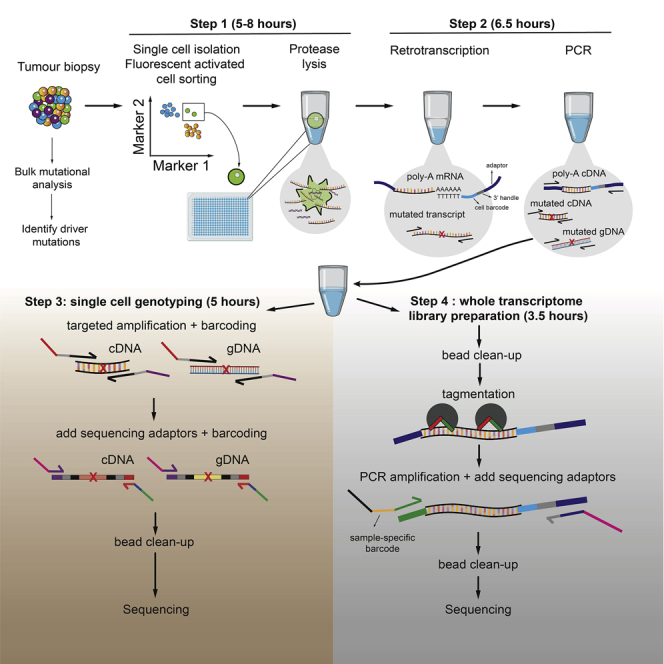
Summary
Single-cell RNA-sequencing technologies are ideally placed to resolve intratumoral heterogeneity. However, the lack of coverage across key mutation hotspots has precluded the correlation of genetic and transcriptional readouts from the same single cell. To overcome this, we developed TARGET-seq, a protocol for TARGETed high-sensitivity single-cell mutational analysis with extremely low allelic dropout rates, parallel RNA SEQuencing, and cell-surface proteomics. Here, we present a detailed step-by-step protocol for TARGET-seq, including troubleshooting tips, approaches for automation, and methods for high-throughput multiplexing of libraries.
For complete details on the use and execution of this protocol, please refer to Rodriguez-Meira et al. (2019).
Graphical Abstract
Highlights
-
•
TARGET-seq provides high-sensitivity single-cell mutational analysis and RNA-sequencing
-
•
TARGET-seq resolves genetic and molecular intratumoral heterogeneity
-
•
Fully automated protocol in robotic platforms for high-throughput analysis
Single-cell RNA-sequencing technologies are ideally placed to resolve intratumoral heterogeneity. However, the lack of coverage across key mutation hotspots has precluded the correlation of genetic and transcriptional readouts from the same single cell. To overcome this, we developed TARGET-seq, a protocol for TARGETed high-sensitivity single-cell mutational analysis with extremely low allelic dropout rates, parallel RNA SEQuencing, and cell-surface proteomics. Here, we present a detailed step-by-step protocol for TARGET-seq, including troubleshooting tips, approaches for automation, and methods for high-throughput multiplexing of libraries.
Before You Begin
Optimization 1: Determine the Number of PCR Cycles Required for Your Specific Cell Type
Timing: 2 days
We strongly recommend that users perform several pilot and optimization experiments to test steps 1–3 of the protocol before proceeding with precious patient samples. These steps should be validated for each cell type and application.
-
1.
Perform an initial test experiment to determine how many cycles of cDNA amplification (step 2, point 44) are required for your specific cell type and/or sample (see note below for specific guidelines on the number of PCR cycles required for each cell type).
-
2.
Due to differences in mRNA content, each cell type will require different degrees of PCR amplification. Ultimately, the aim of this step is to generate enough cDNA for subsequent QC and library preparation. You should aim to generate at least 0.25–0.5 ng/μL per single cell after bead purification of cDNA libraries, and not more than 2 ng/μL.
Note: Generally, cell lines such as K562 (monomyelocytic leukemia cell line) require 18 cycles of amplification, cell lines such as JURKAT (T-cell leukemia cell line; average mRNA 0.35 pg/cell), 20 cycles of amplification and lineage-CD34+ human hematopoietic stem/progenitor cells (HSPCs; average mRNA 0.05 pg/cell), 24 cycles of amplification.
Note: We recommend initially testing at least three different PCR cycling conditions per cell type: the number of cycles estimated using the table below, 2 cycles less and 2 cycles more (i.e., for HSPCs: 22 cycles, 24 cycles, and 26 cycles of PCR amplification).
| Cell Line | Internal mRNA (Average Estimate) | PCR Cycles |
|---|---|---|
| K562 | 0.50 pg | 18 |
| JURKAT, SET2, NALM6 | 0.35 pg | 20 |
| HSPCs | 0.05 pg | 24 |
| HEK293 | 1.6 pg | 18 |
Optimization 2: Design and Validate Target-Specific Primers for RT and PCR Steps
Timing: 2–3 days
CRITICAL: You should always validate the target-specific primers that amplify your amplicons of interest during the initial RT and PCR stages (step 2, see Graphical Abstract). The primers should be validated for (1) successful generation of cDNA libraries and (2) successful specific amplification of target regions. This step is essential because occasionally, addition of primers can generate concatemers that interfere with parallel cDNA amplification and target regions might not be efficiently covered during sequencing if the primers are not specific.
Importantly, the aim of this step is to exclusively assess the successful amplification of target regions of interest, but not the detection of specific mutations in single cells. Therefore, it is not essential to use samples with specific point mutations within the validated amplicons.
-
3.
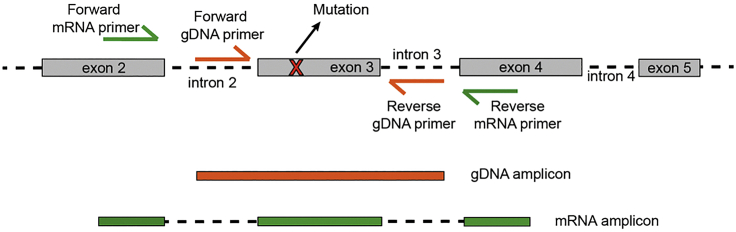
Design your primers using Primer Blast (recommended), Primer3, or an alternative primer design software. An example on how to design TARGET-seq primers using Primer Blast is detailed below (Figure 1).
Figure 1.
Schematic Representation of Primer Design Strategy for mRNA and gDNA Primers for RT and PCR Stages
-
4.You should design your mRNA primers (for the RT step) to meet the following criteria:
-
a.Amplicon size should be in between 250–700 bp. Smaller or larger sizes are not recommended, because they can interfere with cDNA library generation (smaller sizes) or reduced target amplification (longer targets).
-
b.If possible, design your forward primer in the exon before your mutation of interest and your reverse primer, in the exon after. This will allow parallel amplification with gDNA primers.
-
c.Check primer specificity against a transcriptome reference.
-
d.Your primers should have a minimum length of 19 bp and maximum length of 25 bp.
-
e.Design your primers with a melting temperature ranging 57 to 63 C, and a GC content ranging from 20% to 80%.
-
a.
-
5.You should design your gDNA primers (for the PCR step) to meet the following criteria:
-
a.Amplicon size should be in between 250–700 bp. Smaller or larger sizes are not recommended.
-
b.If possible, design your forward primer in the intronic region preceding your mutation of interest and your reverse primer, in the following intron. This will allow parallel amplification with mRNA primers. For exons larger than >700 bp, design a unique primer pair for the amplicon of interest, which will amplify both gDNA and mRNA molecules (combined gDNA+mRNA) and use it exclusively during the PCR step the same concentration as gDNA primers.
-
c.Check primer specificity against a genomic reference and a transcriptomic reference.
-
d.Your primers should have a minimum length of 19 bp and maximum length of 25 bp.
-
e.Design your primers with a melting temperature ranging 57°C to 63°C, and a GC content ranging from 20% to 80%.
-
a.
Optional: cDNA primers for the PCR step contain the same primer sequence used in the RT step, but they also contain and ISPCR adaptor sequence (5′- AAGCAGTGGTATCAACGCAGAGT-3′) in the 5′-end of each primer. Addition of the ISPCR adaptor sequence makes amplification of cDNA specific targets more efficient during the PCR step (Giustacchini et al., 2017), but it is not strictly required for the protocol, and users might choose to use the same target-specific primers as for the RT step.
-
6.
We recommend using PrimerBlast for primer design. A step by step illustration is provided in Figure S1.
-
7.
Validate primer specificity in bulk gDNA (for gDNA primers) and bulk mRNA (for mRNA/cDNA primers). Then, proceed with specificity and amplification efficiency validation in single cells.
-
8.
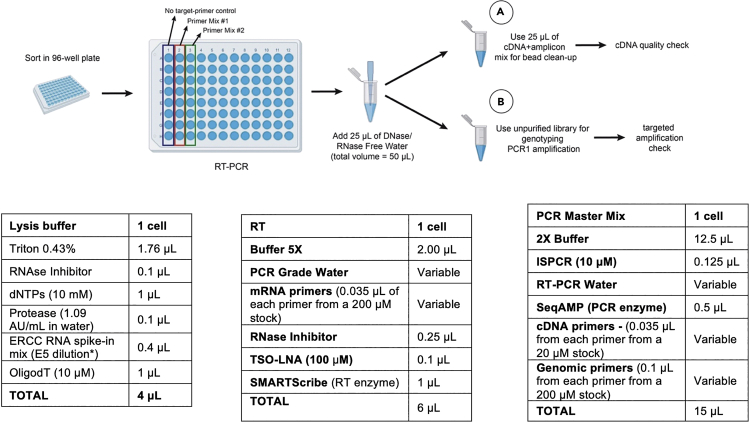
To test the primers, use 8 single cells per condition (Figure 2) in a 96-well plate.
Figure 2.
Schematic Representation of the Experimental Workflow for Primer Testing in 96-Well Plates
-
a.First, sort cells into TARGET-seq buffer in a 96-well plate (step 1 Graphical Abstract, Figure 2), using a total volume of 4 μL of lysis buffer per well. Add RT master mix (without target-specific primers) to 8 cells and RT master mix with target-specific primers for mRNA amplicons to another 8 cells (step 2 Graphical Abstract, Figure 2), using a total volume of 6 μL of RT mix per cell.
-
b.Perform PCR stage (step 2 Graphical Abstract, Figure 2) by adding PCR master mix with (8 cells) or without (8 cells) target-specific primers for cDNA and gDNA amplicons, using a total volume of 15 μL of PCR mix per cell.
-
a.
Note: we recommend using 96-well plates rather than 384-well plates to perform test experiments because they are more easily handled. The lysis, RT and PCR volumes used in 96-well plates are doubled as compared to 384-well plates experiments outlined throughout the protocol.
-
9.
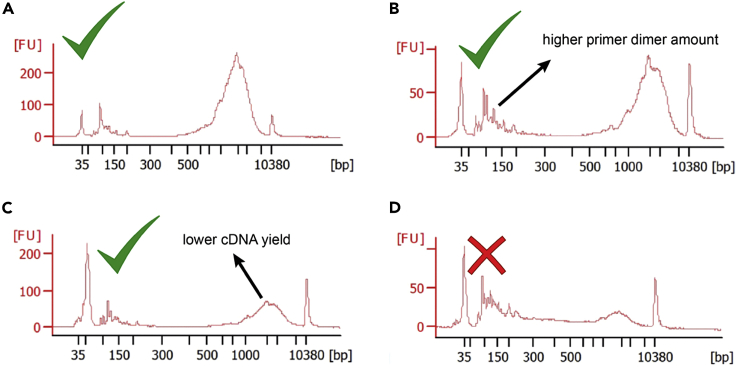
After the RT-PCR stage, add 25 μL of DNase-/RNase-free water to each well (obtaining a total volume of 50 μL per well; Figure 2), purify 25 μL of each library using 0.6:1 ratio of Ampure XP beads (see point 71 for details on Ampure XP purification) and check cDNA quality using a capillary array such as Bioanalyzer or TapeStation D5000. Successful examples of cDNA library generation are shown in Figures 3A–3C.
Figure 3.
Bioanalyzer Traces of Representative Good and Bad Quality cDNA Libraries
(A) Good-quality cDNA library with high yield and low primer dimer amount.
(B) Good-quality cDNA libraries with moderate primer dimer amount.
(C) Good-quality cDNA library with reduced yield (as compared to condition A) and moderate primer dimer amount.
(D) Poor quality cDNA library showing signs of RNA degradation and primer concatemer formation.
Note: A larger amount of low molecular weight fragments (50–300 bp) might appear with certain primer combinations compared to the control condition; these fragments do not typically affect further library preparation if their relative concentration is lower than 25% of the total cDNA amount. Moreover, certain primer combinations might slightly reduce cDNA yield; this does not typically affect library quality if this reduction is lower than 30%–40%.
CRITICAL: Any primer mixes that disrupt cDNA library generation (Figure 3D) should be redesigned and retested. At this stage, testing each primer pair individually rather than combinations of primer pairs is recommended. Usually, less than 5% of primer pairs fail initial testing and would require further optimization.
Optional: mRNA/cDNA primers can generate concatemers and/or affect cDNA library generation more frequently than gDNA primers. When optimizing mRNA/cDNA primers, users might also use mRNA primers for the PCR stage (i.e., target-specific primers which do not include the ISPCR handle sequence) and reduce the primer concentration in the RT stage up to 35 nM. Reducing the concentration of gDNA primers is not recommended.
Optimization 3: Design and Validate Target-Specific Primers for Single-Cell Genotyping
Timing: 1 day
Once successful cDNA library generation is achieved, proceed to validate the amplification of target regions using nested genotyping-PCR1 primers.
-
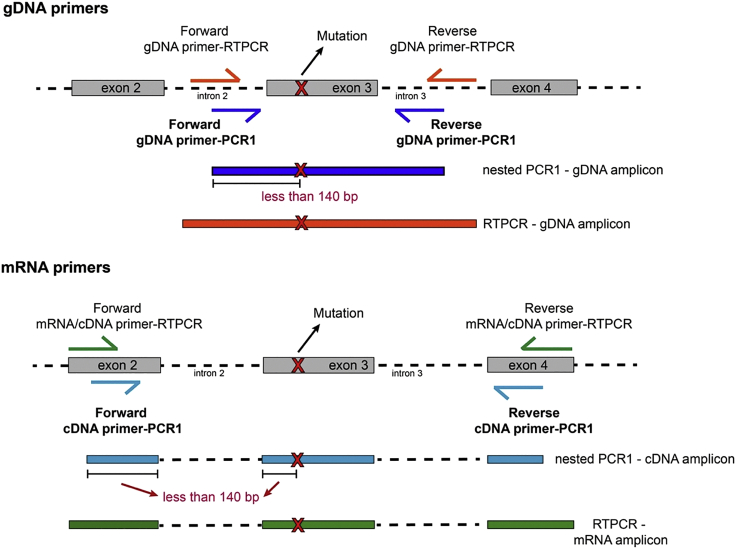
10.
Design nested target-specific primers for genotyping-PCR1 (Figure 4). You should design your cDNA primers (for the PCR1 step) to meet the following criteria:
Figure 4.
Schematic Representation of Primer Design Strategy for mRNA and gDNA Primers for the PCR1 Genotyping Step
-
a.Amplicon size should be in between 150–450 bp. Smaller or larger sizes are not recommended.
-
b.Primers should be nested from the mRNA/cDNA primers used during RT-PCR step.
-
c.Design your primers so that the beginning of the forward or reverse primer is at least within 140 bp of the mutation of interest, so that the mutation will be covered during sequencing.
-
d.If possible, design your forward primer in the exon before your mutation of interest and your reverse primer, in the exon after. This will allow parallel amplification with gDNA primers.
-
e.Check primer specificity against a transcriptome reference.
-
f.Your primers should have a minimum length of 19 bp and maximum length of 25 bp.
-
g.Design your primers with a melting temperature ranging from 57°C to 63°C, and a GC content ranging from 20% to 80%.
-
a.
-
11.You should design your gDNA primers (for the PCR1 step) to meet the following criteria:
-
a.Amplicon size should be in between 150–450 bp. Smaller or larger sizes are not recommended.
-
b.Primers should be nested from the gDNA primers used during the PCR step.
-
c.Design your primers so that the beginning of the forward or reverse primer is at least within 140 bp of the mutation of interest.
-
d.If possible, design your forward primer in the intronic region preceding your mutation of interest and your reverse primer, in the intronic contiguous to it. This will allow parallel amplification with mRNA primers. For certain mutations far away from intronic regions, this might not be possible; in that case, design a unique combined gDNA+mRNA amplicon spanning the mutation of interest.
-
e.Check primer specificity against a genomic reference and a transcriptomic reference.
-
f.Your primers should have a minimum length of 19 bp and maximum length of 25 bp.
-
g.Design your primers with a melting temperature ranging from 57°C to 63°C, and a GC content ranging from 20% to 80%.
-
a.
-
12.
Using the cDNA_amplicon mix libraries generated in the previous RT-PCR step (Figure 2, step B), take a 1.5 μL aliquot as input for genotyping-PCR1:
| Genotyping-PCR1 Master Mix | Per Cell and Amplicon | Storage | Cat. No. | Company |
|---|---|---|---|---|
| KAPA2G Robust HS Ready Mix | 3.125 μL | −20°C | KK5702 | Sigma (Roche) |
| Primer pair 1 (F+R; 20 μM) | 0.375 μL | −20°C | Custom desalted oligos; resuspend in RNase-free TE | Invitrogen |
| Water (DNase-/RNase-free) | 1.25 μL | Room temp | 10977035 | Life Technologies |
| Unpurified cDNA+amplicon mix (Figure 2; step B) | 1.5 μL | |||
| TOTAL | 6.25 μL |
Incubate in a thermocycler using 35 cycles of amplification:
| Temperature | Time | Cycles |
|---|---|---|
| 95°C | 3 min | 1 cycle |
| 95°C | 00:15 | 35 cycles |
| 60°C | 00:20 | |
| 72°C | 1 min | |
| 72°C | 5 min | 1 cycle |
| 4°C | HOLD | 1 cycle |
-
13.
Test each primer pair and cDNA+amplicon mix library individually. Then, run the PCR product from each single cell and amplicon on a gel or a capillary array (such as TapeStation, Agilent Technologies).
-
14.
For gDNA amplicons, you should detect a band of the expected size in all pre-amplified cells in which gDNA primers were added (Figures 5A and 5B). For mRNA/cDNA amplicons, you should detect the amplicon in a larger number of cells in which the mRNA/cDNA target-specific primers were added (Figures 5C and 5D). You should not expect to detect mRNA/cDNA amplicons in every single cell (Figure 5C), specially for lowly expressed genes.
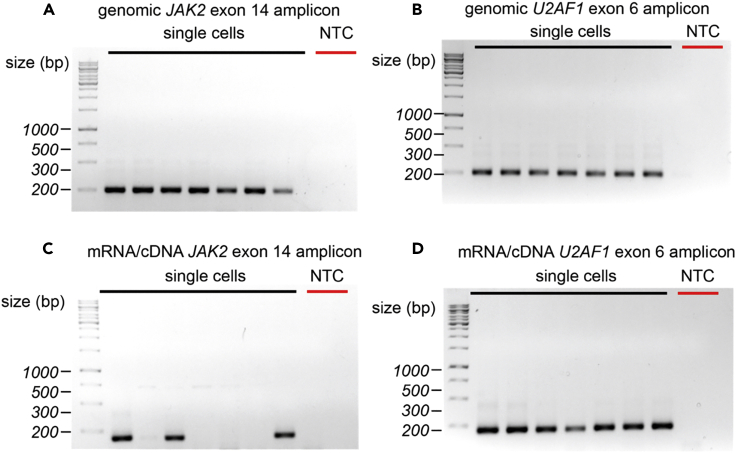
Figure 5.
Representative Agarose Gel Electrophoresis Examples of Genotyping-PCR1 Primer Validation of mRNA/cDNA and gDNA Amplicons for U2AF1 and JAK2 Genes
(A and B) Representative amplification results from JAK2 gDNA (A) or U2AF1 gDNA (B) amplicons in single K562 cells after genotyping-PCR1 using target-specific primers.
(C and D) Representative amplification results from mRNA/cDNA JAK2 (C) and U2AF1 (D) amplicons in single K562 cells after genotyping-PCR1 using target-specific primers. A non-template control condition (NTC) was included for each experimental condition.
Once cDNA generation and target-specific amplification have been successfully completed, primer validation is done.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| See Table 1 for a list of antibodies used for hematopoietic stem and progenitor cell isolation | (Rodriguez-Meira et al., 2019) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease | Qiagen | Cat# 19155 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| dNTPs | Life Technologies | Cat#19155 |
| RNase Inhibitor | Takara (Clontech) | Cat# 2313A |
| SMARTScribe | Takara (Clontech) | Cat# 639537 |
| SeqAMP | Takara (Clontech) | Cat# 638509 |
| Critical Commercial Assays | ||
| Nextera XT DNA Library Preparation Kit | Illumina | Cat# FC-131-1096 |
| Nextera XT Index Kit v2 Set A (See also Table S3) | Illumina | Cat# FC-131-2001 |
| KAPA 2G Robust HS PCR Kit | Sigma (Roche) | Cat# KK5702 |
| FastStart High Fidelity PCR System, dNTPack - Sigma-Aldrich | Roche | Cat# 04-738-292 001 |
| Access Array™ Barcode Library for Illumina® Sequencers-384, Single Direction | Fluidigm | Cat# 100-4876 |
| High Volume MANTIS Chip | FORMULATRIX | Catalog # MCHVSMR6 |
| 12.5 μL GRIPTIP, Sterile, Filter | INTEGRA Biosciences | Catalog Number #6455 |
| P50 tips, filtered, sterile | Beckman Coulter | Cat# A21586 |
| P20 tips, filtered, sterile | Beckman Coulter | Cat# 717256 |
| Ampure XP beads | Beckman Coulter | Cat# A63881 |
| ERCC spike-in | Invitrogen (Ambion) | Cat# 4456740 |
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat# Q32854 |
| Oligonucleotides | ||
| See Table S1 for barcoded OligodT-ISPCR sequences | This paper | N/A |
| TSO-LNA (RNase-free HPLC purification): AAGCAGTGGTATCAACGC AGAGTACATrGrG+G |
(Picelli et al., 2013) | N/A |
| ISPCR (HPLC purification): AAGCAGTGGTATCAACGCAGAGT | (Picelli et al., 2013) | N/A |
| P5_index (HPLC purified): AATGATACGGCGACCACCGAGA TCTACACGCCTGTCCGCGGAAG CAGTGGTATCAACGCAGAGT∗T∗G |
(Rodriguez-Meira et al., 2019) | N/A |
| P5_seq (PAGE purified): GCCTGTCCGCGGAAGCAGT GGTATCAACGCAGAGTTGC∗T |
(Rodriguez-Meira et al., 2019) | N/A |
| CS1 sequencing primer (HPLC purified): A+CA+CTG+ACGACATGGTTCTACA |
N/A | N/A |
| CS2 sequencing primer (HPLC purified): T+AC+GGT+AGCAGAGACTTGGTCT |
N/A | N/A |
| LCS1 sequencing primer (HPLC purified) : GGCGACCACCGAGATCTAC ACTGACGACATGGTTCTACA |
N/A | N/A |
| CS1rc sequencing primer (HPLC purified): T+GT+AG+AACCATGTCGTCAGTGT |
N/A | N/A |
| CS2rc sequencing primer (HPLC purified): A+GAC+CA+AGTCTCTGCTACCGTA |
N/A | N/A |
| See Table S2 for pre-amplification and barcoding PCR1 target-specific primer sequence examples | This paper | Biomers.net, Life Technologies |
| Software and Algorithms | ||
| bcl2fastq (version 2.20) | Illumina | RRID:SCR_015058 |
| STAR (version 2.4.2a) | (Dobin et al., 2012) |
https://github.com/alexdobin/STAR RRID: SCR_015899 |
| TrimGalore (version 0.4.1) | Felix Krueger, The Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| FeatureCounts (version 1.4.5-p1) | (Liao et al., 2014) |
http://subread.sourceforge.net/ RRID: SCR_012919 |
| Samtools (version 1.1) | (Li et al., 2009) |
http://samtools.sourceforge.net/ RRID:SCR_002105 |
| R (version 3.4.3) | CRAN | RRID:SCR_001905 |
| Custom pipelines for TARGET-seq analysis | (Rodriguez-Meira et al., 2019) | N/A https://github.com/albarmeira |
| Other | ||
| Protocols for buffer dispensing using the MANTIS Liquid Handling platform and for genotyping targeted library preparation using the Biomek FxP robotic platform | This Paper | https://doi.org/10.17632/k92cnf2fph.1 |
Materials and Equipment
The day before the sort, prepare media required for sample thawing, staining and sorting (FACS Media, Thaw Media), as well as any antibodies required for sample staining.
Note: do not add the DNase I to the FACS Media and Thaw Media until the day of the sort.
-
•
Prepare FACS Buffer (IMDM + 10% FCS + 1% DNase I) by adding 50 mL of Fetal Calf Serum (Sigma, Cat# F7524) and 5 mL of DNase I (1 mg/mL; Merck) to 445 mL of IMDM (Gibco, Cat# 21056-023).
Note: The DNase I should be added to the media the same day it will be used.
-
•
Prepare Thaw Media (IMDM + 10% FCS + 11% DNase I) by adding 5 mL of DNase I (1 mg/mL; Merck) and 5 mL of FCS to 40 mL of FACS Media. Prepare 10 mL of media per sample to be processed.
Note: The amount of DNase might vary for different tissues and the viability of the samples.
TARGET-seq uses a set of 384 different barcoded oligodT primers (Table S1). These barcodes can be obtained from biomers.net as custom HPLC purified primers. Dilute barcoded oligodT primers 1:10 with RNase-free TE (Invitrogen, Cat# AM9858) to a final concentration of 10 μM, from a 100 μM stock concentration and aliquot them into a 384-well plate (FrameStar, 4titude, 4ti-0384/C). Barcodes can be stored at −20°C for long term storage; however, users should avoid more than 10 freeze-thaw cycles of the stock plate.
TSO-LNA primer used for retrotranscription should be resuspended at a 100 μM concentration in RNase-free TE (Invitrogen, Cat# AM9858) and aliquoted in single-use aliquots, which should be stored at −80°C. The TSO-LNA primer contains RNA bases and therefore will degrade easily by consecutive freeze-thaw cycles or if stored at −20°C for a long period of time. Single-use TSO-LNA aliquots should be kept in dry ice until they are used.
Prepare 0.43% Triton X-100 aliquots. Aliquot 990 μL of RNase-free water (Cat# AM9935, Life Technologies) and carefully pipette pre-warmed 10 μL of Triton X-100. Triton X-100 is extremely viscous; therefore, to facilitate pipetting, pre-warm the Triton X-100 at 50°C–80°C for 5 min. This will generate 1% Triton X-100 dilution. To generate a 0.43% Triton X-100 dilution, mix 430 μL of 1% Triton X-100 with 570 μL of RNase-free water (Cat# AM9935, Life Technologies).
Prepare ERCC aliquots by first diluting the ERCC stock (Cat# 4456740, Invitrogen) in RNase-free TE buffer as indicated below:
| Dilution Name | Transfer (μL) | Mix with (μL of RNase-free TE) | Concentration (Relative to Stock) |
|---|---|---|---|
| Stock (E1) | NA | NA | 1 |
| E2 | 5 μL of E1 | 45 μL of TE | 1:10 |
| E3 | 10 μL of E2 | 990 μL of TE | 1:1,000 |
| E4 | 25 μL of E3 | 975 μL of TE | 1:40,000 |
| E5∗ | 100 μL of E4 | 900 μL of TE | 1:400,000 |
Note: ERCC stocks should be stored in single-use aliquots at −80°C.
Additional Resources
Users are welcome to post any troubleshooting issues found with any steps of method detailed in this protocol in https://groups.google.com/forum/#!forum/target-seq-users-group
Authors will regularly check this space to help other users with optimization of their specific conditions and troubleshooting of any problems that might arise.
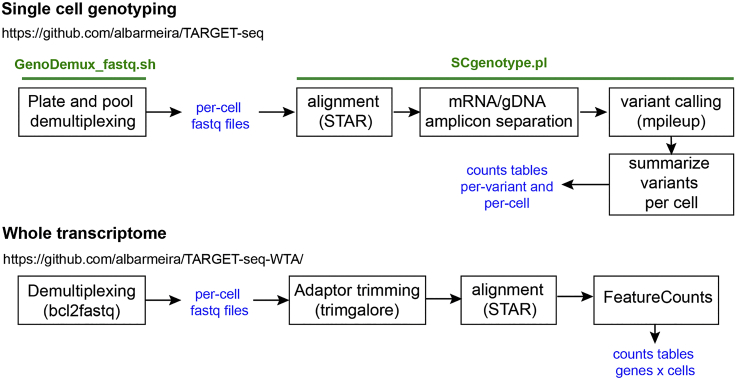
A flowchart of the analytical procedures used to analyze Next Generation sequencing data produced by TARGET-seq can be found at: https://github.com/albarmeira/TARGET-seq and
https://github.com/albarmeira/TARGET-seq-WTA and in Figure 24. Analytical pipelines and example analysis will be regularly updated in the GitHub repository. Users are welcome to post issues and any analytical questions in that same repository.
Figure 24.
Schematic Representation of the Analytical Workflow
Step-By-Step Method Details
Sample Preparation and Single Cell Sorting
Timing: 5–8 h
This step describes the procedures for sample processing, lysis buffer preparation and sorting of single cells into plates.
The protocol below specifies the conditions used for single-cell sorting of hematopoietic stem and progenitor cells (HSPCs) from human bone marrow or peripheral blood.
Note: Different tissues and cell types might require variations of this protocol that should be optimized in advance by the user. Lysis buffer preparation steps are the same for different tissues.
Note: The day before the sort, prepare media required for sample thawing, staining and sorting (FACS Media, Thaw Media), as well as any antibodies required for sample staining. Do not add the DNase I to the FACS Media and Thaw Media until the day of the sort.
CRITICAL: Make sure you have prediluted all reagents required for lysis buffer preparation to the correct concentration: barcoded oligodT primers, 0.43% Triton X-100 and ERCC (E4 or E5 dilution).
Note: The amount of ERCC added to the lysis buffer varies depending on the total mRNA content of each cell type as outlined below. Cell lines usually have mRNA contents ranging from 2 to 8 pg, and therefore we recommend using a 1:8,000,000 dilution ERCC in 2 μL of RT buffer for cell lines (or 0.1 μL from a 1:400,000 E5 stock for cell lines such as K562, Jurkat, NALM6, SET2, or HL60). Primary hematopoietic stem and progenitor cells have less than 0.5 pg of mRNA per cell. Therefore, we use a 1:40,000,000 ERCC dilution in 2 μL of lysis buffer, or 0.02 μL per cell from a 1:400,000 E5 stock. ERCC stocks and aliquots should be stored at −80°C.
| Cell Line | Internal mRNA (Average Estimate) | ERCC in Lysis |
|---|---|---|
| K562 | 0.50 pg | 1:8e6 |
| JURKAT, SET2, NALM6 | 0.35 pg | 1:8e6 |
| HSPCs | 0.05 pg | 1:4e7 |
| HEK293 | 1.6 pg | 1:8e6 |
Sample Preparation
Timing: 2 h
Note: Points 1–3 detail the procedures to follow when using cryopreserved peripheral blood or bone marrow samples. If you are using fresh samples, proceed to point 4. If using other tissues, please refer to relevant literature on how to dissociate each tissue to generate good-quality single cell suspensions (Slyper et al., 2020).
-
1.
Turn on water bath (set at 37°C) and a centrifuge (4°C). Prepare a box of ice and another one of dry ice. Prepare a laminar flow cabinet for sterile work as well as P20, P200, and P1000 filtered tips.
-
2.
Prepare FACS Buffer and Thaw Media (See Materials and Equipment). If they have been prepared in advance, add DNaseI to them.
-
3.
Transfer samples from liquid nitrogen storage in dry ice and thaw each sample in the water bath at 37°C.
Note: thaw 2–3 samples at a time to avoid keeping the samples in freezing media for too long (freezing media will usually contain DMSO, which is toxic to the cells).
-
4.
Add 1 mL of FCS to each sample dropwise using a P1000 and gently transfer it to a 15 mL Falcon tube. Wash the vial with 1 mL of Thaw Media and add 7 mL of Thaw Media to the 15 mL Falcon Tube.
-
5.
Centrifuge at 350 × g for 10 min. Remove the supernatant, gently resuspend in 5 mL of FACS Buffer and count the cells using an hematocytometer.
-
6.
Centrifuge at 350 × g for 10 min. Remove supernatant and resuspend in 105 μL of FACS Buffer (for samples containing less than 25 million cells) or 210 μL (for samples containing more than 25 million cells). At this point, transfer the samples to 1.5 mL microfuge tubes.
Note: One of the samples should be used for single staining and FMO (Fluorescence Minus One) controls, which should be run in every experiment. To prepare single staining and FMO controls, use at least 300,000 cells per condition.
-
7.
Add Human Fc-block (1:200; BD, Cat# 564220) and incubate for 5 min at 4°C.
-
8.
Add antibodies to each sample as well as to single staining and FMO controls, and incubate for 30 min at 4°C. You can find examples of antibody staining to isolate hematopoietic stem and progenitor cells in Table 1.
-
9.
Wash samples with 1 mL of FACS Buffer and spin down at 400 × g during 5 min.
-
10.
Add viability dye (except in the FMO corresponding to such viability dye) and run single staining controls and FMO controls on a cell sorter.
-
11.
Use the single staining and FMO controls to set the right voltages for each fluorochrome and sorting gates. Perform an enrichment sort of your target population and check sorting purity. Purity should be >95% for optimal results.
Table 1.
Antibodies Used for Hematopoietic Stem/Progenitor Isolation
| Antibody-conjugate | Company | Clone | Catalog Number | RRID | Dilution |
|---|---|---|---|---|---|
| CD8-FITC (Lineage) | BioLegend | Clone: RPA-T8 | Cat#: 301006 | RRID: AB_314124 | 1:100 |
| CD20-FITC (Lineage) | BioLegend | Clone: 2H7 | Cat#: 302304 | RRID: AB_314252 | 1:150 |
| CD66b-FITC (Lineage) | BioLegend | Clone: G10F5 | Cat#: 305104 | RRID: AB_314496 | 1:15 |
| CD10-FITC (Lineage) | BioLegend | Clone: HI10a | Cat#: 312208 | RRID: AB_314919 | 1:30 |
| CD127-FITC (Lineage) | eBioscience | Clone eBioRDR5 | Cat#: 11-1278-42 | RRID: AB_1907343 | 1:30 |
| Human Hematopoietic Lineage Cocktail – FITC (Lineage) | eBioscience | NA | Cat# 22-7778-72 | RRID: AB_1311229 | 1:15 |
| CD123-PECy7 | BioLegend | Clone: 6H6 | Cat#: 306010 | RRID: AB_493576 | 1:60 |
| CD38-PETxRed | Invitrogen | Clone: HIT2 | Cat#: MHCD3817 | RRID: AB_10392545 | 1:21 |
| CD90-BV421 | BioLegend | Clone: 5E10 | Cat#: 328122 | RRID: AB_2561420 | 1:30 |
| CD45RA-PE | eBioscience | Clone: HI100 | Cat#: 12-0458-41 | RRID: AB_10717397 | 1:150 |
| CD34-APC-eF780 | eBioscience | Clone: 4H11 | Cat#: 47-0349-42 | RRID: AB_2573956 | 1:150 |
Lysis Buffer Preparation
Timing: 1 h
Lysis buffer plates containing barcoded oligodT primers should be prepared on the same day in which single cell sorting will be performed. To reduce risk of cross-contamination and inconsistencies, this step has been automated on a MANTIS Liquid Handling Platform (FORMULATRIX) and a VIAFLO 96/384 (INTEGRA Biosciences). However, users may opt to adapt this part of the protocol for other liquid handling platforms such as Mosquito HTS (TTP Labtech).
-
12.
First, prepare a 384 well plate containing lysis buffer mix as outlined in the table below. You should prepare enough lysis for the number of cells desired + 13.5% dead volume. For example, to prepare enough lysis buffer for 3,840 cells (10 times 384-well plates), aliquot 17 μL of lysis buffer mix in each well of a 384 well plate (1.5 μL ∗ 3,840 cells = 5,760 μL + 777.6 μL dead volume; 6,537.6 μL divided 384 wells ≈ 17 μL per well).
| Lysis Buffer | 1 Cell | 3,840 Cells (10 Plates) + Dead Volume (13.5%) | Storage | Cat. No | Company |
|---|---|---|---|---|---|
| Triton 0.43% | 0.88 μL | 3,835 μL | −20°C | Cat# T8787, resuspend in UltraPure DNase/RNase-free Distilled Water | Sigma-Aldrich |
| RNase Inhibitor | 0.05 μL | 218 μL | −20°C | Cat# 2313A | TAKARA Clontech |
| dNTPs (10 mM) | 0.5 μL | 2,180 μL | −20°C | Cat# R0192 | Life Technologies |
| Protease (1.09 AU/mL in water) | 0.05 μL | 218 μL | +4°C | Cat#19155; resuspend in UltraPure DNase/RNase-free Distilled Water | Qiagen |
| ERCC RNA spike-in mix (E5 dilution∗) | 0.02 μL | 87 μL | −80°C (single-use aliquot) | Cat# 4456740 | Life Technologies |
| TOTAL | 1.50 μL | 6,538 μL |
-
13.
Once the required amount of lysis buffer has been prepared in microfuge tubes (split in as many tubes as necessary), switch on the MANTIS liquid handling platform. Open the MANTIS software, open the program file “MethodS1.txt” (https://doi.org/10.17632/k92cnf2fph.1). This program will dispense 17 μL of lysis per well into 384 wells of a 384 well plate. This step requires a high volume (HV) chip (FORMULATRIX, Cat# MCHVSMR6), which should be exclusively used for the lysis step.
-
14.
Then, initialize the MANTIS. Clean the chip holder and plastic surfaces with 70% ethanol. Then, prime the chip by placing it into the chip holder and inserting a 1 mL non-filter tip (Biosphere Quality Tips 100 Cat #70.762.200; Figure 6A and Methods Video S1) containing 500 μL of nuclease-free water. Select the reagent name (“lysis buffer”) from the drop down menu, ensure that the correct chip volume next to reagent input is selected (“HV”) and click in the appropriate chip position number (Figure 6B). Hold the icon “Manually Priming Input on MANTIS Arms” (Figure 6C) until the water has gone through the chip. Finally, click the chip position number again to return the chip holder to its position.
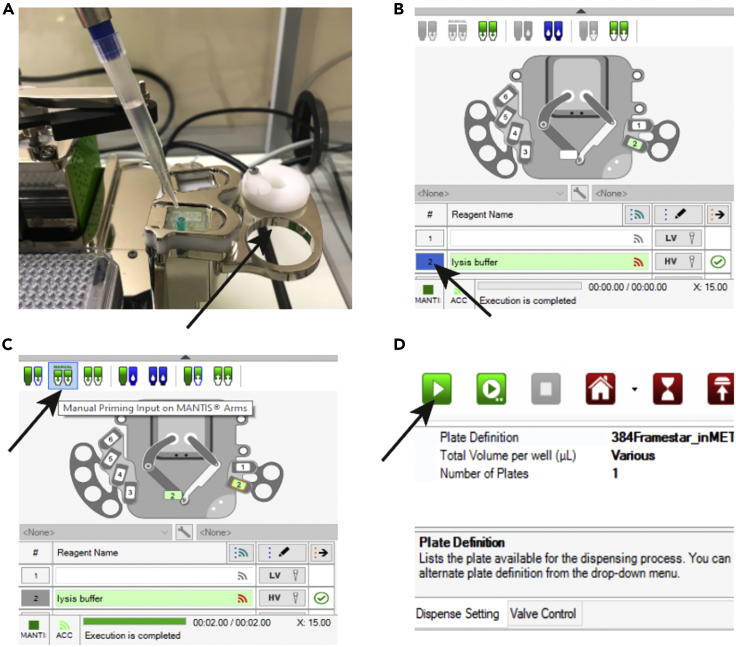
Figure 6.
Usage of MANTIS Liquid Handling Platform for Lysis Buffer Aliquoting
(A) Insert a 1 mL non-filter tip into the high volume chip in the chip holder.
(B) Icon to click to move the chip position number on the software to the MANTIS arms.
(C) Icon to click to manually prime the chip.
(D) Green icon to start the program for dispensing.
-
15.
Place a 384-well cold rack (Cool Rack XT PCR 384, Biocision, Cat# BCS-538) onto the MANTIS holder and place a PCR-clean 384 well plate (FrameStar 384 Well Skirted PCR Plate, 4titude, Cat# 4ti-0384/C) into the cold rack (Methods Video S2). Then, dispense the lysis buffer into a new 1 mL non-filter tip (Biosphere Quality Tips 100 Cat #70.762.200) and insert into the lysis HV chip. Click the green icon (Figure 6D) to start dispensing.
-
16.
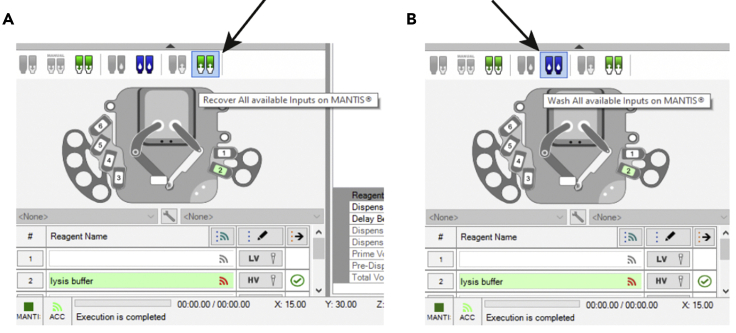
Once dispensing is complete, cover the 384-well plate with a PCR film and spin down briefly at 1,000 × g during 15 s. Click “Recover All Inputs” (Figure 7A) and replace the non-filter tip used for lysis dispensing with an empty tip for washing. Click “Wash All Available Inputs” (Figure 7B) to proceed with a 2-step wash, in which the chip will be flushed with ethanol and water. Place the chip away in a clean RNA/DNA-free 50 mL Falcon tube (Corning, Cat#10291433) and switch off the MANTIS liquid handling platform.
Figure 7.
Cleaning Chips in the MANTIS Liquid Handling Platform
(A) Icon to click to recover remaining liquid in chip.
(B) Icon to click to wash the chip with ethanol followed by nuclease-free water.
-
17.
Next, aliquot each barcoded oligodT-ISPCR primer (prediluted to 10 μM; Table S1) into each well of the 384 well plate containing lysis buffer mix from points 12–16. The barcodes in Table S1 (n=384) provide increased multiplexing capacity as compared to our previously described 96-plex strategy (Rodriguez-Meira et al., 2019). Add the equivalent to 0.5 μL per cell in each well + 13.5% dead volume, as indicated in the table below. For example, to prepare lysis buffer mix + barcoded oligodT stock for 3,840 cells (point 12), add 5.67 μL of each barcoded oligodT-ISPCR primer per well. This step has been automated in a VIAFLO 96/384 INTEGRA electronic pipette (INTEGRA Biosciences) equipped with a 384 pipetting head. This liquid handling platform dispenses 96 or 384 liquids from a source plate into a 384 well plate (one quadrant or full plate, respectively).
| Lysis Buffer Mix + Barcoded OligodT | 1 Cell | Storage | Cat. No | Company |
|---|---|---|---|---|
| Lysis buffer mix (points 12–16) | 1.50 μL | |||
| Oligo-dT-ISPCR (10 μM) barcoded, well-specific | 0.5 μL | −20°C | Custom HPLC primers | Biomers.net |
| TOTAL | 2.00 μL |
-
18.
Switch on the VIAFLO (Figure 8, position 10) and press RUN (Figure 8, position 17) to home the instrument. Press the RUN key a second time to home the pipetting unit.
Figure 8.
Numbered Description of VIAFLO 96/384 Device and Control Unit
-
19.
To load the tips, ensure the plate holder is at the center position, place a tip box (12.5 μL GRIPTIP, Sterile, Filter, Catalog Number #6455, INTEGRA Biosciences) in the place holder (Figure 8, position 8A or 8B), hold the control unit (Figure 8, position 5) and lower the pipetting head onto the tip box until the “Tip load button” (Figure 8, position 4) flashes. The display will then prompt to push the “Tip load button” while at the same time gently push down the control unit (Figure 8, position 5).
-
20.
Place two 384-well cold racks (Cool Rack XT PCR 384, Biocision, Cat# BCS-538) into the plate holders (Figure 8, position 8A and 8B). Use the touch wheel (Figure 8, position 13) to scroll clockwise down the menu list and create a new program to transfer each barcode from the stock plate to a new empty 384-well plate.
-
21.
Place a 384-well plate containing barcoded oligodT-ISPCR primer stock on the top of the cold rack in the source plate position (Figure 8, position 8A) and the 384-well lysis buffer stock plate (from points 12–17; containing c per well) onto the cold rack in the destination plate position (Figure 8, position 8B). To run the program, press the OK button.
-
22.
Once the lysis buffer mix + barcoded oligodT stock plate is aliquoted, cover the plate with a PCR film and briefly spin down at 1,000 × g during 15 s.
-
23.
Next, create a new program to aliquot 2 μL of lysis buffer + barcoded oligodT mix stock plate (point 22) into individual 384-well plates using the VIAFLO 96/384 INTEGRA pipetting robot step. Ensure 384-well cold racks (Cool Rack XT PCR 384, Biocision, Cat# BCS-538) are placed into the plate holders (Figure 8, position 8A and 8B). The lysis buffer + barcoded oligodT mix stock plate should be placed into the source plate holder (Figure 8, position 8A) and an empty sterile 384-well plate should be placed into the destination plate holder (Figure 8, position 8B). Both plates should be place on the top of 384-well cold racks (Cool Rack XT PCR 384, Biocision, Cat# BCS-538). Click OK to run the program. Once dispensing is complete, remove the plate with 2 μL of lysis buffer + barcoded oligodT and replace with another empty sterile 384-well plate and click OK to run the method again (with the same tips). This step can be repeated 10 times with the same tips.
-
24.
Once dispensing is complete, cover the 384-well target plate with a clear PCR film and spin down at 1,000 × g for 15 s. Repeat for further empty 384-well plates until desired number dispensed (the examples volumes for these steps should be sufficient stock for 10∗384-well plates). Keep these plates on ice until the sorting step.
Single Cell Sorting
Timing: 2–5 h
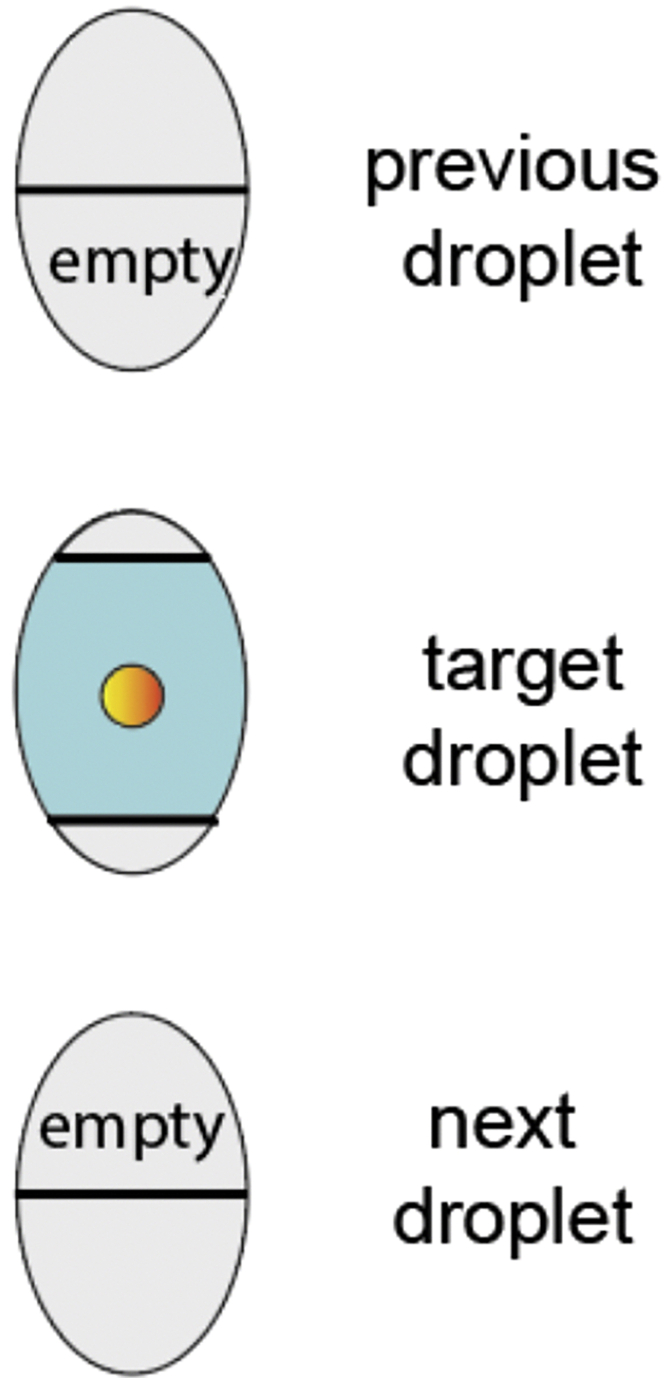
You should now prepare the sorter for single cell sorting. It is essential to choose the right purify mode and singlet gates to ensure that only one single cell is sorted per well. We recommend using a purity mode in which the 50% of the previous and next droplet to the target one are empty, and the target event is within the 75% central part of the target droplet (Figure 9).
Figure 9.
Schematic Representation of the Sorting Purity Settings
You should keep the event rate low (less than 1,000 events per second) to try to maximize cell recovery.
-
25.
Perform an alignment test sort using a 384 well tissue culture plate (Corning® 384 well microplate, CLS3702-100EA) and fluorescent beads (Cat. No). Sort one fluorescent bead per well and check under the microscope that there is only one bead in each well of the 384 well plate.
Note: This can be performed only once when setting up the sorter for single cell sorting for the first time. If previously set up, proceed to point 26.
-
26.
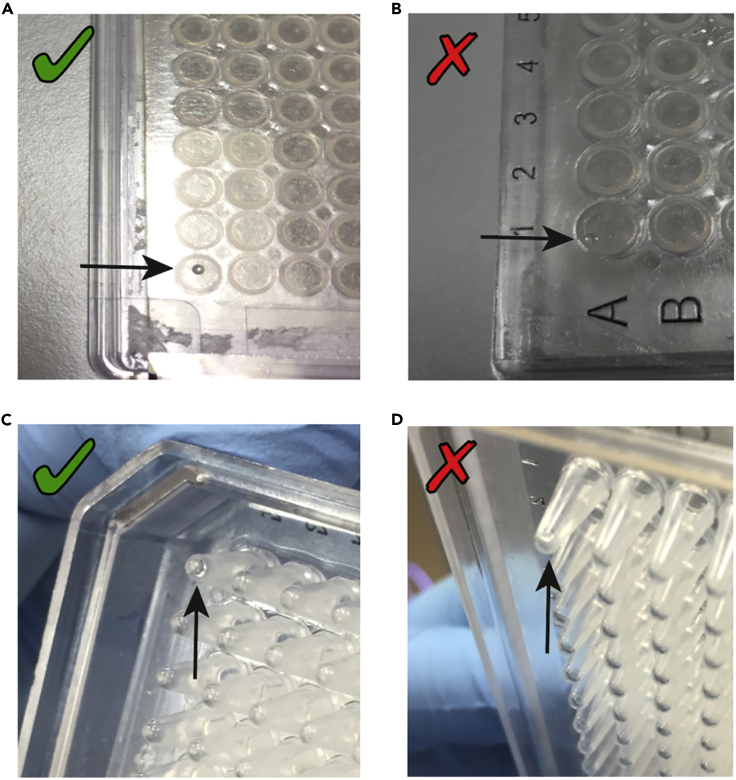
Perform an alignment test sort using a 384 well plate (FrameStar, 4titude, 4ti-0384/C; or the same model in which lysis buffer was aliquoted) covered with a PCR film and sort 50 droplets in the corners of the plate (position 1A, 1P, 24A, 24P). If the plate is correctly aligned (Figure 10A), the droplets should fall perfectly in the center of those well; if not (Figure 10B), re-calibrate the alignment accordingly. If correct, proceed to point 27.
Figure 10.
Representative Examples of 384-Well Plate Sorter Alignment
(A) Drop correctly aligned on center of the well covered with film.
(B) Drop incorrectly aligned at the edge of the well covered with film.
(C) Drop correctly aligned at the center of the bottom of the well.
(D) Drop incorrectly aligned to the side of the well.
-
27.
Take the PCR film off the plate and sort 20 droplets in the same wells are in step 17 (positions 1A, 1P, 24A, 24P). Check that the drops are been deposited on the bottom of each well (Figure 10C) and there is no liquid splashing on the sides of the wells (Figure 10D). If correct, alignment checks are complete. Clean the surfaces around the sorter with RNase away (Thermofisher, Cat# 10328-011).
-
28.
Proceed to single cell sorting of your target populations in plates containing lysis buffer and barcoded oligodT-ISPCR from point 24. Lysis and protease digestion will occur immediately once each cell is sorted into each well. Sort the cells at room temperature (to increase lysis efficiency) for a maximum of 20 min; if your target population if very rare and filling up a plate would take longer than 20 min, you should enrich your target population before sorting into plates to avoid RNA degradation. To obtain cell-surface proteomics data from each single cell, you should record the fluorescence intensity of each cell-surface marker using the “record index sorting” option available in most FACS instruments.
-
29.
Once finished, cover the plates with an aluminum sealing film (StarLab, Cat# E2796-0792), spin down the plate at 1,000 × g for 15 s and snap freeze the plates in dry ice.
Pause Point: Store in −80°C for up to 3 months.
Note: processing plates after 3 months of −80°C storage is associated with higher rates of RNA degradation.
CRITICAL: When freezing plates, it is essential that you use Aluminum Sealing Films (StarLab, Cat# E2796-0792), as plastic films are likely to come off in −80°C freezers.
Heat Inactivation, Retrotranscription, and PCR
Timing: 6.5 h; 75 min hands-on time
This step describes the procedures for retrotranscription and PCR amplification of cDNAs and targeted amplicons from single cells.
This is the most critical step of the method, where most users usually experience difficulties. mRNA degradation can occur easily when handling plates, so you should be extremely careful to keep sterile and PCR-free conditions when performing all of the steps outlined below.
CRITICAL: This step should be performed within 3 months from the start of STEP1.3 (single cell sorting).
CRITICAL: Make sure you have all reagents required for retrotranscription and PCR steps, including pre-made TSO-LNA single-use aliquots and any mRNA, cDNA, and gDNA primers required. It is recommended that you prepare mRNA, cDNA, and gDNA primers mixtures to be used for each patient in advance.
Note: Sorted plates from step 1.3 must be stored at −80°C and kept in dry ice until heat inactivation step. Once the retrotranscription is finished, it is essential that you proceed immediately to PCR.
Note: You should perform heat inactivation, retrotranscription and addition of PCR master mix in a dedicated PCR-free area or clean room, under a laminar flow cabinet free of any amplified material. Once the PCR master mix has been added, the PCR should be performed in a post-PCR area, where all of the steps involving amplified material will be carried out. It is also essential that you use different sets of pipettes for each workspace, to avoid contamination.
Heat Inactivation and Retrotranscription
Timing: 3 h; 30 min hands-on time
-
30.
Take the sorted plate(s) (point 29) from −80°C storage and keep on dry ice until the start of heat inactivation step.
-
31.
Thaw the 5X Buffer (Clontech, Cat#639537), RNase-free PCR Grade Water (Life Technologies, Cat#AM9935) and any mRNA primers that you might add to the reaction. These can be thawed at room temperature.
-
32.
The retrotranscription (RT) step has been automated on the MANTIS Liquid Handling Platform (FORMULATRIX). Initialize the MANTIS as indicated in point 14; cool down a CoolRack® XT PCR384 (Biocision, Cat# BCS-538) to 4°C and clean with RNase away (Thermofisher, Cat# 10328-011).
-
33.
Aliquot the buffer 5X, water and any mRNA primers added to the mix into an RNase-free microfuge tube (Invitrogen, Cat# AM12400) to prepare a master mix for the RT step as indicated in the table below. RNase inhibitor, TSO-LNA, and SMARTScribe enzyme will be added to the mix during heat inactivation step. You should always prepare 15% extra RT mix to account for dead volumes.
| RT | 1 Cell | 384 Cells (+15% Dead Volume) | Storage | Cat. No. | Company |
|---|---|---|---|---|---|
| Buffer 5X | 1.00 μL | 441.6 μL | −20°C | 639537 (delivered with enzyme) | Clontech |
| PCR Grade Water | Variable | Variable | −20°C | AM9935 | Life Technologies |
| mRNA primers (0.0175 μL of each primer from a 200 μM stock) | Variable | Variable | −20°C | Custom HPLC purified primers from; resuspended in RNase-free TE/water | biomers.net |
| TOTAL | 2.325 μL | 1,026.72 μL |
-
34.
Perform heat inactivation of the protease by incubating the plate for 15 min at 72°C in a thermocycler. This step will inactivate the protease within the lysis buffer, so that it does not interfere with any subsequent enzymatic steps.
-
35.
During the heat inactivation step, add the RNase inhibitor, TSO-LNA, and finally, the retrotranscriptase (SMARTScribe) to the retrotranscription mix in that order:
| RT Master Mix | 1 Cell | 384 cells (+15% Dead Volume) | Storage | Cat. No. | Company |
|---|---|---|---|---|---|
| RNase Inhibitor | 0.125 μL | 55.2 μL | −20°C | 2313A | TaKara |
| TSO-LNA (100 μM) | 0.05 μL | 22.08 μL | −80°C | Custom HPLC purified oligo; resuspend in RNase-free TE | Exiqon-Qiagen or biomers.net |
| SMARTScribe (RT enzyme) | 0.5 μL | 220.8 μL | −20°C | 639537 | Clontech |
| TOTAL | 0.675 μL | 298.08 μL | |||
| TOTAL (cumulative) | 3 μL |
Note: Make sure you have finished adding all components to the retrotranscription mix by the time the heat inactivation is finished. It is essential that once heat inactivation is complete, the retrotranscription mix is immediately added to the plate. Otherwise, you might see signs of mRNA degradation.
CRITICAL: The following step must be performed immediately after the heat inactivation is finished.
-
36.
Spin down the plate at 1,000 × g for 15 s, place it on a cold rack (Cool Rack XT PCR 384, Biocision, Cat# BCS-538) and add 3 μL of the retrotranscription mix per well using a MANTIS Liquid Handling Platform or an alternative automated liquid handler such as Mosquito HTS (TTP Labtech). If using a MANTIS, click the Load Dispense list button and open the program file “MethodS2.txt” (https://doi.org/10.17632/k92cnf2fph.1). This step requires a HV chip (FORMULATRIX, catalog # MCHVSMR6), which should be exclusively used for the RT step. Place the HV chip into the chip holder, insert a 1 mL tip (Biosphere Quality Tips 100 Cat #70.762.200) and dispense the RT master mix into the 1 mL tip. Place the plate and cold rack on the MANTIS holder. Click the green “Run Dispense” button (Figure 6D) to start the “MethodS2.txt” program (https://doi.org/10.17632/k92cnf2fph.1).
-
37.
Once the program has finished dispensing the RT mix, carefully seal the plate with 2 PCR films (MicroAMp Clear Adhesive Film, Thermo Fisher Scientific, Cat# 4306311), spin down the plate at 1,000 × g during 15 s, immediately place in the thermocycler and run the RT program:
| Temperature | Time | Cycles |
|---|---|---|
| 42°C | 90 min | 1 |
| 50°C | 2 min | 10 cycles |
| 42°C | 2 min | |
| 70°C | 15 min | 1 |
| 4°C | HOLD | - |
Note: We recommend using two films during this step to facilitate PCR film removal after incubation in the thermocycler, which has proven to be difficult using only one PCR film and is essential to avoid sample cross-contamination.
-
38.
Finally, follow the wash and shut down procedure of the MANTIS outlined in point 16. The RT chip should be placed in a RNA/DNA-free 50 mL Falcon dedicated for the RT chip only.
Note: This is the step where most likely mRNA degradation will occur. Therefore, make sure you are extremely careful in keeping sterile conditions while pipetting reagents and handling plates. After heat inactivation is finished, the plate should be kept at 4°C in a cold rack until the retrotranscription master mix is added and it is placed in the thermocycler. This step should take no longer than 5–6 min; otherwise mRNA degradation will occur!
PCR
Timing: 3 h; 30 min hands-on time
This step has been automated on a MANTIS Liquid Handling Platform (FORMULATRIX), but can be adapted to other liquid handling platforms.
-
39.
15–20 min before the retrotranscription program has finished, start thawing the reagents required for the PCR step. PCR Grade Water (Cat# AM9935) and primer mixes can be thawed at room temperature; the rest of the reagents must be thawed on a cold rack or ice.
-
40.
Open the MANTIS software and open the program file “MethodS3.txt” (https://doi.org/10.17632/k92cnf2fph.1). This step requires a high volume (HV) chip (FORMULATRIX, Cat# MCHVSMR6), which should be exclusively used for the PCR step. Initialize the MANTIS as per point 14 using a HV chip dedicated for PCR only; cool down a CoolRack® XT PCR384 (Biocision, Cat# BCS-538) to 4°C and clean with RNase away (Thermofisher, Cat# 10328-011).
-
41.
Prepare the PCR master mix in a RNase free microfuge tube as indicated in the table below. Add the reagents in the following order: first water, then primers, and lastly, PCR enzyme. Wait until the retrotranscription step has finished to add the PCR enzyme. Prepare 10% additional PCR master mix to account for dead volumes.
| PCR Master Mix | 1 Cell | 384 cells (+10% Dead Volume) | Storage | Cat. No. | Company |
|---|---|---|---|---|---|
| 2X Buffer | 6.5 μL | 2,745.6 μL | −20°C | 638509 | Tech |
| ISPCR (10 μM) | 0.065 μL | 27.46 μL | −20°C | Custom HPLC purified oligo | biomers.net |
| RT-PCR Water | Variable | Variable | −20°C | AM9935 | Life Technologies |
| SeqAMP (PCR enzyme) | 0.25 μL | 105.6 μL | −20°C | 638509 | Clontech |
| cDNA primers - (0.0175 μL from each primer from a 20 μM stock) | Variable | Variable | −20°C | Custom HPLC purified primers; resuspend in RNase-free TE/water | biomers.net |
| Genomic primers (0.05 μL from each primer from a 200 μM stock) | Variable | Variable | −20°C | biomers.net | |
| TOTAL | 8 μL | 3,379.2 μL | |||
| TOTAL (cumulative) | 13 μL |
-
42.
Place the high volume chip into the chip holder, then insert a 1 mL tip (Biosphere Quality Tips 100 Cat #70.762.200) and aliquot the PCR master mix into it. Add a maximum volume of 1.5 mL into the tip; if dispensing larger volumes, stop the program by pressing “Pause,” add extra PCR master mix into the 1 mL tip and continue the program by pressing “Run”.
-
43.
Place a CoolRack® XT PCR384 cold block at 4°C on the MANTIS holder, place the 384 well plate from point 37 on the top of the holder, prime the MANTIS PCR chip and start the “TARGET-seq 384-well PCR step” program.
-
44.
Once dispensing is finished, carefully seal the plate with two MicroAMp Clear Adhesive Films (Thermo Fisher Scientific, Cat# 4306311), spin down the plate at 1,000 × g for 15 s, place the plate on ice and transfer to a dedicated post-PCR area. Then, place the plate on a thermocycler and run the PCR program:
| Temperature | Time | Cycles |
|---|---|---|
| 98°C | 3 min | 1 |
| 98°C | 00:15 | 24 cycles (single HSPCs) |
| 67°C | 00:20 | |
| 72°C | 6 min | |
| 72°C | 5 min | 1 |
| 4°C | HOLD | 1 |
Note: We recommend using two films during this step to facilitate PCR film removal after incubation in the thermocycler, which has proven to be difficult using only one PCR film and is essential to avoid sample cross-contamination.
-
45.
Finally, follow the wash and shut down procedure of the MANTIS outlined in step 16. The PCR chip should be placed in a RNA/DNA-free 50 mL Falcon dedicated for the PCR chip only.
Pause Point: When the PCR finishes, spin down the plate at 1,000 × g for 30 s at room temperature and snap freeze in dry ice. Once the liquid is completely frozen, transfer the plate to −20°C storage for up to 6 months.
Pooling, cDNA+Amplicon Mix Dilution
Timing: 30 min; 15 min hands-on time
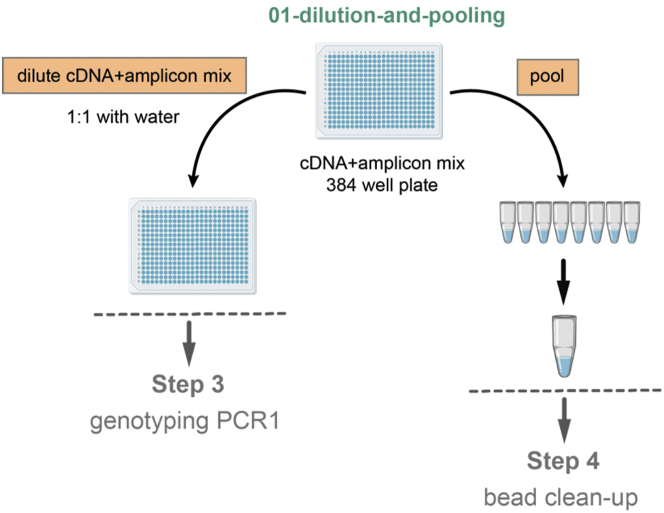
During this step, the cDNA+amplicon mix will be first diluted 1:1 with water, to use as input material in the subsequent single-cell genotyping preparation step, and at the same time, pooled for subsequent bead purification and whole transcriptome library preparation (Figure 11).
Figure 11.
Schematic Representation of the Pooling, cDNA+Amplicon Mix Dilution and Bead Clean-Up Steps
This step has been automated in a Biomek FxP Liquid Handling Platform (Beckman Coulter), and the procedures below detail the steps to follow when using this platform (https://doi.org/10.17632/k92cnf2fph.1). However, users might adapt this part of protocol to other liquid handling platforms such as INTEGRA VIAFLO or Mosquito HTS (TTP Labtech).
Before you start, initialize the Biomek FxP Liquid Handling Platform.
-
46.
Thaw the PCR plate containing the cDNA+amplicon mix and spin it down at 3,000 × g during 1 min at room temperature. When removing the PCR cover, make sure there are no droplets in the PCR film; this might give rise to cross-contamination in between wells.
-
47.
Prepare all the material needed to perform this step:
| Material Needed | Description |
|---|---|
| H20: | 4titude 96-well fully skirted PCR plate (FrameStar, 4titude, Cat# 4ti-0960/C) containing 50–180 μL of RNase-/DNase-free water (Life Technologies, #10977035) per well. This plate might be frozen and used several times |
| cDNA: | cDNA+amplicon mix 384-well plate from point 45, containing 12.5 μL amplified cDNA+amplicon mix after PCR |
| Genotyping_stock | 4 empty 384 well plates (FrameStar, 4titude, Cat# 4ti-0384/C) |
| Pool: | 96-well PCR plate (AB-0900; Thermo Scientific) on the top of a Corning 96-well plate tissue culture flat bottom PCR plate (SLS, Cat# 3595) |
| Tipbox [1-4]: | 4 boxes of P50 tips, sterile, with barrier (Beckman Coulter, Cat# A21586) |
-
48.
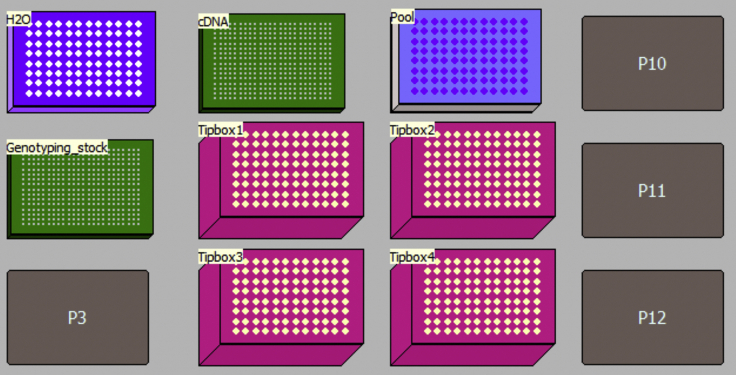
Open the program “MethodS4.bmf” (https://doi.org/10.17632/k92cnf2fph.1). Click Instrument Setup to visualize the method layout and put all required components (plates, tip boxes…) in the right position on the deck (Figure 12). The method can be used with half or entire 384-well plates.
Figure 12.
Schematic Representation of Beckman FxP Deck Layout for Program 01-Dilution-and-Pooling, Using an Entire 384 Well cDNA+Amplicon Mix Plate
-
49.
Click Run, introduce the variable volumes as outlined in the table below (which will be prompted into the screen), and click OK. The program will aliquot 6.25 μL of water and 6.25 μL of cDNA+amplicon mix for each quadrant into a new genotyping_stock plate, which is a 1:1 dilution of the initial cDNA+amplicon mix. This Genotyping_stock plate will be used as the input material for single-cell genotyping PCR1 (step 3). After this, it will pool 1.3 μL from each well of the cDNA+amplicon mix plate into each column of a 96-well plates, which can be subsequently pooled manually in an Eppendorf tube for bead clean-up.
| Variables | Description | Default |
|---|---|---|
| Genotyping_vol: | Ammount of cDNA+amplicon mix to be aliquoted into the Genotyping_stock plate | 6.25 μL |
| Quad: | number of quadrants to be pooled (2 or 4) | 4 |
| Starting_vol: | total volume in each well of the cDNA+amplicon mix plate (from point 45) | 12.5 μL |
| Waterplate_vol: | Volume in each well of the H2O plate | 180 μL |
Pause Point: You might now proceed with bead clean-up of the pooled cDNA+amplicon mix libraries or freeze them and perform bead clean-up later (step 4). You can also store the Genotyping_stock plates until you are ready to proceed with genotyping-PCR1 (step 3).
CRITICAL: The Biomek FxP Liquid Handling Platform has been calibrated to be used with the plasticware specified in this protocol. Any changes to the plasticware might result in suboptimal performance of the method or parts of the robot crashing due to different labware dimensions.
CRITICAL: When freezing plates, it is essential that you use Aluminum Sealing Films (StarLab, Cat# E2796-0792), as plastic films are likely to come off in −20°C freezers. Moreover, it is essential that you spin down and snap freeze the plates in dry ice prior to −20°C freezer storage: this will avoid any liquid freezing in the sides of the wells or cover, which might give rise to cross-contamination in between wells.
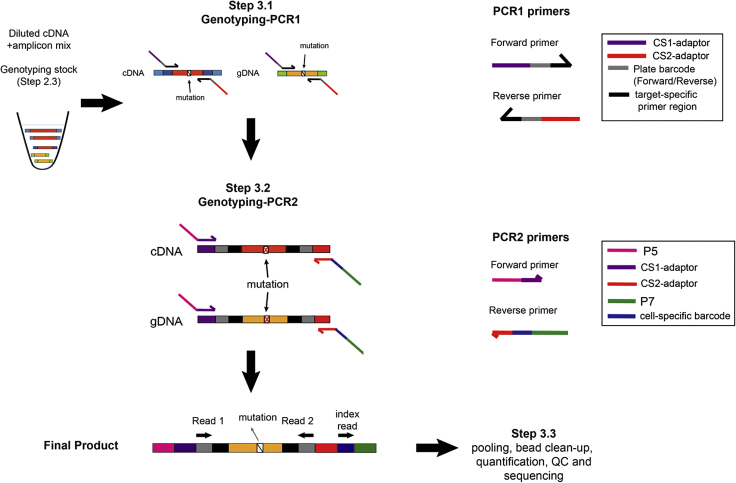
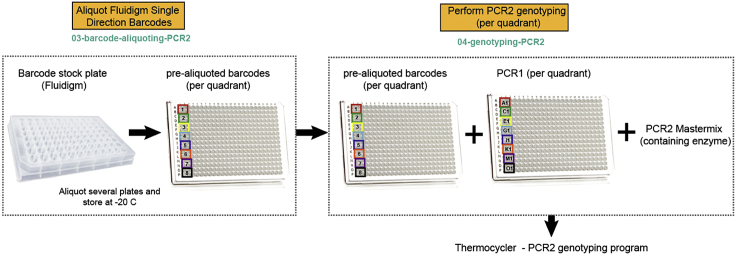
Step 3: Single-Cell Genotyping Library Preparation
Timing: 5 h; 2.5 h hands-on time
This step describes the procedures for single cell genotyping targeted library preparation and sequencing.
During single-cell genotyping library preparation step, two PCR steps will be performed for each single cell to generate Illumina-compatible libraries ready for sequencing (Figure 13). During the first PCR step (genotyping-PCR1), target-specific primers containing CS1 (forward primer) or CS2 (reverse primer) adaptor sequences will amplify specific gDNA and cDNA targets, which have been pre-amplified in the previous RT and PCR step (step 2). At the same time, plate-specific barcode sequences (identifying a set of 96 cells originating from the same plate) will be added (Figure 13). This will increase the multiplexing capacity of the targeted sequencing strategy (Figure 13). Examples on how to design target-specific primers and barcodes for genotyping PCR1 are provided in Table S2. This is an improved higher-multiplexing barcoding strategy as compared to our previously described method (Rodriguez-Meira et al., 2019).
Figure 13.
Schematic Representation of the Targeted Library Preparation Strategy for Single Cell Genotyping
During the subsequent PCR step (genotyping-PCR2), Illumina-compatible adaptors containing a cell-specific barcode are attached to the genotyping-PCR1 products through the CS1 and CS2 adaptor regions. After that, Illumina-compatible libraries are purified using Ampure XP beads and quantified for sequencing (Figure 13).
The steps to perform single-cell genotyping library preparation described here have been automated in a Biomek FxP Liquid Handling Platform (Beckman Coulter). However, users might adapt this protocol to other liquid handling platforms compatible with the method such as Mosquito HTS (TTP Labtech) or VIAFLO (INTEGRA Biosciences).
Genotyping PCR1
Timing: 1.5 h; 30 min hands-on time
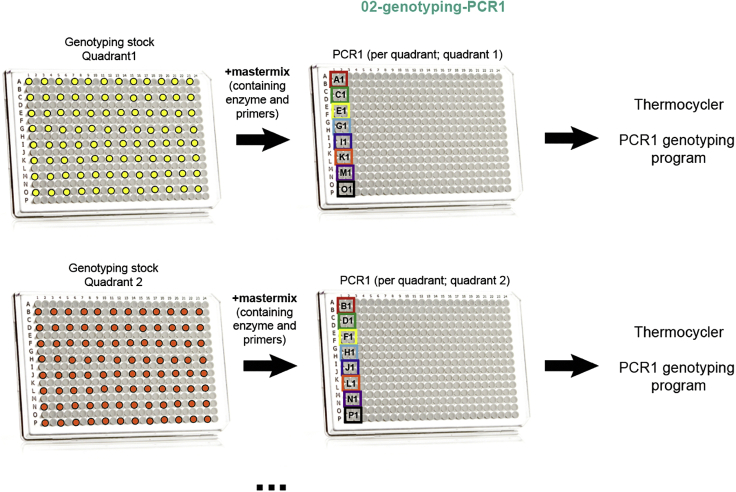
Genotyping-PCR1 is performed individually per each quadrant of a 384-well plate, and up to four different PCR1 reactions are performed per single cell, each reaction containing different primer mixes. The reason behind this is the different amount of input molecules for different gDNA and cDNA amplicons (for example, highly expressed genes will be easily amplified during genotyping PCR1, but lowly expressed ones will be more difficult to amplify). Moreover, different amplification efficiencies of your PCR1 primers will also result in disparate amplification of target regions. Thirdly, some primers targeting gDNA and cDNA from the same target region may not be compatible with parallel amplification in the same reaction. Therefore, compatibility and amplification efficiencies of primers should be established beforehand (see “Optimization and pilot experiments”). Once this has been determined, proceed with Genotyping-PCR1.
Note: The protocol outlined below includes the use of four genotyping PCR1 master mixes per cell, which will be dispensed in the four different quadrants of a 384-well plate (Figure 14); however, the number of mixes required might be reduced depending on the number of targets amplified. Usually, no more than 4 master mixes per cell are required.
Figure 14.
Schematic Representation of Genotyping PCR1 Program
-
50.
Thaw the 384-well plate containing the diluted cDNA+amplicon from “Step 2.3: cDNA+amplicon mix dilution” (genotyping_stock plate). Spin down at 3,000 × g during 1 min at room temperature.
CRITICAL: Make sure the centrifuge is set at room temperature when spinning down the plate; spinning the plate at 4°C will cause condensation on the PCR film covering the plate and as a consequence, potentially cross-contamination in between wells. When removing the PCR cover, make sure there are no droplets on it, as this might also give rise to cross-contamination.
-
51.
Thaw any target-specific genotyping-PCR1 primers to be used in this step and KAPA 2G Ready Mix Enzyme (Roche, Cat# KK5702). Primers might be thawed at room temperature; KAPA enzyme should be thawed and kept on ice or a cold rack at all times.
-
52.
Prepare master mixes containing each primer combination and KAPA 2G enzyme as indicated in the table below. Up to 4 different master mixes might be used per plate.
| Genotyping-PCR1 Master Mix | 1 Cell | 110 Cells∗ (1 Quadrant) | Storage | Cat. No. | Company |
|---|---|---|---|---|---|
| KAPA2G Robust HS Ready Mix | 3.125 μL | 343.75 μL | −20°C | KK5702 | Sigma (Roche) |
| Primer pair 1 (F+R; 20 μM) | 0.375 μL | 41.25 μL | −20°C | Custom desalted oligos; resuspend in RNase-free TE | Invitrogen |
| Primer pair 2 (F+R; 20 μM) | 0.375 μL | 41.25 μL | −20°C | ||
| Primer pair 3 (F+R; 20 μM) | 0.375 μL | 41.25 μL | −20°C | ||
| Water (DNase-/RNase-free) | 0.5 μL | 55 μL | Room temp | 10977035 | Life Technologies |
| TOTAL | 4.75 μL | 522.5 μL |
-
53.
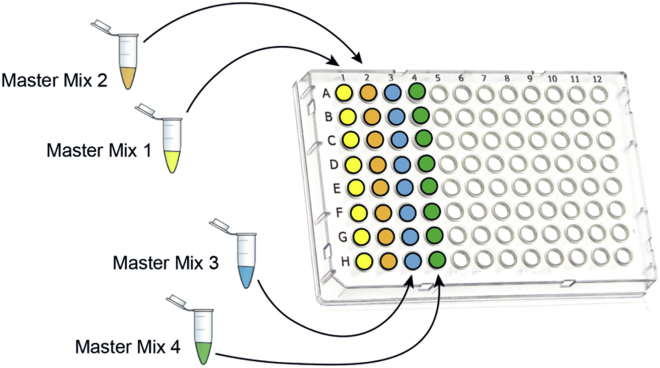
Aliquot each master mix in a column of a 96-well skirted PCR plate (FrameStar, 4titude, Cat# 4ti-0960/C) as indicated in Figure 15.
Figure 15.
Schematic Representation of Mastermix Plate Layout
-
54.
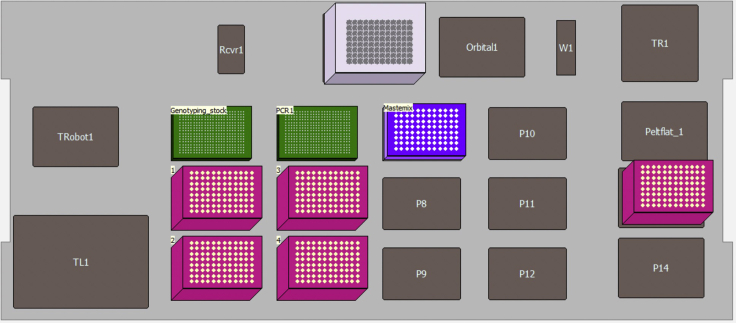
Open the program “MethodS5.bmf” (https://doi.org/10.17632/k92cnf2fph.1). Click Instrument Setup to visualize the method layout. Place the tip boxes, a clean 384-well plate, the genotyping stock plate (step 2.3) and the 96-well skirted plate containing enzyme+primer master mixes (Mastermix plate; Figure 16). The Mastermix plate should be placed on the top of a pre-chilled cold rack.
Figure 16.
Schematic Representation of Deck Layout Used for 02-Genotyping-PCR1 Program in a Biomek FxP Liquid Handling Platform
| Material Needed | Description |
|---|---|
| Genotyping_stock: | Genotyping_stock plate from 01-Dilution-and-pooling (step 2.3) |
| PCR1: | empty 384 well plate to be used for PCR1 (FrameStar, 4titude, 4ti-0384/C) |
| Mastermix | 4titude 96-well skirted PCR plate containing 4.75 μL of master mix per cell per quadrant. The plate should be sitting in a 96-metal rack, which can be chilled prior running the protocol |
| Tip boxes required: | 4 boxes of filtered P50 tips + 32 P50 filtered tips to aliquot master mix |
CRITICAL: The Biomek FxP Liquid Handling Platform has been calibrated to be used with the plasticware specified in this protocol. Any changes to the plasticware might results in suboptimal performance of the method or parts of the robot crashing due to different labware dimensions.
-
55.
Click Run to start the program and introduce the user-defined variables:
| Variables | Description | Default |
|---|---|---|
| Mastermix_vol: | Volume of master mix in each well of the 96-well skirted MasterMix Plate (Figures 15 and 16) | 6.25 μL |
| Number_of_quadrants: | number of quadrants to aliquot master mix and cDNA+amplicon mix into | [1,2,3,4] |
| Starting_vol: | total volume in each well of the genotyping stock plate (point 49) | 12.5 μL |
| Stock_quadrant: | Genotyping stock plate quadrant to use for genotyping PCR1 | [1,2,3,4] |
-
56.
The program will aliquot first 4.75 μL of each Mastermix in each quadrant of the empty 384-well plate (PCR1) and after that, it will aliquot 1.5 μL of prediluted cDNA+amplicon mix from the genotyping stock plate in each quadrant of the same 384-well plate. The entire program will take 10 min to prepare each plate. Once the program is finished, immediately cover the plate with a PCR film, spin it down at 1,000 × g during 15 s and run the following program on a thermocycler:
| Temperature | Time | Cycles |
|---|---|---|
| 95°C | 3 min | 1 cycle |
| 95°C | 00:15 | 20 cycles |
| 60°C | 00:20 | |
| 72°C | 1 min | |
| 72°C | 5 min | 1 cycle |
| 4°C | HOLD | 1 cycle |
Optional: While the robot is aliquoting the first plate, you can start preparing the master mixes for a second plate.
Pause Point: You might now proceed with the second single-cell genotyping PCR (genotyping-PCR2) or spin down the plates, snap freeze them in dry ice and transfer to −20°C storage.
CRITICAL: When spinning down the plates, make sure the centrifuge is set at room temperature; if set at 4°C, the PCR product will condensate on the top of the PCR film and potentially cause contamination between wells (Remember: this PCR does not contain cell-specific barcodes yet!).
Genotyping PCR2
Timing: 1.5 h; 30 min hands-on time
During PCR2 step, Illumina-compatible adaptors containing cell-specific barcodes will be added to the PCR1 products from the previous step (Figure 13). Firstly, Illumina-compatible adaptors with cell-specific barcodes will be aliquoted into each well of a 384-well plate using a Biomek FxP Liquid Handling Platform. These plates can be used immediately after or stored at −20°C for several months. After that, a Mastermix containing PCR enzyme and PCR1 products will be aliquoted in the same plate.
-
57.
Thaw Illumina-compatible adaptors with cell-specific barcodes plates (four 96-well plates in total; Access Array™ Barcode Library for Illumina® Sequencers-384, Single Direction; Fluidigm, Cat# 100-4876) and spin them down at 3,000 × g for 1 min at room temperature.
-
58.
Open the program “MethodS6.bmf” (https://doi.org/10.17632/k92cnf2fph.1). Click Instrument Setup to visualize the method layout (Figure 18). Place the tip boxes (1 P50 box per plate), clean 384-well plates, and the Fluidigm Single-Direction Barcodes plates in the deck, as summarized below:
| Material Required | Description |
|---|---|
| Barcode_stock1, barcode_stock2, barcode_stock3, barcode_stock4 | Fluidigm Access Array Single-Direction Barcodes (plates A1, A2, A3, A4; Fluidigm, Cat# 100-4876) |
| Dest 1, Dest 2, Dest3, Dest4 plates: | Empty 384 well plates (FrameStar, 4titude, Cat# 4ti-0384/C); named prealiquoted-PCR2-barcodes |
| Tip boxes required: | 1 box of filtered P50 tips per plate (Quad1-4) |
-
59.
Click Run, introduce the values of the variables, which will be prompted on the screen, and click OK to start the program. The program will aliquot 2 μL of each barcode four times (if number_of_quadrants=4) in each 384-well plate (Dest1-4). Once finished, seal the plates with a PCR film for further storage or proceed to PCR2 genotyping.
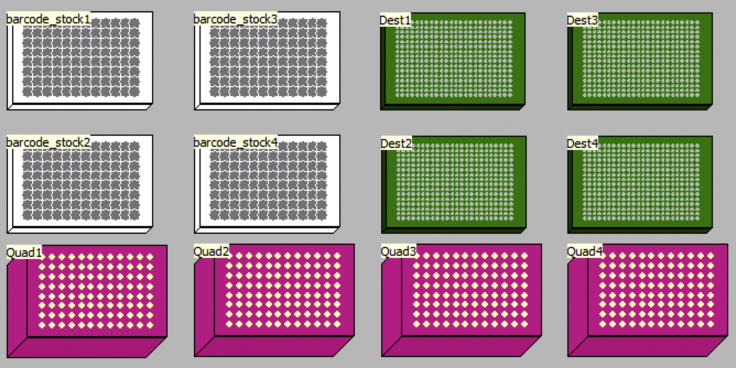
| Variables: | Description: | Default value: |
|---|---|---|
| number_of_plates | Number of barcode_stock plates to aliquot barcodes from. | [1,4] |
| Start_Volume | Starting volume on the Fluidigm Single-Direction Barcodes plates. | 50 μL. Note: the robot will aspirate the liquid from the bottom of the plate, so this volume does not have to be precise |
| number_of_quadrants | Number of quadrants to aliquot barcodes into the destination plates. | [1,4] |
Pause Point:
Figure 18.
Schematic Representation of Deck Layout Used for 03-Barcode-Aliquoting-PCR2 Program in a Biomek FxP Liquid Handling Platform
You might now proceed with the second single-cell genotyping PCR (genotyping-PCR2) or spin down the plates containing cell-specific barcodes, snap freeze them in dry ice and transfer to −20°C storage.
-
60.
Prepare the genotyping-PCR2 master mix in a 5 mL or two 1.5 mL microfuge tubes as per the table below, which is enough for one 384-well plate and takes into account dead volumes of the method (15%).
| Genotyping-PCR2 Master Mix | 1 Reaction | 430 Reactions | Storage | Cat. No. | Company |
|---|---|---|---|---|---|
| Buffer 10 X (labeled 3) | 1 μL | 430 μL | −20°C | FastStart High Fidelity PCR System (Cat# 04738292001) | Roche |
| MgCl2 (labeled 4) | 1.8 μL | 774 μL | |||
| DMSO (labeled 5) | 0.5 μL | 215 μL | |||
| Nucl Mix (labeled 6) | 0.2 μL | 86 μL | |||
| Enzyme (labeled 1) | 0.1 μL | 43 μL | |||
| RNase-/DNase-free Water | 1.9 μL | 817 μL | Room temp | 10977035 | Life Technologies |
| TOTAL | 5.5 μL | 2,365 μL∗ | |||
| cDNA (aliquoted by the robot from PCR1 plate) | 2.5 μL | ||||
| Barcode (prealiquoted in point 59) | 2 μL | ||||
| TOTAL (cumulative) | 10 μL |
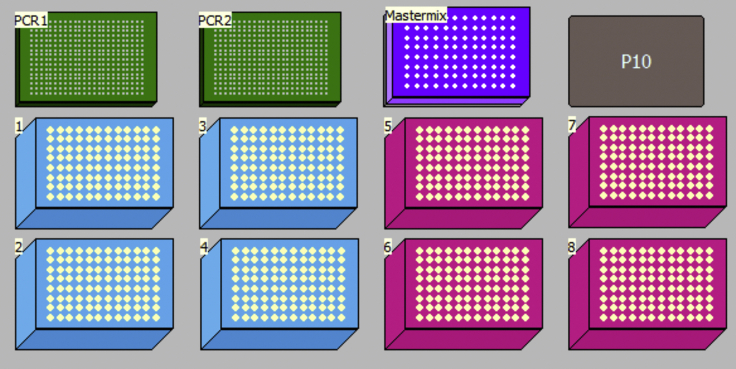
Place the Mastermix, prealiquoted barcode plate (from point 59) and PCR1 genotyping plate (point 56) in the deck as outlined in Figure 19 and in the table below:
| Material Required | Description |
|---|---|
| PCR1 | Genotyping PCR1 plate from point 56 |
| PCR2 | 384 well plate (FrameStar, 4titude, Cat# 4ti-0384/C) containing prealiquoted PCR2 barcodes from point 59 |
| Tip boxes required: | 1 box of filtered P50 tips per plate (Quad1-4; Cat# A21586) and 1 box of filtered P20 tips per plate. |
Note: It is recommended to run several 384-well plates each time to reduce the amount of master mix needed due to dead volumes. If running the method several times, the volume of the genotyping-PCR2 Mastermix plate can be dropped to 400 reactions each time (4% dead volume), rather than 430 reactions (12% dead volume) required for the first run.
-
61.
Start “MethodS7.bmf” program (https://doi.org/10.17632/k92cnf2fph.1) and introduce the user-defined variables (Number of Quads: [1-4], indicating the number of quadrants to aliquot PCR1 product and PCR2 master mix into). Once finished, seal the plate with a PCR film, spin down for 15 s at 1,000 × g and incubate in a thermocycler using the parameters specified in the table below:
| Temperature | Time | Cycles |
|---|---|---|
| 95°C | 10 min | 1 cycle |
| 95°C | 00:15 | 10 cycles |
| 60°C | 00:30 | |
| 72°C | 1 min | |
| 72°C | 5 min | 1 cycle |
| 4°C | HOLD | 1 cycle |
Optional: While the robot is aliquoting the first plate, you can start preparing the master mixes for a second plate. It is recommended to process several plates to reduce dead volumes in the reaction.
Pause Point: You might now proceed with pooling and bead clean-up of the Illumina-compatible single-cell genotyping libraries or freeze the plates at −20°C.
Figure 19.
Schematic Representation of Deck Layout Used for 04-Genotyping-PCR2 Program in a Biomek FxP Liquid Handling Platform
Pooling, Bead Clean-Up, and Library Quantification
Timing: 2 h; 1.5 h hands-on time
-
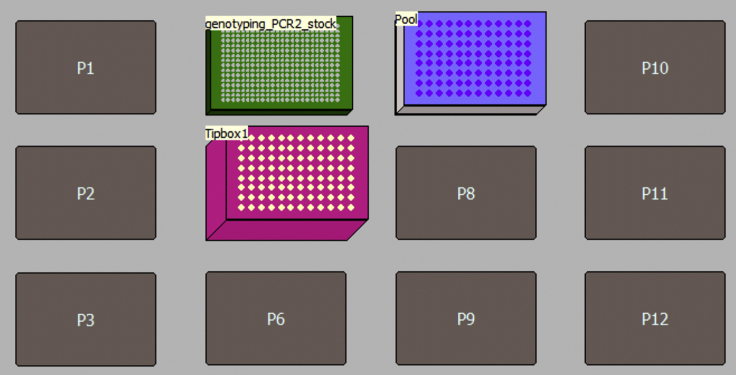
62.
Pool 96 libraries from each genotyping-PCR2 plate quadrant (corresponding to each different primer mix; Figures 14 and 17), using a Biomek FxP Liquid Handling Platform. Start “MethodS8.bmf” program (https://doi.org/10.17632/k92cnf2fph.1), position the material on the deck (Figure 20) and introduce the user-defined variables outlined below. This program will pool 1.5 μL of genotyping-PCR2 reaction from each quadrant. This step can also be easily automated in other liquid handling platforms such as Mosquito HTS (TTP Labtech).
Figure 17.
Schematic Representation of Barcode Aliquoting and Genotyping-PCR2 Steps
Figure 20.
Schematic Representation of Deck Layout Used for 05-Genotyping-Pooling Program in a Biomek FxP Liquid Handling Platform
| Material Required | Description |
|---|---|
| Genotyping_PCR2_stock | PCR2 plate from point 61 |
| Pool | 96-well PCR plate (AB-0900; Thermo Scientific) on the top of a Corning 96-well plate tissue culture flat bottom PCR plate (SLS, Cat# 3595) |
| Tip boxes required: | 1 box of filtered P50 tips |
-
63.
Introduce the user-defined variables and run 05-genotyping-pooling program.
| Variables | Description | Default Value |
|---|---|---|
| Quad | Number of quadrants to pool from genotyping_PCR2_stock plate | [1,4] |
| Starting_Volume | Total volume in each well of genotyping_PCR2_stock plate | 10 μL |
-
64.
Pool the genotyping-PCR2 material from each column of the 96-well plate (“Pool,” Figure 20) into a single 1.5 mL microfuge tube using a P200 tip, as outlined and proceed to bead purification.
-
65.
Aliquot 40 μL of Ampure XP beads (Beckman Coulter; Cat# A63881) into a V-shaped 96 well plate (Axygen, Cat# P-96-450V-C). Add 50 μL of pooled genotyping-PCR2 product from step 49 to the beads (0.8:1 beads to cDNA ratio) and incubate for 5 min at room temperature.
-
66.
Incubate the mixture in a 96-well magnetic stand for 2 min or until the liquid is clear of beads. Remove the liquid and wash the beads twice with 100 μL 80% ethanol, trying not to disturb the bead pellet. Remove any remaining ethanol with P20 tips and let the beads air-dry for 2 min.
-
67.
Resuspend the pellet in 25 μL of EB buffer and check library size distributions and specific target amplification on a microcapillary array (Figure 21), such as Agilent Bioanalyzer (High Sensitivity DNA Kit, Agilent, Cat# 5067-4626), TapeStation (High Sensitivity D1000 ScreenTape and Reagents, Agilent, Cat# 5067-5583 and Cat# 5067-5584) or Fragment Analyzer (High Sensitivity Small Fragment Kit (1–1,500 bp), Advanced Analytical, Cat# DNF-477-0500).
Figure 21.
Representative Traces of Genotyping-PCR2 Pooled Products
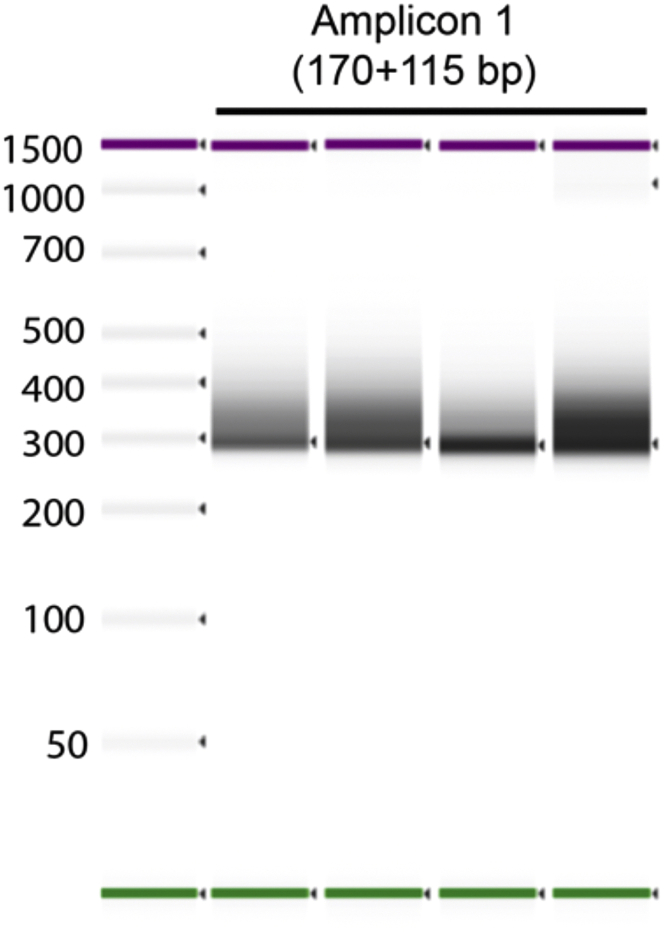
Note: The library sizes would be much shorter than cDNA libraries, so the size range of the microcapillary array should be taken into account in this case.
Note: the barcodes and adaptors add 115 extra bp to the expected amplicon size.
-
68.
Quantify the libraries using Qubit dsDNA HS Assay Kit (Invitrogen, Cat# Q32854).
-
69.
Calculate the molarity of each pool and dilute to 10 nM. Pool each amplicon mix in equimolar concentrations, quantify the final library, and dilute to 4 nM.
-
70.
Sequence the libraries on a MiSeq platform using custom sequencing primers CS1, CS2 (read 1 and read 2; targeted to the CS1/CS2 adaptor sequences added during PCR1) and CS1rc, CS2rc (index read) or in a NextSeq platform using custom sequencing primers LCS1 (read 1), CS2 (read 2) and CS2rc (index read).
If using the MiSeq sequencing platform (Illumina), use 7 μL of 50 μM CS1 and CS2 primers resuspended in 700 μL of HT1 buffer (500 nM final concentration), and 7 μL of 50 μM of CS1rc and CS2rc in a total volume of 700 μL of HT1 buffer (500 nM final concentration). If using the NextSeq platform, use 15 μL of 50 μM LCS1 primer diluted in 2,485 μL of HT1 buffer for read 1 (300 nM final concentration), 15 μL of 50 μM CS2 primer diluted in 2,485 μL of HT1 buffer for read 2 (300 nM final concentration) and 15 μL of 50 μM CS2rc primer diluted in 2,485 μL of HT1 buffer for index read (300 nM final concentration).
| Platform | Primer(s) R1 | Primer(s) R2 | Index Read Primer(s) | Custom Primer Total Volume |
|---|---|---|---|---|
| MiSeq | CS1+CS2 (500 nM) | CS1+CS2 (500 nM) | CS1rc+CS2rc (500 nM) | 700 μL |
| NextSeq | LCS1 (300 nM) | CS2 (300 nM) | CS2rc (300 nM) | 2,500 μL |
Primer sequences can be found in the Key Resources Table. Use the following sequencing configuration: 151 cycles (read1) + 10 cycles (index read) + 151 cycles (read2).
Note: CS1/CS2 and CS1rc/CS2rc sequencing primers contain LNA modifications as compared to CS1/CS2 tags used for PCR1 target-specific primers. Primers should be aliquoted and stored in single-use aliquots. Primer stocks should be stored at −80°C.
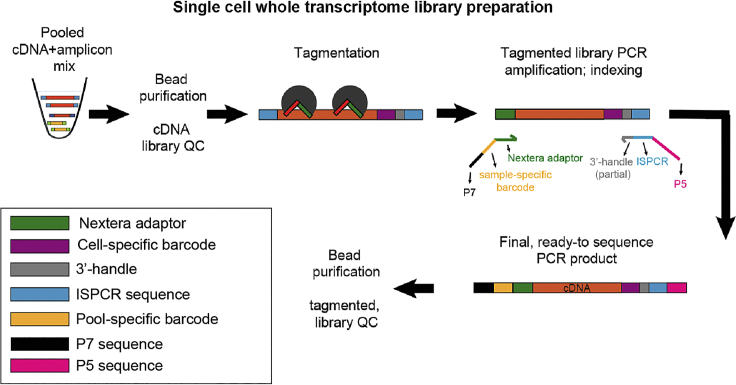
Whole Transcriptome Library Preparation
Timing: 3.5 h; 2.5 h hands-on time
This step describes the procedures for whole transcriptome library preparation from pooled cDNA material using a tagmentation-based approach specifically tailored to TARGET-seq libraries (Rodriguez-Meira et al., 2019)
Whole transcriptome library preparation consists of three steps: first, the pooled cDNA libraries from step 2.3 are bead-purified; secondly, the libraries are tagmented using a commercial Tn5 enzyme (Illumina). Thirdly, the tagmented libraries will be amplified and indexed using a custom P5 primer binding to the ISPCR and 3′-handle adaptor sequence (introduced during the RT and PCR steps, which will allow specific amplification of the 3′-ends), as well as a P7 primer containing a sample-specific index. In this way, only the tagmented fragments corresponding to the 3′-end of each cDNA molecule will be amplified. Lastly, tagmented and amplified libraries are purified using Ampure XP beads, quantified and sequenced.
CRITICAL: Pre-warm an aliquot of Ampure XP beads (Beckman Coulter; Cat# A63881) at room temperature for 30 min.
-
71.
Pool the cDNA material from the 96-well plate (step 2.3, point 49) into a single 1.5 mL microfuge tube using a P200 tip and proceed to cDNA purification.
-
72.
Aliquot 66 μL of Ampure XP beads (Beckman Coulter; Cat# A63881) into a V-shaped 96 well plate (Axygen, Cat# P-96-450V-C). Add 110 μL of pooled cDNA+amplicon mix from step 2.3 to the beads (0.6:1 beads to cDNA ratio) and incubate for 5 min at room temperature.
-
73.
Incubate the mixture in a 96-well magnetic stand for 2 min or until the liquid is clear of beads. Remove the liquid and wash the beads twice with 100 μL 80% ethanol, trying not to disturb the bead pellet. Remove any remaining ethanol with P20 tips and let the beads air-dry for 2 min.
-
74.
Resuspend the beads in 110 μL of EB buffer (Qiagen Cat# 19086) and repeat the bead purification step. Add 66 μL of Ampure XP beads to the 110 μL of pre-purified cDNA mix, incubate 5 min at room temperature, then incubate the mixture for 2 min in the 96-well magnetic stand. Once clear of beads, remove the liquid and what twice with 100 μL 80% ethanol, trying not to disrupt the beads. Remove any remaining ethanol using P20 tips.
-
75.
Resuspend the pellet in 25 μL of EB buffer and check cDNA traces on a microcapillary array, such as Agilent Bioanalyzer (High Sensitivity DNA Kit, Agilent, Cat# 5067-4626), TapeStation (High Sensitivity D5000 ScreenTape and Reagents, Agilent, Cat# 5067-5592 and Cat# 5067-5593) or Fragment Analyzer (High Sensitivity NGS Fragment Analysis Kit (1–6,000 bp), Advanced Analytical, Cat# DNF-474-1000). An example of good-quality libraries can be found in Figure 3.
-
76.
Quantify the libraries using Qubit dsDNA HS Assay Kit (Invitrogen, Cat# Q32854).
-
77.
Prepare one PCR tube (StarLab; Cat# I1402-2908) per each pool of libraries from point 71 and add 10 μL of Tagmentation Buffer (Nextera XT Kit, Illumina, Cat# FC-131-1096) into each tube.
-
78.
Add 1 ng of purified cDNA from each pool from point 71, up to a total volume of 5 μL, and add 5 μL of Amplicon Tagmentation Mix (Nextera XT Kit) on ice as per the table below.
Note: keep the Amplicon Tagment Mix on ice at all times.
| Reagents | 1 Reaction (μL) | Catalog Number | Company |
|---|---|---|---|
| Tagmentation Buffer | 10 μL | Nextera XT Kit, Cat# FC-131-1096 | Illumina |
| ATM (Amplicon Tagment Mix) | 5 μL | ||
| Bead-purified cDNA from point 75 (0.2 ng/μL) | 5 μL | NA | NA |
| TOTAL | 20 μL |
-
79.
Briefly vortex the mix, spin down and incubate at 55°C for 6 min.
-
80.
Immediately after, add 5 μLof Neutralization Buffer (Nextera XT Kit, Illumina); pipet up and down to mix the liquid.
-
81.
Prepare PCR master mix as per the table below. i7 index primers can be obtained commercially (Illumina) or as custom HPLC purified oligonucleotides (Table S3). P5_index is a custom primer binding to the ISPCR and 3′-handle sequence (Table S3) as outlined in Figure 22.
Figure 22.
Schematic Representation of the Tagmentation-Based Library Preparation Strategy for 3′-Biased Whole Transcriptome Sequencing
| Reagents | 1 Reaction | Catalog Number | Company |
|---|---|---|---|
| i7 index (2 μM) | 5 μL | Nextera XT Index Kit v2 (Set A,B,C or D), FC-131-2001/2002/2003/2004 | Illumina |
| P5_index (2 μM) | 5 μL | Custom HPLC purified oligonucletide | biomers.net |
| NPM (PCR master mix) | 15 μL | Nextera XT Kit, Cat# FC-131-1096 | Illumina |
| TOTAL | 25 μL | ||
| TOTAL (cumulative) | 50 μL |
-
82.
Incubate in a thermocycler with the following program:
| Temperature | Time | Cycles |
|---|---|---|
| 95°C | 30 s | 1 |
| 95°C | 10 s | 14 cycles |
| 55°C | 30 s | |
| 72°C | 30 s | |
| 72°C | 5 min | 1 |
| 4°C | HOLD | 1 |
-
83.
Purify the tagmented and amplified libraries using Ampure XP beads. Add 35 μL of beads to 50 μL (0,7:1 beads to cDNA ratio) of tagmented library in a V-shaped 96 well plate (Cat. No. P-96-450V-C, Axygen) and incubate for 5 min at room temperature.
-
84.
Incubate in a 96-well magnetic stand for 2 min or until the liquid is clear of beads. Remove the liquid and wash twice with 100 μL of 80% ethanol. Remove any remaining ethanol with a P20 tip and resuspend in 35 μL of EB buffer.
-
85.
Repeat this step by adding 24.5 μL of beads to the 35 μL of tagmented library and incubate for 5 min at room temperature. Remove the liquid, wash twice with ethanol, and air-dry the pellet for at least 5 min. Resuspend in 20 μL of EB buffer.
CRITICAL: It is essential that the beads are clear of ethanol at this point before resuspending in EB buffer; these libraries are now ready to sequence and any remaining traces of ethanol might affect sequencer performance.
Pause Point: You might now proceed with quantification and sequencing or store the bead-purified tagmented and indexed libraries at −20°C.
-
86.
Check tagmented library size distributions (Figure 23) on a microcapillary array such as Agilent Bioanalyzer (High Sensitivity DNA Kit, Agilent, Cat# 5067-4626), TapeStation (High Sensitivity D5000 ScreenTape and Reagents, Agilent, Cat# 5067-5592 and Cat# 5067-5593) or Fragment Analyzer (High Sensitivity NGS Fragment Analysis Kit (1–6,000 bp), Advanced Analytical, Cat# DNF-474-1000). The average size of this library should be between 500 and 750 bp (range 300–1000 bp); if there are traces of primer dimers (<200 bp fragments), repeat bead purification and re-check fragment size distributions.
Figure 23.
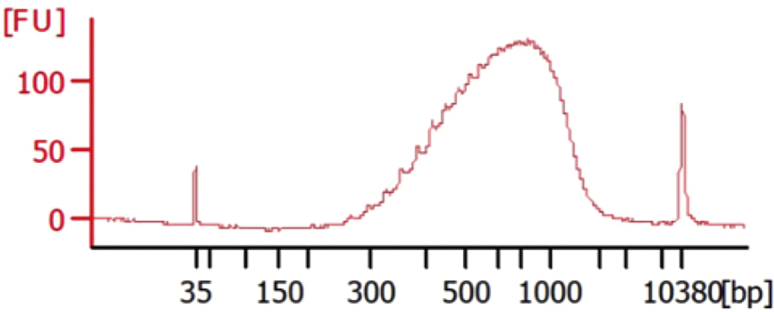
Representative Traces of a Tagmented and Purified 3′-TARGET-seq Whole Transcriptome Library (Average Size = 600 bp)
-
87.
Quantify the libraries using Qubit and calculate library molarity.
-
88.
Dilute libraries to 4 nM and pool equimolar concentrations of each library in a microfuge tube. Libraries are now ready for sequencing!
-
89.
Sequence the libraries on an Illumina platform (NextSeq, HiSeq or NovaSeq platforms) using a custom primer for read1 (P5_SEQ; Key Resources Table). If using the NextSeq platform, load a 3 pM library diluted in 1.3 mL of HT1 Buffer (Illumina) and 900 nM of P5_SEQ primer in a total volume of 3 mL of HT1 buffer. Use the following sequencing configuration: 15 cycles (read1) + 8 cycles (index read) + 65 cycles (or more, read2).
Expected Outcomes
After step 2, you should expect to generate cDNA libraries such as the one presented in Figures 3A–3C, with a concentration ranging from 0.5 ng/μL to 5 ng/μL.
After step 3, you should expect to generate targeted libraries such as the ones presented in Figure 21, with a yield ranging from 5 ng/μL up to 100 ng/μL. If sequencing is performed on the NextSeq, you should expect over 85% of bases over Q30. In a good-quality run, all amplicons should be detected in over 85% of the cells and the allelic dropout rate should be approximately 5% for genomic DNA amplicons. The allelic dropout rate for mRNA amplicons is variable and depends on the level of expression of each transcript.
After step 4, you should expect to generate tagmented libraries such as the ones presented in Figure 23, with a yield ranging from 0.5 ng/ μL to 5 ng/μL. After whole transcriptome sequencing, you should expect at least 40%–50% of reads mapping to exonic regions in the vast majority of cells, and less than 5% of reads mapping to ERCC molecules on average. QC thresholds for HSPC TARGET-seq libraries are usually set at library size >2,000, number of genes >500, mitochondrial reads <20%, and ERCC <50%; these parameters might vary depending on the cell type of interest.
Single-cell genotyping libraries and whole transcriptome libraries from the same single cell are assigned different barcodes during the experimental workflow. Mapping of genotyping and whole transcriptome barcodes to their original cell is achieved through the initial well location in which each cell was sorted. For example, plate 1 Well A1 is assigned identifier Plate1_A1, and the barcodes that are subsequently assigned to this position are tracked through the experimental design with this cell identifier.
Limitations
TARGET-seq relies on targeted mutational analysis to achieve high-sensitivity mutation detection in single cells. Consequently, only a limited number of target regions (we have tested up to 12 in total) can be profiled per cell, as opposed to a less sensitive whole-genome amplification approach. In our workflow, we characterize tumors through bulk mutational analysis using whole-genome sequencing, whole-exome sequencing, or a targeted gene panel for the regions of interest. We then select a list of candidate driver mutations to be profiled using TARGET-seq.
TARGET-seq is a plate-based protocol which has been completely automated using robotic platforms, so that a few thousand cells (1,500–2,000) can be easily processed per week. Moreover, through novel optimized barcoding and multiplexing strategies implemented in this protocol, we improve the throughput as compared on our previously described 3′-TARGET-seq approach. Despite this, processing hundreds of thousands of cells using within a few months could be challenging, and the throughput of the platform is likely to be limited to tens of thousands of cells.
Other techniques achieving higher throughput are currently available (Nam et al., 2019, van Galen et al., 2019). However, the sensitivity of mutation detection is considerably lower and therefore, these technologies cannot be used to confidently reconstruct clonal hierarchies, determine loss of wild-type allele or profile mutations in lowly expressed genes.
Troubleshooting
Problem 1
You are unable to generate cDNA libraries; all of your samples seem to be blank (steps 1 and 2; Figure S2).
Potential Solution
Make sure your sorter alignment is optimal and single cells are been efficiently sorted in each well containing lysis buffer (step 1.3, points 25–27) . Some fluorescent-activated cell sorters cannot be easily calibrated to sort into 384-well plates and therefore, it is recommended to first generate libraries into 96-well plates.
Alternatively, some of your reagents might be degraded and/or contaminated with RNase/DNase. Change your reagents and always keep sterile and RNase-/DNase-free conditions when handling reagents used for lysis buffer preparation, RT, and PCR steps.
Problem 2
You are unable to generate good-quality cDNA libraries; your library size distributions are skewed to lower ranges (steps 1 and 2; Figure S3).
Potential Solution
The patterns presented in Figure S3 correspond to RNA degradation in your samples. There are several reasons which might be causing this degradation: sorter alignment, reagent contamination with RNases, high evaporation during the RT step or sample viability.
Check first that your sorted alignment is accurate (step 1.3, points 25–27): if you are sorting cells on the sides rather than center of the wells, RNA will be partially degraded. Secondly, make sure that none of your reagents are contaminated with RNases and that when you handle the samples, this is done in a RNase-free environment. Thirdly, make sure that the duration of the sort does not exceed 20 min for each plate (step 1.3, point 28); longer times might cause sample degradation. High evaporation rates during the RT step (step 2.1, point 37) will also give rise to concatemer formation/RNA degradation; you should carefully seal your plates with strongly adhesive PCR films prior the RT step and if necessary, two sealing films. Finally, poor cell viability in your samples will also give rise to RNA degradation.
Problem 3
You are unable to generate libraries when you include the protease on the lysis buffer, or you are experiencing library degradation when you include the protease (steps 1 and 2; Figures S2 and S3).
Potential Solution
Protease should be efficiently inactivated by incubating samples at 72°C for 15 min (step 2.1, point 34). If addition of protease is associated with lower yields or signs of RNA degradation (Figure S3), it is likely that the protease itself is contaminated with DNases or RNases. Resuspend the protease using DNase-/RNase-free water. Re-order the protease and avoid the presence of potential contaminants when resuspending the lyophilized solution.
Problem 4
You are unable to generate libraries when you include target-specific primers in your RT or PCR mix (step 2)
Potential Solution
Primers should be HPLC purified to minimize the presence of any contaminants that might affect cDNA library generation. Always resuspend RT/PCR primers using DNase-/RNase-free water in a PCR-clean environment. Re-order the target-specific primers and avoid the presence of potential contaminants when resuspending the lyophilized solution. Alternative, redesign primers (Optimization and pilot experiments; points 2 and 3) or reduce the concentration of primers during the RT step (step 2.1, point 33), down to 35 nM. mRNA primers used during the RT step are more likely to interfere with successful cDNA library generation than gDNA primers used during the PCR step.
Problem 5
You are generating good-quality cDNA libraries, but you cannot amplify your target regions of interest with high efficiency (Optimization and pilot experiments; points 2 and 3)
Potential Solution
It is likely that RT/PCR or genotyping-PCR1 primers are not specific for your target region of interest. Redesign the primers (Optimization and pilot experiments) trying to minimize binding to off-target regions, 3′ self-complementarity and high number of adenine and thymidine bases at the 3′-end of the primer (Figures 1 and S1).
Problem 6
You are generating good-quality libraries from cell lines but you are struggling to generate libraries from primary cells
Potential Solution
Cell lines amplify more easily than primary cells because they (usually) have more mRNA. So, it is not unusual to obtain good libraries from cell lines but not from primary cells. You should increase the number of PCR cycles that you perform on the cDNA amplification step accordingly (Optimization and pilot experiments) and reduce the ERCC amount used in the lysis buffer (step 1.2, point 12). The minimum number of PCR cycles recommended is 18, and the maximum, 28. If cDNA is not successfully amplified after 28 cycles of PCR amplification, it is unlikely that further amplification will generate cDNA libraries.
One major complication when handling primary cells is cell viability, which might have major implications in successfully generating cDNA libraries. Make sure your viability gate is stringent enough to avoid sorting dead cells, and that the viability dye used for sorting is bright enough to allow clear separation of viable versus non-viable cells (we do not recommend using Aqua Dead Cell Stain dyes or similar; 7-AAD and DAPI provide better results). If the viability of your sample is 50% or lower, you might not be able to generate good-quality cDNA libraries from single cells.
Problem 7
You can only see two big peaks with characteristic size distributions around 600 bp and 1,300 bp but you are not generating bell-shaped cDNA libraries (step 1; Figure S4)
Potential Solution
It is very likely that the peaks you are detecting are the ERCC spike-in mix transcripts (Figure S4), which corresponds to the size distributions of the most abundant ERCC molecules. You should reduce the input ERCC amount (step 1.2, point 12). Also, you should take into consideration that the ERCC amount used for primary cells as compared to cell lines is usually a lot lower (5 to 10-fold difference; Optimization and pilot experiments; point 1).
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Adam J Mead (adam.mead@imm.ox.ac.uk)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The code required to analyze datasets produced using this protocol are available at GitHub https://github.com/albarmeira/. An schematic workflow of the analysis is presented in Figure 24.
Acknowledgments
The authors thank Dr. Sally-Ann Clark at the WIMM Flow Cytometry Facility, Dr. Neil Ashley at the WIMM Single Cell Facility, Dr. Affaf Aliouat, and Dr. Asger Jakobsen for helpful comments. The authors also thank Gemma Buck for input and initial development of automation protocols and Wei Wen for input on library demultiplexing for high-throughput single-cell genotyping. This work was funded by a CRUK DPhil Prize DPhil Prize Studentship (C5255/A20936) to A.R.-M., a senior CRUK Senior Cancer Research Fellowship to A.J.M. and an MRC Clinical Research Training Fellowship (J.O’S.; MR/S001190/1). Some of the figures in this manuscript were created using BioRender.
Author Contributions
A.R.-M. and A.J.M. conceived the protocol. A.R.-M., J.O.S., and H.R. performed and analyzed experiments. A.R.-M. wrote the manuscript with contributions from all authors. A.J.M. supervised the study and acquired funding.
Declaration of Interests
A.J.M. and A.R.-M. are cited as inventors in two patent applications related to the method described in this manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100125.
Contributor Information
Alba Rodriguez-Meira, Email: alba.rodriguezmeira@imm.ox.ac.uk.
Adam J. Mead, Email: adam.mead@imm.ox.ac.uk.
Supplemental Information
References
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustacchini A., Thongjuea S., Barkas N., Woll P.S., Povinelli B.J., Booth C.A.G., Sopp P., Norfo R., Rodriguez-Meira A. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017;23:692–702. doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Nam A.S., Kim K.T., Chaligne R., Izzo F., Ang C., Taylor J., Myers R.M., Abu-Zeinah G., Brand R., Omans N.D. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature. 2019;571:355–360. doi: 10.1038/s41586-019-1367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Bjorklund A.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Meira A., Buck G., Clark S.A., Povinelli B.J., Alcolea V., Louka E., Mcgowan S., Hamblin A., Sousos N., Barkas N. Unravelling Intratumoral Heterogeneity through High-Sensitivity Single-Cell Mutational Analysis and Parallel RNA Sequencing. Mol. Cell. 2019;73:1292–1305.e8. doi: 10.1016/j.molcel.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyper M., Porter C.B.M., Ashenberg O., Waldman J., Drokhlyansky E., Wakiro I., Smillie C., Smith-Rosario G., Wu J., Dionne D. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med. 2020;26:792–802. doi: 10.1038/s41591-020-0844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P., Hovestadt V., Wadsworth M.H., II, Hughes T.K., Griffin G.K., Battaglia S., Verga J.A., Stephansky J., Pastika T.J., Lombardi Story J. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell. 2019;176:1265–1281 e24. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code required to analyze datasets produced using this protocol are available at GitHub https://github.com/albarmeira/. An schematic workflow of the analysis is presented in Figure 24.