6.1. The rise of computational pathology
Synergistic technological developments in the areas of slide scanning, digital storage and high‐performance computing have enabled the uptake of digital pathology for routine diagnostics, resulting in the generation of several terabytes of information‐rich, high‐resolution whole slide images from individual clinical histopathology departments (the so‐called “big image data”) on a daily basis. The raw pixel information in whole slide images is ripe for AI‐based automated or computer‐assisted diagnosis and subtyping of cancer in a fast, accurate, objective and reproducible manner. Whole slide image pixel data with linked clinicopathological data are also an invaluable resource for mining and discovery of deep spatial patterns and novel image‐based markers of direct benefit to patients in terms of prediction of recurrence, progression and response to therapy, paving the way for personalized treatment. Some of the notable developments in the area of computational pathology include the discovery of stromal nuclear features shown to be more strongly associated with survival of breast cancer than tumour cell features, 43 the Immunoscore method for predicting outcome in Stage I colon cancer, 44 superhuman performance of AI algorithms in the detection of lymph node metastasis in breast cancer, 45 nuclear shape and orientation features for predicting oncotype DX‐risk categories, 46 and digital scoring of the abundance of tumour‐infiltrating lymphocytes in oral cancers. 47
6.2. Benchmark datasets for AI R&D and validation
Modern deep learning algorithms are known to be generally “data hungry”. These algorithms are also quite vulnerable to a data bias problem where assurance is needed that the dataset used for prediction is as fully representative as possible of the phenomenon to be predicted. In future, the construction of large multicentric benchmark datasets will be key to the research and development (R&D) of novel AI algorithms that are robust and reliable in addition to international standards. Such benchmark datasets can also serve as a vehicle for the rapid evaluation and validation of AI algorithms for regulatory approvals. With these objectives in mind and to achieve higher reproducibility in computational pathology, the PathLAKE (Pathology Image Data Lake for Analytics Knowledge & Education) (https://www.pathlake.org) Centre of Excellence in the United Kingdom is putting together a massive repository of multicentric pathology images linked with diagnostics and clinical outcome data.
6.3. AI‐based evidence synthesis
In recent years, AI algorithms have demonstrated significant potential for synthesis of evidence—one of the key objectives of IC3R—in particular, tools for identifying randomized controlled trials 48 and assessment of bias in their reporting. 49 , 50 These tools are still in the early stages of development and will need to be rigorously validated. Nevertheless, they can be used to assist with systematic reviews for evidence synthesis, saving time and costs in the synthesis. It will also be of paramount importance to ensure that key principles of evidence synthesis (eg, inclusivity, transparency, rigor and accessibility, as recommended by the UK Royal Society and Academy of Medical Sciences), are upheld when AI algorithms are deployed for the purposes of evidence synthesis.
6.4. Deep and integrative mining for better classification
AI‐based deep and integrative mining of vast amounts of raw pixel information, together with linked clinical outcomes and other types of linked data (eg, genomic and physiological data), can also be used to revisit the existing taxonomies. In the genomic space, AI algorithms can be employed to infer the effects of DNA mutations on gene splicing and to predict the effects of genetic mutations on disease risk or drug response. 51 As a specific example, there are >30 different subtypes of salivary gland tumours in the current WHO classification. However, the classification may not be of enough prognostic relevance or sufficiently informative on its own to drive patient management. AI‐based integrated analysis has the potential to drill down into the data, pick out deep subvisual and integrative patterns, and form disease subgroups directly related to outcome. Although there are clear benefits to the detailed subtyping of tumours (such as avoiding suboptimal treatment of patients due to excessively broad classifications), it would perhaps be better to have fewer subtypes that are more useful in determining the best course of action for a given patient than an overly detailed subtyping that is not as informative for patient management.
7. UPDATING THE EVIDENCE
The available evidence must be reviewed periodically in order to keep tumour classifications up to date, but it is not always clear how or when such reviews should be performed. 52 Literature reviews produce variable results depending on the expertise and perspective of the reviewer and the methodology used, with the potential for relevant studies to be missed and interpretations of results to be misleading. 53 In addition, not every statement needs to be informed by complex systematic reviews. Often, traditional background or scoping reviews provide sufficient information to evaluate a subject, and available resources can be saved to be used to inform pressing issues of tumour classification—these topics need to be assessed by systematic reviews due to their potential controversy. Evidence‐based medicine has been providing recommendations and promoting the performance of systematic reviews as the cornerstone of its methods, encouraging comprehensive literature searching, transparency in methods and rigorous study appraisal. 54 However, best practice guidelines, such as the Cochrane Handbook, 55 and related guidelines, such as PRISMA, 56 are closely aligned with meta‐analytic reviews for medical interventions evaluated by randomized controlled trials—methods that are not necessarily appropriate for pathology. Evidence‐based medicine principles and practices need to be adapted to be useful in tumour classification, and an evidence‐based pathology approach that adjusts methods, provides recommendations and offers training is greatly needed. 57
There are additional challenges for an evidence‐based pathology approach to the classification of tumours. Evidence levels and study design considerations are applied with great variations within the field, and if we consider the known evidence hierarchy, 58 most pathology studies examine associations with the lowest levels of evidence. These studies usually do not consider the limitations of their study design and the consequent risk of bias needing to be considered in drawing conclusions and planning research. Pathology as a specialty would benefit from adding research methodology to the curriculum and continuous professional development for its specialists. Methodological discussions are also needed, as well as an initiative for expert consensus on principles and best practices in the field that would allow more consistent interpretation and comparison of studies in pathology. This would also identify areas where new solutions and underpinning methodological research are required.
Thus, evidence‐based pathology will have to deal with the need for timely assessments of controversial decisions, the exponential rise in the number of scientific publications and the management of new types of information such as evidence from genetics or big data sources in bioinformatics. How much subgrouping of uncommon tumours can be sustained before the size of the groups worldwide becomes too small to prove that differences in behaviour or outcome could have occurred by chance alone? To address these challenges, new methodologies will have to be developed, a collective of skilled experts trained and an extended network of evidence‐based pathology hubs created. Such an evidence‐based pathology movement, with sufficient resources and adequate coordination, could respond with evidence‐based assessments to the predicted large number of decisions that will have to be addressed in the near future for tumour classification as the number of potential classifiers continues to increase rapidly. IC3R is ideally positioned to efficiently promote and coordinate such an endeavour, working with other partners in pathology and drawing on the expertise of the more widespread evidence‐based movements.
8. COPYRIGHT AND OPEN SCIENCE
Copyright law is somewhat at variance with the principles of open science. The restrictions that certain copyright transfer provisions impose on scientists remain a hindrance to information dissemination. 59 For journals, restricting access to their content has been traditionally acceptable and seen as necessary for their financial survival. However, this amounts to securing profit from work financed by others—often by public funds—and it delays the potential impact of translating discoveries into patient care. Open science is a movement that aims to make scientific research and data available to all, from professionals to citizens. Open science advocates for practices such as publishing open research and open access to publications in order to facilitate scientific growth, with the ultimate aim of making it easier to publish and communicate scientific knowledge. 60 However, the transition towards effective open science is slow and challenging, and although journals do offer authors the option of submitting articles as open access, this is often accompanied by increased financial burden on the authors.
Open access is not an infringement on copyright, because authors or their institutions still own the original copyright for their publications. The problem is that they are often asked to transfer these rights to the publisher, allowing publishers to set the terms for providing open access. Ideally, an open license would be used to publish, with clearly defined access and reuse rights, and there are certain journals that do offer the option of publishing under a Creative Commons license (eg, CC BY 4.0 for publications and CC0 for data) or GNU (for software and code). 61 , 62 In addition, some large institutions, such as the National Institutes of Health (NIH), impose open access requirements for articles arising from their funding, requiring that the articles be made publicly available on PubMed Central. 59 Moreover, movements such as OpenAIRE 63 encourage researchers to choose publishers who allow them to retain their authors' rights, attempting to change practices that limit knowledge exchange. Working to improve access to information, through various means, is something that IC3R is committed to examining.
9. CONCLUSIONS
The IC3R has been founded to address the need of coordination and collaboration in the multidimensional environment of cancer research. The collaboration will provide essential guidance for involved parties, set standards in the field and provide evidence and evidence synthesis considering priorities in tumour classification.
The IC3R work plan includes five main actions that will ensure the future of the consortium and the implementation of appropriate measures to meet the objectives (see supplementary material):
1. The establishment of a stable managerial structure to facilitate the planning and production of standards well into the future, as well as to design, coordinate and fund projects that address the identified challenges (harmonization of standards, information system development, evidence‐based data, etc.) at the initiative of a steering group.
Action: A consortium has been founded to provide the managerial structure needed to facilitate the design and implementation of appropriate measures to meet the objectives detailed in our study. In the initial year, the structure and working organigram will be set up, including the coordination team (composed by a project manager and assistant part of the secretariat) and a communications strategy. In parallel, workgroups will be created to address the identified challenges (harmonization of standards, information system development, evidence‐based data, etc.) at the initiative of the Steering Group.
2. Drafting of recommendations for quality management in research laboratories are one of the most pressing needs. Options for formal accreditations and best practices need to be provided
Action: The adoption of an ISO standard for cancer research laboratories is in preparation.
3. Promotion and development of evidence‐based pathology to provide the methodological and technical support needed in the field is also relevant and should be initiated as soon as possible.
Action: A project for the promotion and development of evidence‐based pathology will be launched that will include deliverables such as evidence levels adapted for the field of Pathology, new systematic review methods, training in evidence‐based pathology, adapted systematic review tools and a network of systematic reviewers (Figure 3).
FIGURE 3.
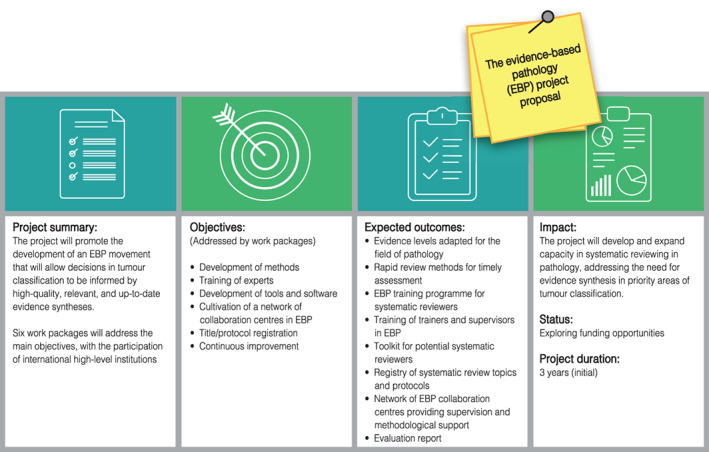
The evidence‐based pathology project proposal [Color figure can be viewed at wileyonlinelibrary.com]
4. Identification of tumour types for which there is insufficient information about their characteristics are clearly stated within the WHO Blue Books, but there is no organized database of the research needs identified or coordinated response to these needs. As part of IC3R, IARC should maintain an organized record of current research needs and allow other organizations to add to it.
Action: A database of current research needs will be established based on IARC publications and others, including a proposal for the prioritization of issues.
One of the key advancements provided by such a consortium and its activities would be the establishment of an open and interactive network of experts that describe for tumour classification relevant features, participate in the identification of needs and assess trends. Importantly, one might envisage the development of a community around this network that participates actively in the continuous improvement of the initiative.
CONFLICT OF INTEREST
Dr Nicola Normanno reports grants and/or personal fees from Amgen, MSD, Quiagen, Roche, BMS, Merck, Thermofisher, Astrazeneca, Zanofi, ELI Lilly, Archer Dx and Illumina outside the submitted work and being President of the International Quality Network for Pathology (IQN Path) and the Italian Cancer Society (SIC). Dr Richard L. Schilsky reports his institution (ASCO) receives funding on his behalf from Astra‐Zeneca, Bayer, Boehringer‐Ingelheim, Bristol Myers Squibb, Eli Lilly & Co., Genentech, Merck, Pfizer. Dr Stefan Holdenrieder reports grants from Roche Diagnostics and Volition RX, as well as being a Scientific Advisory Board Member and consultant of Volition RX. The other authors declare that they have no conflict of interest. The other authors indicated no financial relationships.
DISCLAIMER
The content of this article represents the personal views of the authors and does not necessarily represent the views of the authors' employers or associated institutions. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and these views do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Supporting information
Appendix S1. Supporting Information.
ACKNOWLEDGEMENTS
We are grateful to Dr Louise Jones and Professor Mark Caulfield for their participation in the preliminary draft of the manuscript, to Asiedua Asante for her assessment and to Jessica Cox and Catarina Marques for language and image editing.
Cree IA, Indave Ruiz BI, Zavadil J, et al. The International Collaboration for Cancer Classification and Research. Int. J. Cancer. 2021;148:560–571. 10.1002/ijc.33260
REFERENCES
- 1. Douet LJ, Preedy D, Thomas V, Cree IA. An exploratory investigation of the influence of publication on translational medicine research. J Transl Med. 2010;8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edelman ER, FitzGerald GA. A decade of science translational medicine. Sci Transl Med. 2019;11(489):eaax4327. [DOI] [PubMed] [Google Scholar]
- 3. Sartor RB. Translational research: bridging the widening gap between basic and clinical research. Gastroenterology. 2003;124(5):1178. [DOI] [PubMed] [Google Scholar]
- 4. (EMBL‐EBI) TEBI . Experimental Factor Ontology The European Molecular Biology Laboratory (EMBL); 2020. https://www.ebi.ac.uk/
- 5. Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29(4):365‐371. [DOI] [PubMed] [Google Scholar]
- 6. Genomic Data Commons NCI . Genomic Data Harmonization National Health Institute. https://gdc.cancer.gov/about-data/publications/HG38QC
- 7. Cieslik M, Chinnaiyan AM. Global genomics project unravels cancer's complexity at unprecedented scale. Nature. 2020;578(7793):39‐40. [DOI] [PubMed] [Google Scholar]
- 8. Regier AA, Farjoun Y, Larson DE, et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat Commun. 2018;9(1):4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Danos AM, Ritter DI, Wagner AH, et al. Adapting crowdsourced clinical cancer curation in CIViC to the ClinGen minimum variant level data community‐driven standards. Hum Mutat. 2018;39(11):1721‐1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Federal Ministry for Economic Affairs and Energy . Export Initiative for the German Healthcare Industry. Mittelstand Global. Health Made in Germany. Germany: Federal Ministry for Economic Affairs and Energy; 2020. https://www.exportinitiative-gesundheitswirtschaft.de/EIG/Navigation/EN/Home/home.html. [Google Scholar]
- 11. Bramesfeld AAM, Deandrea S, Gusmeroli C, et al. Plenary of ECIBC 2015: Improving Breast Cancer Screening, Diagnosis and Care in Europe. Luxembourg: European Commission; 2016. [Google Scholar]
- 12. Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6(11):e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Fam Pract. 2004;21(1):4‐10. [DOI] [PubMed] [Google Scholar]
- 14. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ (Clinical Research Ed). 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heus P, Damen J, Pajouheshnia R, et al. Poor reporting of multivariable prediction model studies: towards a targeted implementation strategy of the TRIPOD statement. BMC Med. 2018;16(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heus P, Damen J, Pajouheshnia R, et al. Uniformity in measuring adherence to reporting guidelines: the example of TRIPOD for assessing completeness of reporting of prediction model studies. BMJ Open. 2019;9(4):e025611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crowl A, Sharma A, Sorge L, Sorensen T. Accelerating quality improvement within your organization: applying the model for improvement. J Am Pharm Assoc (2003). 2015;55(4):e364‐e374. quiz e75‐6. [DOI] [PubMed] [Google Scholar]
- 18. Hastings RJ, Howell RT. The importance and value of EQA for diagnostic genetic laboratories. J Community Genet. 2010;1(1):11‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. International Organization for Standardization . Medical laboratories—Requirements for quality and competence (ISO 15189: 2012). https://www.iso.org/standard/56115.html [DOI] [PubMed]
- 20. Deans ZC, Bilbe N, O'Sullivan B, et al. Improvement in the quality of molecular analysis of EGFR in non‐small‐cell lung cancer detected by three rounds of external quality assessment. J Clin Pathol. 2013;66(4):319‐325. [DOI] [PubMed] [Google Scholar]
- 21. van Krieken JH, Normanno N, Blackhall F, et al. Guideline on the requirements of external quality assessment programs in molecular pathology. Virchows Archiv. 2013;462(1):27‐37. [DOI] [PubMed] [Google Scholar]
- 22. Schneider F, Maurer C, Friedberg RC. International Organization for Standardization (ISO) 15189. Ann Lab Med. 2017;37(5):365‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agrawal L, Engel KB, Greytak SR, Moore HM. Understanding preanalytical variables and their effects on clinical biomarkers of oncology and immunotherapy. Semin Cancer Biol. 2018;52(Pt 2):26‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deans S. DNA extraction and DNA quantification EQAs Performance Criteria.; 2018.
- 25. Hastings R; Rack K; Fairley J; Morgan F; Sales M; Deans S. External Quality Assessment (EQA) for Genomics and Clinical Genetics ensuring the quality of the entire genetics service. Abstract of the 12(th) European Cytogenomics Conference 2019: Salzburg, Austria. 6–9 July 2019. Mol Cytogenet. 2019;12(Suppl 1):30.
- 26. Schrijver I, Aziz N, Farkas DH, et al. Opportunities and challenges associated with clinical diagnostic genome sequencing: a report of the Association for Molecular Pathology. J Mol Diagnost. 2012;14(6):525‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cree IA, Uttley L, Buckley Woods H, et al. The evidence base for circulating tumour DNA blood‐based biomarkers for the early detection of cancer: a systematic mapping review. BMC Cancer. 2017;17(1):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keppens C, Dequeker EMC, Patton SJ, et al. International pilot external quality assessment scheme for analysis and reporting of circulating tumour DNA. BMC Cancer. 2018;18(1):804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aziz N, Zhao Q, Bry L, et al. College of American Pathologists' laboratory standards for next‐generation sequencing clinical tests. Arch Pathol Lab Med. 2015;139(4):481‐493. [DOI] [PubMed] [Google Scholar]
- 30. Endrullat C, Glökler J, Franke P, Frohme M. Standardization and quality management in next‐generation sequencing. Appl Transl Genom. 2016;10:2‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hume S, Nelson TN, Speevak M, et al. CCMG practice guideline: laboratory guidelines for next‐generation sequencing. J Med Genet. 2019;56(12):792‐800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rehm HL, Bale SJ, Bayrak‐Toydemir P, et al. ACMG clinical laboratory standards for next‐generation sequencing. Genetics in Medicine: Official Journal of the American College of Medical Genetics. 2013;15(9):733‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schrijver I, Aziz N, Jennings LJ, Richards CS, Voelkerding KV, Weck KE. Methods‐based proficiency testing in molecular genetic pathology. J Mol Diagnos. 2014;16(3):283‐287. [DOI] [PubMed] [Google Scholar]
- 34. Zhang R, Ding J, Han Y, et al. The reliable assurance of detecting somatic mutations in cancer‐related genes by next‐generation sequencing: the results of external quality assessment in China. Oncotarget. 2016;7(36):58500‐58515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simera I, Altman DG. Writing a research article that is “fit for purpose”: EQUATOR network and reporting guidelines. Evidence Based Med. 2009;14(5):132‐134. [DOI] [PubMed] [Google Scholar]
- 36. Retamero JA, Aneiros‐Fernandez J, Del Moral RG. Complete digital pathology for routine histopathology diagnosis in a multicenter hospital network. Arch Pathol Lab Med. 2020;144(2):221‐228. [DOI] [PubMed] [Google Scholar]
- 37. Williams B, Hanby A, Millican‐Slater R, et al. Digital pathology for primary diagnosis of screen‐detected breast lesions—experimental data, validation and experience from 4 centres. Histopathology. 2020;76:968‐975. [DOI] [PubMed] [Google Scholar]
- 38. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402‐2410. [DOI] [PubMed] [Google Scholar]
- 40. Hinton G. Deep learning‐a technology with the potential to transform health care. JAMA. 2018;320(11):1101‐1102. [DOI] [PubMed] [Google Scholar]
- 41. Silver D, Schrittwieser J, Simonyan K, et al. Mastering the game of go without human knowledge. Nature. 2017;550(7676):354‐359. [DOI] [PubMed] [Google Scholar]
- 42. Salto‐Tellez M, Maxwell P, Hamilton P. Artificial intelligence‐the third revolution in pathology. Histopathology. 2019;74(3):372‐376. [DOI] [PubMed] [Google Scholar]
- 43. Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3(108):108ra13. [DOI] [PubMed] [Google Scholar]
- 44. Pages F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet (London, England). 2018;391(10135):2128‐2139. [DOI] [PubMed] [Google Scholar]
- 45. Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199‐2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu C, Romo‐Bucheli D, Wang X, et al. Nuclear shape and orientation features from H&E images predict survival in early‐stage estrogen receptor‐positive breast cancers: laboratory investigation. J Tech Methods Pathol. 2018;98(11):1438‐1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shaban M, Khurram SA, Fraz MM, et al. A novel digital score for abundance of tumour infiltrating lymphocytes predicts disease free survival in oral squamous cell carcinoma. Sci Rep. 2019;9(1):13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marshall IJ, Noel‐Storr A, Kuiper J, Thomas J, Wallace BC. Machine learning for identifying randomized controlled trials: an evaluation and practitioner's guide. Res Synth Methods. 2018;9(4):602‐614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marshall IJ, Kuiper J, Wallace BC. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. J Am Med Informat Assoc. 2016;23(1):193‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marshall IJ, Wallace BC. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Syst Rev. 2019;8(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24‐29. [DOI] [PubMed] [Google Scholar]
- 52. Uttley L, Indave BI, Hyde C, White V, Lokuhetty D, Cree I. Invited commentary—WHO classification of tumors: how should tumours be classified? Expert consensus, systematic reviews or both? Int J Cancer. 2020;146:3516‐3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jadad AR, Cook DJ, Browman GP. A guide to interpreting discordant systematic reviews. Canadian Med Assoc J. 1997;156(10):1411‐1416. [PMC free article] [PubMed] [Google Scholar]
- 54. Mulrow CD. Rationale for systematic reviews. BMJ (Clinical Research Ed). 1994;309(6954):597‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1.0. Chichester, West Sussex; Hoboken NJ: John Wiley; 2008. [Google Scholar]
- 56. Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group . Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marchevsky AM, Wick MR. Evidence‐based pathology: systematic literature reviews as the basis for guidelines and best practices. Arch Pathol Lab Med. 2015;139(3):394‐399. [DOI] [PubMed] [Google Scholar]
- 58. Murad MH, Asi N, Alsawas M, Alahdab F. New evidence pyramid. Evid Based Med. 2016;21(4):125‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reichman JH, Okediji RL. When copyright law and science collide: empowering digitally integrated research methods on a global scale. Minnesota Law Review. 2012;96(4):1362‐1480. [PMC free article] [PubMed] [Google Scholar]
- 60. (ORION) ORraItfOK . ORION Open Science. What is Open Science? 2017. https://www.orion-openscience.eu/resources/open-science
- 61. Hagedorn G, Mietchen D, Morris RA, et al. Creative commons licenses and the non‐commercial condition: implications for the re‐use of biodiversity information. ZooKeys. 2011;150:127‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maggio LA, Stranack K. Understanding creative commons. Acad Med. 2020;95(2):322. [DOI] [PubMed] [Google Scholar]
- 63. OpenAIRE‐Advance . OpenAIRE Science set free. https://www.openaire.eu/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.


