Summary
We present a reproducible protocol to prepare droplet-embedded vesicles (DEVs) consisting of an oil droplet embedded within a phospholipid bilayer. This model system mimics a cellular lipid droplet (LD) in physical contact with the endoplasmic reticulum (ER) bilayer. It has the advantage that the lipid composition and the biophysical properties of the droplet and the bilayer are controlled and tunable. DEVs can be used to study LD biogenesis factors and determinants of protein binding between ER and LD interfaces.
For complete details on the use and execution of this protocol, please refer to Chorlay and Thiam (2020) and Santinho et al. (2020).
Graphical Abstract
Highlights
-
•
Droplet-embedded vesicles (DEVs) model lipid droplet (LD) and ER contiguity
-
•
Biophysical properties of model LD and membrane of the DEV can be modulated
-
•
DEVs are a powerful tool for testing and predicting regulatory mechanisms of LDs
We present a reproducible protocol to prepare droplet-embedded vesicles (DEVs) consisting of an oil droplet embedded within a phospholipid bilayer. This model system mimics a cellular lipid droplet (LD) in physical contact with the endoplasmic reticulum (ER) bilayer. It has the advantage that the lipid composition and the biophysical properties of the droplet and the bilayer are controlled and tunable. DEVs can be used to study LD biogenesis factors and determinants of protein binding between ER and LD interfaces.
Before You Begin
-
1.
The day before making the droplet-embedded vesicles (DEVs), make 10 mL of Bovine Serum Albumin solution 10% (w/w) in Milli-Q water. In the protocol, you only need a few thousand microliters of the BSA solution. However, we recommend preparing a volume of 10 mL which can be stored at 4°C and reused for a few weeks.
-
2.
Prepare a HEPES-potassium acetate-magnesium chloride buffer (HKM): 50 mM HEPES, 120 mM KOAc, and 1 mM MgCl2 in Milli-Q water and adjust pH to 7.4 with diluted HCl or KOH. Osmolarity should be around 270 ± 10 mOsm.
-
3.
Prepare 50 mL of a sucrose solution at 5 g/L in Milli-Q water. Check that the osmolarity is around 270 ± 10 mOsm.
-
4.
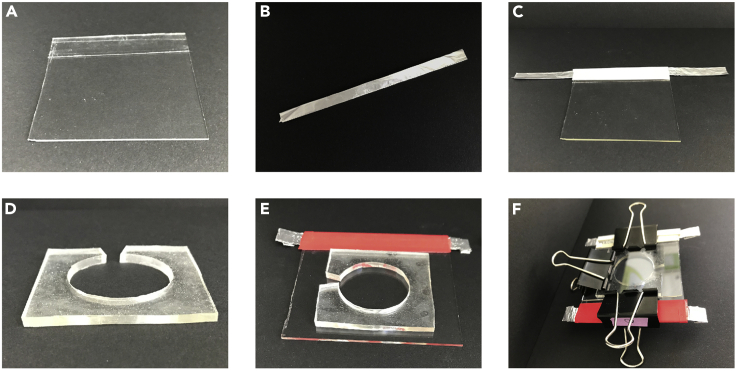
Prepare the electroformation chamber of the giant unilamellar vesicles (GUVs) (Dimova and Marques, 2019). Make two 6 cm square indium tin oxide (ITO) coated glass plate (Figure 1A). Cut two aluminum foil rectangles of 15 cm × 5 cm and fold them each six times along their length (Figure 1B). Place each folded foil on the border of the two ITO coated glass plates. Make sure to place them it on the electrically conductive border. Use adhesive tape to fix them (Figure 1C). The aluminum foils must protrude on both sides. Prepare a 3 mm thick polydimethylsiloxane polymer slab (10 × 10 cm), and punch in it a large hole that will represent the GUV preparation chamber (Figure 1D). This chamber will be sandwiched between the conductive faces of the ITO glass plates (Figures 1E and 1F).
Figure 1.
Preparing the Electroformation Chamber
Related to the “Before You Begin” section.
(A) Square plate of Indium Tin Oxide coated glass (6 × 6 cm).
(B) Aluminum strip (15 cm long) serving as a contact electrode.
(C) Aluminum strip taped on the conductive face at one extremity of the ITO coated glass plate.
(D) Picture of the PDMS spacer used to form the electroformation chamber (3 mm width).
(E) Positioning of the PDMS spacer on the ITO coated glass plate.
(F) Picture whole electroformation chamber: second coated glass plate is clipped to the first one with the PDMS spacer in between.
Note that GUVs containing sucrose can be made by other methods that are not described here, such as by the inverted emulsion approach (Pautot et al., 2003). The latter for instance enables to generate GUVs faster but with a lower yield and with oil molecules being present in the GUV membrane, already before DEVs are generated.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, and Recombinant Proteins | ||
| DOPC (1,2-dioleoyl-sn-3-glycero-3-phosphocholine) | Avanti Polar Lipids | 850375C |
| Rhodamine-DOPE 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) | Avanti Polar Lipids | 810150P |
| Chloroform containing 0.5%–1.0% ethanol as stabilizer | Sigma Aldrich | 288306 |
| Triolein | Sigma Aldrich | T7140 |
| BODIPY 493/503 | Thermo Fisher | D3922 |
| HCS LipidTox Deep Red | Thermo Fisher | H34477 |
| Bovine Serum Albumin (BSA) | Sigma Aldrich | A7906-100G |
| HEPES | Sigma Aldrich | 54457–250-F |
| CH3COOK (potassium acetate) | Sigma Aldrich | P1190 |
| MgCl2 | Sigma Aldrich | M8266-100G |
| Sucrose | Sigma Aldrich | 59378-500G |
| Other | ||
| Coverslip | Menzel | P35G-0-20-C |
| Indium Tin Oxide coated glass (150 × 150 mm) | Delta-Technologies | CB-90IN-1511 |
| PDMS | DOW CORNING | SYLGARD 184 / 1.1kg |
| Eppendorf tubes 0.5 and 2 mL | Eppendorf | N°0030121023 and N°0030125150 |
| Sonicator | Electris | DVELUC500CH |
| Vortex | Scientific Industries | Vortex Genie 2 |
| Dessicator | Deltalab Spain | 19232 |
| Tube rotator | Ika loopster basics | n/a |
| Microsyringe 10 μL, fixed needle | Hamilton | 701N |
| Osmometer | Roebling | Micro-osmometer Autocal Type 13 D-14129 |
| Signal generator | Rigol | DG1022 |
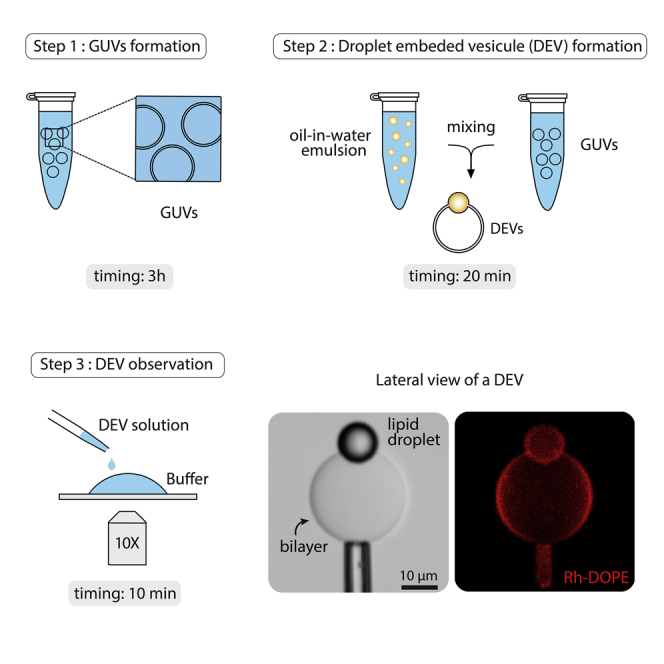
Step-By-Step Method Details
Formation of Giant Unilamellar Vesicles (GUVs)
Timing: 3 h
Electroformation of GUVs which will serve as the necessary bilayers for the production of DEV.
-
1.
Wash the two ITO coated glass plate and the PDMS spacer with water.
-
2.
Dry the two ITO coated glass plate and the PDMS spacer and wash it with ethanol.
CRITICAL: When chloroform is used, work under a laminar flow hood and wear gloves.
-
3.
Wash the two ITO coated glass plate with chloroform.
Note: ITO coated glasses and PDMS chamber can be reused dozens of times to generate GUVs.
-
4.
Dilute a DOPC 99.5% (w/w) and Rh-DOPE 0.5% (w/w) mixture in 200 μL of chloroform at 2 mg/mL for DOPC.
-
5.
Homogenize the phospholipid mixture by vortexing.
-
6.
Collect 20 μL with a microsyringe and make little droplet spots (2 μL) on the ITO coated glass inside the PDMS chamber (Figure 2A).
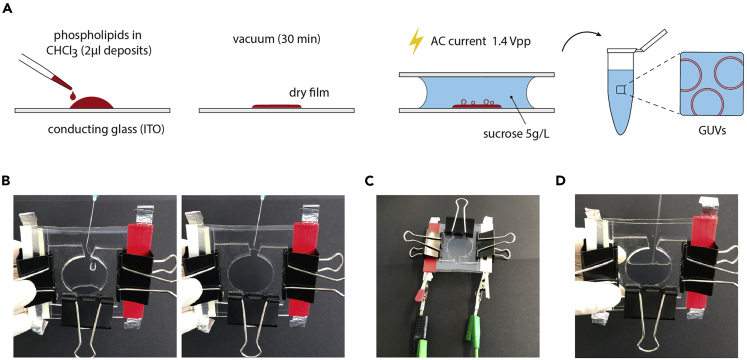
Figure 2.
GUV Formation
(A) Step by step schematic illustration of the GUV formation protocol. From the left to the right: Deposit of the phospholipid mixture diluted in chloroform on the conductive face of the ITO coated glass plate (step 6); Lipid mixture is dried under vacuum during 30 min (step 8); Electroformation chamber is formed and sucrose solution is added (step 9), then submitted to a AC current of 1.4 Vpp (step 10); GUVs are collected into Eppendorf tube (step 11).
(B) Picture of the sucrose solution injection into the electroformation chamber. Left: beginning of the injection. Right: end of the injection (step 9).
(C) Electroformation chamber connected to the signal generator in order to apply AC current (step 10).
(D) Illustration of the collect of the sucrose solution containing the GUV with a Pasteur pipette (step 11).
Note: The phospholipid mixture can be stored at −20°C by saturating the upper volume of the tube with argon before closing the tube. If there is no evaporation or oxidation, the mixture can be reused for a few weeks.
-
7.
Seal the electroformation chamber with the other ITO coated glass plate as shown in (Figure 1F).
-
8.
Desiccate the lipid film during 1h in a vacuum chamber (Figure 2A).
-
9.
Clip the chamber with fold back clips as shown in (Figure 1F) and add very slowly 3 mL of sucrose solution in the chamber (Figures 2A and 2B, Methods Video S1).
CRITICAL: While injecting sucrose into the chamber, try to minimize the number of air bubbles that form in the sucrose volume. This may alter the formation of GUV.
-
10.
Apply 100 Hz AC voltage at 1.4 Vpp by using conductive alligator clip between the generator and the aluminum strips. Maintain it for at least 2h (Figures 2A and 2C).
-
11.
Collect GUVs gently with a Pasteur pipette in two Eppendorf tubes of 1,5mL (Figures 2A and 2D, Methods Video S2).
-
12.
Store them at 4°C.
Note: GUVs stored at 4°C can be used during approximately two weeks.
Note: To check if the electroformation process is successful, one can observe the GUVs solution under confocal microscopy (see section observation of DEVs for more details) by using the following dilution: 20 μL of the sucrose-GUV solution into 80 μL of HKM buffer on a Menzel coverslip. This dilution allows GUVs to settle down on the coverslip, since the HKM buffer is lighter than the sucrose encapsulated by the GUVs. By using a 10× objective dozens of GUVs, between 5 μm and 40 μm in size, can be observed per target region.
Formation of Droplet-Embedded Vesicles (DEVs)
Timing: 20 min
Production of DEVs by contacting the bilayers of the GUVs with an oil emulsion.
-
13.
Prepare two empty Eppendorf tubes of 0.5 mL cleaned with air stream.
-
14.
Add 70 μL of HKM buffer in the first Eppendorf tube (Figure 3A). Make sure your HKM buffer is clean and dust-free by filtering it with 0.45μm filter. To double check if the buffer does not contain huge dusts, one can place a small volume of buffer (e.g., 5 μL) on a Menzel coverslip and observe it under the microscope.
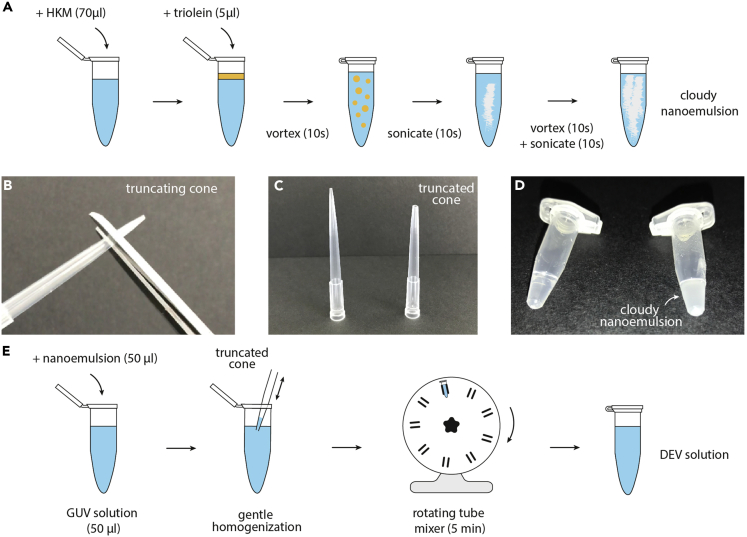
Figure 3.
DEVs Formation by Mixing an Oil Nano-Emulsion with a GUV Solution
(A) Step by step schematic illustration of how to make the nano-emulsion with triolein and HKM. From the left to the right: HKM and oil are added in an Eppendorf tube (steps 14 and 15); the Eppendorf tube is vortexed resulting in a coarse emulsion (step 18); the solution is sonicated until a cloudy nano-emulsion is obtained and then homogenized by vortexing (steps 19–21).
(B) The micropipette tip cone is truncated, used in (step 17).
(C) Picture of a truncated micropipette tip cone (right) compared to a normal tip cone (left), (step 17).
(D) Oil-buffer solution before (left, step 15) and after sonication (right, step 21), resulting in a cloudy solution indicating the formation of the nano-emulsion.
(E) Step by step protocol of the mixing of GUVs solution with the nano-emulsion to form DEVs. From left to right: the nano-emulsion is injected into the GUV solution with the truncated cone (steps 22 and 23). The double arrow heads designate the back and forth mixing with the micropipette to gently mix the emulsion with the GUVs (step 24). Before observation, the previous solution is homogenized with a rotating tube mixer to allow further mixing (step 25). After 5 min, the DEV solution is ready for observation. (step 26).
-
15.
Add 5 μL of triolein oil into the HKM volume and close the Eppendorf tube (Figure 3A).
Optional: Different oil types can be used and be fluorescently stained for subsequent imaging by fluorescence microscopy.
-
16.
In the other Eppendorf tube, add gently 50 μL of previously produced GUVs solution (from step 12) and keep it within easy reach.
-
17.
Prepare a 200 μL micropipette to collect the 50 μL and cut the cone of the micropipette 5 mm from the tip to obtain a large aperture. This modified cone is further referred as truncated cone. (Figures 3B and 3C, Methods Video S3).
-
18.
Vortex the first Eppendorf tube for 10 s to make a coarse emulsion (Figure 3A).
-
19.
Sonicate this emulsion with the water bath sonicator for 10 s until the solution starts to be cloudy. Do not sonicate too long to avoid the formation of a gel phase. (Figure 3A).
-
20.
Vortex again during 10 s (Figure 3A).
-
21.
Sonicate again during 10 s to obtain a nano-emulsion (Figure 3A). At this stage, you are supposed to get a milky solution (Figure 3D), if necessary, vortex and sonicate it again.
-
22.
Collect rapidly a volume of 50 μL of the nano-emulsion with the truncated cone (Figures 3B, 3C, and 3E).
-
23.
Pour this volume very gently into the second Eppendorf tube containing the GUVs solution (Figure 3E, Methods Video S4).
-
24.
Mix back and forth very gently with the same truncated pipette tip to homogenize the GUVs and nano-droplets mixture (Figure 3E, Methods Video S4).
Note: This step brings GUVs into contact with the nano-oil droplets. It promotes the formation of DEV and it is a crucial step for a high yield of DEVs.
-
25.
Then, immediately place the Eppendorf tube on a rotating tube mixer during 5 min. This allows further mixing of the GUVs and the nano-emulsion while avoiding precipitation and aggregation of the DEVs and GUVs at the bottom of the Eppendorf tube. (Figure 3E).
-
26.
DEV solution is ready for observation.
Observation of Droplet-Embedded Vesicles (DEVs)
Timing: 10 min
How to observe the DEVs produced in the previous step using a confocal microscope
-
27.
Take a Menzel glass coverslip for confocal microscopy observation.
-
28.
With a pipette, place an 80 μL of the BSA solution on the coverslip and wait for 10 s (Figure 4A).
Figure 4.
Observation of DEVs under the Confocal Microscope
(A) Step by step illustration of the DEV observation by microscopy. From left to right: the BSA solution is placed on the glass coverslip (step 28) and then discarded after 10 s. The HKM buffer solution is used to rinse the BSA-treated spot; rinsing is performed three times for the efficient removal of the excess BSA (step 30). An 80 μL HKM solution is then deposited on the washed coverslip and an 20 μL of DEV solution added to it (step 31). The coverslip is placed under the microscope for observation (step 35).
(B) Picture of the deposited droplet of HKM (step 31) on the coverslip before adding the DEVs and the observation.
(C) Picture of the coverslip of the DEVs in the HKM solution ready for observation (end of step 31).
(D) Representative microscope image of DEVs at 10 min, after DEVs settled down (end of step 35) on the glass coverslip (brightfield 10× objective). DEVs and GUVs without droplet can be seen in the same area. Nano-emulsion oil droplets, not incorporated in GUVs, are indicated by black arrowheads.
(E) Confocal brightfield and fluorescence images of a DEV, side view. The DEV is maintained thanks to a micropipette. Rh-PE is used to report for phospholipids. Objective 10×. Scale bar is 10 μm.
(F) Brightfield and fluorescence side view of a DEV obtained with 63× oil objective. See that the droplet is embedded in between the two leaflets of the bilayer. Note that the phospholipid signal on the droplet monolayer is dimmed because light refraction due to index mismatch between the oil and the buffer (Chorlay and Thiam, 2020); this observation mainly happens with a 63× oil objective. Scale bar is 10 μm.
-
29.
Remove the BSA solution with the pipette. (Figure 4A).
-
30.
To remove the excess of BSA from the coverslip, rinse the BSA spot on the coverslip with 80 μL of HKM buffer and discard this solution (Figure 4A, Methods Video S5). Repeat this process three times.
-
31.
Deposit again 80 μL of the HKM buffer on the glass coverslip, on the BSA-treated spot (Figures 4A and 4B) and place the coverslip on the microscope for observation (Figure 4C).
-
32.
Prepare a 20 μL micropipette to collect 10 μL of the DEV solution with pipette tip also cut 5 mm from the tip.
-
33.
Collect gently 10 μL of the DEVs solution with the micropipette and pour it in the HKM solution on the glass coverslip (Figure 4A).
Optional: You can add oil markers such as Bodipy to the DEV solution to visualize the oil droplets by fluorescence microscopy.
-
34.
Wait 5 min for the DEVs to settle down on the coverslip and for the excess non-GUV-incorporated nano-droplets to float at the top of the sample, cleared from the observation regions of the DEVs (Chorlay and Thiam, 2018; Thiam et al., 2013).
-
35.
DEVs that settle down on the coverslip can be pre-visualized by an 10× objective (Figure 4D).
Note: DEVs can be easily recognized thanks to the dark contrast in brightfield of their oil droplets (due to their refractive index) (Figure 4E).
Expected Outcomes
At the end of the protocol, you can expect to have at least one vesicle over two with droplets incorporated. It corresponds to the formation of 50% of DEVs from the total GUVs. A suitable DEV is a vesicle with a radius of 10 μm with an over-micrometric droplet. (Figures 4D–4F). By using a 63× objective, we determine the DEV radius as the radius of the equatorial plane of the DEV bilayer.
Limitations
This protocol allows the production of DEVs in a reproducible and robust manner. However, there is some variability in the stability of DEVs depending on the physical chemistry of their molecular components. For example, the combination of triolein and diolein for the oil phase may result in less stable DEVs. This can be problematic because stability is essential if one wants to manipulate DEVs with micropipettes or to use them for long-term experiments such as reconstituting biological reactions.
In addition, the production efficiency of DEVs can be modified depending on the type of oil used. For example, viscous oils seem to be more difficult to be incorporate in GUVs.
In conclusion, the ability of an oil to produce a nano-emulsion is crucial for this protocol, especially in step 24. In fact, the smaller the size of the oil droplets (nanometric) is, the higher the efficiency to incorporate them into the bilayer will be.
Troubleshooting
Problem 1
You do not visualize any DEV but only GUV in step 35.
Potential Solutions
It is possible that the GUVs have not been sufficiently brought into contact with the nanodrops of the emulsion. It is therefore necessary to mix the emulsion better with the GUVs by making more back and forth mixing (step 24). If this does not work, check that the nano-emulsion is milky, a sign of the presence of many nanodrops. If necessary, re-sonicate the emulsion.
Problem 2
DEVs seem to be normal when observed from the top, but when positioned in lateral view, droplets are not incorporated into the bilayer as in (Figure 5A).
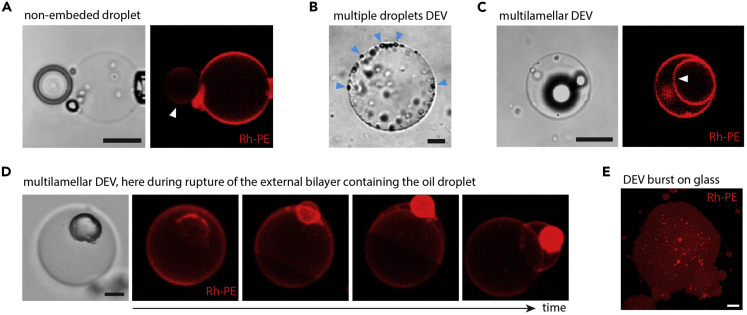
Figure 5.
Illustration of Troubleshooting during DEVs Formation
(A) Representative image of an oil droplet NOT embedded in a bilayer (white arrowhead). If observed in a top view, this system could be mistaken for a DEV, related to problem 2. Scale bar is 10 μm.
(B) Representative image of a DEV presenting multiple small droplets (blue arrowheads), related to problem 3. 63× objective, scale bar is 10 μm.
(C) Representative image (brightfield and fluorescence) of a DEV which exhibit multilamellar bilayers, see arrowhead in fluorescence image. Related to problem 4 section. 63× objective, scale bar is 10 μm.
(D) Time-lapse of the rupture of the external bilayer of a multilamellar DEV. After a pore opening, the external bilayer containing the droplet is ejected. Note that prior to the rupture of the external bilayer, the multilamellar DEV looked similar to a unilamellar DEV, except for the phospholipid fluorescence that was more pronounced inside the vesicle. Related to problem 4. 63× objective, scale bar is 10 μm.
(E) Representative fluorescence image of a DEV which had burst and spread at the surface of a glass coverslip not enough treated with BSA. Related to problem 5 section, 63× objective, scale bar is 10 μm.
Potential Solutions
Check that your HKM buffer is clean (step 14). If some contaminating molecules have polluted your solution, they may act as surfactants recruited to the surface of the nano-oil droplets. This will interfere with the incorporation of the droplets between the two phospholipid leaflets of the GUV, and the droplets may simply adhere to the membrane. Prepare a new HKM buffer solution.
Make more back and forth when mixing the nano-emulsion with the GUVs (step 24), to allow better incorporation of the nano-droplets in the bilayer of the GUVs
Problem 3
DEVs presents a lot of droplets in their bilayer (Figure 5B).
Potential Solutions
Wait longer after deposition of the DEV on the observation coverslip (step 34) to allow the droplets to laterally fuse ant gather as one bigger droplet (Salo et al., 2019). Lateral fusion is helped by higher bilayer surface tension. Thus, you can slightly increase the tension of the bilayer by adding a few microliters of MQ water in the medium. You can also repeat step 21 by sonicating your emulsion for a shorter time. This will result in fewer and larger droplets.
Problem 4
Many DEVs present multilamellar structures more (Figure 5C) or less visible (Figure 5D).
Potential Solutions
After electroformation (step 11), Ensure that GUVs are not multilamellar. If too many are multilamellar, repeat the GUVs electroformation process from the beginning.
Problem 5
You do not visualize any DEV or GUV in step 35.
Potential Solutions
Make sure that after the electroformation step you have enough GUVs in the solution, step 11.
If the mixing of the emulsion with the GUVs is too abrupt (step 24), the vesicles may burst due to the high shear rate. Mix the emulsion with the GUVs more gently.
It is possible for the DEVs to burst and spread over the glass coverslip if the glass slide is not well covered with BSA (step 28). If this is the case and the phospholipids are fluorescently labeled, you should see vesicles burst and spread on the glass slide (Figure 5E).
Be sure to deposit BSA on the glass coverslip used for observation (step 28). Coating the glass with BSA creates a steric barrier between the bilayer of the vesicles and the glass, which prevents bursting.
Resource Availability
Lead Contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact Abdou Rachid Thiam (thiam@ens.fr).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets and code generated or analyzed for this study.
Acknowledgments
This work was supported by grants from the Emergence program of Ville de Paris, ANR-Nanodrop, ANR-17-CE11-0003 and ANR-MOBIL, ANR-18-CE11-0012-01 to A.R.T. We are thankful to all the group members for their valuable comments and critical discussions.
Author Contributions
A.S. and A.C. drafted the manuscript, edited by A.R.T. A.S., A.C., and A.R.T. developed the protocol.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100116.
References
- Chorlay A., Thiam A.R. An asymmetry in monolayer tension regulates lipid droplet budding direction. Biophys. J. 2018;114:631–640. doi: 10.1016/j.bpj.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A., Thiam A.R. Neutral lipids regulate amphipathic helix affinity for model lipid droplets. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201907099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova R., Marques C. CRC Press; 2019. The Giant Vesicle Book. [Google Scholar]
- Pautot S., Frisken B.J., Weitz D.A. Production of unilamellar vesicles using an inverted emulsion. Langmuir. 2003;19:2870–2879. [Google Scholar]
- Salo V.T., Li S., Vihinen H., Hölttä-Vuori M., Szkalisity A., Horvath P., Belevich I., Peränen J., Thiele C., Somerharju P. Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev. Cell. 2019;50:478–493. doi: 10.1016/j.devcel.2019.05.016. [DOI] [PubMed] [Google Scholar]
- Santinho A., Salo V.T., Chorlay A., Li S., Zhou X., Omrane M., Ikonen E., Thiam A.R. Membrane curvature catalyzes lipid droplet assembly. Curr. Biol. 2020;30:2481–2494.e6. doi: 10.1016/j.cub.2020.04.066. [DOI] [PubMed] [Google Scholar]
- Thiam A.R., Antonny B., Wang J., Delacotte J., Wilfling F., Walther T.C., Beck R., Rothman J.E., Pincet F. COPI buds 60-nm lipid droplets from reconstituted water–phospholipid–triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. U S A. 2013;110:13244–13249. doi: 10.1073/pnas.1307685110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets and code generated or analyzed for this study.