Summary
Studying brown and brite adipose tissue requires precise and reliable quantification of cellular thermogenesis. This protocol describes the isolation of primary murine pre-adipocytes, differentiation into thermogenic brown and brite adipocytes, and subsequent oxygen consumption analysis. Commonly applied procedures only measure basal and maximal proton leak-linked oxygen consumption but not explicitly uncoupling protein 1 (UCP1)-dependent respiration. Meaningful oxygen consumption analyses require (1) the activation of UCP1, (2) control over intracellular free-fatty-acid levels, and (3) inhibition of ATP-consuming futile cycles.
For complete details on the use and execution of this protocol, please refer to Li et al. (2014, 2017, 2018) and Schweizer et al. (2018).
Graphical Abstract

Highlights
-
•
Culture and bioenergetic phenotyping of UCP1-expressing thermogenic adipocytes
-
•
Inactive UCP1 protein does not generate heat
-
•
Meaningful assay requires ignition, control over FFA levels, and blocked ATP synthase
-
•
ECAR does not reflect the glycolytic rate of adipocytes during active lipolysis
Studying brown and brite adipose tissue requires precise and reliable quantification of cellular thermogenesis. This protocol describes the isolation of primary murine pre-adipocytes, differentiation into thermogenic brown and brite adipocytes, and subsequent oxygen consumption analysis. Commonly applied procedures only measure basal and maximal proton leak-linked oxygen consumption but not explicitly uncoupling protein 1 (UCP1)-dependent respiration. Meaningful oxygen consumption analyses require (1) the activation of UCP1, (2) control over intracellular free-fatty-acid levels, and (3) inhibition of ATP-consuming futile cycles.
Before You Begin
Note: Primary cultures of brown and brite adipocytes are a valuable tool to study UCP1-mediated thermogenesis. Microplate-based cellular respirometry provides a convenient, high-throughput system for the characterization of cellular bioenergetics with high resolution.
CRITICAL: The most critical step prior to bioenergetic analysis is to differentiate precursor cells into UCP1-expressing adipocytes in microplates specifically designed for the Seahorse XF technology.
Because primary cultures of brown and brite adipocytes reliably and reproducibly express high levels of UCP1, they are well suited to respiration measurements centered on UCP1 functionality. This protocol describes the isolation as well as differentiation of precursor cells from murine adipose tissue into UCP1-expressing mature adipocytes and subsequent microplate-based respirometry.
Note: This protocol can be combined with siRNA-based gene silencing and microplate-based respirometry to facilitate the investigation of gene function in UCP1-dependent thermogenesis.
The protocol below describes the use of primary cultures of pre-adipocytes, which can be differentiated into UCP1-expressing adipocytes. However, if you are using established brown or brite adipocyte cell lines expressing UCP1, you can simply skip steps 1–11 and proceed from step 12.
CRITICAL: Inactive UCP1 protein does not generate heat. Meaningful oxygen consumption analysis requires (1) activation of UCP1, (2) control over free fatty acid levels, and (3) inhibition of ATP-consuming futile cycles. Extracellular acidification rate (ECAR) does not reflect glycolytic rate of adipocytes during active lipolysis.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Albumin Fraction V | Carl Roth | Cat# 8076 |
| Amphotericin B solution | Sigma-Aldrich | Cat# A2942 |
| Antimycin A | Sigma-Aldrich | Cat# A8674 |
| Bovine serum albumin (BSA) Fatty Acid Free | Sigma-Aldrich | Cat# A3803 |
| Collagenase, Type I | Fisher Scientific | Cat# 10114532 |
| Dexamethasone | Sigma-Aldrich | Cat# D4902 |
| DMEM | Sigma-Aldrich | Cat# D5796 |
| DMEM | Sigma-Aldrich | Cat# D5030 |
| EDTA | N/A | N/A |
| FBS Superior | Sigma-Aldrich | Cat# S0615 |
| FCCP | Sigma-Aldrich | Cat# C2920 |
| Gentamicin solution | Sigma-Aldrich | Cat# G1272 |
| Glucose | N/A | N/A |
| GlutaMAX™ | Fisher Scientific | Cat# 13462629 |
| HBSS | Fisher Scientific | Cat# 11540476 |
| HEPES solution (1 M) | N/A | N/A |
| Indomethacin | Sigma-Aldrich | Cat# I7378 |
| Insulin | Sigma-Aldrich | Cat# I9278 |
| Isobutylmethylxanthine | Sigma-Aldrich | Cat# I5879 |
| Isoproterenol | Sigma-Aldrich | Cat# I6504 |
| K2HPO4 | N/A | N/A |
| Lipofectamine™ RNAiMAX (optional) | Fisher Scientific | Cat# 13778 |
| NaCl | N/A | N/A |
| NH4Cl | N/A | N/A |
| Oligomycin | Sigma-Aldrich | Cat# O4876 |
| PBS | N/A | N/A |
| Penicillin-Streptomycin | Sigma-Aldrich | Cat# P4333 |
| Phenol Red | N/A | N/A |
| Rosiglitazone | Biomol | Cat# Cay71740 |
| T3 | Sigma-Aldrich | Cat# T6397 |
| Trypsin-EDTA Solution | Sigma-Aldrich | Cat# 59417C |
| Critical Commercial Assays | ||
| Seahorse XF96 fluxPak | Agilent | Cat# 102416-100 |
| Experimental Models: Cells | ||
| Primary murine brown and white cells | N/A | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse | N/A | N/A |
| Oligonucleotides | ||
| siRNA (optional) | IDT | N/A |
| Software and Algorithms | ||
| Seahorse Wave Desktop | Agilent | Online |
| Spreadsheet software | N/A | N/A |
| Other | ||
| 0.22 μm filter | N/A | N/A |
| 250 μm nylon mesh | N/A | N/A |
| 40 μm cell strainer | N/A | N/A |
| 6-well cell culture plate (optional) | N/A | N/A |
| Benchtop centrifuge | N/A | N/A |
| Biosafety cabinet | N/A | N/A |
| Disposable reagent reservoir | N/A | N/A |
| Multichannel pipette | N/A | N/A |
| Surgical scissors | N/A | N/A |
| Temperature-controlled orbital shaker (for 15 mL tubes) | N/A | N/A |
| Thermoshaker (microtubes) | N/A | N/A |
| Water bath | N/A | N/A |
| XF96 Extracellular Flux Analyzer | Agilent | N/A |
Materials and Equipment
Preparation of Buffers and Solutions for the Isolation of Pre-adipocytes from Murine Adipose Tissue and Differentiation into Mature Adipocytes
Note: This part of the protocol can be skipped, if using established brown or brite adipocyte cell lines expressing UCP1.
Prepare PBS-A (PBS supplemented with antibiotic and antifungal agents), collagenase solution, wash medium, and red blood cell lysis buffer. Sterilize collagenase solution, wash medium, and red blood cell lysis buffer using a 0.22 μm filter. PBS-A and red blood cell lysis buffer are stored at 4°C. Collagenase solution and wash medium are stored at −20°C.
Refer to Key Resources Table and Materials and Equipment for a complete list of materials, equipment, and recipes.
CRITICAL: Avoid repeatedly freezing and thawing the collagenase solution.
Preparation of Respiration Base Medium and Injection Stock Solutions for Respiration Analysis
Prepare respiration base medium, adjust pH to 7.4, and sterilize using a 0.22 μm filter. Respiration base medium is stored at 4°C.
Optional: Agilent offers a range of liquid media specially formulated for use in oxygen consumption assays (Seahorse XF media), which eliminate the liabilities connected with hydrating powdered medium and sterile filtration of solutions.
Prepare injection stock solutions including Oligomycin, Isoproterenol, carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (FCCP) and Antimycin A using the appropriate solvent, and aliquot stock solutions into small volumes. Injection stock solutions are stored at −20°C.
CRITICAL: Protect Isoproterenol from light. Avoid repeatedly freezing and thawing stock solutions.
Note: Recipes are sorted alphabetically.
Collagenase Solution
| Reagent | Stock Concentration | Final Concentration | Volume/Weight |
|---|---|---|---|
| HBSS | n/a | n/a | 100 mL |
| Albumin Fraction V | n/a | 3.5% (w/v) | 3.5 g |
| Glucose | n/a | 0.55 mM | 1 mg |
| Collagenase Type I | n/a | 0.1% (w/v) | 100 mg |
| Penicillin-Streptomycin | 10,000 units/mL, 10 mg/mL | 40 units/mL, 40 μg/mL | 400 μL |
| Gentamicin | 10 mg/mL | 40 μg/mL | 400 μL |
| Amphotericin B | 250 μg/mL | 500 ng/mL | 200 μL |
| Total | n/a | n/a | 100 mL |
Sterilize using a 0.22 μm filter, store at −20°C, do not store for more than 3 months, avoid repeated freeze and thaw cycles.
Differentiation Medium
| Reagent | Stock Concentration | Final Concentration | Volume |
|---|---|---|---|
| DMEM (D5796) | n/a | n/a | 8.9 mL |
| FBS | n/a | 10% (v/v) | 1 mL |
| Penicillin-Streptomycin | 10,000 units/mL, 10 mg/mL | 40 units/mL, 40 μg/mL | 40 μL |
| Gentamicin | 10 mg/mL | 40 μg/mL | 40 μL |
| Insulin | 1.72 mM | 850 nM | 5 μL |
| T3 | 2 μM | 1 nM | 5 μL |
| Rosiglitazone | 10 mM | 1 μM | 1 μL |
| Total | n/a | n/a | 10 mL |
Always prepare freshly, do not store.
Growth Medium
| Reagent | Stock Concentration | Final Concentration | Volume |
|---|---|---|---|
| DMEM (D5796) | n/a | n/a | 7.9 mL |
| FBS | n/a | 20% (v/v) | 2 mL |
| Penicillin-Streptomycin | 10,000 units/mL, 10 mg/mL | 40 units/mL, 40 μg/mL | 40 μL |
| Gentamicin | 10 mg/mL | 40 μg/mL | 40 μL |
| Amphotericin B | 250 μg/mL | 500 ng/mL | 20 μL |
| Total | n/a | n/a | 10 mL |
Store at 4°C, do not store for more than 5 days.
Induction Medium
| Reagent | Stock Concentration | Final Concentration | Volume |
|---|---|---|---|
| DMEM (D5796) | n/a | n/a | 8.9 mL |
| FBS | n/a | 10% (v/v) | 1 mL |
| Penicillin-Streptomycin | 10,000 units/mL, 10 mg/mL | 40 units/mL, 40 μg/mL | 40 μL |
| Gentamicin | 10 mg/mL | 40 μg/mL | 40 μL |
| Insulin | 1.72 mM | 850 nM | 5 μL |
| T3 | 2 μM | 1 nM | 5 μL |
| Rosiglitazone | 10 mM | 1 μM | 1 μL |
| Dexamethasone | 10 mM | 1 μM | 1 μL |
| IBMX | 500 mM | 500 μM | 10 μL |
| Indomethacine | 125 mM | 125 μM | 10 μL |
| Total | n/a | n/a | 10 mL |
Always prepare freshly, do not store.
Injection Stock Solutions
| Compound | Stock Concentration (mM) | Solvent |
|---|---|---|
| Oligomycin | 2.5 | DMSO |
| Isoproterenol | 10 | Ethanol |
| FCCP | 2.5 | DMSO |
| Antimycin A | 2.5 | DMSO |
Store at −20°C, avoid repeated freeze and thaw cycles, protect Isoproterenol from light.
Injection Volumes and Concentrations
| Final Concentration in Well (μM) | Stock Solution Volume (μL) | Respiration Base Medium Volume (μL) | 10× Concentration in Port (μM) | Volume Added to Port (μL) | |
|---|---|---|---|---|---|
| Port A Oligomycin |
5 | 52 | 2548 | 50 | 20 |
| Port B Isoproterenol |
0.5 | 1.4 | 2798.6 | 5 | 22 |
| Port C FCCP |
7.5 | 90 | 2910 | 75 | 24 |
| Port D Antimycin A |
5 | 64 | 3136 | 50 | 26 |
Prepare injection working solutions in respiration base medium, do not store injection working solutions. The volume of injection working solution added to each port depends on the initial volume of respiration assay medium per well. Here we calculate the volume of injection working solution based on a starting volume of 180 μL respiration assay medium per well.
PBS-A
| Reagent | Stock Concentration | Final Concentration | Volume |
|---|---|---|---|
| PBS | n/a | n/a | 9.9 mL |
| Penicillin-Streptomycin | 10,000 units/mL, 10 mg/mL | 40 units/mL, 40 μg/mL | 40 μL |
| Gentamicin | 10 mg/mL | 40 μg/mL | 40 μL |
| Amphotericin B | 250 μg/mL | 500 ng/mL | 20 μL |
| Total | n/a | n/a | 10 mL |
Store at 4°C.
Red Blood Cell Lysis Buffer
| Reagent | Stock Concentration | Final Concentration | Volume/Weight |
|---|---|---|---|
| NH4Cl | n/a | 154 mM | 824 mg |
| K2HPO4 | n/a | 10 mM | 174 mg |
| EDTA | 500 mM | 0.1 mM | 20 μL |
| ddH2O | n/a | n/a | 100 mL |
| Total | n/a | n/a | 100 mL |
Adjust pH to 7.3, sterilize using a 0.22 μm filter, store at 4°C.
Respiration Assay Medium
| Reagent | Stock Concentration | Final Concentration | Volume/Weight |
|---|---|---|---|
| Respiration base medium | n/a | n/a | 10 mL |
| BSA fatty acid-free | n/a | 2% (w/v) | 200 mg |
| Total | n/a | n/a | 10 mL |
Adjust pH to 7.4, always prepare freshly, do not store.
Respiration Base Medium
| Reagent | Stock Concentration | Final Concentration | Volume/Weight |
|---|---|---|---|
| DMEM (D5030) | n/a | n/a | 1 Package |
| Glucose | n/a | 25 mM | 4.5 g |
| NaCl | n/a | 31 mM | 1.85 g |
| Phenol Red | n/a | 15 mg/l | 15 mg |
| GlutaMAX™ | 200 mM | 2 mM | 10 mL |
| HEPES | 1 M | 5 mM | 5 mL |
| ddH2O | n/a | n/a | 985 mL |
| Total | n/a | n/a | 1000 mL |
Adjust pH to 7.4, sterilize using a 0.22 μm filter, store at 4°C.
Transfection Mix
| Reagent | Stock Concentration | Final Concentration | Volume for 1 Well | Volume for 16 Wells | With 10% Extra |
|---|---|---|---|---|---|
| OptiMEM | n/a | n/a | 24.1 μL | 385.6 μL | 424.1 μL |
| Lipofectamine™ RNAiMAX | n/a | 5 μL/mL (in well) | 0.75 μL | 12 μL | 13.2 μL |
| siRNA | 50 μM | 50 nM (in well) | 0.15 μL | 2.4 μL | 2.7 μL |
| Total | n/a | n/a | 25 μL | 400 μL | 440 μL |
Always prepare freshly, do not store.
Wash Medium
| Reagent | Stock Concentration | Final Concentration | Volume/Weight |
|---|---|---|---|
| HBSS | n/a | n/a | 100 mL |
| Albumin Fraction V | n/a | 3.5% (w/v) | 3.5 g |
| Penicillin-Streptomycin | 10,000 units/mL, 10 mg/mL | 40 units/mL, 40 μg/mL | 400 μL |
| Gentamicin | 10 mg/mL | 40 μg/mL | 400 μL |
| Amphotericin B | 250 μg/mL | 500 ng/mL | 200 μL |
| Total | n/a | n/a | 100 mL |
Sterilize using a 0.22 μm filter, store at −20°C.
Step-By-Step Method Details
Isolation of Stromal Vascular Fraction from Murine Adipose Tissue and Differentiation into Mature Adipocytes
Timing: 4 h + differentiation
Note: Skip the part on isolating stromal vascular fraction, if you are using established brown or brite adipocyte cell lines. For a detailed description regarding seeding pre-adipocytes into Seahorse XF96 cell culture microplates and the differentiation procedure jump to step 12. If you already have an established differentiation protocol, directly jump to step 19 of the respirometry part for instructions on how to perform the oxygen consumption assay.
For one whole Seahorse XF96 cell culture microplate (corresponding to one measurement) you require either iBAT depots from 1 to 2 mice or iWAT depots from 1 mouse.
Optional: For siRNA-based gene silencing prior to oxygen consumption analysis follow the cell culture protocol until step 10. For 4 individual siRNA treatment groups with 16 wells per condition on one Seahorse XF96 cell culture microplate you require either at least iBAT depots from 2 to 3 mice or iWAT depots from 1 mouse; the number of depots can be scaled accordingly.
Note: The initial number of fat depots is critical, as it will dictate the seeding density later on. However, the cell yield can be influenced by a variety of factors and may consequently slightly differ between labs depending on chemicals and equipment used. Therefore, the numbers stated above are guidelines and may require further optimization. Troubleshooting 1 (Cells do not differentiate well or have low/no UCP1 expression)
-
1.
Euthanize mice (young mice aged 5–6 weeks are to be preferred) by CO2 asphyxiation.
-
2.
Dissect interscapular brown adipose tissue and/or inguinal white adipose tissue. Try not to damage major blood vessels during the dissection, as this will help minimizing the contamination with erythrocytes.
-
3.
Rinse adipose tissue with PBS-A and remove remaining connective tissue and muscle tissue (when dissecting iWAT, remove the inguinal lymph node). Cover with PBS-A to prevent the tissue from drying out; process as quickly as possible in a sterile manner.
-
4.
Mince adipose tissue into small pieces (<1–2 mm3) using scissors. Large pieces will negatively affect the digestion process and result in a low cell yield.
-
5.
Transfer tissue into 15 mL tube and add 7 mL of collagenase solution to start the digestion reaction.
CRITICAL: One digestion reaction (7 mL) is sufficient for iBAT or iWAT depots of up to 3 mice. If the experimental design requires more depots to be processed at the same time, separate digestion reactions can be combined at step 9 (after the first two centrifugation steps or after the optional red blood cell lysis step); volumes need to be adjusted accordingly. The “7 mL of collagenase solution” stated above and the following volumes refer to one digestion reaction.
Avoid adding significantly more than 7 mL of collagenase solution into a 15 mL tube, since an increased liquid amount results in impaired mixing and less mechanical force during the digestion process. Otherwise, the incubation time will drastically increase.
-
6.
Place the tube horizontally into an orbital shaker. Digest by incubating at 37°C (120–180 rpm) for 45 min to 1 h 15 min. Vigorous shaking by hand for 10 s every 15 min facilitates digestion. Digestion is completed when almost no visible pieces of adipose tissue are left and the mixture becomes homogenous.
-
7.
Filter through a 250 μm nylon mesh and capture flow through in a new 15 mL tube. Rinse the tube, in which the digestion was carried out, with 7 mL of wash buffer, filter through the same nylon mesh, and combine flow through.
-
8.
Centrifuge at 250 × g for 5 min at 18°C–26°C (room temperature). Carefully invert the tube several times to break up the fat layer floating on top. Centrifuge at 250 × g for 5 min at 18°C–26°C (room temperature).
-
9.
Completely remove fat layer and supernatant (do not disturb the cell pellet), add 1 mL of wash medium, and break up the cell pellet by carefully pipetting up and down (separate digestion reactions can be combined at this point).
Optional: Erythrocytes can be removed by using a buffer designed to lyse red blood cells. Add 1–2 mL of red blood cell lysis buffer instead of wash medium to break up the cell pellet. Incubate for 3–5 min at 18°C–26°C (room temperature; separate digestion reactions can be combined at this point).
-
10.
Add 13 mL of wash buffer and centrifuge at 500 × g for 5 min at 18°C–26°C (room temperature). Completely remove the supernatant (do not disturb the cell pellet), add 1 mL of growth medium, and resuspend cells by carefully pipetting up and down.
Optional: For the siRNA-based knockdown procedure jump to the chapter “siRNA-based gene silencing in murine adipocytes with oxygen consumption analysis (optional)” below.
-
11.
Add 9 mL of growth medium and filter the suspension through a 40 μm cell strainer into a reagent reservoir or a 10 cm petri dish.
CRITICAL: Before seeding the cells and throughout the seeding procedure repeatedly mix the cells in the reservoir by gently pipetting up and down (or carefully shaking the reservoir) to ensure that cells are thoroughly resuspended and to prevent the cells from settling down.
Note: We recommend seeding at least 8–16 wells per condition/treatment.
-
12.
Seed 80 μL of cell suspension per well into a Seahorse XF96 cell culture microplate using a multichannel pipette. Do not seed cells in the four background correction wells (A1, A12, H1, H12), but add growth medium (without cells) in these wells. If your experimental design does not require a whole 96-well plate, add PBS or growth medium in the empty wells to reduce the microplate edge effect. Troubleshooting 2 (The position of a well on the cell culture plate systematically influences oxygen consumption rates)
-
13.
Let the plate rest at 18°C–26°C (room temperature) in the cell culture hood for 1 h after seeding. Place the cell culture plate toward the back of the incubator to minimize changes in temperature and humidity caused by frequently accessing the incubator.
Note: As counting stromal vascular fraction or differentiated adipocytes after the trypsination procedure is problematic due to the sheer amount of cell debris, the presence of cell aggregates, and the heterogeneous size distribution, we explicitly did not state cell numbers nor provide a seeding density. Instead, we simply stick to a defined number of fat depots or wells of a cell culture plate, which is simple and astonishingly reproducible. Following our strategy, at the latest 72 h after seeding the SVF cells should have reached 30%–40% confluence. When working with established brown or brite adipocyte cell lines, seed 10,000 cells per well into a Seahorse XF96 cell culture microplate.
-
14.
On the next day or at least 6 h after seeding carefully remove the growth medium using a vacuum manifold adapter and add 150 μL of fresh growth medium with a multichannel pipette. From this point on, treat the background correction wells exactly as you treat the remaining wells. The differentiation procedure is schematically summarized in Figure 1.
Figure 1.
Graphical Timeline Illustrating the Differentiation Procedure from Pre-adipocytes into Mature Adipocytes
CRITICAL: To prevent the cells from drying out only process one half or even one third of the microplate at a time. Remove as much of the medium as possible but do not completely remove the medium (leaving around 20 μL behind), as this may cause the cells to detach. Changing the medium will require some practice.
-
15.
Change growth medium every other day until the cells reach 80%–100% confluency (termed “day 0”).
Note: We recommend 80%–100% confluency, because pre-adipocytes will still proliferate even after the induction/differentiation process is initiated. Therefore, pre-adipocytes/differentiating adipocytes will reach 100% confluence during the 48 h induction phase, which lowers the chances of obtaining overgrown cells. Ensuring that the cells only grow in a monolayer may help to slightly reduce the inter-assay variability. Images of pre-adipocytes and adipocytes at all stages during the differentiation process can be found in the section “Expected outcomes: Cultivation of Murine Pre-adipocytes and Differentiation into Mature Adipocytes.”
-
16.
On day 0 remove growth medium and add 150 μL of induction medium per well.
CRITICAL: Induction medium and differentiation medium have to be prepared freshly prior to usage. We do not recommend freezing, and/or storing medium for several days.
-
17.
On day 2 remove induction medium and add 150 μL of differentiation medium per well.
Note: In this context brite adipocytes are defined as cells derived from inguinal white adipose tissue and differentiated in the presence of the peroxisome proliferator-activated receptor γ (PPAR-γ) agonist rosiglitazone. Rosiglitazone induces a thermogenic gene signature promoting UCP1 expression. We specifically recommend working with pre-adipocytes isolated from adipose tissue of a “129 mouse” (any of the substrains), as they differentiate into adipocytes, which are characterized by high UCP1 levels.
-
18.
Change differentiation medium every other day until the cells are fully differentiated on day 8.
Optional: Chemicals that supposedly affect UCP1 expression levels can be added to the differentiation medium at this point.
siRNA-Based Gene Silencing in Murine Adipocytes with Oxygen Consumption Analysis (Optional)
Timing: 2 h
The cell culture protocol can be combined with siRNA-based gene silencing prior to respirometry (Isidor et al., 2016). This allows studying the effect of a specific gene on UCP1-dependent respiration (Li et al., 2019). Isolated pre-adipocytes are cultured and differentiated on a 6-well cell culture plate prior to reverse transfection onto a Seahorse XF96 cell culture microplate containing the siRNA transfection mix. One decisive advantage of this protocol is that a gene of interest can be knocked down at three different time points: 1) during proliferation, 2) directly after induction (early phase of differentiation), and 3) at a late stage of differentiation (after 5 days of differentiation) to test the role of a certain gene at different time points during adipogenesis. Nevertheless, it should be mentioned that siRNAs only cause a transient knockdown.
Note: This procedure starts after step 10 of the “Isolation of stromal vascular fraction from murine adipose tissue” process. The volumes below are calculated for four individual siRNA treatment groups with 16 wells per condition (1 negative control, 3 different siRNAs per target gene) on one Seahorse XF96 cell culture microplate and can be scaled accordingly.
-
1.
Add 8 mL of growth medium (iBAT) or 6 mL of growth medium (iWAT) and filter suspension through a 40 μm cell strainer into a 15 mL tube.
CRITICAL: Before seeding the cells and throughout the seeding procedure repeatedly mix the cells in the reservoir by gently pipetting up and down (or carefully shaking the reservoir) to ensure that cells are thoroughly resuspended and to prevent the cells from settling down.
-
2.
Seed 2 mL of cell suspension per well into a 6-well cell culture plate (4 wells for iBAT and 3 wells for iWAT). Troubleshooting 3 (Basal oxygen consumption rate of cells is too high or too low following the reverse transfection procedure)
-
3.
Carefully shake the plate backward and forward, then left and right, and let the plate rest at 18°C–26°C (room temperature) in the cell culture hood for 10 min after seeding to allow equal distribution of the cells.
Note: See Note above (Note after step 13). When working with established brown or brite adipocyte cell lines, seed 60,000 cells per well into a 6-well cell culture plate.
-
4.
On the next day or at least 6 h after seeding carefully remove the growth medium and wash the cells twice with prewarmed PBS (the washing steps help to remove cell debris and erythrocytes). Completely remove PBS and add 2 mL of fresh growth medium.
Note: Depending on the experimental question the reverse transfection procedure can be carried out at different time points: day 0 (commitment to the adipogenic lineage), day 2 (early differentiation), or day 7 (fully differentiated) of the differentiation procedure. Follow this protocol until the cells have reached the desired stage (step 6, step 7, or step 8), then continue with step 9 (“Reverse transfection”)
-
5.
Change growth medium every other day until the cells reach 80%–100% confluency (termed “day 0”).
Note: See Note above (Note after step 15).
-
6.
On day 0 remove growth medium and add 2 mL of induction medium per well.
CRITICAL: Induction medium and differentiation medium have to be prepared freshly prior to usage. We do not recommend freezing, and/or storing medium for several days.
-
7.
On day 2 remove induction medium and add 2 mL of differentiation medium per well.
-
8.
Change differentiation medium every other day until cells are considered as differentiated on day 7.
Reverse Transfection
-
9.
Use OptiMEM, Lipofectamine RNAiMAX, and siRNA stock solution to make a transfection mix for each siRNA treatment group. Troubleshooting 4 (Knockdown efficiency is low)
Transfection Mix for siRNA-mediated knockdown. When preparing the transfection mix, at least 10% extra volume should be added, so that even the last well can be filled properly.
| Volume OptiMEM | Volume RNAiMax | Volume siRNA (50μM stock) | Volume cell suspension | |
|---|---|---|---|---|
| Per well of a XF96 cell culture microplate | 24.1 μL | 0.75 μL | 0.15 μL | 125 μL |
-
10.
Mix gently. Add 25 μL of transfection mix per well into a Seahorse XF96 cell culture plate and incubate for 20 min at 18°C–26°C (room temperature) in the cell culture hood.
-
11.
Retrieve the 6-well cell culture plate from the cell culture incubator.
CRITICAL: Differentiating and especially fully differentiated adipocytes are very fragile. Handle with care; reduce the number of pipetting, resuspension, and mixing steps to the absolute minimum, and always pipette slowly and gently.
-
12.
Wash cells twice with prewarmed PBS.
-
13.
Remove PBS and add Trypsin-EDTA solution (cells processed at day 0 and day 2), or Trypsin-EDTA-Collagenase solution (cells processed at day 5). Return 6-well cell culture plate to the cell culture incubator and leave for 2–10 min. View cells under the microscope to ensure that all cells are detached and floating.
CRITICAL: Cells should only be exposed to Trypsin-EDTA-(Collagenase) long enough to detach the cells. Prolonged exposure may cause damage or even excess cell death.
-
14.
Once cells are detached add growth medium (cells processed at day 0) or differentiation medium (cells processed at day 2 and day 5) without antibiotics to inactivate trypsin.
Optional: DMEM supplemented with at least 10% FBS is essentially sufficient to inactivate trypsin.
-
15.
Centrifuge at 250 × g for 5min at 18°C–26°C (room temperature). Completely remove medium.
Note: We recommend plating at least 8 - 16 wells per negative control siRNA/siRNA treatment group. Knockdown efficiency can be directly assessed in cells that were subjected to oxygen consumption analysis (pooled RNA isolated from the content of 16 wells of a XF96 cell culture microplate is sufficient to perform qPCR; various suppliers offer kits designed to isolate RNA from small samples). Alternatively, an additional XF96 cell culture microplate/multiwell plate specifically designated to quantify knockdown efficiency can be plated and treated in parallel.
-
16.
Resuspend the cells in an appropriate volume (125 μL per well) of growth medium without antibiotics and antifungal agents (cells processed at day 0) or differentiation medium without antibiotics (cells processed at day 2 and day 7). For four different groups with 16 wells of a Seahorse XF96 cell culture microplate per group resuspend in 10 mL of medium. After at least 20 min incubation of the transfection mix at 18°C–26°C (room temperature) add 125 μL of cell suspension per well into the Seahorse XF96 cell culture plate. The necessity of counting cells is avoided by using a defined number of confluent wells, which corresponds to a defined constant number of cells, and seeding this given number of cells into a certain number of wells in a new plate again. Troubleshooting 3 (Basal oxygen consumption rate of cells is too high or too low following the reverse transfection procedure)
Re-seeding of Cultured Adipocytes during the Reverse Transfection
| Cell type | Number of Wells of a 6-Well Plate Required | Final Number of Wells on a Seahorse XF96 Cell Culture Plate after Re-seeding |
|---|---|---|
| iBAT | 4 wells | 64 wells (4 groups, each group 16 wells) |
| iWAT | 3 wells | 64 wells (4 groups, each group 16 wells) |
CRITICAL: Before seeding the cells and throughout the seeding procedure gently mix the cells in the reservoir by pipetting up and down (or carefully shaking the reservoir) to ensure that the cells are thoroughly resuspended and to prevent the cells from settling down.
-
17.
Place the cell culture plate toward the back of the incubator to minimize changes in temperature and humidity caused by frequently accessing the incubator.
-
18.
24 h after seeding carefully remove the medium and add 150 μL of fresh induction medium (cells processed at day 0) or differentiation medium (cells processed at day 2 and day 7).
-
19.
After another 48 h the cells can be subjected to oxygen consumption analysis. After bioenergetic phenotyping, cells can be harvested for assessing knockdown efficiency if needed.
Microplate-Based Respirometry: Hydrating the Sensor Cartridge and Creating a Seahorse Assay Template (Day −1)
Timing: 30 min + overnight hydration
Note: The Seahorse XF technology is offered as a 24 or 96-well microplate-based assay platform. This protocol is tailored for the analysis of cultures of primary murine brown and beige adipocytes in a 96-well microplate format. Nevertheless, the rationale behind this protocol applies to all adipocytes expressing UCP1, and it can be extended to the 24-well microplate platform.
On the day before the assay the Seahorse XFe96 sensor cartridge has to be hydrated. Furthermore, the Seahorse assay template can be created in advance.
-
20.
On the day prior to the oxygen consumption assay hydrate the SeahorseXFe96 sensor cartridge according to the manufacturer’s recommendations (instructions can be found online; “How to Hydrate an Agilent SeahorseXFe96 Sensor Cartridge”). In brief: Aliquot 22 mL of Seahorse XF Calibrant into a 50 mL tube. Incubate tube at 37°C in a laboratory (non-CO2) incubator overnight. Remove sensor cartridge from the utility plate. Add 200 μl of sterile water per well into the utility plate. Lower sensor cartridge onto utility plate (do not apply force). Incubate sensor cartridge and utility plate at 37°C in a laboratory (non-CO2) incubator overnight.
Alternatives: Outdated hydration method: Remove sensor cartridge from the utility plate. Add 200 μl of XF Calibrant per well into the utility plate. Lower sensor cartridge onto utility plate (do not apply force). Incubate sensor cartridge and utility plate at 37°C in a laboratory (non-CO2) incubator overnight.
Note: The sensor cartridge hydration procedure has slightly changed over the years. This assay was initially developed using a now outdated hydration method. However, based on our experience the “old” hydration method does not perform differently compared to the “new” method in the oxygen consumption assay. Thus, we definitely recommend following the updated version of the hydration procedure.
-
21.
Create a Seahorse assay template including the following instrument protocol:
We recommend assigning at least eight wells of a Seahorse XF96 cell culture microplate to each experimental group.
Protocol Commands
| Steps | Time | Cycles |
|---|---|---|
| Calibration | ||
| Equilibration | ||
| Mix | 4 min | 4 |
| Wait | 0 min | |
| Measure | 2 min | |
| Injection, Port A (Oligomycin) | ||
| Mix | 4 min | 4 |
| Wait | 0 min | |
| Measure | 2 min | |
| Injection, Port B (Isoproterenol) | ||
| Mix | 4 min | 4 |
| Wait | 0 min | |
| Measure | 2 min | |
| Injection, Port C (FCCP) | ||
| Mix | 4 min | 3 |
| Wait | 0 min | |
| Measure | 2 min | |
| Injection, Port D (Antimycin A) | ||
| Mix | 4 min | 4 |
| Wait | 0 min | |
| Measure | 2 min | |
Note: Mix time is set to 4 min to ensure proper reoxygenation between two “Measure” commands. This protocol intentionally leaves out a “Wait” command, since during periods of high oxygen consumption even 1–2 min between a “Mix” and “Measure” command can be sufficient to significantly lower the prevailing oxygen partial pressure at the beginning of a “Measure” period. The “Measure” command is set to the shortest possible time of 2 min to prevent hypoxia in the transient microchamber, which is created during a “Measure” command (Gerencser et al., 2009).
Microplate-Based Respirometry: Preparation of Respiration Media (Day 1)
Timing: 45 min
On the day of the assay the hydration procedure is completed, and bovine serum albumin (BSA)-buffered respiration assay medium is prepared freshly. All volumes regarding medium and injections below are sufficient for one assay (a whole Seahorse XF96 cell culture microplate).
-
22.
Complete the Agilent SeahorseXFe96 sensor cartridge hydration procedure (instructions can be found online; “How to Hydrate an Agilent SeahorseXFe96 Sensor Cartridge”). In brief: Separate sensor cartridge from the utility plate. Remove water from utility plate. Add 200 μL of warm XF Calibrant per well into the utility plate. Lower the sensor cartridge onto utility plate (do not apply force). Incubate at 37°C in a laboratory (non-CO2) incubator for around 1 h prior to loading the injection ports.
Outdated hydration method: Prior to loading the injection ports (step 12) separate sensor cartridge from the utility plate and lower it again onto utility plate (do not apply force).
-
23.
Aliquot 45 mL of respiration base medium into a 50 mL tube (used for wash steps later on), 22 mL of respiration base medium into a 50 mL tube (used for preparation of respiration assay medium later on), and 13 mL of respiration base medium into a 15 mL tube (used for injections later on). Incubate respiration base medium at 37°C until ready for use.
-
24.
Prepare respiration assay medium by supplementing respiration base medium with essentially fatty acid free BSA to a final concentration of 2% (w/v). Add 440 mg of essentially fatty acid free BSA to 22 mL of respiration base medium. Bring respiration assay medium to 37°C (ensure that BSA is completely dissolved) and adjust pH to 7.4. Incubate the final respiration assay medium at 37°C until ready for use. Troubleshooting 5 (I do not want to include BSA in the respiration assay medium)
Note: Fatty acid-free BSA is employed to control intracellular free fatty acid (FFA) levels and to prevent unspecific FFA-induced uncoupling (Li et al., 2017; Li et al., 2014). However, BSA can bind significant amounts of any given substance depending on the molecular nature of the compound. Thus, the presence of BSA in the respiration assay medium may affect the unbound and available concentration of some chemicals (e.g., FCCP). In some cases you will have to significantly increase the amount of a substance used to exert the same effect as in the absence of BSA. Vice versa, there are examples of BSA facilitating the import of compounds into cells leading to an increased availability.
Optional: Chemicals or inhibitors that supposedly affect UCP1-dependent respiration can be added to the respiration assay medium at this point. However, some chemicals may significantly alter the pH of the respiration assay medium. In doubt, measure the pH after adding your compound of interest again and adjust if necessary.
Microplate-Based Respirometry: Washing the Cells and Adding Respiration Assay Medium (Day 1)
Timing: 30 min
Regular cell culture medium very often contains bicarbonate as a buffering agent. As the oxygen consumption analysis is conducted under ambient atmosphere, a bicarbonate-based buffering agent would significantly alter the pH of the medium over time. Additionally, extracellular acidification rate (ECAR), the second readout of the Seahorse XF technology, can only be detected in a medium with low buffering capacity. Thus, culture medium is replaced with a weakly HEPES-buffered medium containing a defined amount of BSA. BSA is a mandatory addition to the respiration assay medium due to its ability to buffer excess free fatty acids, which prevents non-specific uncoupling. UCP1-dependent respiration can only be unequivocally quantified in the presence of a certain amount of BSA (Li et al., 2017; Li et al., 2014).
-
25.
Retrieve the Seahorse XF96 cell culture microplate from the cell culture incubator.
-
26.
Carefully remove all but 20 μL of the differentiation medium using a vacuum manifold adapter or multichannel pipette and add 200 μL of fresh respiration base medium with a multichannel pipette. Repeat this step one more time, leaving 20 μL behind after each wash.
CRITICAL: To prevent the cells from drying out only process one half or even one third of the microplate at a time. Changing the medium will require some practice.
-
27.
After the second wash, add 160 μL of respiration assay medium to each well for a final volume of 180 μL per well. A detailed instruction how to properly wash adherent cells in Seahorse XF96 cell culture microplates can be found online (“Washing Adherent Cells in Agilent Seahorse XF96 Cell Culture Microplates”).
-
28.
View cells under the microscope to ensure that cells were not damaged or washed away.
-
29.
Incubate the Seahorse XF96 cell culture microplate at 37°C in a laboratory (room air or non-CO2) incubator for 60 min prior to the assay.
Microplate-Based Respirometry: Loading the Injection Ports and Starting the Assay (Day 1)
Timing: 30 min
CRITICAL: A defined set of compounds is required to specifically measure oxygen consumption attributable to the action of UCP1. Many publications use a combination of Oligomycin and FCCP without Isoproterenol or any other pro-lipolytic stimulant. This setup stimulates the flow of electrons along the electron transport chain, but UCP1 is inherently inactive and it will remain inactive in this scenario. Therefore, solely using Oligomycin and FCCP is sufficient to determine electron transport chain capacity, but conclusions about UCP1-dependent respiration must on no account be drawn. Only the combination of Oligomycin and Isoproterenol allows a separate look at the action of UCP1 (Li et al., 2017; Li et al., 2014). Oligomycin shuts down all oxygen-dependent ATP-consuming processes and thereby prevents any compensatory ATP-dependent futile cycle, which may lead to an overestimation of UCP1-dependent oxygen consumption (Schweizer et al., 2018), whereas Isoproterenol (or any other substance stimulating lipolysis) activates UCP1 through the induction of lipolysis. The working solutions of compounds that are loaded into the sensor cartridge’s injection ports have to be prepared freshly on the day of the assay. Injection working solutions have to be prepared at a 10× working concentration, since the volume of injection working solution per injection port (1 part) and the volume of respiration assay medium per well (9 parts) are defined in a way that each compound finally reaches a 1× concentration in the well of the microplate. Therefore, the volume of injection working solution per injection port has to increase from port “A” to port “D,” as the volume of respiration assay medium per well simultaneously increases after the addition of each injection.
-
30.
Thaw appropriate volume of Oligomycin, FCCP, and Antimycin A. Isoproterenol stock solution does not freeze, as it is solved in ethanol.
CRITICAL: Protect Isoproterenol from light.
-
31.
Use Oligomycin, Isoproterenol, FCCP, and Antimycin A stock solutions to make compound working solutions for loading into the injection ports of the sensor cartridge. Prepare working solutions for each compound in respiration base medium (no BSA added) and load the sensor cartridge ports using a multichannel pipette and reagent reservoir. A detailed explanation on how to properly load the injection ports can be found online (“Loading the Agilent Seahorse XFe96 Sensor Cartridge Injection Ports”). Troubleshooting 6 (Oxygen consumption is not different between WT and UCP1KO adipocytes, or between groups with supposedly high and low UCP1 protein levels.)
Optional: Isoproterenol can be replaced with a specific set of chemicals, which reliably lead to the activation of UCP1: Norepinephrine, any β3-adenergic receptor agonist, dibutyryl-cAMP, or Forskolin. Therefore, any compound that supposedly activates UCP1 directly or indirectly can be injected instead of Isoproterenol (Li et al., 2018). Vice versa, putative compounds negatively affecting UCP1 activation or activity (e.g., lipolysis inhibitors) can be injected either after Oligomycin and prior to Isoproterenol or after Isoproterenol depending on the experimental question.
-
32.
Place the utility plate with the loaded sensor cartridge onto the instrument tray and start the assay around 40 min after the Seahorse XF96 cell culture microplate was placed in a laboratory (room air or non-CO2) incubator; calibration takes approximately 20 min.
CRITICAL: Before placing the sensor cartridge including the utility plate, and later on the cell culture plate onto the instrument tray remove all loading guides and plastic lids. Make sure that the utility plate and also the cell culture plate fits properly on the instrument tray. Check if everything is oriented in the correct direction.
-
33.
When prompted, replace the utility plate with the cell culture microplate then click Start. Assay duration is around 2 h.
-
34.
Once the final measurement is completed, an “Unload Sensor Cartridge dialog” will be displayed.
-
35.
Click “Eject” to eject the sensor cartridge and cell culture microplate.
-
36.
Separate the sensor cartridge from the cell culture microplate. Check the injection ports of the sensor cartridge for failed injections. Remaining injection medium, still stuck in the injection ports, can be easily identified at this point. Note down ports that were not completely emptied for subsequent data analysis Troubleshooting 7 (Injection ports are not completely empty after the measurement).
Optional: If desired, the cell culture microplate can be set aside or stored at −80°C for later analysis, such as determination of protein or DNA content for normalization, isolation of RNA for qPCR, or isolation of protein for immunodetection. Troubleshooting 8 (Large variation between groups. How can I normalize my data?)
Expected Outcomes
Cultivation of Murine Pre-adipocytes and Differentiation into Mature Adipocytes
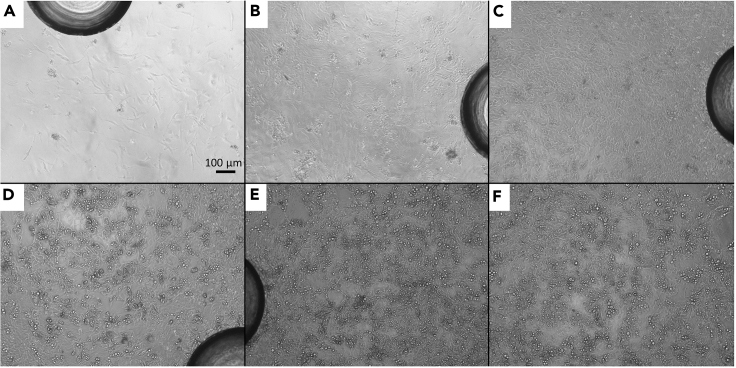
Upon adipogenic induction pre-adipocytes will drastically alter their morphological appearance (Figures 2A–2C). Starting from day 3 to day 4, first small lipid droplets will appear (Figures 2C and 2D). Over the course of the differentiation process the number and size of lipid droplets will further increase until adipocytes are densely packed with lipids (Figures 2D–2F). Differentiation efficiency, the percentage of cells on the plate storing triglycerides in the form of lipid droplets, ranges between 70%–90%. Differentiated cells will express significant (brown adipocytes) to moderate (brite adipocytes) amounts of UCP1.
Figure 2.
Bright-Field Images Illustrating the Differentiation Process from Pre-adipocytes into Mature Adipocytes
Scale bar represents 100 μm. The large spheroid-like structures in (A)–(E) are the molded stops on the bottom of a Seahorse XF96 cell culture microplate.
(A) Proliferating pre-adipocytes.
(B) Pre-adipocytes at the start of the induction phase (day 0).
(C) Differentiating adipocytes after the 48 h induction phase (day 2).
(D) Differentiating adipocytes at day 5.
(E and F) Mature adipocytes at day 7.
siRNA-Based Gene Silencing in Murine Adipocytes
Expected knockdown efficiency on mRNA level 72 h after treatment is 75%–95% compared to the negative control with no signs of cell damage or cytotoxicity.
Oxygen Consumption Analysis
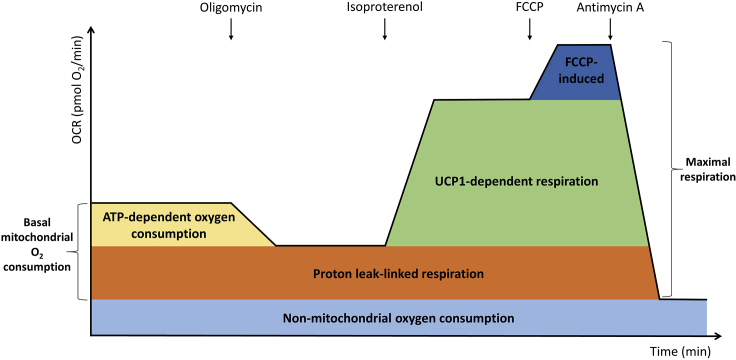
The different states of respiration in thermogenic adipocytes are graphically illustrated in Figure 3.
-
•
Basal mitochondrial oxygen consumption: Value prior to the injection of Oligomycin minus non-mitochondrial oxygen consumption.
-
•
Basal proton leak-linked respiration: Lowest recorded rate after the injection of Oligomycin, but prior to the addition of Isoproterenol, minus non-mitochondrial oxygen consumption.
-
•
ATP-dependent oxygen consumption: Basal oxygen consumption minus proton leak-linked respiration.
-
•
UCP1-dependent respiration: Highest recorded value after the injection of Isoproterenol, but prior to the addition of FCCP, minus lowest recorded rate after the injection of Oligomycin, but prior to the addition of Isoproterenol.
-
•
Maximal respiration: Highest recorded value after the injection of FCCP, but prior to the addition of Antimycin A, minus non-mitochondrial oxygen consumption.
-
•
Non-mitochondrial oxygen consumption: Lowest recorded rate after the injection of Antimycin A.
Figure 3.
Schematic OCR Trace Depicting the Different Raw Oxygen Consumption Metrics and Their Contribution to Oxygen Consumption during a Respiration Measurement
Arrows indicate injections.
Basal oxygen consumption during the first four measurements is generally between 150–250 pmol O2/min (Figure 4A). The first two rates are sometimes unstable, but at least the last two basal measurements should be stable. After the injection of Oligomycin respiration rates will immediately drop. The magnitude of this decrease depends on the basal ATP-demand, which is met by the mitochondrial ATP synthase. Therefore, the effect of Oligomycin on oxygen consumption can vary between different cell types, treatments, or conditions. However, we usually observe a pronounced decrease in respiration rates of at least 30%–50% after the injection of Oligomycin. Oxygen consumption rates are rather stable in this state. Cellular respiration increases again upon the injection of Isoproterenol Troubleshooting 9 (Oxygen consumption does not increase following the addition of Isoproterenol). As Isoproterenol treatment causes an activation of UCP1, respiration rates are largely determined by the amount of UCP1 present; compare group A, high UCP1 expression, and group B, significantly lower UCP1 expression (Figure 4A). Oxygen consumption usually reaches a plateau after the first two rate measurements following the addition of Isoproterenol. Cells with a high UCP1 expression, such as brown adipocytes, will show a 2–3-fold increase in oxygen consumption over leak respiration Troubleshooting 10 (Oxygen consumption rates rapidly peak in response to the Isoproterenol injection followed by a gradual decrease). FCCP, a chemical uncoupler, is the third substance added, which will cause cells to further increase their oxygen consumption until they have reached their maximal uncoupled respiration rate. Generally, there is an inverse relationship between respiration prior to the FCCP injection and respiration after the FCCP injection. The lower the oxygen consumption is after the Isoproterenol injection, the stronger is the effect of FCCP on respiration rates and vice versa. FCCP-stimulated oxygen consumption rates are almost always characterized by a sharp peak and very unstable rate measurements Troubleshooting 11 (Treatment groups with lower lipolysis rates do not reach the same level of maximal respiratory capacity in response to FCCP as control cells). The last injection, Antimycin A, shuts down the electron transport chain by inhibiting cytochrome c reductase, complex III of the respiration chain, which causes oxygen consumption to immediately drop to residual non-mitochondrial oxygen consumption. Again, rate measurements are quite stable at this stage.
Figure 4.
This Scenario Depicts a Potential Outcome Showing the OCR Traces of a Group with High UCP1 Expression (Group A) and a Group with Lower UCP1 Expression (Group B)
Data are presented as means ± SEM.
(A) Raw OCR traces of an exemplary respiration measurement using cultures of primary differentiated brown adipocytes. Arrows indicate injections. Compounds were added in the following order: Oligomycin (Oligo), Isoproterenol (Iso), FCCP, Antimycin A (AA).
(B) Corresponding OCR metrics calculated from the raw rate measurements.
Below we further discuss additional outcomes, highlight common pitfalls, and present ways to avoid them.
Outcome 1: A Representative Difference in UCP1-Dependent Respiration
The oxygen consumption traces of Group A and Group B show a significant overlap over large parts of the measurement (Figure 4A). Basal mitochondrial oxygen consumption and proton leak-linked respiration are comparable (Figure 4B). Both groups reach approximately the same maximally uncoupled oxygen consumption rate after the injection of FCCP suggesting that oxidative capacity is not markedly different. The only obvious difference between groups is the response to Isoproterenol. Cells in group A drastically increase oxygen consumption, whereas cells in Group B respond modestly (Figure 4A). This altered response translates into a markedly reduced UCP1-dependent respiration and a more than 2.5-fold decreased ratio of UCP1-dependent respiration to proton leak-linked respiration (Figure 4B).
Data can be explained by the following phenomena:
-
•
Cells in group B have lower UCP1 levels compared to cells in group A (confirm by immunoblotting).
-
•
Cells in group B have a reduced lipolytic capacity (the ability to release fatty acids in response to adrenergic stimulation) compared to cells in group A (check ECAR as a “quick and dirty” surrogate for lipolysis, which should be further confirmed by measuring the release of glycerol or fatty acids).
Note: ECAR (extracellular acidification rate), the second readout of the Extracellular Flux Analyzer, can be used to measure glycolytic flux and the anaerobic conversion of pyruvate to lactate. Catabolism of glucose to lactate generates protons, which contribute to the acidification of the extracellular medium. Consequently, the change in pH can serve as a surrogate marker for lactate production. However, glycolysis is not the only source of protons, as respiratory production of CO2 can significantly contribute to ECAR depending on the cell type. Mookerjee et al. developed a sophisticated protocol to solve this problem and to dissect glycolytic acidification from respiratory acidification of the extracellular medium facilitating an accurate determination of lactate production (Mookerjee et al., 2016). Unfortunately, this protocol cannot be applied to adipocytes, because release of free fatty acids into the surrounding medium during active lipolysis is a third and major factor leading to medium acidification (Schweizer et al., 2018). Until now, no solution was found to distinguish between ECAR deriving from anaerobic glycolysis, respiratory CO2 production, and release of free fatty acids. Furthermore, determining glycolytic flux and conversion of glucose to pyruvate in a quantitative manner requires prior knowledge of the respiratory quotient, i.e., the substrate metabolized in a certain state, which cannot be easily measured in cultured adipocytes. In conclusion, ECAR is not a suitable measure of glycolytic flux of adipocytes during active lipolysis.
Outcome 2: The Shifted Curve
The oxygen consumption traces of Group A and Group B show a very similar behavior during the whole measurement (Figure 5A). However, the oxygen consumption rates of group B are systematically lower than the rates of group A across all rate measurements. The difference almost completely disappears after calculating raw oxygen consumption metrics. Basal mitochondrial oxygen consumption, proton leak-linked respiration, and the ratio of UCP1-dependent respiration to proton leak-linked respiration are almost identical between groups (Figure 5B). This highlights the importance of calculating certain metrics during the analysis and interpretation of oxygen consumption data in addition to a careful analysis of respiration traces.
Figure 5.
A Frequently Observed Outcome: Two Different Groups Respond Almost Identical to the Injection Regimen, but the Oxygen Consumption of One Group Is Consistently Higher/Lower Compared to the Other Group
OCR traces of both groups would completely overlap, if the OCR trace of one group was shifted slightly upward/downward. Data are presented as means ± SEM.
(A) Raw OCR traces. Arrows indicate injections. Compounds were added in the following order: Oligomycin (Oligo), Isoproterenol (Iso), FCCP, Antimycin A (AA).
(B) Corresponding OCR metrics calculated from the raw rate measurements.
Data can be explained by the following phenomenon:
-
•
Most likely there are less cells per well in group B. This is an observation we frequently make. If one curve appears to be shifted upward or downward and all rate measurements are systematically higher or lower, this strongly suggests a difference in cell number. Although raw oxygen consumption rates are different between groups, we conclude that the treatment does not affect cellular respiration, if none of the oxygen consumption metrics is affected (confirm by measuring DNA content, protein content, number of nuclei, cell count). Troubleshooting 8 (Large variation between groups. How can I normalize my data?)
Outcome 3: Altered Proton Leak
The oxygen consumption traces of Group A and Group B show a significant overlap over large parts of the measurement (Figure 6A, left). Nevertheless, oxygen consumption rates after the Oligomycin injection are slightly higher in group B compared to group A. Accordingly, basal mitochondrial oxygen consumption is not different between groups, but proton leak-linked respiration is 2-fold higher in group B (Figure 6B). Raw oxygen consumption rates in both groups are around 400–420 pmol O2/min after the addition of Isoproterenol (Figure 6A). Yet, UCP1-dependent respiration is almost 10% higher in group A than in group B and the ratio of UCP1-dependent respiration to proton leak-linked respiration is 2.5-fold decreased in group B (Figure 6B).
Figure 6.
An Exaggerated Scenario, Which Very Often Entails Wrong Interpretations
Two groups only differing in oxygen consumption following the addition of Oligomycin, whereas the remaining rate measurements are almost identical. Data are presented as means ± SEM.
(A) Raw OCR traces and the same traces scaled to the rate measurement prior to the addition of Isoproterenol using the baseline feature. Arrows indicate injections. Compounds were added in the following order: Oligomycin (Oligo), Isoproterenol (Iso), FCCP, Antimycin A (AA).
(B) Corresponding OCR metrics calculated from the raw rate measurements.
This particular outcome cannot be interpreted in a straightforward fashion, potential explanations are:
-
•
Only proton leak-linked respiration is higher in group B than in group A, while assuming that the number of cells per well is similar across groups. UCP1-dependent respiration in group A is slightly higher than in group B and the ratio of UCP1-dependent respiration to proton leak-linked respiration is drastically altered, but these differences are only caused by an increase in proton leak-linked respiration, as raw oxygen consumption values after the addition of Isoproterenol are comparable. Consequently, UCP1-dependent oxygen consumption, which is a function of UCP1 expression and/or activity, is most likely not significantly different between groups (confirm by measuring UCP1 expression and quantifying cell number/mass). This interpretation may be rather applicable to an experiment, in which different treatments are applied to the same cell type, as basal mitochondrial OCR, maximal respiration, and non-mitochondrial O2-consumption of Group A and B are comparable, while the treatment merely affects basal proton leak and mitochondrial ATP-synthesis.
-
•
Proton leak-linked respiration is in fact not different between groups, but the number of cells per well is higher in group B than in group A. Thus, UCP1-dependent respiration is indeed different between groups and the difference is most likely underestimated, as comparable cell numbers would cause raw OCR traces to clearly deviate from each other following the addition of Isoproterenol. Consequently, UCP1-dependent oxygen consumption is as a matter of fact lower in group B than in group A (confirm by measuring UCP1 expression and quantifying cell number/mass). This interpretation may be rather applicable to an experiment, in which different cell types are assayed, as the interpretation assumes parallel alterations of basal mitochondrial OCR, oxygen consumption linked to ATP-synthesis, maximal respiration, and non-mitochondrial O2-consumption, which is not plausibly caused by a single treatment variable, but more likely by cell-type-intrinsic characteristics. After all, if a treatment is applied to more than one cell type, different cells have to be compared in a treated and untreated manner to dissect treatment-dependent and cell-type-dependent effects.
This example highlights the pitfalls that may arise from only looking at single metrics (UCP1-dependent respiration) or data expressed as ratios (UCP1-dependent respiration to proton leak-linked respiration), which would give a completely wrong impression, if raw oxygen consumption traces were not presented.
The problem becomes even more prominent, when oxygen consumption rate traces are scaled using the “Baseline” function (Figure 6A, right). In this example the raw traces of Figure 6A were scaled by selecting the rate measurement prior to the Isoproterenol injection. Transforming the data suddenly implies a dramatic difference regarding basal oxygen consumption and UCP1-dependent respiration between groups, which may be in fact not true and only an artifact caused by scaling the data.
Quantification and Statistical Analysis
-
1.
Open assay result file (∗.asyr) with Wave Desktop.
-
2.
Go to the “Overview” analysis view.
-
3.
Switch to “Well mode” to display individual traces for each well. Check each and every well for extreme outliers (wells in row “A” and “H,” and wells in column “1” and “12” generally tend to show slightly higher oxygen consumption rates than the remaining wells) and incomplete/failed injections. Exclude these wells from the analysis. We want to stress once more the importance of looking at single wells and not only at group averages.
-
4.
Export assay result data to Microsoft Excel.
CRITICAL: In general, we strongly advise to calculate certain metrics, such as basal mitochondrial oxygen consumption, proton leak-linked respiration, and non-mitochondrial oxygen consumption, only from rather stable respiration rates.
An excellent more general framework regarding the execution of microplate-based respirometry and analysis of oxygen consumption data can be found here (Divakaruni et al., 2014).
An alternative way to present the increase in UCP1-dependent oxygen consumption after the addition of Isoproterenol is to calculate a ratio between UCP1-dependent respiration and proton leak-linked respiration.
Similarly, oxygen consumption rate traces can be scaled using the “Baseline” function of the Wave software. The “Baseline” function enables rate data to be expressed as a percent of a selected rate measurement. This feature can help to minimize variability between experimental groups, and to visualize the acute effect of an injection in a clear and simple manner. We recommend selecting either the rate measurement prior to the addition of Oligomycin or the rate measurement prior to the injection of Isoproterenol.
Nevertheless, we want to emphasize that using the “Baseline” function or calculating ratios very often reduces the amount of information gained from an experiment, and can sometimes even lead to serious misinterpretations. Therefore, we want to encourage researchers to not publish scaled data, ratios, or fold changes without presenting the original raw oxygen consumption traces.
To illustrate the analysis process we calculated the raw oxygen consumption metrics (Table 2) from the raw rate measurements (Table 1) corresponding to the OCR traces in Figure 4A.
Table 2.
OCR Metrics Calculated from the Raw Rate Measurements According to the Guidelines Stated Above “Calculation of Raw Oxygen Consumption Metrics”
| Group A |
Group B |
|||
|---|---|---|---|---|
| Well |
B3 |
B4 |
C3 |
C4 |
| Metric | OCR (pmol O2/min) | OCR (pmol O2/min) | ||
| Non-mitochondrial oxygen consumption | 85.92 | 84.32 | 93.84 | 95.07 |
| Basal mitochondrial oxygen consumption | 131.39 | 131.04 | 125.76 | 129.05 |
| Proton leak-linked respiration | 24.42 | 22.44 | 22.92 | 24.21 |
| ATP-dependent oxygen consumption | 106.97 | 108.60 | 102.84 | 104.84 |
| UCP1-dependent respiration | 311.55 | 313.77 | 122.13 | 115.49 |
| Maximal respiration | 345.31 | 350.13 | 311.00 | 313.29 |
| UCP1-dependent respiration/Proton leak-linked respiration | 12.76 | 13.98 | 5.33 | 4.77 |
Table 1.
Raw Rate Measurements of Representative Wells B3, B4, C3, and C4
| Group A |
Group B |
|||
|---|---|---|---|---|
| Well |
B3 |
B4 |
C3 |
C4 |
| Time (min) | OCR (pmol O2/min) | OCR (pmol O2/min) | ||
| 0.80 | 218.71 | 216.88 | 226.50 | 229.61 |
| 7.48 | 217.69 | 215.91 | 223.07 | 225.45 |
| 14.15 | 217.28 | 215.46 | 221.21 | 223.90 |
| 20.82 | 217.31 | 215.36 | 219.60 | 224.12 |
| Injection: Oligomycin | ||||
| 27.73 | 128.20 | 110.43 | 129.06 | 124.69 |
| 34.37 | 120.95 | 108.85 | 118.79 | 119.28 |
| 41.07 | 118.41 | 113.78 | 117.85 | 123.35 |
| 47.73 | 110.34 | 106.76 | 116.76 | 119.28 |
| Injection: Isoproterenol | ||||
| 54.62 | 311.19 | 306.97 | 218.38 | 207.12 |
| 61.27 | 402.26 | 406.83 | 214.66 | 219.97 |
| 67.93 | 416.32 | 418.52 | 219.28 | 223.29 |
| 74.63 | 421.89 | 420.53 | 238.89 | 234.77 |
| Injection: FCCP | ||||
| 81.47 | 431.22 | 434.44 | 327.99 | 336.25 |
| 88.13 | 406.96 | 401.83 | 365.76 | 361.12 |
| 94.87 | 390.60 | 392.62 | 404.84 | 408.36 |
| Injection: Antimycin A | ||||
| 101.67 | 90.60 | 86.62 | 100.60 | 101.98 |
| 108.33 | 87.12 | 84.33 | 98.64 | 98.44 |
| 115.02 | 85.92 | 85.48 | 96.17 | 97.48 |
| 121.67 | 86.24 | 84.32 | 93.84 | 95.07 |
Limitations
Among the disadvantages are that the price of the Seahorse XFe Analyzer platform itself and the corresponding consumables are quite high (especially in comparison to ordinary Clark-type oxygen sensors), which clearly limits the number of laboratories and scientists having access to this technology.
Another distinct disadvantage is the limited number of four injections per well and measurement, which effectively excludes titrating injection chemicals until a maximal response has been achieved and using different substances at the same time.
Furthermore, the Seahorse XFe Analyzer platform can measure oxygen consumption only discontinuously, as against continuous monitoring of respiration using high-resolution respirometry. This can be considered as only a slight disadvantage, since the assay protocol can be modified in a way to achieve almost continuous readings, which effectively allows capturing fast and transient events.
Another limitation of this technology is the fact that oxygen consumption is measured as a surrogate marker for UCP1 activity. Nevertheless, as the protonophoric activity of UCP1 cannot be routinely and directly recorded in intact cells, oxygen consumption still represents the best indirect readout for the reliable approximation of UCP1 activity. However, we want to emphasize that oxygen consumption is a complex function of various processes. Especially the variable non-linear nature of proton leak and substrate oxidation kinetics contributes a significant portion of uncertainty to cellular respirometry centered on UCP1-dependent thermogenesis. Conclusively, UCP1-knockout adipocytes or cells with very low UCP1 expression, such as white adipocytes, are valuable tools to demonstrate beyond a shadow of doubt that a given treatment or condition specifically affects UCP1-dependent respiration and not any of the other factors.
The last limitation is the extraordinarily high oxidative capacity of brown and brite adipocytes, which may result in oxygen consumption rates beyond the dynamic range of the Seahorse XFe96 Analyzer. Insufficient reoxygenation between rate measurements, hypoxic periods during a measurement command, or an inadequate recovery of the oxygen-sensitive fluorescent probe can substantially distort respiration measurements during phases of high oxygen consumption causing sharp peaks in respiration rates followed by declining and implausible recordings (discussed in the Troubleshooting section “Problem 10”). Fortunately, incorrect rate measurements can easily be identified by checking the raw oxygen partial pressure values using the Wave Desktop software. Moreover, Agilent offers the Seahorse XFe24 Analyzer as an alternative to the XFe96 platform. Seahorse XF24 V28 PS cell culture plates have a higher microchamber volume, which facilitates characterizing highly oxidative cells, such as thermogenic adipocytes.
Troubleshooting
Problem 1
Cells do not differentiate well or have low/no UCP1 expression.
Potential Solutions
Time between seeding and pre-adipocytes reaching 80%–100% confluency (start of the induction phase) is too long. A high number of cell divisions during the proliferation phase can negatively affect the differentiation potential. The proliferation phase should not exceed 5–6 days. Increasing the seeding density by using more fat depots per experiment will reduce the time pre-adipocytes require to reach the desired cell density.
Induction phase was initiated too late and cells were heavily overgrown. Start the induction phase earlier.
Pre-adipocytes are heavily contaminated with erythrocytes. Erythrocytes do not proliferate but still occupy growth surface. After several medium changes a portion of red blood cells will be washed away, which will cause a patchy appearance of the cell monolayer. Add more red blood cell lysis buffer and/or increase the incubation time.
Primary cultures of brown adipocytes are strongly recommended.
Problem 2
The position of a well on the cell culture plate systematically influences oxygen consumption rates.
Potential Solution
Differentiation into adipocytes can depend on the position on the plate (the outer wells may show an altered differentiation rate). Mix the cells well prior to plating, ensure that cells are thoroughly resuspended, distribute groups symmetrically across the plate, or even exclude outer wells from the analysis.
Problem 3
Basal oxygen consumption rate of cells is too high or too low following the reverse transfection procedure
Potential Solutions
Ensure seeding an appropriate number of cells per well onto the Seahorse XF96 cell culture microplate during reverse transfection procedure. Optimal cell number depends on the cell type or condition and should be determined experimentally. Basal oxygen consumption rates should not exceed 250 - 275 pmol O2/min.
Excess cell death following the siRNA treatment. Transfection reagent is cytotoxic. Lower the concentration of Lipofectamine RNAiMAX to 2.5 μg/mL.
High amount of siRNA causing cytotoxic effects. Lower the concentration of the siRNA to 10, or 25 nM.
Gene of interest is coding for a protein that is essential for cell survival. Use less siRNA to decrease knockdown efficiency.
Pipetting and/or resuspension of cells was too harsh. Adipocytes are fragile, reduce the number of pipetting steps and always treat gently. Use pipette tips with a wider opening to reduce shear forces.
Problem 4
Knockdown efficiency is low
Potential Solution
Titrate the concentration of Lipofectamine RNAiMAX (2.5–5 μg/mL) and the siRNA (10–50 nM), try different combinations of concentrations. Sometimes less is more.
Problem 5
I do not want to include BSA in the respiration assay medium.
Potential Solution
Adding BSA to the respiration assay medium has a certain disadvantage due to BSA’s characteristic of very efficiently binding some compounds. Nevertheless, adding at least a certain amount of BSA to the respiration assay medium is a prerequisite for measuring UCP1-dependent respiration (Li et al., 2017; Li et al., 2014).
Problem 6
Oxygen consumption is not different between WT and UCP1KO adipocytes, or between groups with supposedly high and low UCP1 protein levels.
Potential Solution
A) Oligomycin Injection Is Mandatory
Both UCP1 wild-type (WT) and knockout (UCP1KO) brown fat cells drastically increase oxygen consumption in response to the addition of Isoproterenol (Figure 7A). In contrast to the in vivo situation (Kazak et al., 2017), primary cultures of brown UCP1KO adipocytes do not experience a global mitochondrial dysfunction, as maximal FCCP-stimulated respiration values of cultured WT and UCP1KO adipocytes are identical (oxygen consumption following the addition of FCCP is largely driven by electron transport chain capacity, which can serve as a general surrogate marker for electron transport chain complex protein levels) (Li et al., 2014). Oxygen consumption rates of both groups only differ after the Oligomycin injection. Oxygen consumption of UCP1KO adipocytes immediately drops down to basal respiration rates (similar to OCR prior to the injection of Isoproterenol), whereas WT adipocytes still consume almost twice as much oxygen as compared to basal respiration. This example clearly demonstrates that only the combination of Oligomycin and Isoproterenol enables a discrimination between WT and UCP1KO adipocytes, when microplate-based respirometry is employed to quantify UCP1 activity. In other words, if the measured Isoproterenol-stimulated respiration can be inhibited by the addition of Oligomycin, then it is not UCP1-dependent.
Figure 7.
Highlighting the Importance of Oligomycin in Determining UCP1-Dependent Respiration
Data are presented as means ± SEM.
(A) OCR traces of cultures of primary brown wild-type (WT) and UCP1-knockout (UCP1KO) adipocytes. Arrows indicate injections. Compounds were added in the following order: Isoproterenol (Iso), Oligomycin (Oligo), FCCP, Antimycin A (AA).
(B) Comparing different injection regimens. OCR traces of cultures of primary brown adipocytes measured on a Seahorse XFe24 Analyzer. Arrows indicate injections. Compounds were added in the following order: ●, Isoproterenol (Iso), Oligomycin (Oligo), FCCP, Antimycin A (AA). ■, Oligomycin (Oligo), Isoproterenol (Iso), FCCP, Antimycin A (AA).
ATP-consuming futile cycles, such as lipid cycling (lipolysis and simultaneous re-esterification), can contribute to Isoproterenol-stimulated respiration, when the ATP synthase is still operating. Thus, the addition of Oligomycin prevents a significant overestimation of UCP1-dependent respiration (Li et al., 2017; Li et al., 2014). We compared two different injection regimes: Oligomycin followed by Isoproterenol vs. Isoproterenol followed by Oligomycin (Figure 7B). Oxygen consumption of WT cells is not different after the addition of both substances independent of the order of addition. A decisive advantage of the first injection regime (Oligomycin followed by Isoproterenol) is that maximal observed rates after the addition of Isoproterenol are overall slightly lower, as the ATP synthase is already inhibited at this point, which reduces the chances of oxygen consumption rates being out of the dynamic range, i.e., preventing hypoxia.
B) Wild-Type Cells Lose UCP1 Expression during Cultivation/Differentiation
Some immortalized brown fat cell lines tend to have relatively low or even no UCP1 expression, when cultured in microplates tailored for bioenergetic analyses. Confirm UCP1 expression on protein level.
Problem 7
Injection ports are not completely empty after the measurement.
Potential Solutions
Do not add BSA to the injection working solutions.
Avoid empty injection ports. Fill all ports irrespective of use.
Loading the injection ports has to be performed at an appropriate speed using a certain pressure to ensure that the injection solution reaches the bottom part of the port. Nevertheless, applying too much pressure may overcome the surface tension keeping the injection solution in the injection ports, which may cause premature emptying of the ports, or lead to injection solution partially leaking into the well at an undesired time.
Problem 8
Large variation between groups. How can I normalize my data?
Potential Solution
Oxygen consumption data can be normalized to various readouts, such as protein content, DNA content, nuclei count, and cell number. The most accurate method is counting nuclei or cells, but this approach is very time-consuming and without an automated process not feasible (Divakaruni et al., 2014). However, based on our experience normalization very often does not reduce variability but in turn introduces even more variation into the dataset. Optimizing the isolation, seeding and differentiation procedure and the execution of the oxygen consumption assay itself can reduce variation to an absolute minimum.
Moreover, scaling data with the “Baseline” function can sometimes aid in interpreting respiration data. Scaling data to different respiration states may serve as an approximate surrogate marker for certain parameters: basal respiration correlates with cell number, whereas basal leak respiration and FCCP-induced maximal respiration correlate with mitochondrial content. However, this measure has to be treated with caution.
Problem 9
Oxygen consumption does not increase following the addition of Isoproterenol (UCP1-dependent oxygen consumption)
Potential Solutions
UCP1 expression is low. Confirm UCP1 expression on protein level.
Isoproterenol concentration was too low or too high. We observe effects starting at concentrations as low as 50 nM. However, very high concentrations of Isoproterenol can cause an excess release of fatty acids, which may exert cytotoxic effects. Isoproterenol concentration should not exceed 500 nM–1 μM.
Isoproterenol was degraded, protect from light.
BSA concentration was too high. Cells with a low lipolytic capacity may be assayed in the presence of less BSA. If the release of free fatty acids is low, an excess amount of BSA may prevent UCP1 activation. Titrate BSA concentration.
Problem 10
Oxygen consumption rates rapidly peak in response to the Isoproterenol injection followed by a gradual decrease (frequently observed with brown adipocytes).
Potential Solutions
Reduce/titrate Isoproterenol concentration.
Inject Oligomycin prior the addition of Isoproterenol.
Do not additionally add pyruvate to the respiration assay medium. Pyruvate favors the occurrence of this phenomenon.
Switch to the Seahorse XFe24 Analyzer. Especially V28 PS cell culture plates offer an increased microchamber volume, which reduces the occurrence of hypoxic phases during a measurement command.
Problem 11
Treatment groups with lower lipolysis rates do not reach the same level of maximal respiratory capacity in response to FCCP as control cells despite no expected difference in mitochondrial number.
Potential Solution
BSA in the Seahorse respiration assay medium can interact with FCCP negatively affecting its uncoupling properties. In treatment groups with lower lipolysis rates, less free fatty acids are released, which would be scavenged by BSA, resulting in a lower occupation of BSA’s binding sites and this may in turn favor the binding of FCCP to BSA (Figure 8).
Figure 8.
Potential Connection between Lipolysis Rate and the Magnitude of FCCP-Induced Uncoupling
Data are presented as means ± SEM. OCR traces of cultures of primary brown adipocytes. Inhibitors targeting ATGL (Ai) and HSL (Hi) were used to block lipolysis. The effect of different concentrations of FCCP on OCR was compared during active or pharmacologically blocked lipolysis.
Titrate FCCP to higher concentrations to ensure enough free FCCP to reach maximal uncoupling.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Dr. Yongguo Li (yongguo.li@tum.de).
Materials Availability
This study did not generate any unique materials or reagents.
Data and Code Availability
We did not generate any unique datasets or code.
Acknowledgments
This work was supported by the German Research Foundation (KL 973/12-1, LI 3716/1-1) and the Else Kröner-Fresenius-Stiftung (EKFS). The graphical abstract was created with BioRender.com.
Author Contributions
Conceptualization, J.O, A.B.-H., and Y.L.; Investigation, J.O., A.B.-H., and Y.L.; Writing – Original Draft, J.O., A.B.-H. and Y.L.; Writing – Review & Editing, J.O., A.B.-H., and Y.L.; Funding Acquisition, M.K and Y.L.; Supervision, J.O., T.F., M.K., and Y.L.
Declaration of Interests
The authors declare no competing interests.
Contributor Information
Josef Oeckl, Email: josef.oeckl@tum.de.
Yongguo Li, Email: yongguo.li@tum.de.
References
- Divakaruni A.S., Paradyse A., Ferrick D.A., Murphy A.N., Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309–354. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- Gerencser A.A., Neilson A., Choi S.W., Edman U., Yadava N., Oh R.J., Ferrick D.A., Nicholls D.G., Brand M.D. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal. Chem. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidor M.S., Winther S., Basse A.L., Petersen M.C.H., Cannon B., Nedergaard J., Hansen J.B. An siRNA-based method for efficient silencing of gene expression in mature brown adipocytes. Adipocyte. 2016:175–185. doi: 10.1080/21623945.2015.1111972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L., Chouchani E.T., Stavrovskaya I.G., Lu G.Z., Jedrychowski M.P., Egan D.F., Kumari M., Kong X., Erickson B.K., Szpyt J. UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proc. Natl. Acad. Sci. U S A. 2017;114:7981–7986. doi: 10.1073/pnas.1705406114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fromme T., Klingenspor M. Meaningful respirometric measurements of UCP1-mediated thermogenesis. Biochimie. 2017;134:56–61. doi: 10.1016/j.biochi.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Li Y., Fromme T., Schweizer S., Schöttl T., Klingenspor M. Taking control over intracellular fatty acid levels is essential for the analysis of thermogenic function in cultured primary brown and brite/beige adipocytes. EMBO Rep. 2014;15:1069–1076. doi: 10.15252/embr.201438775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Schnabl K., Gabler S.M., Willershauser M., Reber J., Karlas A., Laurila S., Lahesmaa M., U Din M., Bast-Habersbrunner A. Secretin-activated brown fat mediates prandial thermogenesis to induce satiation. Cell. 2018;175:1561–1574.e12. doi: 10.1016/j.cell.2018.10.016. [DOI] [PubMed] [Google Scholar]
- Li Y., Schwalie P.C., Bast-Habersbrunner A., Mocek S., Russeil J., Fromme T., Deplancke B., Klingenspor M. Systems-genetics-based inference of a core regulatory network underlying white fat browning. Cell Rep. 2019;29:4099–4113.e5. doi: 10.1016/j.celrep.2019.11.053. [DOI] [PubMed] [Google Scholar]
- Mookerjee S.A., Nicholls D.G., Brand M.D. Determining maximum glycolytic capacity using extracellular flux measurements. PLoS One. 2016;11:e0152016. doi: 10.1371/journal.pone.0152016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer S., Oeckl J., Klingenspor M., Fromme T. Substrate fluxes in brown adipocytes upon adrenergic stimulation and uncoupling protein 1 ablation. Life Sci. Alliance. 2018;1:e201800136. doi: 10.26508/lsa.201800136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We did not generate any unique datasets or code.