Abstract
Heme, as a hydrophobic iron-containing organic ring, is lipid soluble and can interact with biological membranes. The very same properties of heme that nature exploits to support life also renders heme potentially cytotoxic. In order to utilize heme, while also mitigating its toxicity, cells are challenged to tightly control the concentration and bioavailability of heme. On the bright side, it is reasonable to envision that, analogous to other transition metals, a combination of membrane-bound transporters, soluble carriers, and chaperones coordinate heme trafficking to subcellular compartments. However, given the dual properties exhibited by heme as a transition metal and lipid, it is compelling to consider the dark side: the potential role of non-proteinaceous biomolecules including lipids and nucleic acids that bind, sequester, and control heme trafficking and bioavailability. The emergence of inter-organellar membrane contact sites, as well as intracellular vesicles derived from various organelles, have raised the prospect that heme can be trafficked through hydrophobic channels. In this review, we aim to focus on heme delivery without deliverers - an alternate paradigm for the regulation of heme homeostasis through chaperone-less pathways for heme trafficking.
Keywords: Heme, Tetrapyrrole, Porphyrin, Iron, Trafficking
1. Introduction
Heme (iron protoporphyrin IX) is an essential but potentially cytotoxic iron-containing organic ring that functions as a cofactor, signaling molecule, and nutrient [1–9]. As a cofactor, heme endows proteins with the ability to mediate electron transfer, catalysis, and gas binding and transport [2, 7]. As a signaling molecule, heme binding to or dissociation from a target protein can alter its expression, activity, or both [4, 10–16]. For instance, heme regulation of transcription factors, translational activators, ion channels, kinases, and micro-RNA processing factors collectively act to control a multitude of pathways, including the antioxidant stress response, mitochondrial respiration and biogenesis, mitophagy, translation, apoptosis, circadian rhythms, and cell proliferation [5, 17–33]. As a nutrient, heme-iron is a major source of dietary iron for many microbes and animals, being more bioabsorbable than non-heme iron [34]. However, the very same properties of heme that nature exploits to support life, including its redox activity, Lewis acidity, and hydrophobicity, also renders heme potentially cytotoxic [11]. Indeed, if inappropriately handled by cells, heme can catalyze deleterious redox reactions and/or become misincorporated into proteins, nucleic acids, or lipid bilayers, thereby disrupting biomolecular structure. Unsurprisingly, defects in heme homeostasis are associated with numerous diseases, including cardiovascular, neurodegenerative, and hemolytic disorders, anemias, porphyrias, and certain cancers [6]. As a consequence of the duality of heme being an “essential toxin”, cells and organisms must safely mobilize heme from sites of synthesis or uptake to heme-dependent proteins and pathways throughout the cell. Here, we review current knowledge and discuss future paradigms in eukaryotic heme transport and trafficking.
In order to utilize heme, while also mitigating its toxicity, cells are challenged to tightly control the concentration and bioavailability of heme. Cellular heme concentration is governed by the relative rates of synthesis, degradation, import, and export. The synthesis and degradation of heme are well understood processes and have been reviewed extensively elsewhere [4, 5, 35–39]. Indeed, all eight eukaryotic heme biosynthetic enzymes and heme degrading heme oxygenases (HMOX)s are structurally characterized to atomic resolution and their molecular mechanisms have been studied in depth [36, 40–42]. By contrast, the cellular import and export of heme, as well as the factors governing heme trafficking, remain poorly understood and have been an area of active research over the past 15 years since the discovery of the first eukaryotic heme transporters [43–46]. Analogous to other transition metals, such as copper and iron, it is thought that a combination of membrane embedded transporters, soluble heme carriers, and chaperones coordinate the distribution and insertion of heme into hemoproteins residing in every subcellular compartment [6, 47–50]. Certainly, as discussed herein and summarized in Figure 1, this is borne out by the discovery of a number of proteinaceous factors that fall under each of the aforementioned categories. However, far less attention has been given to the potential role of non-proteinaceous biomolecules in controlling heme trafficking and bioavailability, including lipids and nucleic acids [11].
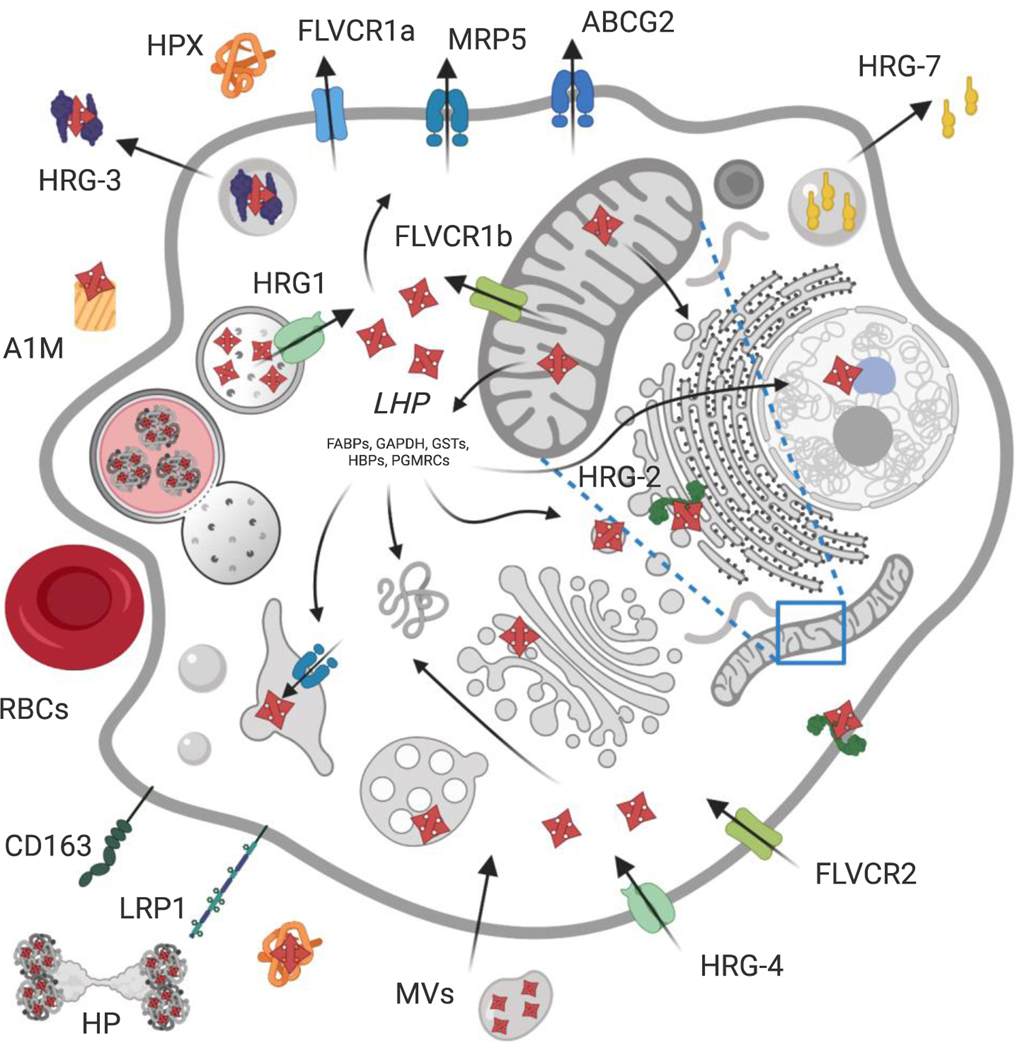
Figure 1.
Consensus model of known proteinaceous mechanisms for metazoan heme transport and trafficking within and between cells. Intracellular heme is either produced via de novo synthesis in the mitochondria or imported via transporters or endocytosis (green components). The efflux of heme to exoplasmic spaces is mediated by transporters or exocytosis (blue components). The labile heme pool (LHP) consists of heme that is buffered by a network of proteins and facilitates its trafficking to other organelles throughout the cell. In mammals, FLVCR2 has been implicated as a plasma membrane heme importer, whereas LRP1 and CD163 are cell surface scavenger receptors for hemopexin (HPX) and the haptoglobin-hemoglobin (HP) complex (or hemoglobin alone), respectively. Heme is also transported via endocytosis of senescent red blood cells (RBCs) and shed microvesicles (MVs) or exosomes in vertebrates. In worms, HRG-4 homologs are dedicated plasma membrane importers and HRG-2 aids in heme utilization by functioning as an oxidoreductase. Finally, heme is imported into the cytosol from exoplasmic compartments via HRG1 and FLVCR1b at the endolysosome and mitochondria, respectively. Conversely, heme efflux is primarily achieved via plasma membrane exporters FLVCR1a, ABCG2 and MRP5 in mammals, with MRP5 also having been localized to the secretory pathway for heme efflux from the cytosol across metazoans. In worms, heme additionally is secreted from intestinal cells for intercellular heme transport via the heme binding protein HRG-3, while interorgan heme homeostasis is modulated via secretion of HRG-7. Created with BioRender - see text for additional details and Table 1.
Heme, as a hydrophobic molecule, is lipid soluble and can interact with biological membranes. The emergence of inter-organellar membrane contact sites (MCSs), as well as intracellular vesicles derived from various organelles, have raised the prospect that heme can be trafficked through hydrophobic channels present at MCSs or within vesicles [4, 11, 51]. Additionally, the π-conjugated electron system of the heme porphyrin can interact with other aromatics, including certain nucleobases, through π-stacking interactions. For instance, guanine-tracts in RNA and DNA, can stack to form guanine-quadruplexes (G4s) and bind to heme via π-π interactions, potentially regulating heme bioavailability in vivo [52]. In this review, we aim to focus on an alternate but emerging paradigm for the regulation of heme homeostatic pathways through “chaperone-less” pathways for heme trafficking.
2. Protein-based heme trafficking mechanisms.
2.1. Heme Import and Export Pathways.
Leading into the last decade, the canonical viewpoint of metazoan heme homeostasis was that every cell makes and utilizes its own mitochondrial-derived heme. Thus, the concept of heme transport was limited to intestinal heme absorption as a means of iron acquisition, and scavenging circulating heme and hemoglobin (Hb) released due to hemolysis via hemopexin and haptoglobin, respectively [53–60]. Internalized heme from hemolysis was largely thought to be degraded by heme oxygenases, releasing iron that can then be recycled for new heme synthesis, stored, or distributed to other cells and tissues. However, with the discovery of heme exporters and importers, new paradigms for cellular heme detoxification and new hypotheses involving the exchange of heme between cells for nutritional purposes or intercellular communication were born [4, 5, 8]. These concepts still remain controversial [61]. For instance, the need for a heme exporter to detoxify excessive heme clashes with current notions that heme degrading heme oxygenases are sufficient to protect cells from heme toxicity [61]. The need for a cell to import heme is obviated by the fact that heme is biosynthesized by most eukaryotic cells [61]. Nevertheless, life evolved heme transporters and in this section, we review heme import and export pathways.
The earliest examples of metazoan heme internalization involved pathways for scavenging cell-free heme and Hb. Hemolysis during diseases such as malaria, sickle cell anemia, autoimmune hemolytic anemia, and cancers causes rupturing of red blood cells which releases toxic heme and Hb into circulation [62–69]. The proteins required for scavenging heme and Hb include hemopexin and haptoglobin [70, 71]. Circulating heme is bound by hemopexin (HPX) and the complex is endocytosed via the LRP1 (CD91) receptor at many different tissues including macrophages, hepatocytes, placental syncytiotrophoblasts and neurons where the hemopexin is recycled and the heme is degraded [72–76]. Likewise, cell-free Hb is bound by haptoglobin and subsequently endocytosed, via binding to the CD163 receptors on macrophages, and degraded for iron recycling [77–81]. Additionally, other serum proteins such as human serum albumin (HSA) have been shown to participate in heme binding and scavenging, albeit with much lower affinities. For reference, the HPX-heme Kd is 5 fM, whereas the HSA-heme Kd values have been measured to be as low as 20 pM or as high as 40 μM [82–85]. Alpha 1 microglobulin (A1M) and high density lipoproteins can also requisition heme in circulation and are thought to serve as additional scavengers [86–90]. Though not the focus of this review, these examples highlight the importance of proteins that bind and limit circulating heme to prevent cellular and tissue damage [57, 91].
Another pathway for cellular heme uptake is through phagocytosis of red blood cells (RBCs) by reticuloendothelial system (RES) macrophages [92–95]. Erythrophagocytosis (EP) eliminates RBCs from the circulation as they become damaged or senescent, and serves as the primary method for recycling the iron within heme [96–99]. Hb accounts for > 90% of a mature red cell protein, which translates to a staggering one billion heme molecules [92, 100–102]. After RBCs are broken down in the endophagolysosome, this bolus of potentially cytotoxic heme must be transported out of the lysosomal lumen and trafficked to the ER-tethered HMOX1 to be broken down for its iron [103–105]. The mechanism underlying how heme reaches HMOX1, particularly the trafficking to HMOX1’s active site in the cytosol is not known [4, 93, 106, 107].
The notion that all free-living metazoans made their own heme was challenged by the discovery that the microscopic nematode, Caenorhabditis elegans, lacked all eight enzymes for heme biosynthesis [108]. However, this heme auxotroph still requires hemoproteins and acquires environmental heme through diet and then disseminates it to other cells throughout its body for survival [4, 108, 109]. Though clearly this mechanism is an outlier amongst eukaryotes, C. elegans’ unique heme auxotrophy therefore allowed for the study of heme transport and trafficking without the confounding contributions from endogenous heme synthesis [8, 108, 110]. It was also possible that the pathways that enabled the distribution of heme to different tissues and cells in C. elegans may also be conserved in the vast majority of animals that can synthesize heme.
As has been reviewed elsewhere, genetic screens and subsequent characterization of heme-regulated genes have identified novel importers, exporters, chaperones and signaling molecules [4–6, 8, 9, 12, 39, 50, 110, 111]. HRG-1 and its homologs HRG-4, −5, −6 were the first bona fide eukaryotic heme transporters to be discovered, whose critical function of heme import into the cytosol is evolutionarily conserved [44]. The worm-specific HRG-4 localizes exclusively to the plasma membrane and helps import heme directly from the intestinal lumen, whereas HRG1 typically resides in endo-lysosomal recycling compartments and has orthologs from arthropods to vertebrates including humans [44]. HRG1’s function has been demonstrated to be closely coupled with V-type ATPases and the lumenal pH of the organelle, as acidic conditions increase free heme solubility and a decrease in pH also raises HRG1’s affinity for heme by transport from endosomes in cell lines [44, 112]. Early studies of this four transmembrane domain (TMD) importer showed transport by experimentation with yeast reporter assays, Xenopus oocytes, zebrafish embryos, fluorescent heme analogs and genetic manipulations in worms and cell lines but recent genetic knockouts in zebrafish and mouse models have now characterized HRG1’s essential role in heme/iron homeostasis at the organismal level [44, 107, 113–115]. Upon EP, HRG1 is highly upregulated in RES macrophages where it is recruited to erythrophagosomal membranes and aids in recycling degraded RBCs by importing heme into the cytosol for HMOX1 or subsequent trafficking [107]. In zebrafish this recycling occurs in the kidney marrow, the primary adult hematopoietic organ, where fish lacking HRG1 display aberrant histological and heme-iron metabolism gene expression [114, 116]. The loss of HRG1 in mice is even more striking, with a hyper accumulation of HMOX1 inaccessible heme in RES macrophages resulting in enlarged lysosomes and the endogenous production of hemozoin, a phenomenon never seen before in mammals [115]. Critically the organs where these macrophages reside turn visibly dark from the accrual of inert heme crystals which confer the mammalian cells tolerance to heme toxicity, the same mechanism employed by malarial parasites [58, 115, 117]. HRG1 is highly expressed in other non-RES tissues as well, including brain, kidney, intestine, lung, and smooth muscle though its role in these organs remains to be elucidated.
The reasoning that since mammalian cells are capable of synthesizing, utilizing and degrading their own heme calls into question as to why these cells would export heme across the plasma membrane in the first place [61]. This notion has been challenged with the discovery of heme exporters, including FLVCR and MRP5. The feline leukemia virus subgroup C receptor-related proteins (FLVCR) are members of the major facilitator superfamily proteins containing 12 TMDs and were first discovered for their role in non-regenerative anemia in cats resembling pure red cell aplasia in humans [118, 119]. Normally localized to the cell surface, these transporters act as receptors for feline leukemia retroviruses, forming the point of entry for infection and subsequent cellular disruption [119]. Later characterization of their endogenous function revealed their role in heme homeostasis, with FLCVR1 demonstrated to be a heme exporter while its closely related homolog FLVCR2 has been implicated as a heme importer into the cell [43, 45, 120]. Lines of evidence that support this role for FLVCR2 include: binding to hemin-agarose, expression in Xenopus oocytes and CHO cells resulted in accumulation of exogenous heme and heme sensitivity, and depletion by siRNA reduced the transport of the fluorescent heme analog zinc mesoporphyrin (ZnMP) [45]. On the other hand, mutations in the FLVCR2 gene in humans is the cause of the autosomal recessive Fowler syndrome, a proliferative vasculopathy of the brain, which despite associated defects in respiratory complexes, has no obvious dysfunction in heme homeostasis [45, 121]. Furthermore, ectopic expression of FLVCR2 in the yeast reporter system cannot rescue heme-deficient strain’s growth from exogenous heme [113]. For these reasons, FLVCR2 heme import function may need further investigation.
MRP5/ABCC5 was identified as an essential heme exporter in C. elegans and for regulating systemic heme homeostasis [122]. MRP5 belongs to the family of multidrug resistance-associated protein (MRP) within the type C subfamily of the ATP Binding Cassette (ABC) transporters, which have been investigated extensively over the past 20-plus years for their role in cancer drug resistance [123–126]. Closely related to MRPs 4, 8 and 9, MRP5 falls in the “short MRP clade” with 12 TMDs split into two membrane spanning domains each with a nucleotide binding cassette, and is typically localized to the plasma membrane and endosomal compartments [122, 126, 127]. Prior to the discovery in C. elegans, the role of MRP5 appeared to be that of cyclic nucleotide export from cells as demonstrated by transport of guanosine 3’,5’-cyclic monophosphate (cGMP) in membrane vesicles expressing human MRP5 in vitro [128, 129]. However, the generation of Mrp5−/− mice demonstrated little to no contribution in cGMP efflux and no observable overt phenotypes, leaving the physiological relevance of this transporter unclear [130, 131]. However, genetic studies in C. elegans showed that intestinal heme acquired from the importers HRG-1/4 is transported by MRP-5 from the basolateral surface of the gut intestinal epithelium for utilization by extra-intestinal tissues [122]. Key lines of evidence for this model include functional heme transport assays in yeast, accumulation of fluorescent ZnMP in the intestine, survival in the presence of the toxic heme analog Ga protoporphyrin IX, high labile heme accumulation in the intestine with a concomitant reduction in extra-intestinal tissues, and embryonic lethality due to mrp-5 deficiency that is rescued by excess heme supplementation [122, 132]. In vertebrates, knockdown of mrp5 in zebrafish resulted in severe anemia, indicating a critical role in erythropoiesis for this transporter which is also highly expressed in human RBCs [122, 133]. Furthermore in mammalian cell culture experiments, MRP5 knockout mouse embryonic fibroblasts demonstrated reduced heme export into the secretory pathway, as measured by heme incorporation into a Golgi-targeted horseradish peroxidase reporter [122]. Once in the secretory pathway, the notion is that heme can then be trafficked for incorporation into hemoproteins or directed to other subcellular locations, including the plasma membrane / extracellular space [134].
ABCG2, also known as the breast cancer resistance protein or BRCP, is another family member of the ATP-binding cassette transporters that has been implicated in heme efflux from the cell. ABCG2 is one of the three well-known major multidrug resistance proteins to be discovered (along with MRP1 and P-gp) and consequently its mechanistic function in cancer cells and pathophysiology has been studied exhaustively for over two decades [135–138]. As is common amongst the ABC transporters, ABCG2 is quite promiscuous and has been shown to efflux myriads of drugs/xenobiotics, in particular mitoxantrone, estrone sulfate (E1S), dehydroepiandrosterone sulfate (DHEA), methotrexate, cGMP, topotecan, imatinib, irinotecan and 9-(2-phosphonyl-methoxyethyl) adenine (PMEA) [135–137, 139–144]. Although, a true consensus on ABCG2’s endogenous substrate has yet to be identified, studies have shown that it can efflux heme, protoporphyrin IX, and other porphyrins from the cell [145–150]. ABCG2 is expressed in many different tissues, including the erythroid lineage like MRP5, and in particular ABCG2 has been shown to have a role in blood barrier tissues such as the placenta, mammary epithelium and the blood brain barrier [146, 147]. A key study demonstrated that ABCG2 requires an extracellular-binding loop to export heme to circulating albumin [148].
While FLVCR1 was initially described as a plasma membrane heme exporter in developing erythroid cells to efflux heme and reduce toxicity it was determined that FLVCR1 actually has two different isoforms: FLVCR1a and FLVCR1b with different subcellular localizations [46, 151, 152]. FLVCR1a, the full-length 12 TMD protein, which is targeted to the plasma membrane and transports heme from the cytosol, has an alternate start site in its first intron which generates the FLVCR1b isoform. This truncated six TMD protein localizes to the mitochondria and is thought to be responsible for heme transport after the completion of heme synthesis [46]. Strikingly, FLVCR1 null mice which lack both a and b isoforms die in utero during mid gestation due to a block in erythropoiesis [151]. In an effort to untangle the roles of each isoform, a mouse model still expressing the mitochondrial FLVCR1b but lacking the full length FLVCR1a was generated. This mutant mouse showed in utero lethality due to hemorrhaging and skeletal deformations [46]. The restoration of erythroid maturation in these mice indicated that FLVCR1a is dispensable for definitive erythropoiesis, and the role for heme transport in the block of erythroid maturation was associated with the mitochondrial isoform [46]. The logical explanation is that rather than accumulation of excess cytosolic heme inducing toxicity, the cell is starved for heme that is trapped in the mitochondria [8, 153, 154].
These findings beg the question - what in fact is the role of FLVCR1a at the plasma membrane, and why does heme need to be exported from the red cell to prevent cell toxicity? This subject is discussed at length in other reviews, and undoubtedly additional questions still remain regarding FLVCR1 as a heme transporter [39, 61, 134]. In particular, gaps in mechanistic biochemical details as to how it functions require further elucidation. It is known that FLVCR1a requires an extracellular heme carrier protein in the media for heme export to be detected [155]. Furthermore, in the presence of a dedicated carrier, hemopexin, this efflux rate is increased over 100-fold versus albumin, in keeping with their relative affinities [155]. Interestingly, this has led to the theory that heme may be channeled from the cytosol through FLVCR1a, docked at critical histidine residues, and subsequently released to the heme carrier protein [153, 155]. Furthermore, this logic implicates extracellular HPX as more than just a traditional scavenger, with a designated role for facilitating heme efflux from the cell for intercellular trafficking. This notion is of particular interest within the scope of this review, as the key residues which are implicated in the interaction of FLVCR1a with hemopexin for facilitating heme transport and loading are missing from FLCVR1b [153]. How then is the heme exported from the mitochondria without a dedicated chaperone? This is a question that has been brought up repeatedly in recent reviews [8, 39, 50, 111, 134].
Long awaited biochemical characterizations of efflux notwithstanding, lacking a traditional chaperone-based export mechanism should not be viewed as a detractor for mitochondrial export, as conceivably heme transported by FLVCR1b or another yet to be identified mitochondrial exporter may be directly pumped into another organelle and subsequently trafficked to other subcellular compartments. Additional questioning lies in where FLVCR1b localizes within the mitochondria, as this has implications for how it acquires and subsequently transports heme. Is the transporter in the inner or outer mitochondrial membrane? Although it is possible that FLVCR1b could interact with the heme biosynthetic machinery clustered around ferrochelatase (FECH) on the inner mitochondrial membrane (IMM), data from affinity purification studies of the complex do not identify FLVCR1b in the complex [156]. If FLVCR1b is in fact on the IMM but not associated with the FECH complex, one possibility is that this may facilitate heme incorporation into certain types of mitochondrial-derived vesicles. Thus, FLVCR1b may constitute one way of contributing to labile heme in the cytosol, however it is likely not the only mechanism.
FLVCR1a makes up the third putative heme exporter described herein which has been shown to localize to the plasma membrane of the erythroid lineage and implicated in the efflux of heme to reduce toxicity [43]. In addition to MRP5 and ABCG2, eight other dedicated ABC transporters have now been shown to be expressed specifically on the plasma membrane of red cells, including the putative coproporphyrinogen III transporter, ABCB6 [157–159]. Originally thought to be yet another heme transporter, ABCB6 is normally localized to the mitochondria but has an isoform which is directed to the plasma membrane [160, 161]. Critically, overexpression of this plasma membrane isoform was unable to efflux heme, but did reduce the cellular accumulations of another tetrapyrrole, pheophorbide A [161]. Though ABCB6 functions are still not well-understood, it recently was shown to be involved with cadmium detoxification and likely has affinity for multiple substrates [162].
Why are there so many transporters at the surface of RBCs, particularly those capable of effluxing porphyrins? This is perplexing, given the established cytotoxic nature of free heme in circulation and the concerted mechanisms for actively avoiding free heme by scavenging and recycling it. Indeed, it is hard to imagine that RBCs, which need to synthesize a billion heme molecules for incorporation into globins may contain multiple heme or porphyrin exporters. Part of the explanation however may lie with the compromise that is required for higher levels of specialization. In simplistic terms, mammalian red cells become specialized hemoglobin-producing cells to maximize oxygen delivery and systematically strip themselves of all intracellular organelles during differentiation [163]. Therefore a current model posits that FLVCR1a is essential during early stages of red cell differentiation to maintain stoichiometric amounts of heme and globin, and that excess heme causes cell death [164]. Thus, it is reasonable to assume that the exporters orchestrate a delicate balance between ridding the intracellular milieu of a toxic redox-active catalyst versus the physiologic disadvantage of exposing the extracellular milieu, which harbors heme scavenging proteins. Heme import and export may be highly dynamic, thereby mitigating the risk of both intracellular and extracellular heme toxicity. However, the physiological and pathological contexts in which various heme import and export pathways are activated and regulated remain to be determined. Moreover, it is unclear at this time if heme transporters work independently of each other or operate in a synergistic manner. The determination of a temporospatial hierarchy for heme transporters during cell and tissue development will be critical for helping to define heme transport dynamics in various contexts.
2.2. Intercellular heme trafficking.
The discovery that a wide range of cell types in metazoans can both import and export heme raises the possibility that cells and tissues can share their heme quota for a systemic control of heme homeostasis. In C. elegans, HRG-3 was discovered to be an essential 8 kDa intercellular heme chaperone protein which binds and delivers maternal heme to developing oocytes [165]. Originally secreted by the maternal intestine, HRG-3 binds heme with a stoichiometry of 2:1 respectively and is released into the interstitial fluid for transport to extra-intestinal cells, including oocytes [165, 166]. Loss of HRG-3 resulted either in death during embryogenesis or developmental arrest immediately post-hatching, which was completely rescued by maternal expression of HRG-3 [165]. Though there are no obvious homologs for HRG-3 in vertebrates, these results demonstrate that heme can be transferred between different tissues and cell types as a nutritional heme source [110, 165]. Another C. elegans protein, HRG-2, facilitates heme import and utilization in the hypodermis. HRG-2 is a predicted 32 kDa single-pass type I transmembrane protein and localizes to both the apical plasma membrane and the endoplasmic reticulum [167]. It is thought to function as a heme binding oxidoreductase, as it contains both a thioredoxin-like fold and a glutathione S-transferase (GST) domain, which have been recently demonstrated to bind heme in the blood-feeding nematode Haemonchus contortus [168]. Based on these dual functions and localizations, HRG-2 likely reduces heme at the cell surface to facilitate its entry into the cell, and assists with sequestration or redistribution of intracellular heme at the ER, possibly transferring bound heme to membrane-bound or maturing luminal hemoproteins [110, 167].
The fact that heme can be transported between cells and distal tissues imply that systemic signals may be able to communicate and regulate its trafficking and usage accordingly [110, 165]. Such systems have already been demonstrated for other critical metallo-nutrients, such as hepcidin’s regulation of systemic iron homeostasis and the yet to be identified factor that communicates copper deficiency from the heart to copper storage organs [169, 170]. Indeed, in C. elegans, a secreted signaling factor termed HRG-7 communicates intestinal heme status to extra-intestinal tissues [171]. HRG-7 belongs to the family of A1 aspartic proteases and is released from the intestine during heme starvation and signals to sensory neurons, which in-turn regulates hrg-7 expression in the intestine through DBL-1, a worm homolog of bone morphogenetic protein 5 (BMP5), via the “small” worm phenotype “Mothers Against Decapentaplegic” family (SMAD) homolog SMA-9 [171]. Loss of HRG-7 causes a heme-dependent growth defect and a striking upregulation of heme deficiency signal, as the worms are unable to cell non-autonomously regulate intestinal heme import [171]. It remains to be determined whether similar pathways modulate systemic heme homeostasis in mammals as homologs for HRG-7, DBL-1, and SMA-9 exist in mammals. These long-range inter-organ communication systems allow for a concerted physiologic response by the whole organism in response to nutrient fluctuations.
2.3. Intracellular heme trafficking.
Once heme is synthesized on the matrix side of the mitochondrial inner-membrane, or transported into the cell, it must be trafficked to hemoproteins located throughout the cell [4, 5, 8, 39, 48, 173, 174]. To mitigate the potential for heme toxicity, there must be proteins that bind and buffer heme, deliver and insert it into downstream hemoproteins, sequester it into a chemically inert form until it is needed, and transport it into and out of various subcellular compartments [4, 5, 8, 12, 39, 134]. Here, we discuss proteins and processes implicated in these delivery systems.
Due to the high concentration of biomolecules in cells, there is likely no free unbound heme. Intracellular heme can be partitioned between exchange inert and labile heme binding sites. The former comprises proteins that have high affinity heme binding sites with considerable kinetic barriers for exchange. Labile heme (LH) comprises proteins with relatively modest heme binding affinities and sites that are accessible for facile heme exchange between various biomolecules. The network of proteins that bind and buffer LH, which are largely unknown, represent the source of heme for heme-requiring proteins and pathways. Recently, tools for measuring LH using fluorescence and activity-based sensors in yeast and non-erythroid mammalian cell lines have found that LH is in the nM regime, spanning ~20 – 430 nM [12, 132, 175–177]. This implies that the proteins that buffer LH exhibit heme dissociation constants of similar magnitude, providing a thermodynamic signature for proteins that comprise the LH pool.
Overall, while little is known about the speciation of the LH pool, certain proteins have been implicated in buffering heme, including glutathione S-transferases (GSTs), fatty acid binding proteins (FABPs), various heme binding proteins (HBPs), and glyceraldehyde phosphate dehydrogenase (GAPDH). Glutathione S-transferases are a family of Phase II detoxification enzymes that have been shown to bind heme in the cell and play a role in modulating heme homeostasis [178–180]. Highly abundant in the cytosol, the canonical function of GST is to catalyze the conjugation of glutathione (GSH) to electrophilic compounds in order to facilitate their clearance [181]. Though the scope of GST’s impact on heme binding and trafficking in mammalian cells is still poorly understood, it is clearly critical in blood-feeding parasites which have redundant means to buffer cytotoxic heme from their diet [182–184]. FABP, whose function (as the name would suggest) involves the trafficking of fatty acids and lipids, can also bind heme, albeit its exact physiological role in maintaining heme homeostasis has yet to be determined [185, 186]. The FABP affinity for heme has been shown to be 10-fold greater than that of some fatty acids such as oleic acid, prompting speculation that it could traffic heme [4, 8, 185]. The heme binding proteins p22HBP and HBP23 also known as heme binding protein 1 (HEBP1) and peroxiredoxin I (PRDX1) respectively, are highly expressed in the liver and have been comparatively well-studied [187, 188]. Originally named for their molecular weights, HBP heme binding affinities (p22HBP/HEBP1: Kd = 26 nM, HBP23/PRDXI: Kd = 55 nM) are greater than those of GSTs and FABPs (Kd = 100–200 nM) [187, 189]. Additionally, their expression has been shown to be inducible by heme-related stimuli; p22HBP by erythroid differentiation in mouse erythroleukemia cells, and HBP23 by heme, protoporphyrin IX, and other metalloporphyrins in hepatocytes [187, 189, 190]. p22HBP bound to both heme and its precursor’s crystal structures show tetrapyrrole binding [187, 189, 191]. In summary GSTs, FABPs, and HBPs all bind heme, though evidence for their specific roles in heme homeostasis is lacking. While all these factors exhibit heme binding affinities within the estimated concentration range of the LH pool, suggesting that these proteins may be components of a network of heme buffering proteins, experiments probing their individual and cumulative roles in heme homeostasis are currently lacking.
GAPDH is a canonical glycolytic enzyme that was recently found to play key roles in heme homeostasis as both a heme buffering molecule and a chaperone [192–194]. Of note, GAPDH is able to regulate heme delivery and insertion into hemoproteins such as nitric oxide synthase (NOS) and guanylate cyclase [193, 195, 196]. The stoichiometry and mechanisms of binding for GAPDH have been elucidated, with one heme bound per GAPDH tetramer and coordinated by a highly-conserved histidine, His51 [197, 198]. His51, which is conserved from yeast to human, appears to be essential for heme binding and trafficking to NOS and other hemoproteins, but also for heme delivery to transcription factors in the nucleus [175, 197, 198]. Furthermore, GAPDH has been shown to be an important component of the heme buffering system, with a heme binding affinity of 24 nM similar to estimates of labile heme in the cytosol [175, 199].
The progesterone receptor membrane component (PGRMC) proteins were recently classified as intracellular heme chaperones. Although PGRMCs have been known to be capable of heme binding, they now have been discovered to have essential roles in the distribution of heme from the mitochondria [156, 200–203]. Indeed, both PGRMC1 and PGRMC2 can be localized to the mitochondria, where they were confirmed to be interacting with the heme biosynthetic machinery clustered around FECH, the final step in heme production [156]. In probing this relationship further, it was determined that PGRMC1 is in fact capable of regulating heme synthesis via interactions with FECH and was suggested to be a chaperone for efflux from the mitochondria [202]. Key supporting lines of evidence for this included: inhibition resulted in observed decrease in hemoglobinization in erythroid differentiation model; purified PGRMC1 was able to donate heme to apo-cytochrome b5; and in vitro measured FECH activity decreased in a PGRMC1 dose-dependent manner [202]. The ER-based PGRMC2 was required for heme delivery to transcription factors in the nucleus [203]. Specifically, deletion of PGMRC2 in brown fat adipocytes reduced LH in the nucleus and altered heme regulated gene expression resulting in severe mitochondrial defects and a failure to activate adaptive thermogenesis, while obese-diabetic mice treated with a small-molecule activator of PGRMC2 showed substantial improvement of diabetic features [203]. Though additional studies are required to determine their exact roles in trafficking, these PGRMCs serve as an outlet for newly synthesized heme to reach distinct compartments, such as hemylating ER-localized cytochromes or distributing to nuclear transcription factors [39, 134].
Unlike iron that is encased in ferritin for storage, there are no known heme storage proteins that sequester heme until it is needed. However, recently, transient absorption imaging of heme in C. elegans and mammalian cell lines revealed the presence of heme containing granules [204]. These compartments of heme appear to be highly dynamic and can be broken down and mobilized within the cell [204]. While the physiological function and physical composition of these granules are unknown, it is tempting to speculate that they represent novel heme storage sites. Though these heme granules were initially attributed to membrane-enclosed puncta-like structures, it is also possible that they may be maintained in distinct biocondensates, or “membrane-less” compartments akin to p-granules in the cell cytosol [205]. This liquid-liquid phase separation may facilitate its accumulation and rapid dissemination as a heme store for hemoproteins and signaling in an aqueous environment [205, 206]. Biocondensate formation may be driven by scaffolding or by heme binding / chaperoning proteins. Elucidation of these heme granules will be an exciting new avenue for future research in understanding cellular heme homeostasis and trafficking.
3. Membrane centered heme trafficking mechanisms.
3.1. Heme trafficking through membrane contact sites.
Inter-organelle membrane contact sites have emerged as being critical for signaling and metabolite exchange, including the transfer of various phospholipids [207–220]. MCSs are physical contacts between two membrane-bound organelles and have key characteristics, including structural proteins that physically tether two membranes, functional proteins that fulfill specific role(s) for a given MCS, and a defined lipid composition that supports MCS function [51]. While the type, frequency, function, and composition of MCSs are still under active investigation and a matter of debate, virtually all MCSs identified to date involve the ER, including ER-Mitochondria, ER-Plasma Membrane, ER-Golgi, ER-Peroxisome, ER-Lysosome/Vacuole, and ER-Lipid Droplet contact sites [190] (Table 2 and Figure 2). More recently, mitochondrial-nuclear contact sites have been purported to be identified [221]. MCSs are also highly dynamic and remodeled in response to various physiologic stimuli and perturbations to other contact sites [222, 223]. Given that the ER forms contact sites with the mitochondria, where heme is synthesized, and many other subcellular compartments that house hemoproteins, we previously proposed that the ER network may serve as a highway for intracellular heme distribution [4, 11]. Indeed, we recently found that mitochondrial-ER contact sites, the first junction in this proposed heme distribution network, is important for regulating heme trafficking to the nucleus [224].
Table 2.
Inter-organellar contact sites in yeast and mammals. Mito = Mitochondria; N/A = Not Available.
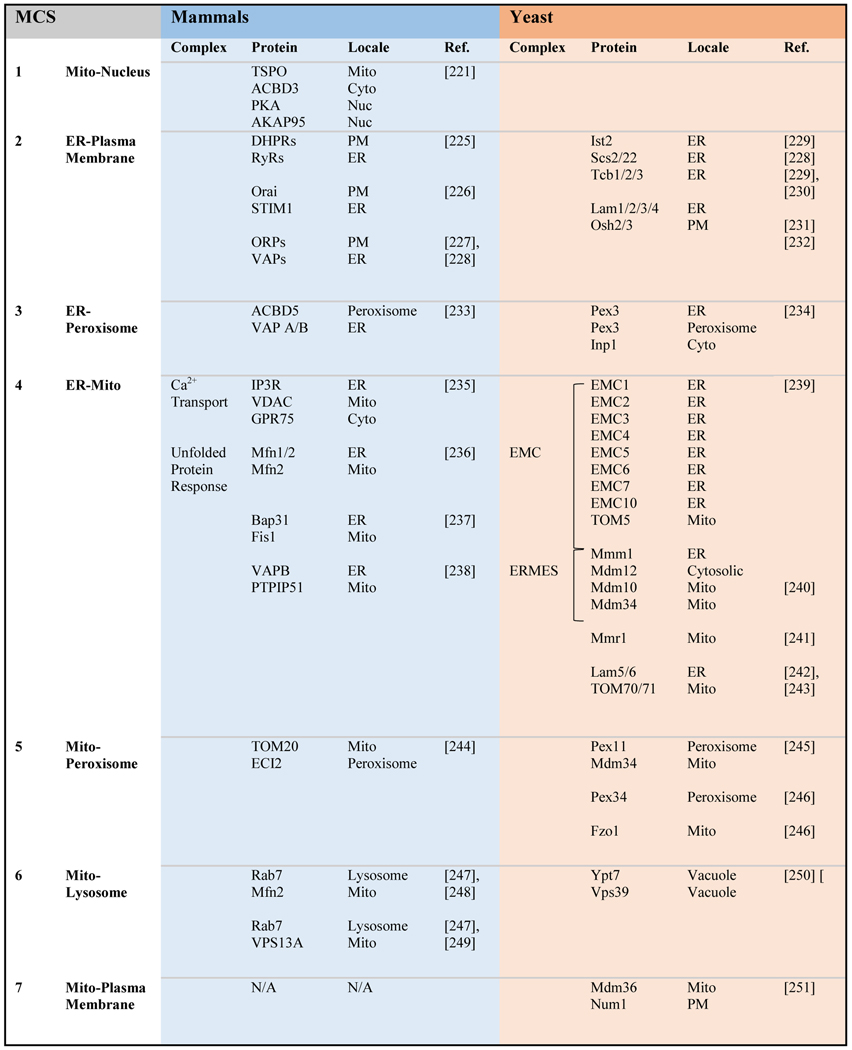 |
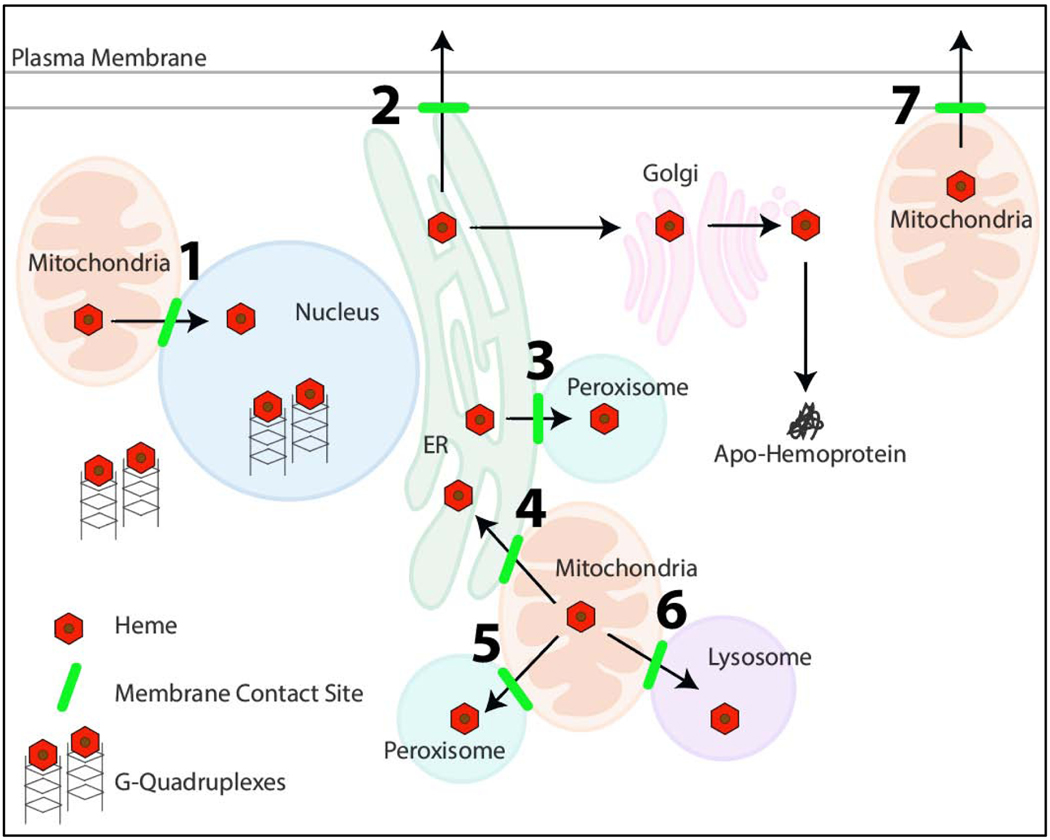
Figure 2.
Proposed model for “chaperone-less” heme trafficking via inter-organelle membrane contact sites (highlighted in green) and G-quadruplexes. The specific organellar contact sites, numbered 1–7, are described in Table 2. RNA and DNA G-quadruplexes are also hypothesized to regulate intracellular heme bioavailability.
ER-mitochondrial contact sites are arguably the most well understood in Saccharomyces cerevisiae. In Baker’s yeast, two types of ER-mitochondrial contact sites have been identified, the ER-Mitochondria Encounter Structure (ERMES) and ER Membrane Protein Complexes (EMCs) [51, 239, 252]. In humans, ER-mitochondrial contact sites are less well defined and are referred to as Mitochondria-Associated Membranes (MAMs). In yeast, ERMES facilitates calcium signaling, phospholipid exchange, protein import, and mitochondrial dynamics [204, 205]. In order to identify factors that regulate heme distribution, including the role of ER-mitochondrial contact sites, heme distribution dynamics were probed in yeast using genetically-encoded heme sensors targeted to different cellular locales, including the mitochondrial matrix, cytosol, and nucleus. Interestingly, upon initiation of heme synthesis, heme is trafficked from the matrix side of the IMM, where the last step of heme synthesis occurs, to the nucleus ~25% faster than to the cytosol or mitochondrial matrix [224]. The faster rate of heme trafficking to the nucleus suggested there is a pathway that bypasses export into the cytosol prior to import into the nucleus and raised the prospects of a more direct mechanism for heme transfer between the mitochondria and nucleus. One possibility is that there are mitochondrial-nuclear contact sites, which has only recently been proposed [221]. Another possibility is that heme is transferred via the ER network which forms contact sites with the mitochondria and is contiguous with the nuclear envelope [132].
In order to test the role of ERMES in nuclear heme distribution, Saccharomyces cerevisiae deletion mutants with defects in ERMES were screened for alterations in nuclear heme trafficking rates [224]. As indicated in Figure 3 and Table 2, there are four structural proteins, Mdm12, Mmm1, Mdm34, and Mdm10, that tether the ER and mitochondrial outer membrane in the ERMES complex [239, 240, 253, 254]. Moreover, ERMES is highly dynamic and responsive to mitochondrial dynamics. ERMES is required for the ER-mediated constriction of mitochondrial tubules to help facilitate mitochondrial division by the GTPase, Dnm1. After division, the GTPase Gem1 disengages ERMES, thereby releasing the ER from the mitochondrial tubule [240, 255]. There is an increased prevalence of ERMES if mitochondrial division cannot occur or occurs at a lower rate than fusion since the completion of a division cycle initiates the release of the ER from the mitochondria. Conversely, there is a decreased prevalence in ERMES if mitochondrial fusion cannot occur or occurs at a faster rate than division. Thus, deletion of core components of ERMES or mitochondrial dynamics factors are expected to alter the frequency of ERMES, thereby providing an excellent test bed to probe the role of ER-mitochondrial contact sites in heme trafficking. Consistent with a role for ERMES in mediating mitochondrial-nuclear heme transfer, deletion of GEM1 and DNM1, which increase ERMES when deleted, increase mitochondrial-nuclear heme trafficking rates [224]. Moreover, deletion of MGM1, a GTPase required for mitochondrial fusion that decreases ERMES when deleted, decreases mitochondrial-nuclear heme trafficking rates [224]. However, deletion of core ERMES components, Mdm10 and Mdm34, did not affect heme trafficking rates [224]. The lack of a phenotype in ERMES tethers might suggest that other mitochondrial-ER contact sites provide alternative routes for heme trafficking. Indeed, other studies have also reported that phosphatidylserine transfer is not affected in cells lacking ERMES [239, 256, 257]. A major experimental challenge to contend with is the fact that perturbation of one ER-mitochondrial contact site alters the distribution of other types of mitochondrial contact sites, thereby masking heme trafficking phenotypes. Future work should involve multi-genic knockouts of multiple contact sites to address how these pathways act to mobilize heme to distinct cellular locations or if these pathways are uni- or bi-directional. In humans, mitochondrial-ER contact sites have composition that is distinct from yeast and is less well understood [258]. Future studies will be needed to determine if ER-mitochondrial contact sites in mammalian cells influence heme trafficking and mobilization.
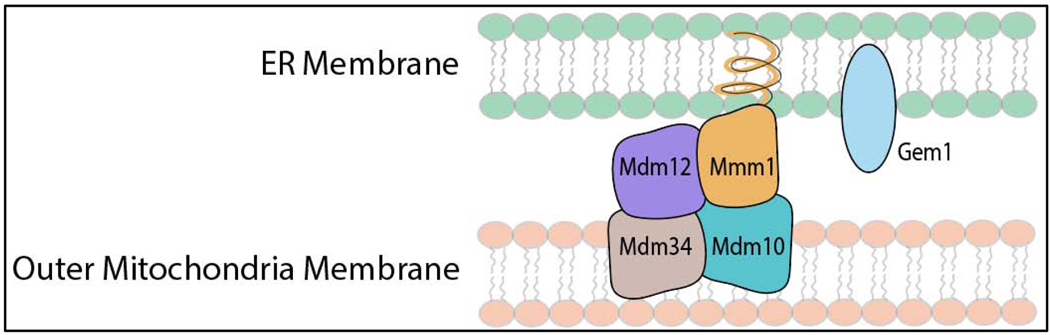
Figure 3.
Cartoon of the ER-mitochondrial encounter structure (ERMES) in yeast, which facilitates calcium and phospholipid exchange, as well as possibly heme trafficking. ERMES consists of four proteins, Mdm12, Mmm1, Mdm34, and Mdm10, that physically tether the ER and outer mitochondrial membranes. Gem1 is a GTPase that disengages ERMES after mitochondrial fission. Altering the frequency of ERMES, by deletion of Gem1, or GTPases that control mitochondrial fusion, e.g. Mgm1, and fission, e.g. Dnm1, has been shown to affect mitochondrial-nuclear heme trafficking rates. See text for details.
Mitochondria-lysosomal/vacuolar contact sites are a particularly intriguing axis with respect to heme homeostasis [250, 259–263]. Lysosomes are critical for mitochondrial recycling, fusing with autophagosomes to engulf, and degrade entire mitochondria in a process called mitophagy [264–267]. Furthermore, mitochondrial derived vesicles (MDVs) bud off from mitochondria and can fuse with distant lysosomes, likely facilitating more specific mitochondrial protein turnover [268]. We envision heme may be recycled in such processes, i.e. stored in the lysosome/vacuole upon degradation of mitochondrial material and subsequently re-routed to the mitochondria when needed via mitochondria-lysosome/vacuolar contact sites. Alternatively, a mitochondrial-lysosome/vacuolar contact site may serve as an exit valve if heme builds up in the mitochondria to excessive levels. Given the compendium of other MCSs (Table 2), systemic perturbation of these contact sites along with measurements of bioavailable heme distribution will reveal novel heme trafficking networks.
3.2. Heme trafficking through membrane vesicles.
Another mode of heme transport and trafficking involves the use of membrane vesicles. Heme can be exported, imported, and trafficked intracellularly using such vesicles. For instance, RBCs are known to shed heme-loaded microparticles, especially during pathophysiological conditions such as sickle cell disease and acute lung injuries / respiratory distress syndromes [269, 270]. These submicron vesicles have also been described in great detail as a challenge associated with the erythrocyte storage lesion, a term used to describe the structural, biochemical, and metabolic changes associated with prolonged aging of stored red blood cell units for transfusions [271]. RBCs suffer from a loss of structural integrity in accelerated senescence and as a result have increased shedding of exosomes loaded with hemoglobin and heme [272–274]. These microvesicles can be highly inflammatory to the vasculature, particularly to pulmonary endothelial cells which have been shown to endocytose them in Rab5-dependent manner [274]. Heme transport via microparticles or exosomes in circulation is a relatively new field of study and warrants additional investigation, particularly to determine if this mechanism of heme uptake by cells is really only relevant in pathophysiologic contexts and if other cell types are capable of releasing heme in exosomes, microparticles, and other membranous components.
Heme can also be imported into the cell via vesicular endocytosis / phagocytosis. As discussed earlier, Hb and HPX receptors in vertebrates have well established mechanisms to bind their extracellular heme sources and internalize them via traditional endocytosis [70, 71]. Likewise, eukaryotic microbes such as S. pombe can also bind extracellular heme via cell surface heme receptors, e.g. Shu1, for import by endocytosis and subsequent trafficking to the vacuole for storage [275–277]. Intracellular movement of heme-loaded membrane vesicles already inside the cell could be facilitated via a similar process, with endosomal sorting driving mobilization to other organelles. Aligned with this, prior studies have found that iron can be transferred from the endosome to the mitochondria through a “kiss and run” mechanism [278, 279]. Heme could be trafficked to the mitochondria in a similar endosome-dependent manner. Moreover, the endosomal-based vesicular transport would allow for direct connection to the ER, as ER-endosome contact sites have been reported and function in mediating cholesterol transport [280, 281]. Heme trafficking to the ER highway via the endosome would allow heme to be mobilized throughout the entire cell.
An additional mechanism that may compliment this endosomal vesicle heme trafficking is the intracellular distribution of mitochondrial-sourced heme within MDVs, which are single- or double-membrane bound vesicles approximately 70 – 100 nm in diameter that are released from the mitochondria [282]. Initially, these MDVs were characterized as a specific form of vesicular transport to the peroxisome, as some vesicles that contained the mitochondrial marker mitochondria-anchored protein ligase (MAPL) were shown to be able to fuse with peroxisomes; likewise a subset of peroxisomes also possessed matching MDV markers [282]. Furthermore, this novel vesicular transport was determined to be cargo-specific as MDVs that were positive for MAPL lacked another mitochondrial outer-membrane marker, TOM20, while the MDVs that contained TOM20 excluded MAPL from their membranes [282]. This distinct vesicle population labeled with TOM20 did not fuse with peroxisomes, but rather was later determined to be selectively transported to the lysosome [283]. Critically, MDV formation targeted to the lysosome is distinct from mitophagy-dependent markers and results in the transport of proteins from healthy, non-fragmented mitochondria [283]. Taken together, these findings indicated that MDVs are able to discriminate between types of cargo loaded within them and likely the incorporation of specific cargo is a primary factor for their requisite trafficking [282–284]. Transfer of heme via such vesicles would not only minimize the release of cytotoxic heme, but could also facilitate regulated movement of sequestered heme from its “depot” for rapid availability [4]. Additionally, if it is determined that these MDVs can be targeted to additional organelles, it is easy to visualize mitochondria effectively becoming their own mini-secretory pathway for the distribution of heme as well as other mitochondrial components and products by cargo vesicles [285]. The trafficking of heme via MDVs may not even require heme crossing the IMM, as a subset of the MDVs are stainable by membrane-potential dyes (MitoTracker Red and tetramethylrhodamine, ethyl ester TMRE), indicating both their membranes and electrochemical potential are intact [282, 283]. Additionally, subsets of MDVs were shown to contain matrix protein markers such as pyruvate dehydrogenase (PDH) as well as complexes II, III, and IV of the IMM but would not incorporate complexes I and V or nucleoids, further demonstrating their specificity of cargo incorporation [283, 286].
What are the major cargo for MDVs? It has been proposed that the primary function of MDVs is the selective removal of oxidized proteins, complexes and lipids, as oxidized species were found to be highly enriched in their cargo [286]. Ultimately, the contents being sequestered by these MDVs and their purpose still remain to be fully elucidated, and may very well include heme [286, 287]. What is further reinforced by these studies however is the longstanding intimacy already understood between peroxisomes and the mitochondria [288, 289]. With shared major roles in fatty acid metabolism, reactive oxygen species (ROS) production / scavenging, and now clear physical and metabolic interactions, it has long been postulated that these organelles are evolutionarily intertwined [287, 288, 290, 291]. Indeed, they are more than just distant relatives, as recent studies have now demonstrated the essential role for mitochondria in the de novo generation of peroxisomes in mammalian cells via the budding of vesicles coined pre-peroxisomes [292]. As there is a critical need for heme to be trafficked between these two organelles (incorporation into key antioxidant heme binding protein catalase), it may make both physiological and evolutionary sense for heme to be trafficking via this highly specific and conserved vesicular route.
4. Nucleic Acids and Heme
Nucleic acid polymers have been shown to form various secondary structures. DNA forms double stranded helices and guanine quadruplexes (G4), while RNA has a more diverse repertoire consisting of hairpins, internal loops, pseudoknots, bulges, multiloops, and G4s. One common distinct structure between DNA and RNA is G4s, formed at guanine-rich sequences via Hoogsteen hydrogen bonding of four guanine bases to create a square planar guanine tetrad (G-quartet or G-tetrad). Two or more of these G-quartets stacked together form a G4, which are typically folded in parallel, antiparallel, or hybrid structures. This nucleic acid folding results in various topologies as G4 formation is primarily determined by strand polarities and loop orientation. Whether these different topologies influence G4 function is yet to be elucidated.
4.1. Heme association with guanine quadruplexes.
G4 structures are enriched at promoter and telomere regions and have been shown to affect transcription, reverse transcription, translation, and epigenetics [293–295]. For example, during transcription elongation DNA G4 formation in the template strand can inhibit RNA polymerases and impede transcription. The presence of G4s have also been shown to contribute to double-stranded breaks inducing DNA instability [296]. As a protective mechanism, helicases can unfold these G4s to limit DNA damage [297]. In fact, many cancer cells have mutated, dysfunctional helicases allowing for G4 formation, resulting in the progression of DNA damage and transcription stalling [298]. Apart from transcription, it was reported that RNA and DNA G4 interacts with porphyrins, including heme, enabling catalysis and protection against heme-related oxidative damage [299, 300]. Specifically, G4s associated with hemin exhibited peroxidase activity [301, 302]. These interesting findings represent yet another unique heme interaction in the cell that may affect its bioavailability and mobilization.
The earliest evidence for porphyrin interactions with G4s was nearly 25 years ago, with the discovery of “DNAzymes” capable of catalyzing the metalation of mesoporphyrin IX [301]. Since then, heme itself has been shown to interact with nucleic acids, as its planar and hydrophobic characteristics enable stacking with the G4 structure, corroborated by specific docking and molecular dynamics studies [303, 304]. Recently, a new role for G4s regarding heme bioavailability was reported [300]. This study showed that G4s in mammalian cell culture sequester heme and, when G4s are disrupted, genes related to heme and iron homeostasis are dysregulated, including over a 30-fold induction of HMOX1. These results highlight G4s’ potential ability to regulate heme availability and gene expression. However, it is unclear at this point if the displaced heme from G4s directly or indirectly affect HMOX1 expression. Indeed, other genes found to be upregulated by heme and synthetic G4 ligands included ferritin heavy chain FTH1, Glutamate-Cysteine Ligase Modifier Subunit GCLM, and NAD(P)H Quinone Dehydrogenase 1 NQO1, which all share nuclear factor erythroid 2-related factor 2 (NRF2) as a common regulator [300]. Therefore, the release of heme could be indirectly activating HMOX1 through the stimulation of oxidative stress and subsequent stabilization of NRF2 [300].
G4 formation in rRNA and human ribosomes in vivo have also been reported [305]. rRNA is the most abundant RNA in cells and the greatest source of cytosolic G4s. Mestre-Fos et al. reported rRNA G4-hemin interactions in vivo and established their role in buffering cytosolic heme [305]. Inducing rRNA G4s using PhenDC3, a small molecule that stabilizes G4s, resulted in lower cytosolic heme as measured by fluorescent heme sensors [305]. These results suggest that, upon induction, rRNA G4s can bind and sequester heme, thereby limiting its availability in the cytosol. Overall, these results provide evidence for G4s in regulating cellular heme levels. However, the precise physiological roles and context of G4-heme binding remain to be determined. Are G4s simply a storage site for heme? Or are there specific heme-dependent reactions catalyzed by G4s that are physiologically important? For instance, could the catalase activity of G4-heme defend nucleic acids against peroxide stress? Could rRNA G4s participate in catalyzing hemylation of nascent hemoproteins? Much work still remains to be done to characterize the significance of G4-heme interactions.
5. Conclusion
We envision that the area of heme trafficking and homeostasis will flourish as more discoveries, either planned or serendipitous, will fill in the enormous gaps in our knowledge. Clearly, in the past 15 years new molecules and pathways have been identified through directed genetic screening in model systems, creating sensitive reporters and sensors for dynamic heme monitoring, and establishing new platforms for label-free imaging modalities. Emerging concepts from membrane trafficking, some that were considered radical just 30 years ago, have further contributed to our understanding of how heme that exhibits properties of both, transition metals and lipids, can move within organelles [212]. We envision that this movement will not be restricted to between organelles but extends to the myriad of cell types that constitute whole organs. It is now time to rethink heme beyond a static macrocycle contained within a protein inside a cell to one that is dynamic and available on-demand at the intercellular and organismal level.
Table 1.
Metazoan Heme Transporters.
| Directionality of Transport | Transporter | Localization | Species | Ref. |
|---|---|---|---|---|
| Import | HRG1 | Endosome / Lysosomal | All metazoans | [115] |
| FLVCR1b | Mitochondria | Vertebrates | [46] | |
| HRG-4 | Plasma Membrane | Worms | [44] | |
| FLVCR2 | Plasma Membrane | Mammals | [45] | |
| Export | MRP5 | Endosome / Plasma Membrane | All metazoans | [122] |
| FLVCR1a | Plasma Membrane | Vertebrates | [172] | |
| ABCG2 | Plasma Membrane | Vertebrates | [148] | |
| HRG-3 | Secretory | Worms | [165] |
Highlights.
Heme is an essential cofactor, signaling molecule and metallonutrient.
The mechanisms underlying heme transport and trafficking are not well understood.
Heme trafficking is often thought to be mediated by transporters and chaperones.
The mobilization of heme may also be mediated by nucleic acids and lipid membranes.
Acknowledgements
This work was supported in part by funding from the National Institutes of Health - DK85035 (IH), DK125740 (IH), ES025661 (ARR, IH) - the National Science Foundation (MCB- 1552791 to ARR), and the Blanchard Professorship from the Georgia Institute of Technology (to ARR).
I.G.C and M.M.W, I.H., and A.R.R. were involved in the conceptualization and writing of the manuscript. I.H. and A.R.R. supervised the work and were responsible for funding acquisition.
Footnotes
Declaration of competing interest
IH is the President and Founder of Rakta Therapeutics Inc. (College Park, MD), a company involved in the development of heme transporter-related diagnostics. He declares no other competing financial interests.
Author Roles
Declaration of interest
Iqbal Hamza is the President and Founder of Rakta Therapeutics Inc. (College Park, MD), a company involved in the development of heme transporter-related diagnostics. He declares no other competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CITATIONS
- [1].Sassa S, Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide?, Antioxid Redox Signal, 6 (2004) 819–824. [DOI] [PubMed] [Google Scholar]
- [2].Reedy CJ, Gibney BR, Heme protein assemblies, Chem Rev, 104 (2004) 617–649. [DOI] [PubMed] [Google Scholar]
- [3].Kumar S, Bandyopadhyay U, Free heme toxicity and its detoxification systems in human, Toxicol Lett, 157 (2005) 175–188. [DOI] [PubMed] [Google Scholar]
- [4].Severance S, Hamza I, Trafficking of heme and porphyrins in metazoa, Chem Rev, 109 (2009) 4596–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hamza I, Dailey HA, One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans, Biochim Biophys Acta, 1823 (2012) 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chiabrando D, Vinchi F, Fiorito V, Mercurio S, Tolosano E, Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes, Front Pharmacol, 5 (2014) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Poulos TL, Heme Enzyme Structure and Function, Chemical Reviews, 114 (2014) 3919–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Reddi AR, Hamza I, Heme Mobilization in Animals: A Metallolipid’s Journey, Accounts of Chemical Research, 49 (2016) 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chiabrando D, Fiorito V, Petrillo S, Tolosano E, Unraveling the Role of Heme in Neurodegeneration, FrontNeurosci, 12 (2018) 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mense SM, Zhang L, Heme: a versatile signaling molecule controlling the activities of diverse regulatorsranging from transcription factors to MAP kinases, Cell research, 16 (2006) 681–692. [DOI] [PubMed] [Google Scholar]
- [11].Reddi AR, Hamza I, Heme Mobilization in Animals: A Metallolipid’s Journey, Acc Chem Res, 49 (2016) 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanna DA, Martinez-Guzman O, Reddi AR, Heme Gazing: Illuminating Eukaryotic Heme Trafficking, Dynamics, and Signaling with Fluorescent Heme Sensors, Biochemistry, 56 (2017) 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shimizu T, Lengalova A, Martínek V, Martínková M, Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres, Chem Soc Rev, 48 (2019) 5624–5657. [DOI] [PubMed] [Google Scholar]
- [14].Donegan RK, Moore CM, Hanna DA, Reddi AR, Handling heme: The mechanisms underlying the movement of heme within and between cells, Free Radic Biol Med, 133 (2019) 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang L, Heme Biology, Place Published, 2020. [Google Scholar]
- [16].Pradhan P, Vijayan V, Gueler F, Immenschuh S, Interplay of Heme with Macrophages in Homeostasis and Inflammation, International Journal of Molecular Sciences, 21 (2020) 740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, Kamenecka TM, Schaart G, Lefebvre P, Neviere R, Burris TP, Schrauwen P, Staels B, Duez H, Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy, Nature medicine, 19 (2013) 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA, Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha, Genes Dev, 23 (2009) 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM, p53 regulates mitochondrial respiration, Science, 312 (2006) 1650–1653. [DOI] [PubMed] [Google Scholar]
- [20].Dohi Y, Ikura T, Hoshikawa Y, Katoh Y, Ota K, Nakanome A, Muto A, Omura S, Ohta T, Ito A, Yoshida M, Noda T, Igarashi K, Bach1 inhibits oxidative stress-induced cellular senescence by impeding p53 function on chromatin, Nature structural & molecular biology, 15 (2008) 1246–1254. [DOI] [PubMed] [Google Scholar]
- [21].Pfeifer K, Kim KS, Kogan S, Guarente L, Functional dissection and sequence of yeast HAP1 activator, Cell, 56 (1989) 291–301. [DOI] [PubMed] [Google Scholar]
- [22].Zhang L, Guarente L, Heme binds to a short sequence that serves a regulatory function in diverse proteins, EMBO J, 14 (1995) 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Igarashi K, Sun J, The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation, Antioxid Redox Signal, 8 (2006) 107–118. [DOI] [PubMed] [Google Scholar]
- [24].Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H, Igarashi K, Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1, EMBO J, 20 (2001) 2835–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shen J, Sheng X, Chang Z, Wu Q, Wang S, Xuan Z, Li D, Wu Y, Shang Y, Kong X, Yu L, Li L, Ruan K, Hu H, Huang Y, Hui L, Xie D, Wang F, Hu R, Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function, Cell Rep, 7 (2014) 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F, Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta, Nature structural & molecular biology, 14 (2007) 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Burton MJ, Kapetanaki SM, Chernova T, Jamieson AG, Dorlet P, Santolini J, Moody PC, Mitcheson JS, Davies NW, Schmid R, Raven EL, Storey NM, A heme-binding domain controls regulation of ATP-dependent potassium channels, Proc Natl Acad Sci U S A, 113 (2016) 3785–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Barr I, Smith AT, Chen Y, Senturia R, Burstyn JN, Guo F, Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing, Proc Natl Acad Sci U S A, 109 (2012) 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Faller M, Matsunaga M, Yin S, Loo JA, Guo F, Heme is involved in microRNA processing, Nature structural & molecular biology, 14 (2007) 23–29. [DOI] [PubMed] [Google Scholar]
- [30].Quick-Cleveland J, Jacob JP, Weitz SH, Shoffner G, Senturia R, Guo F, The DGCR8 RNA-binding heme domain recognizes primary microRNAs by clamping the hairpin, Cell Rep, 7 (2014) 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weitz SH, Gong M, Barr I, Weiss S, Guo F, Processing of microRNA primary transcripts requires heme in mammalian cells, Proc Natl Acad Sci U S A, 111 (2014) 1861–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Barrientos A, Zambrano A, Tzagoloff A, Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae, EMBO J, 23 (2004) 3472–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fontanesi F, Soto IC, Horn D, Barrientos A, Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis, Mol Cell Biol, 30 (2010) 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Korolnek T, Hamza I, Like iron in the blood of the people: the requirement for heme trafficking in iron metabolism, Front Pharmacol, 5 (2014) 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ryter SW, Tyrrell RM, The heme synthesis and degradation pathways: role in oxidant sensitivity: Heme oxygenase has both pro- and antioxidant properties, Free Radical Biology and Medicine, 28 (2000) 289–309. [DOI] [PubMed] [Google Scholar]
- [36].Ajioka RS, Phillips JD, Kushner JP, Biosynthesis of heme in mammals, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1763 (2006) 723–736. [DOI] [PubMed] [Google Scholar]
- [37].Bonkovsky HL, Guo JT, Hou W, Li T, Narang T, Thapar M, Porphyrin and heme metabolism and the porphyrias, Compr Physiol, 3 (2013) 365–401. [DOI] [PubMed] [Google Scholar]
- [38].Sun F, Cheng Y, Chen C, Regulation of heme biosynthesis and transport in metazoa, Science China Life Sciences, 58 (2015) 757–764. [DOI] [PubMed] [Google Scholar]
- [39].Swenson SA, Moore CM, Marcero JR, Medlock AE, Reddi AR, Khalimonchuk O, From Synthesis to Utilization: The Ins and Outs of Mitochondrial Heme, Cells, 9 (2020) 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bianchetti CM, Yi L, Ragsdale SW, Phillips GN Jr., Comparison of apo- and heme-bound crystal structures of a truncated human heme oxygenase-2, J Biol Chem, 282 (2007) 37624–37631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL, Crystal structure of human heme oxygenase-1, Nat Struct Biol, 6 (1999) 860–867. [DOI] [PubMed] [Google Scholar]
- [42].Unno M, Matsui T, Ikeda-Saito M, Crystallographic studies of heme oxygenase complexed with an unstable reaction intermediate, verdoheme, J Inorg Biochem, 113 (2012) 102–109. [DOI] [PubMed] [Google Scholar]
- [43].Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL, Identification of a Human Heme Exporter that Is Essential for Erythropoiesis, Cell, 118 (2004) 757–766. [DOI] [PubMed] [Google Scholar]
- [44].Rajagopal A, Rao AU, Amigo J, Tian M, Upadhyay SK, Hall C, Uhm S, Mathew MK, Fleming MD, Paw BH, Krause M, Hamza I, Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins, Nature, 453 (2008) 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Duffy SP, Shing J, Saraon P, Berger LC, Eiden MV, Wilde A, Tailor CS, The Fowler Syndrome-Associated Protein FLVCR2 Is an Importer of Heme, Molecular and Cellular Biology, 30 (2010) 5318–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chiabrando D, Marro S, Mercurio S, Giorgi C, Petrillo S, Vinchi F, Fiorito V, Fagoonee S, Camporeale A, Turco E, Merlo GR, Silengo L, Altruda F, Pinton P, Tolosano E, The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation, Journal of Clinical Investigation, 122 (2012) 4569–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hamza I, Gitlin JD, Copper chaperones for cytochrome c oxidase and human disease, J Bioenerg Biomembr, 34 (2002) 381–388. [DOI] [PubMed] [Google Scholar]
- [48].Hamza I, Intracellular Trafficking of Porphyrins, ACS Chemical Biology, 1 (2006) 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Robinson NJ, Winge DR, Copper Metallochaperones, Annual Review of Biochemistry, 79 (2010) 537–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chiabrando D, Mercurio S, Tolosano E, Heme and erythropoieis: more than a structural role, Haematologica, 99 (2014) 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnoczky G, Kornmann B, Lackner LL, Levine TP, Pellegrini L, Reinisch K, Rizzuto R, Simmen T, Stenmark H, Ungermann C, Schuldiner M, Coming together to define membrane contact sites, Nat Commun, 10 (2019) 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gray LT, Puig Lombardi E, Verga D, Nicolas A, Teulade-Fichou M-P, Londoño-Vallejo A, Maizels N, G-quadruplexes Sequester Free Heme in Living Cells, Cell Chemical Biology, 26 (2019) 1681–1691.e1685. [DOI] [PubMed] [Google Scholar]
- [53].Bunn HF, Jandl JH, Exchange of heme among hemoglobins and between hemoglobin and albumin, J Biol Chem, 243 (1968) 465–475. [PubMed] [Google Scholar]
- [54].Halliwell B, Gutteridge JMC, The antioxidants of human extracellular fluids, Archives of Biochemistry and Biophysics, 280 (1990) 1–8. [DOI] [PubMed] [Google Scholar]
- [55].Balla G, Vercellotti GM, Muller-Eberhard U, Eaton J, Jacob HS, Exposure of endothelial cells to free heme potentiates damage mediated by granulocytes and toxic oxygen species, Laboratory investigation; a journal of technical methods and pathology, 64 (1991) 648–655. [PubMed] [Google Scholar]
- [56].Vercellotti GM, Balla G, Balla J, Nath K, Eaton JW, Jacob HS, Heme and the vasculature: an oxidative hazard that induces antioxidant defenses in the endothelium, Artif Cells Blood Substit Immobil Biotechnol, 22 (1994) 207–213. [DOI] [PubMed] [Google Scholar]
- [57].Jeney V, Balla J, Yachie A, Varga Z, Vercellotti GM, Eaton JW, Balla G, Pro-oxidant and cytotoxic effects of circulating heme, Blood, 100 (2002) 879–887. [DOI] [PubMed] [Google Scholar]
- [58].Kumar S, Bandyopadhyay U, Free heme toxicity and its detoxification systems in human, Toxicology Letters, 157 (2005) 175–188. [DOI] [PubMed] [Google Scholar]
- [59].Fibach E, Rachmilewitz E, The role of oxidative stress in hemolytic anemia, Curr Mol Med, 8 (2008) 609–619. [DOI] [PubMed] [Google Scholar]
- [60].Dutra FF, Alves LS, Rodrigues D, Fernandez PL, De Oliveira RB, Golenbock DT, Zamboni DS, Bozza MT, Hemolysis-induced lethality involves inflammasome activation by heme, Proceedings of the National Academy of Sciences, 111 (2014) E4110–E4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ponka P, Sheftel AD, English AM, Scott Bohle D, Garcia-Santos D, Do Mammalian Cells Really Need to Export and Import Heme?, Trends in Biochemical Sciences, 42 (2017) 395–406. [DOI] [PubMed] [Google Scholar]
- [62].Rytting M, Worth L, Jaffe N, HEMOLYTIC DISORDERS ASSOCIATED WITH CANCER, Hematology/Oncology Clinics, 10 (1996) 365–376. [DOI] [PubMed] [Google Scholar]
- [63].Beutler E, Luzzatto L, Hemolytic anemia, Semin Hematol, 36 (1999) 38–47. [PubMed] [Google Scholar]
- [64].Ashley-Koch A, Yang Q, Olney RS, Sickle Hemoglobin (Hb S) Allele and Sickle Cell Disease: A HuGE Review, American Journal of Epidemiology, 151 (2000) 839–845. [DOI] [PubMed] [Google Scholar]
- [65].Dhaliwal G, Cornett PA, Tierney LM Jr., Hemolytic anemia, Am Fam Physician, 69 (2004) 2599–2606. [PubMed] [Google Scholar]
- [66].Roseff SD, Sickle cell disease: a review, Immunohematology, 25 (2009) 67–74. [PubMed] [Google Scholar]
- [67].Buehler PW, Abraham B, Vallelian F, Linnemayr C, Pereira CP, Cipollo JF, Jia Y, Mikolajczyk M, Boretti FS, Schoedon G, Alayash AI, Schaer DJ, Haptoglobin preserves the CD163 hemoglobin scavenger pathway by shielding hemoglobin from peroxidative modification, Blood, 113 (2009) 2578–2586. [DOI] [PubMed] [Google Scholar]
- [68].Ratanasopa K, Chakane S, Ilyas M, Nantasenamat C, Bulow L, Trapping of Human Hemoglobin by Haptoglobin: Molecular Mechanisms and Clinical Applications, Antioxidants & Redox Signaling, 18 (2013) 2364–2374. [DOI] [PubMed] [Google Scholar]
- [69].White NJ, Anaemia and malaria, Malar J, 17 (2018) 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Schaer DJ, Buehler PW, Alayash AI, Belcher JD, Vercellotti GM, Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins, Blood, 121 (2013) 1276–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Smith A, McCulloh RJ, Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders, Frontiers in Physiology, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hrkal Z, Vodrazka Z, Kalousek I, Transfer of Heme from Ferrihemoglobin and Ferrihemoglobin Isolated Chains to Hemopexin, European Journal of Biochemistry, 43 (1974) 73–78. [DOI] [PubMed] [Google Scholar]
- [73].Hvidberg V, Maniecki MB, Jacobsen C, Højrup P, Møller HJ, Moestrup SK, Identification of the receptor scavenging hemopexin-heme complexes, Blood, 106 (2005) 2572–2579. [DOI] [PubMed] [Google Scholar]
- [74].Vinchi F, Gastaldi S, Silengo L, Altruda F, Tolosano E, Hemopexin Prevents Endothelial Damage and Liver Congestion in a Mouse Model of Heme Overload, The American Journal of Pathology, 173 (2008) 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hahl P, Davis T, Washburn C, Rogers JT, Smith A, Mechanisms of neuroprotection by hemopexin: modeling the control of heme and iron homeostasis in brain neurons in inflammatory states, Journal of Neurochemistry, 125 (2013) 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mehta NU, Reddy ST, Role of hemoglobin/heme scavenger protein hemopexin in atherosclerosis and inflammatory diseases, Current Opinion in Lipidology, 26 (2015) 384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hwang PK, Greer J, Interaction between hemoglobin subunits in the hemoglobin. haptoglobin complex, J Biol Chem, 255 (1980) 3038–3041. [PubMed] [Google Scholar]
- [78].Wejman JC, Hovsepian D, Wall JS, Hainfeld JF, Greer J, Structure and assembly of haptoglobin polymers by electron microscopy, 174 (1984) 343–368. [DOI] [PubMed] [Google Scholar]
- [79].Okazakia T, Yanagisawa Y, Nagai T, Analysis of the affinity of each haptoglobin polymer for hemoglobin by two-dimensional affinity electrophoresis, Clinica Chimica Acta, 258 (1997) 137–144. [DOI] [PubMed] [Google Scholar]
- [80].Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman H-J, Law SKA, Moestrup SK, Identification of the haemoglobin scavenger receptor, Nature, 409 (2001) 198–201. [DOI] [PubMed] [Google Scholar]
- [81].Graversen JH, Madsen M, Moestrup SK, CD163: a signal receptor scavenging haptoglobin–hemoglobin complexes from plasma, The International Journal of Biochemistry & Cell Biology, 34 (2002) 309–314. [DOI] [PubMed] [Google Scholar]
- [82].Hrkal Z, Vodrazka Z, Kalousek I, Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin, Eur J Biochem, 43 (1974) 73–78. [DOI] [PubMed] [Google Scholar]
- [83].Hargrove MS, Barrick D, Olson JS, The association rate constant for heme binding to globin is independent of protein structure, Biochemistry, 35 (1996) 11293–11299. [DOI] [PubMed] [Google Scholar]
- [84].Kamal JK, Behere DV, Binding of heme to human serum albumin: steady-state fluorescence, circular dichroism and optical difference spectroscopic studies, Indian J Biochem Biophys, 42 (2005) 7–12. [PubMed] [Google Scholar]
- [85].Amoorahim M, Ashrafi-Kooshk MR, Esmaeili S, Shahlaei M, Moradi S, Khodarahmi R, Physiological changes in the albumin-bound non-esterified free fatty acids critically influence heme/bilirubin binding properties of the protein: A comparative, in vitro, spectroscopic study using the endogenous biomolecules, Spectrochimica Acta Part A: Molecular Spectroscopy, 235 (2020) 118298. [DOI] [PubMed] [Google Scholar]
- [86].Akerström B, Lögdberg L, Berggård T, Osmark P, Lindqvist A, alpha(1)-Microglobulin: a yellow-brown lipocalin, Biochim Biophys Acta, 1482 (2000) 172–184. [DOI] [PubMed] [Google Scholar]
- [87].Karnaukhova E, Rutardottir S, Rajabi M, Wester Rosenlöf L, Alayash AI, Åkerström B, Characterization of heme binding to recombinant α1-microglobulin, Front Physiol, 5 (2014) 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zager RA, Alpha 1 Microglobulin: A Potentially Paradoxical Anti-Oxidant Agent, Adv Tech Biol Med, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Miller YI, Shaklai N, Kinetics of hemin distribution in plasma reveals its role in lipoprotein oxidation, Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1454 (1999) 153–164. [DOI] [PubMed] [Google Scholar]
- [90].Zunszain PA, Ghuman J, Komatsu T, Tsuchida E, Curry S, Crystal structural analysis of human serum albumin complexed with hemin and fatty acid, BMC Struct Biol, 3 (2003) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Jeney VR, Balla GR, Balla JZ, Red blood cell, hemoglobin and heme in the progression of atherosclerosis, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bratosin D, Mazurier J, Tissier JP, Estaquier J, Huart JJ, Ameisen JC, Aminoff D, Montreuil J, Cellular and molecular mechanisms of senescent erythrocyte phagocytosis by macrophages. A review, Biochimie, 80 (1998) 173–195. [DOI] [PubMed] [Google Scholar]
- [93].Knutson M, Wessling-Resnick M, Iron Metabolism in the Reticuloendothelial System, Critical Reviews in Biochemistry and Molecular Biology, 38 (2003) 61–88. [DOI] [PubMed] [Google Scholar]
- [94].Ganz T, Macrophages and Systemic Iron Homeostasis, Journal of Innate Immunity, 4 (2012) 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Klei TRL, Meinderts SM, Van Den Berg TK, Van Bruggen R, From the Cradle to the Grave: The Role of Macrophages in Erythropoiesis and Erythrophagocytosis, Frontiers in Immunology, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M, Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages, Blood, 102 (2003) 4191–4197. [DOI] [PubMed] [Google Scholar]
- [97].Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M, Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin, Proceedings of the National Academy of Sciences, 102 (2005) 1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Delaby C, Pilard N, Hetet G, Driss F, Grandchamp B, Beaumont C, Canonne-Hergaux F, A physiological model to study iron recycling in macrophages, Exp Cell Res, 310 (2005) 43–53. [DOI] [PubMed] [Google Scholar]
- [99].Winter WE, Bazydlo LAL, Harris NS, The Molecular Biology of Human Iron Metabolism, Laboratory Medicine, 45 (2014) 92–102. [DOI] [PubMed] [Google Scholar]
- [100].Korolnek T, Hamza I, Macrophages and iron trafficking at the birth and death of red cells, Blood, 125 (2015) 2893–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Knutson MD, Iron transport proteins: Gateways of cellular and systemic iron homeostasis, J Biol Chem, 292 (2017) 12735–12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].D’Alessandro A, Dzieciatkowska M, Nemkov T, Hansen KC, Red blood cell proteomics update: is there more to discover?, Blood Transfus, 15 (2017) 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yoshida T, Sato M, Posttranslational and direct integration of heme oxygenase into microsomes, Biochem Biophys Res Commun, 163 (1989) 1086–1092. [DOI] [PubMed] [Google Scholar]
- [104].Yanatori I, Tabuchi M, Kawai Y, Yasui Y, Akagi R, Kishi F, Heme and non-heme iron transporters in non-polarized and polarized cells, BMC Cell Biology, 11 (2010) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gottlieb Y, Truman M, Cohen LA, Leichtmann-Bardoogo Y, Meyron-Holtz EG, Endoplasmic reticulum anchored heme-oxygenase 1 faces the cytosol, Haematologica, 97 (2012) 1489–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ponka P, Cell biology of heme, Am J Med Sci, 318 (1999) 241–256. [DOI] [PubMed] [Google Scholar]
- [107].White C, Yuan X, Paul E Bresciani, Tamika, D. Campagna, C. Hall, K. Bishop, Monica, A. Lapierre, Diane, P. Liu, Mark, I. Hamza, HRG1 Is Essential for Heme Transport from the Phagolysosome of Macrophages during Erythrophagocytosis, Cell Metabolism, 17 (2013) 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Rao AU, Carta LK, Lesuisse E, Hamza I, Lack of heme synthesis in a free-living eukaryote, Proceedings of the National Academy of Sciences, 102 (2005) 4270–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chung J, Chen C, Paw BH, Heme Metabolism and Erythropoiesis, Current opinion in hematology, 19 (2012) 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Sinclair J, Hamza I, Lessons from bloodless worms: heme homeostasis in C. elegans, Biometals, 28 (2015) 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chiabrando D, Bertino F, Tolosano E, Hereditary Ataxia: A Focus on Heme Metabolism and Fe-S Cluster Biogenesis, Int J Mol Sci, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].O’Callaghan KM, Ayllon V, O’Keeffe J, Wang Y, Cox OT, Loughran G, Forgac M, O’Connor R, Heme-binding protein HRG-1 is induced by insulin-like growth factor I and associates with the vacuolar H+-ATPase to control endosomal pH and receptor trafficking, J Biol Chem, 285 (2010) 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yuan X, Protchenko O, Philpott CC, Hamza I, Topologically Conserved Residues Direct Heme Transport in HRG-1-related Proteins, Journal of Biological Chemistry, 287 (2012) 4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhang J, Chambers I, Yun S, Phillips J, Krause M, Hamza I, Hrg1 promotes heme-iron recycling during hemolysis in the zebrafish kidney, PLOS Genetics, 14 (2018) e1007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pek RH, Yuan X, Rietzschel N, Zhang J, Jackson L, Nishibori E, Ribeiro A, Simmons W, Jagadeesh J, Sugimoto H, Alam MZ, Garrett L, Haldar M, Ralle M, Phillips JD, Bodine DM, Hamza I, Hemozoin produced by mammals confers heme tolerance, eLife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhang J, Hamza I, Zebrafish as a model system to delineate the role of heme and iron metabolism during erythropoiesis, Molecular Genetics and Metabolism, 128 (2019) 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Coronado LM, Nadovich CT, Spadafora C, Malarial hemozoin: From target to tool, Biochimica et Biophysica Acta (BBA) - General Subjects, 1840 (2014) 2032–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Abkowitz JL, Holly RD, Grant CK, Retrovirus-induced feline pure red cell aplasia. Hematopoietic progenitors are infected with feline leukemia virus and erythroid burst-forming cells are uniquely sensitive to heterologous complement, Journal of Clinical Investigation, 80 (1987) 1056–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL, Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia, Blood, 95 (2000) 1093–1099. [PubMed] [Google Scholar]
- [120].Brown JK, Fung C, Tailor CS, Comprehensive Mapping of Receptor-Functioning Domains in Feline Leukemia Virus Subgroup C Receptor FLVCR1, Journal of Virology, 80 (2006) 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Meyer E, Ricketts C, Morgan NV, Morris MR, Pasha S, Tee LJ, Rahman F, Bazin A, Bessières B, Déchelotte P, Yacoubi MT, Al-Adnani M, Marton T, Tannahill D, Trembath RC, Fallet-Bianco C, Cox P, Williams D, Maher ER, Mutations in FLVCR2 Are Associated with Proliferative Vasculopathy and Hydranencephaly-Hydrocephaly Syndrome (Fowler Syndrome), The American Journal of Human Genetics, 86 (2010) 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Korolnek T, Zhang J, Beardsley S, Scheffer GL, Hamza I, Control of metazoan heme homeostasis by a conserved multidrug resistance protein, Cell Metab, 19 (2014) 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kool M, de Haas M, Scheffer GL, Scheper RJ, van Eijk MJ, Juijn JA, Baas F, Borst P, Analysis of expression of cMOAT (MRP2), MRP3, MRP4, and MRP5, homologues of the multidrug resistance-associated protein gene (MRP1), in human cancer cell lines, Cancer Res, 57 (1997) 3537–3547. [PubMed] [Google Scholar]
- [124].McAleer MA, Breen MA, White NL, Matthews N, pABC11 (Also Known as MOAT-C and MRP5), a Member of the ABC Family of Proteins, Has Anion Transporter Activity but Does Not Confer Multidrug Resistance When Overexpressed in Human Embryonic Kidney 293 Cells, Journal of Biological Chemistry, 274 (1999) 23541–23548. [DOI] [PubMed] [Google Scholar]
- [125].Borst P, Evers R, Kool M, Wijnholds J, A family of drug transporters: the multidrug resistance-associated proteins, J Natl Cancer Inst, 92 (2000) 1295–1302. [DOI] [PubMed] [Google Scholar]
- [126].Slot AJ, Molinski SV, Cole SP, Mammalian multidrug-resistance proteins (MRPs), Essays Biochem, 50 (2011) 179–207. [DOI] [PubMed] [Google Scholar]
- [127].Deeley RG, Westlake C, Cole SP, Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins, Physiol Rev, 86 (2006) 849–899. [DOI] [PubMed] [Google Scholar]
- [128].Ritter CA, Jedlitschky G, Meyer zu Schwabedissen H, Grube M, Kock K, Kroemer HK, Cellular export of drugs and signaling molecules by the ATP-binding cassette transporters MRP4 (ABCC4) and MRP5 (ABCC5), Drug Metab Rev, 37 (2005) 253–278. [DOI] [PubMed] [Google Scholar]
- [129].Jedlitschky G, Burchell B, Keppler D, The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides, J Biol Chem, 275 (2000) 30069–30074. [DOI] [PubMed] [Google Scholar]
- [130].Jansen RS, Mahakena S, de Haas M, Borst P, van de Wetering K, ATP-binding Cassette Subfamily C Member 5 (ABCC5) Functions as an Efflux Transporter of Glutamate Conjugates and Analogs, J Biol Chem, 290 (2015) 30429–30440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Reid G, Wielinga P, Zelcer N, de Haas M, van Deemter L, Wijnholds J, Balzarini J, Borst P, Characterization of the Transport of Nucleoside Analog Drugs by the Human Multidrug Resistance Proteins MRP4 and MRP5, Molecular Pharmacology, 63 (2003) 1094–1103. [DOI] [PubMed] [Google Scholar]
- [132].Yuan X, Rietzschel N, Kwon H, Walter Nuno AB, Hanna DA, Phillips JD, Raven EL, Reddi AR, Hamza I, Regulation of intracellular heme trafficking revealed by subcellular reporters, Proc Natl Acad Sci U S A, 113 (2016) E5144–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Jedlitschky G, Burchell B, Keppler D, The Multidrug Resistance Protein 5 Functions as an ATP-dependent Export Pump for Cyclic Nucleotides, Journal of Biological Chemistry, 275 (2000) 30069–30074. [DOI] [PubMed] [Google Scholar]
- [134].Donegan RK, Moore CM, Hanna DA, Reddi AR, Handling heme: The mechanisms underlying the movement of heme within and between cells, Free Radical Biology and Medicine, 133 (2019) 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Austin Doyle L, Ross DD, Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2), Oncogene, 22 (2003) 7340–7358. [DOI] [PubMed] [Google Scholar]
- [136].Szakács G, Váradi A, Özvegy-Laczka C, Sarkadi B, The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME–Tox), Drug Discovery Today, 13 (2008) 379–393. [DOI] [PubMed] [Google Scholar]
- [137].Robey RW, To KK, Polgar O, Dohse M, Fetsch P, Dean M, Bates SE, ABCG2: a perspective, Adv Drug Deliv Rev, 61 (2009) 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Khunweeraphong N, Szöllősi D, Stockner T, Kuchler K, The ABCG2 multidrug transporter is a pump gated by a valve and an extracellular lid, Nature Communications, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Suzuki M, Suzuki H, Sugimoto Y, Sugiyama Y, ABCG2 Transports Sulfated Conjugates of Steroids andXenobiotics, Journal of Biological Chemistry, 278 (2003) 22644–22649. [DOI] [PubMed] [Google Scholar]
- [140].Takenaka K, Morgan JA, Scheffer GL, Adachi M, Stewart CF, Sun D, Leggas M, Ejendal KFK, Hrycyna CA, Schuetz JD, Substrate Overlap between Mrp4 and Abcg2/Bcrp Affects Purine Analogue Drug Cytotoxicity and Tissue Distribution, Cancer Research, 67 (2007) 6965–6972. [DOI] [PubMed] [Google Scholar]
- [141].De Wolf C, Jansen R, Yamaguchi H, De Haas M, Van De Wetering K, Wijnholds J, Beijnen J, Borst P,Contribution of the drug transporter ABCG2 (breast cancer resistance protein) to resistance against anticancer nucleosides, Molecular Cancer Therapeutics, 7 (2008) 3092–3102. [DOI] [PubMed] [Google Scholar]
- [142].Nagai S, Takenaka K, Nachagari D, Rose C, Domoney K, Sun D, Sparreboom A, Schuetz JD, DeoxycytidineKinase Modulates the Impact of the ABC Transporter ABCG2 on Clofarabine Cytotoxicity, 71 (2011) 1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Sodani K, Patel A, Kathawala RJ, Chen Z-S, Multidrug resistance associated proteins in multidrug resistance, Chin J Cancer, 31 (2012) 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Fukuda Y, Schuetz JD, ABC transporters and their role in nucleoside and nucleotide drug resistance,Biochem Pharmacol, 83 (2012) 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Krishnamurthy P, Ross DD, Nakanishi T, Bailey-Dell K, Zhou S, Mercer KE, Sarkadi B, Sorrentino BP, Schuetz JD, The Stem Cell Marker Bcrp/ABCG2 Enhances Hypoxic Cell Survival through Interactions with Heme, Journal of Biological Chemistry, 279 (2004) 24218–24225. [DOI] [PubMed] [Google Scholar]
- [146].Zhou S, Zong Y, Ney PA, Nair G, Stewart CF, Sorrentino BP, Increased expression of the Abcg2 transporter during erythroid maturation plays a role in decreasing cellular protoporphyrin IX levels, Blood, 105 (2005) 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Krishnamurthy P, Schuetz JD, ROLE OF ABCG2/ BCRP IN BIOLOGY AND MEDICINE, 46 (2006) 381–410. [DOI] [PubMed] [Google Scholar]
- [148].Desuzinges-Mandon E, Arnaud O, Martinez L, Huché F, Di Pietro A, Falson P, ABCG2 Transports and Transfers Heme to Albumin through Its Large Extracellular Loop, Journal of Biological Chemistry, 285 (2010) 33123–33133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Saison C, Helias V, Ballif BA, Peyrard T, Puy H, Miyazaki T, Perrot S, Vayssier-Taussat M, Waldner M, Le Pennec P-Y, Cartron J-P, Arnaud L, Null alleles of ABCG2 encoding the breast cancer resistance protein define the new blood group system Junior, Nature Genetics, 44 (2012) 174–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Kitajima Y, Ishii T, Kohda T, Ishizuka M, Yamazaki K, Nishimura Y, Tanaka T, Dan S, Nakajima M, Mechanistic study of PpIX accumulation using the JFCR39 cell panel revealed a role for dynamin 2-mediated exocytosis, Scientific Reports, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL, A heme export protein is required for red blood cell differentiation and iron homeostasis, Science, 319 (2008) 825–828. [DOI] [PubMed] [Google Scholar]
- [152].Mercurio S, Petrillo S, Chiabrando D, Bassi ZI, Gays D, Camporeale A, Vacaru A, Miniscalco B, Valperga G, Silengo L, Altruda F, Baron MH, Santoro MM, Tolosano E, The heme exporter Flvcr1 regulates expansion and differentiation of committed erythroid progenitors by controlling intracellular heme accumulation, Haematologica, 100 (2015) 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Khan AA, Quigley JG, Heme and FLVCR-related transporter families SLC48 and SLC49, Molecular Aspects of Medicine, 34 (2013) 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Kafina MD, Paw BH, Intracellular iron and heme trafficking and metabolism in developing erythroblasts, Metallomics, 9 (2017) 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Yang Z, Philips JD, Doty RT, Giraudi P, Ostrow JD, Tiribelli C, Smith A, Abkowitz JL, Kinetics and Specificity of Feline Leukemia Virus Subgroup C Receptor (FLVCR) Export Function and Its Dependence on Hemopexin, Journal of Biological Chemistry, 285 (2010) 28874–28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, Phillips JD, Dailey HA, Identification of the Mitochondrial Heme Metabolism Complex, PLoS One, 10 (2015) e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Prenni JE, Vidal M, Olver CS, Preliminary characterization of the murine membrane reticulocyte proteome, Blood Cells, Molecules, and Diseases, 49 (2012) 74–82. [DOI] [PubMed] [Google Scholar]
- [158].Pesciotta EN, Lam H-S, Kossenkov A, Ge J, Showe LC, Mason PJ, Bessler M, Speicher DW, In-Depth, Label-Free Analysis of the Erythrocyte Cytoplasmic Proteome in Diamond Blackfan Anemia Identifies a Unique Inflammatory Signature, PLOS ONE, 10 (2015) e0140036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Bryk AH, Wiśniewski JR, Quantitative Analysis of Human Red Blood Cell Proteome, Journal of Proteome Research, 16 (2017) 2752–2761. [DOI] [PubMed] [Google Scholar]
- [160].Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, Mercer KE, Wang J, Sosa-Pineda B, Murti KG,Schuetz JD, Identification of a mammalian mitochondrial porphyrin transporter, Nature, 443 (2006) 586–589. [DOI] [PubMed] [Google Scholar]
- [161].Paterson JK, Shukla S, Black CM, Tachiwada T, Garfield S, Wincovitch S, Ernst DN, Agadir A, Li X, Ambudkar SV, Szakacs G, Akiyama S-I, Gottesman MM, Human ABCB6 Localizes to Both the Outer Mitochondrial Membrane and the Plasma Membrane†, Biochemistry, 46 (2007) 9443–9452. [DOI] [PubMed] [Google Scholar]
- [162].Rakvács Z, Kucsma N, Gera M, Igriczi B, Kiss K, Barna J, Kovács D, Vellai T, Bencs L, Reisecker JM, Szoboszlai N, Szakács G, The human ABCB6 protein is the functional homologue of HMT-1 proteins mediating cadmium detoxification, Cellular and Molecular Life Sciences, 76 (2019) 4131–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Moras M, Lefevre SD, Ostuni MA, From Erythroblasts to Mature Red Blood Cells: Organelle Clearance inMammals, Frontiers in Physiology, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Yang Z, Keel SB, Shimamura A, Liu L, Gerds AT, Li HY, Wood BL, Scott BL, Abkowitz JL, Delayed globin synthesis leads to excess heme and the macrocytic anemia of Diamond Blackfan anemia and del(5q) myelodysplastic syndrome, Sci Transl Med, 8 (2016) 338ra367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Chen C, Tamika J Sinclair Harry, Hamza I, An Intercellular Heme-Trafficking Protein Delivers Maternal Heme to the Embryo during Development in C. elegans, Cell, 145 (2011) 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Marciano O, Moskovitz Y, Hamza I, Ruthstein S, Histidine residues are important for preserving the structure and heme binding to the C. elegans HRG-3 heme-trafficking protein, J Biol Inorg Chem, 20 (2015) 1253–1261. [DOI] [PubMed] [Google Scholar]
- [167].Chen C, Samuel TK, Krause M, Dailey HA, Hamza I, Heme Utilization in theCaenorhabditis elegansHypodermal Cells Is Facilitated by Heme-responsive Gene-2, Journal of Biological Chemistry, 287 (2012) 9601–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Zhou JR, Bu DR, Zhao XF, Wu F, Chen XQ, Shi HZ, Yao CQ, Du AF, Yang Y, Hc-hrg-2, a glutathione transferase gene, regulates heme homeostasis in the blood-feeding parasitic nematode Haemonchus contortus, Parasit Vectors, 13 (2020) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J, Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization, Science, 306 (2004) 2090–2093. [DOI] [PubMed] [Google Scholar]
- [170].Kim B-E, Turski ML, Nose Y, Casad M, Rockman HA, Thiele DJ, Cardiac Copper Deficiency Activates a Systemic Signaling Mechanism that Communicates with the Copper Acquisition and Storage Organs, Cell Metabolism, 11 (2010) 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Sinclair J, Pinter K, Samuel T, Beardsley S, Yuan X, Zhang J, Meng K, Yun S, Krause M, Hamza I, Inter-organ signalling by HRG-7 promotes systemic haem homeostasis, Nature Cell Biology, 19 (2017) 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Mercurio S, Petrillo S, Chiabrando D, Bassi ZI, Gays D, Camporeale A, Vacaru A, Miniscalco B, Valperga G, Silengo L, Altruda F, Baron MH, Santoro MM, Tolosano E, The heme exporter Flvcr1 regulates expansion and differentiation of committed erythroid progenitors by controlling intracellular heme accumulation, Haematologica, 100 (2015) 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Schultz IJ, Chen C, Paw BH, Hamza I, Iron and porphyrin trafficking in heme biogenesis, J Biol Chem, 285 (2010) 26753–26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Yuan X, Fleming MD, Hamza I, Heme transport and erythropoiesis, Current Opinion in Chemical Biology, 17 (2013) 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Hanna DA, Harvey RM, Martinez-Guzman O, Yuan X, Chandrasekharan B, Raju G, Outten FW, Hamza I, Reddi AR, Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors, Proceedings of the National Academy of Sciences, 113 (2016) 7539–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Atamna H, Brahmbhatt M, Atamna W, Shanower GA, Dhahbi JM, ApoHRP-based assay to measure intracellular regulatory heme, Metallomics, 7 (2015) 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Song Y, Yang M, Wegner SV, Zhao J, Zhu R, Wu Y, He C, Chen PR, A Genetically Encoded FRET Sensor for Intracellular Heme, ACS Chemical Biology, 10 (2015) 1610–1615. [DOI] [PubMed] [Google Scholar]
- [178].Ketley JN, Habig WH, Jakoby WB, Binding of nonsubstrate ligands to the glutathione S-transferases, Journal of Biological Chemistry, 250 (1975) 8670–8673. [PubMed] [Google Scholar]
- [179].Senjo M, Ishibashi T, Imai Y, Purification and characterization of cytosolic liver protein facilitating heme transport into apocytochrome b5 from mitochondria. Evidence for identifying the heme transfer protein as belonging to a group of glutathione S-transferases, J Biol Chem, 260 (1985) 9191–9196. [PubMed] [Google Scholar]
- [180].Perally S, Lacourse EJ, Campbell AM, Brophy PM, Heme transport and detoxification in nematodes: subproteomics evidence of differential role of glutathione transferases, J Proteome Res, 7 (2008) 4557–4565. [DOI] [PubMed] [Google Scholar]
- [181].Townsend DM, Tew KD, The role of glutathione-S-transferase in anti-cancer drug resistance, Oncogene, 22 (2003) 7369–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Harwaldt P, Rahlfs S, Becker K, Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target, Biol Chem, 383 (2002) 821–830. [DOI] [PubMed] [Google Scholar]
- [183].Van Rossum AJ, Jefferies JR, Rijsewijk FAM, Lacourse EJ, Teesdale-Spittle P, Barrett J, Tait A, Brophy PM, Binding of Hematin by a New Class of Glutathione Transferase from the Blood-Feeding Parasitic Nematode Haemonchus contortus, Infection and Immunity, 72 (2004) 2780–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Zhan B, Liu S, Perally S, Xue J, Fujiwara R, Brophy P, Xiao S, Liu Y, Feng J, Williamson A, Wang Y, Bueno LL, Mendez S, Goud G, Bethony JM, Hawdon JM, Loukas A, Jones K, Hotez PJ, Biochemical Characterization and Vaccine Potential of a Heme-Binding Glutathione Transferase from the Adult Hookworm Ancylostoma caninum, Infection and Immunity, 73 (2005) 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Vincent SH, Muller-Eberhard U, A protein of the Z class of liver cytosolic proteins in the rat that preferentially binds heme, J Biol Chem, 260 (1985) 14521–14528. [PubMed] [Google Scholar]
- [186].Stewart JM, Slysz GW, Pritting MA, Muller-Eberhard U, Ferriheme and ferroheme are isosteric inhibitors of fatty acid binding to rat liver fatty acid binding protein, Biochemistry and Cell Biology, 74 (1996) 249–255. [DOI] [PubMed] [Google Scholar]
- [187].Iwahara S, Satoh H, Song DX, Webb J, Burlingame AL, Nagae Y, Muller-Eberhard U, Purification, characterization, and cloning of a heme-binding protein (23 kDa) in rat liver cytosol, Biochemistry, 34 (1995) 13398–13406. [DOI] [PubMed] [Google Scholar]
- [188].Jacob Blackmon B, Dailey TA, Lianchun X, Dailey HA, Characterization of a human and mouse tetrapyrrole-binding protein, Archives of Biochemistry and Biophysics, 407 (2002) 196–201. [DOI] [PubMed] [Google Scholar]
- [189].Taketani S, Adachi Y, Kohno H, Ikehara S, Tokunaga R, Ishii T, Molecular Characterization of a Newly Identified Heme-binding Protein Induced during Differentiation of urine Erythroleukemia Cells, Journal of Biological Chemistry, 273 (1998) 31388–31394. [DOI] [PubMed] [Google Scholar]
- [190].Immenschuh S, Nell C, Iwahara S, Katz N, Muller-Eberhard U, Gene regulation of HBP 23 by metalloporphyrins and protoporphyrin IX in liver and hepatocyte cultures, Biochem Biophys Res Commun, 231 (1997) 667–670. [DOI] [PubMed] [Google Scholar]
- [191].Micaelo NM, Macedo AL, Goodfellow BJ, Félix V, Tetrapyrrole binding affinity of the murine and human p22HBP heme-binding proteins, J Mol Graph Model, 29 (2010) 396–405. [DOI] [PubMed] [Google Scholar]
- [192].Barber RD, Harmer DW, Coleman RA, Clark BJ, GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues, Physiological Genomics, 21 (2005) 389–395. [DOI] [PubMed] [Google Scholar]
- [193].Chakravarti R, Aulak KS, Fox PL, Stuehr DJ, GAPDH regulates cellular heme insertion into inducible nitric oxide synthase, Proceedings of the National Academy of Sciences, 107 (2010) 18004–18009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Eisenberg E, Levanon EY, Human housekeeping genes, revisited, Trends in Genetics, 29 (2013) 569–574. [DOI] [PubMed] [Google Scholar]
- [195].Boradia VM, Raje M, Raje CI, Protein moonlighting in iron metabolism: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Biochem Soc Trans, 42 (2014) 1796–1801. [DOI] [PubMed] [Google Scholar]
- [196].Dai Y, Sweeny EA, Schlanger S, Ghosh A, Stuehr DJ, GAPDH delivers heme to soluble guanylyl cyclase, Journal of Biological Chemistry, 295 (2020) 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Hannibal L, Collins D, Brassard J, Chakravarti R, Vempati R, Dorlet P, Santolini J, Dawson JH, Stuehr DJ, Heme Binding Properties of Glyceraldehyde-3-phosphate Dehydrogenase, Biochemistry, 51 (2012) 8514–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Sweeny EA, Singh AB, Chakravarti R, Martinez-Guzman O, Saini A, Haque MM, Garee G, Dans PD, Hannibal L, Reddi AR, Stuehr DJ, Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells, Journal of Biological Chemistry, 293 (2018) 14557–14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Hannibal L, Collins D, Brassard J, Chakravarti R, Vempati R, Dorlet P, Santolini J, Dawson JH, Stuehr DJ, Heme binding properties of glyceraldehyde-3-phosphate dehydrogenase, Biochemistry, 51 (2012) 8514–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Hughes AL, Powell DW, Bard M, Eckstein J, Barbuch R, Link AJ, Espenshade PJ, Dap1/PGRMC1 Binds and Regulates Cytochrome P450 Enzymes, Cell Metabolism, 5 (2007) 143–149. [DOI] [PubMed] [Google Scholar]
- [201].Thompson AM, Reddi AR, Shi X, Goldbeck RA, Moënne-Loccoz P, Gibney BR, Holman TR, Measurement of the Heme Affinity for Yeast Dap1p, and Its Importance in Cellular Function†, Biochemistry, 46 (2007) 14629–14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Piel RB, Shiferaw MT, Vashisht AA, Marcero JR, Praissman JL, Phillips JD, Wohlschlegel JA, Medlock AE, A Novel Role for Progesterone Receptor Membrane Component 1 (PGRMC1): A Partner and Regulator of Ferrochelatase, Biochemistry, 55 (2016) 5204–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Galmozzi A, Kok BP, Kim AS, Montenegro-Burke JR, Lee JY, Spreafico R, Mosure S, Albert V, Cintron-Colon R, Godio C, Webb WR, Conti B, Solt LA, Kojetin D, Parker CG, Peluso JJ, Pru JK, Siuzdak G, Cravatt BF, Saez E, PGRMC2 is an intracellular haem chaperone critical for adipocyte function, Nature, 576 (2019) 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Chen AJ, Yuan X, Li J, Dong P, Hamza I, Cheng J-X, Label-Free Imaging of Heme Dynamics in Living Organisms by Transient Absorption Microscopy, Analytical Chemistry, 90 (2018) 3395–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Zhang C, Rabouille C, Membrane-Bound Meet Membraneless in Health and Disease, Cells, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Chong PA, Forman-Kay JD, Liquid–liquid phase separation in cellular signaling systems, Current Opinion in Structural Biology, 41 (2016) 180–186. [DOI] [PubMed] [Google Scholar]
- [207].Schrader M, Islinger M, Editorial: Molecular Mechanisms and Physiological Significance of Organelle Interactions and Cooperation, Front Cell Dev Biol, 4 (2016) 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].van Vliet AR, Verfaillie T, Agostinis P, New functions of mitochondria associated membranes in cellular signaling, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1843 (2014) 2253–2262. [DOI] [PubMed] [Google Scholar]
- [209].Sano R, Annunziata I, Patterson A, Moshiach S, Gomero E, Opferman J, Forte M, d’Azzo A, GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca(2+)-dependent mitochondrial apoptosis, Mol Cell, 36 (2009) 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Pinton P, Mitochondria-associated membranes (MAMs) and pathologies, Cell Death Dis, 9 (2018) 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Vance JE, MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond, Biochim Biophys Acta, 1841 (2014) 595–609. [DOI] [PubMed] [Google Scholar]
- [212].Vance JE, Phospholipid synthesis in a membrane fraction associated with mitochondria, J Biol Chem, 265 (1990) 7248–7256. [PubMed] [Google Scholar]
- [213].Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M, A Tether Is a Tether Is a Tether: Tethering at Membrane Contact Sites, Dev Cell, 39 (2016) 395–409. [DOI] [PubMed] [Google Scholar]
- [214].van der Veen JN, Kennelly JP, Wan S, Vance JE, Vance DE, Jacobs RL, The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease, Biochimica et Biophysica Acta (BBA) - Biomembranes, 1859 (2017) 1558–1572. [DOI] [PubMed] [Google Scholar]
- [215].Pinto MCX, Kihara AH, Goulart VAM, Tonelli FMP, Gomes KN, Ulrich H, Resende RR, Calcium signaling and cell proliferation, Cellular Signalling, 27 (2015) 2139–2149. [DOI] [PubMed] [Google Scholar]
- [216].Helle SCJ, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B, Organization and function of membrane contact sites, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1833 (2013) 2526–2541. [DOI] [PubMed] [Google Scholar]
- [217].van Meer G, Voelker DR, Feigenson GW, Membrane lipids: where they are and how they behave, Nature Reviews Molecular Cell Biology, 9 (2008) 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Wakai T, Vanderheyden V, Fissore RA, Ca2+ signaling during mammalian fertilization: requirements, players, and adaptations, Cold Spring Harb Perspect Biol, 3 (2011) a006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Szent-Györgyi AG, Calcium regulation of muscle contraction, Biophys J, 15 (1975) 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].de la Fuente S, Fonteriz RI, Montero M, Alvarez J, Ca2+ homeostasis in the endoplasmic reticulum measured with a new low-Ca2+-affinity targeted aequorin, Cell Calcium, 54 (2013) 37–45. [DOI] [PubMed] [Google Scholar]
- [221].Desai R, East DA, Hardy L, Crosby J, Rigon M, Faccenda D, Alvarez MS, Singh A, Mainenti M, Hussey LK, Bentham R, Szabadkai G, Zappulli V, Dhoot G, Romano LE, Dong X, Coppens I, Hamacher-Brady A, Chapple JP, Abeti R, Fleck RA, Vizcay-Barrena G, Smith K, Campanella M, Mitochondria form contact sites with the nucleus to couple pro-survival retrograde response, bioRxiv, (2020) 445411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M, A dynamic interface between vacuoles and mitochondria in yeast, Dev Cell, 30 (2014) 95–102. [DOI] [PubMed] [Google Scholar]
- [223].Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J, ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast, Elife, 2 (2013) e00422.23682313 [Google Scholar]
- [224].Martinez-Guzman O, Willoughby MM, Saini A, Dietz JV, Bohovych I, Medlock AE, Khalimonchuk O, Reddi AR, Mitochondrial–nuclear heme trafficking in budding yeast is regulated by GTPases that control mitochondrial dynamics and ER contact sites, Journal of Cell Science, 133 (2020) jcs237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Rebbeck RT, Karunasekara Y, Gallant EM, Board PG, Beard NA, Casarotto MG, Dulhunty AF, The β(1a) subunit of the skeletal DHPR binds to skeletal RyR1 and activates the channel via its 35-residue C-terminal tail, Biophys J, 100 (2011) 922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Soboloff J, Rothberg BS, Madesh M, Gill DL, STIM proteins: dynamic calcium signal transducers, NatureReviews Molecular Cell Biology, 13 (2012) 549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P, PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts, Science, 349 (2015) 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Murphy SE, Levine TP, VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis ofFFAT-like motifs in the VAPome, Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 1861 (2016) 952–961. [DOI] [PubMed] [Google Scholar]
- [229].Manford Andrew G., Stefan Christopher J., Yuan Helen L., Jason A. MacGurn, Scott D. Emr, ER-to-PlasmaMembrane Tethering Proteins Regulate Cell Signaling and ER Morphology, Developmental Cell, 23 (2012) 1129–1140. [DOI] [PubMed] [Google Scholar]
- [230].Toulmay A, Prinz WA, Lipid transfer and signaling at organelle contact sites: the tip of the iceberg, Curr Opin Cell Biol, 23 (2011) 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Gatta AT, Wong LH, Sere YY, Calderón-Noreña DM, Cockcroft S, Menon AK, Levine TP, A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport, eLife, 4 (2015) e07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Levine TP, Munro S, Dual Targeting of Osh1p, a Yeast Homologue of Oxysterol-binding Protein, to both the Golgi and the Nucleus-Vacuole Junction, Molecular Biology of the Cell, 12 (2001) 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Schuldiner M, Zalckvar E, Incredibly close-A newly identified peroxisome-ER contact site in humans, The Journal of cell biology, 216 (2017) 287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Knoblach B, Sun X, Coquelle N, Fagarasanu A, Poirier RL, Rachubinski RA, An ER-peroxisome tether exerts peroxisome population control in yeast, The EMBO Journal, 32 (2013) 2439–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].de Brito OM, Scorrano L, Mitofusin 2 tethers endoplasmic reticulum to mitochondria, Nature, 456 (2008) 605–610. [DOI] [PubMed] [Google Scholar]
- [236].Szabadkai G.r., Bianchi K, rnai P.t., De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T.s., Rizzuto R, Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels, Journal of Cell Biology, 175 (2006) 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Iwasawa R, Mahul-Mellier A-L, Datler C, Pazarentzos E, Grimm S, Fis1 and Bap31 bridge the mitochondria–ER interface to establish a platform for apoptosis induction, The EMBO Journal, 30 (2011) 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].De Vos KJ, Mórotz GM, Stoica R, Tudor EL, Lau K-F, Ackerley S, Warley A, Shaw CE, Miller CCJ, VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis, Human Molecular Genetics, 21 (2011) 1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, Loewen CJ, Prinz WA, A conservedendoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria, PLoS Biol, 12 (2014) e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P, An ER-mitochondriatethering complex revealed by a synthetic biology screen, Science, 325 (2009) 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Swayne TC, Zhou C, Boldogh IR, Charalel JK, McFaline-Figueroa JR, Thoms S, Yang C, Leung G, McInnes J, Erdmann R, Pon LA, Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast, Curr Biol, 21 (2011) 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Murley A, Sarsam RD, Toulmay A, Yamada J, Prinz WA, Nunnari J, Ltc1 is an ER-localized steroltransporter and a component of ER–mitochondria and ER–vacuole contacts, Journal of Cell Biology, 209 (2015) 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Elbaz-Alon Y, Eisenberg-Bord M, Shinder V, Sebastian B Stiller, E. Shimoni, N. Wiedemann, T. Geiger, M. Schuldiner, Lam6 Regulates the Extent of Contacts between Organelles, Cell Reports, 12 (2015) 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Fan J, Li X, Issop L, Culty M, Papadopoulos V, ACBD2/ECI2-Mediated Peroxisome-Mitochondria Interactions in Leydig Cell Steroid Biosynthesis, Molecular Endocrinology, 30 (2016) 763–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Mattiazzi Ušaj M, Brložnik M, Kaferle P, Žitnik M, Wolinski H, Leitner F, Kohlwein SD, Zupan B, Petrovič U, Genome-Wide Localization Study of Yeast Pex11 Identifies Peroxisome-Mitochondria Interactions through the ERMES Complex, Journal of molecular biology, 427 (2015) 2072–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Shai N, Schuldiner M, Zalckvar E, No peroxisome is an island — Peroxisome contact sites, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1863 (2016) 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Wong YC, Kim S, Peng W, Krainc D, Regulation and Function of Mitochondria–LysosomeMembrane Contact Sites in Cellular Homeostasis, Trends in Cell Biology, 29 (2019) 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, Maria V Schiaffino, Mitochondria andMelanosomes Establish Physical Contacts Modulated by Mfn2 and Involved in Organelle Biogenesis, Current Biology, 24 (2014) 393–403. [DOI] [PubMed] [Google Scholar]
- [249].Muñoz-Braceras S, Tornero-Écija AR, Vincent O, Escalante R, VPS13A is closely associated withmitochondria and is required for efficient lysosomal degradation, Dis Model Mech, 12 (2019) dmm036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C, Cellular Metabolism Regulates Contact Sites between Vacuoles and Mitochondria, Developmental Cell, 30 (2014) 86–94. [DOI] [PubMed] [Google Scholar]
- [251].Ping HA, Kraft LM, Chen W, Nilles AE, Lackner LL, Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities, The Journal of cell biology, 213 (2016) 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Lackner LL, The Expanding and Unexpected Functions of Mitochondria Contact Sites, Trends Cell Biol, 29 (2019) 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Youngman MJ, Hobbs AEA, Burgess SM, Srinivasan M, Jensen RE, Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids, The Journal of cell biology, 164 (2004) 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Jeong H, Park J, Jun Y, Lee C, Crystal structures of Mmm1 and Mdm12–Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites, Proceedings of the National Academy of Sciences, 114 (2017) E9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Kornmann B, Osman C, Walter P, The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections, Proc Natl Acad Sci U S A, 108 (2011) 14151–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Nguyen TT, Lewandowska A, Choi J-Y, Markgraf DF, Junker M, Bilgin M, Ejsing CS, Voelker DR, Rapoport TA, Shaw JM, Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance, Traffic, 13 (2012) 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA, ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in <em>S. cerevisiae</em>, Journal of Cell Science, 125 (2012) 4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Xu L, Wang X, Tong C, Endoplasmic Reticulum–Mitochondria Contact Sites and Neurodegeneration, Frontiers in Cell and Developmental Biology, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J, Applying systems-level spectral imaging and analysis to reveal the organelle interactome, Nature, 546 (2017) 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Wong YC, Ysselstein D, Krainc D, Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis, Nature, 554 (2018) 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Aston D, Capel RA, Ford KL, Christian HC, Mirams GR, Rog-Zielinska EA, Kohl P, Galione A, Burton RAB, Terrar DA, High resolution structural evidence suggests the Sarcoplasmic Reticulum forms microdomains with Acidic Stores (lysosomes) in the heart, Scientific Reports, 7 (2017) 40620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Fermie J, Liv N, ten Brink C, van Donselaar EG, Müller WH, Schieber NL, Schwab Y, Gerritsen HC, Klumperman J, Single organelle dynamics linked to 3D structure by correlative live-cell imaging and 3D electron microscopy, Traffic, 19 (2018) 354–369. [DOI] [PubMed] [Google Scholar]
- [263].Chen Q, Jin C, Shao X, Guan R, Tian Z, Wang C, Liu F, Ling P, Guan J-L, Ji L, Wang F, Chao H, Diao J, Super-Resolution Tracking of Mitochondrial Dynamics with An Iridium(III) Luminophore, Small, 14 (2018) 1802166. [DOI] [PubMed] [Google Scholar]
- [264].Lemasters JJ, Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging, Rejuvenation Res, 8 (2005) 3–5. [DOI] [PubMed] [Google Scholar]
- [265].Yamashita SI, Jin X, Furukawa K, Hamasaki M, Nezu A, Otera H, Saigusa T, Yoshimori T, Sakai Y, Mihara K, Kanki T, Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy, J Cell Biol, 215 (2016) 649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Biel TG, Rao VA, Mitochondrial dysfunction activates lysosomal-dependent mitophagy selectively in cancer cells, Oncotarget, 9 (2018) 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Rodger CE, McWilliams TG, Ganley IG, Mammalian mitophagy - from in vitro molecules to in vivo models, Febs j, 285 (2018) 1185–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Sugiura A, McLelland G-L, Fon EA, McBride HM, A new pathway for mitochondrial quality control: mitochondrial-derived vesicles, The EMBO Journal, 33 (2014) 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Kim-Shapiro DB, Lee J, Gladwin MT, Storage lesion: role of red blood cell breakdown, Transfusion, 51 (2011) 844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Gaggar A, Patel RP, There is blood in the water: hemolysis, hemoglobin, and heme in acute lung injury, American Journal of Physiology-Lung Cellular and Molecular Physiology, 311 (2016) L714–L718. [DOI] [PubMed] [Google Scholar]
- [271].Hoehn RS, Jernigan PL, Chang AL, Edwards MJ, Pritts TA, Molecular mechanisms of erythrocyte aging, Biological Chemistry, 396 (2015) 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Donadee C, Raat NJH, Kanias T, Tejero J, Lee JS, Kelley EE, Zhao X, Liu C, Reynolds H, Azarov I, Frizzell S, Meyer EM, Donnenberg AD, Qu L, Triulzi D, Kim-Shapiro DB, Gladwin MT, Nitric Oxide Scavenging by Red Blood Cell Microparticles and Cell-Free Hemoglobin as a Mechanism for the Red Cell Storage Lesion, Circulation, 124 (2011) 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Camus SM, De Moraes JA, Bonnin P, Abbyad P, Le Jeune S, Lionnet F, Loufrani L, Grimaud L, Lambry J-C, Charue D, Kiger L, Renard J-M, Larroque C, Le Clésiau H, Tedgui A, Bruneval P, Barja-Fidalgo C, Alexandrou A, Tharaux P-L, Boulanger CM, Blanc-Brude OP, Circulating cell membrane microparticles transfer heme to endothelial cells and trigger vasoocclusions in sickle cell disease, Blood, 125 (2015) 3805–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Kim Y, Abplanalp WA, Jung AD, Schuster RM, Lentsch AB, Gulbins E, Caldwell CC, Pritts TA, Endocytosis of Red Blood Cell Microparticles by Pulmonary Endothelial Cells is Mediated By Rab5, SHOCK, 49 (2018) 288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Labbé S, Mourer T, Brault A, Vahsen T, Machinery for fungal heme acquisition, Curr Genet, (2020). [DOI] [PubMed] [Google Scholar]
- [276].Mourer T, Jacques JF, Brault A, Bisaillon M, Labbe S, Shu1 is a cell-surface protein involved in iron acquisition from heme in Schizosaccharomyces pombe, J Biol Chem, 290 (2015) 10176–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Mourer T, Brault A, Labbe S, Heme acquisition by Shu1 requires Nbr1 and proteins of the ESCRT complex in Schizosaccharomyces pombe, Mol Microbiol, 112 (2019) 1499–1518. [DOI] [PubMed] [Google Scholar]
- [278].Todkar K, Ilamathi HS, Germain M, Mitochondria and Lysosomes: Discovering Bonds, Front Cell Dev Biol, 5 (2017) 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Das A, Nag S, Mason AB, Barroso MM, Endosome–mitochondria interactions are modulated by iron release from transferrin, Journal of Cell Biology, 214 (2016) 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Raiborg C, Wenzel EM, Stenmark H, ER–endosome contact sites: molecular compositions and functions, The EMBO Journal, 34 (2015) 1848–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Eden ER, The formation and function of ER-endosome membrane contact sites, Biochimica et biophysica acta, 1861 (2016) 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM, Cargo-Selected Transport from the Mitochondria to Peroxisomes Is Mediated by Vesicular Carriers, Current Biology, 18 (2008) 102–108. [DOI] [PubMed] [Google Scholar]
- [283].Soubannier V, McLelland G-L, Zunino R, Braschi E, Rippstein P, Edward, Heidi, A Vesicular Transport Pathway Shuttles Cargo from Mitochondria to Lysosomes, Current Biology, 22 (2012) 135–141. [DOI] [PubMed] [Google Scholar]
- [284].Schumann U, Subramani S, Special delivery from mitochondria to peroxisomes, Trends in Cell Biology, 18 (2008) 253–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Spinelli JB, Haigis MC, The multifaceted contributions of mitochondria to cellular metabolism, Nature Cell Biology, 20 (2018) 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM, Reconstitution of Mitochondria Derived Vesicle Formation Demonstrates Selective Enrichment of Oxidized Cargo, PLoS ONE, 7 (2012) e52830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Eisenberg-Bord M, Schuldiner M, Mitochatting – If only we could be a fly on the cell wall, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1864 (2017) 1469–1480. [DOI] [PubMed] [Google Scholar]
- [288].Mcbride H, Mohanty A, Emerging roles of mitochondria in the evolution, biogenesis, and function of peroxisomes, Frontiers in Physiology, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Demarquoy J, Crosstalk between mitochondria and peroxisomes, World Journal of Biological Chemistry, 6 (2015) 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Bolte K, Rensing SA, Maier U-G, The evolution of eukaryotic cells from the perspective of peroxisomes, 37 (2015) 195–203. [DOI] [PubMed] [Google Scholar]
- [291].Gould SB, Garg SG, Martin WF, Bacterial Vesicle Secretion and the Evolutionary Origin of the Eukaryotic Endomembrane System, Trends in Microbiology, 24 (2016) 525–534. [DOI] [PubMed] [Google Scholar]
- [292].Sugiura A, Mattie S, Prudent J, McBride HM, Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes, Nature, 542 (2017) 251–254. [DOI] [PubMed] [Google Scholar]
- [293].Huppert JL, Balasubramanian S, G-quadruplexes in promoters throughout the human genome, Nucleic Acids Res, 35 (2007) 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM Jr., Sen D, Lansdorp PM, Detection of G-quadruplex DNA in mammalian cells, Nucleic Acids Res, 42 (2014) 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S, The regulation and functions of DNA and RNA G-quadruplexes, Nat Rev Mol Cell Biol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Puget N, Miller KM, Legube G, Non-canonical DNA/RNA structures during Transcription-Coupled Double-Strand Break Repair: Roadblocks or Bona fide repair intermediates?, DNA Repair, 81 (2019) 102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Técher H, Koundrioukoff S, Nicolas A, Debatisse M, The impact of replication stress on replication dynamics and DNA damage in vertebrate cells, Nature Reviews Genetics, 18 (2017) 535–550. [DOI] [PubMed] [Google Scholar]
- [298].Neidle S, Quadruplex Nucleic Acids as Novel Therapeutic Targets, Journal of Medicinal Chemistry, 59 (2016) 5987–6011. [DOI] [PubMed] [Google Scholar]
- [299].Sen D, Poon LCH, RNA and DNA complexes with hemin [Fe(III) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean?, Critical Reviews in Biochemistry and Molecular Biology, 46 (2011) 478–492. [DOI] [PubMed] [Google Scholar]
- [300].Gray LT, Puig Lombardi E, Verga D, Nicolas A, Teulade-Fichou MP, Londono-Vallejo A, Maizels N, G-quadruplexes Sequester Free Heme in Living Cells, Cell Chem Biol, 26 (2019) 1681–1691 e1685. [DOI] [PubMed] [Google Scholar]
- [301].Li Y, Sen D, A catalytic DNA for porphyrin metallation, Nature Structural Biology, 3 (1996) 743–747. [DOI] [PubMed] [Google Scholar]
- [302].Travascio P, Li Y, Sen D, DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex, Chemistry & Biology, 5 (1998) 505–517. [DOI] [PubMed] [Google Scholar]
- [303].Poon LCH, Methot SP, Morabi-Pazooki W, Pio F, Bennet AJ, Sen D, Guanine-Rich RNAs and DNAs That Bind Heme Robustly Catalyze Oxygen Transfer Reactions, Journal of the American Chemical Society, 133 (2011) 1877–1884. [DOI] [PubMed] [Google Scholar]
- [304].Ghahremani Nasab M, Hassani L, Mohammadi Nejad S, Norouzi D, Interaction of hemin with quadruplex DNA, J Biol Phys, 43 (2017) 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Mestre-Fos S, Ito C, Moore CM, Reddi AR, Williams LD, Human Ribosomal G-Quadruplexes Regulate Heme Bioavailability, J Biol Chem, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]