Abstract
Oncogenic Bcr-Abl kinase mimics pre-B cell receptor (pre-BCR) survival signals in BCR-ABL1-positive B-cell acute lymphoblastic leukemia (BCR-ABL1+ B-ALL), driving B-cell progenitor malignant transformation; thus, defining a particularly unfavorable prognosis for patients. During B-cell development, pre-BCR differentiation signaling components terminate proliferative expansion and promote B-cell maturation. To study whether pre-BCR differentiation signaling components regulate the initiation and development of BCR-ABL1+ B-ALL, the tumor suppression mechanism of differentiation-related signaling molecules in BCR-ABL1-transformed pro-B cells were analyzed. The results demonstrated that Bcr-Abl kinase activated the PI3K/Akt pathway, promoting cell growth, and upregulated Aid expression, increasing genomic instability in pro-B cells. These findings suggest that Bcr-Abl kinase mediates pro-B cell malignant transformation. Furthermore, the present data revealed that BCR-ABL1 oncogenic stress triggered enhanced expression of B-cell differentiation components B-cell linker (Blnk) and forkhead box protein O1 (Foxo1) in BCR-ABL1 transformed pro-B cells. Using the CRISPR/Cas9-mediated Blnk or Foxo1 knockout BCR-ABL1-transformed pro-B cells, it was identified that, in BCR-ABL1-transformed pro-B cells, Blnk and Foxo1 reduced Bcr-Abl kinase activity to induce cell cycle arrest and decrease genomic instability. In addition, Blnk suppressed the PI3K/Akt pathway to reduce Foxo1 phosphorylation and heighten the Foxo1 activity, indicating that, in BCR-ABL1-transformed pro-B cells, Foxo1 participated in the regulation of Bcr-Abl kinase by Blnk. The present data highlighted the antitumor mechanisms of Blnk and Foxo1 in the regulation of Bcr-Abl kinase, and thus, may offer an alternative therapeutic strategy to Bcr-Abl kinase regulation in BCR-ABL1+ B-ALL.
Keywords: BCR-ABL1+ B-ALL, Bcr-Abl kinase activity, B-cell linker, forkhead box protein O1, activation-induced cytidine deaminase
Introduction
The BCR-ABL1 fusion gene, derived from the t(9;22) translocation, is the most common cytogenetic abnormality found in adult B-acute lymphoblastic leukemia (B-ALL), accounting for approximately 25% of all B-ALL (1–3). BCR-ABL1 rearrangement results in a fusion protein with constitutively active tyrosine kinase activity that fosters B-cell progenitor malignant transformation (4). Bcr-Abl kinase activates a series of intracellular signaling pathways, resulting in increased cell survival and limited growth factor dependence (5–8). Numerous studies have suggested that additional genomic alterations are the hallmark of BCR-ABL1+ B-ALL (9,10). These genomic alterations could be created by two classes of enzymes required for the diversity of immunoglobulins in B-cell development: the Rag proteins, encoded by recombination-activating genes (RAG1 and RAG2), which introduce DNA rearrangement of the variable (V), diversity (D), and joining (J) segments of immunoglobulin (Ig) genes in pro-B and pre-B cells, and activation-induced cytidine deaminase (Aid) that initiate somatic hypermutation (SHM) and class-switch recombination (CSR) on Ig genes in activated B cells (11). Mistargeting of Rag mediates the deletion of IKZF1, leading to the loss of Ikaros function, which is highly relevant for the leukemogenesis of BCR-ABL1+ B-ALL (12). Studies in mice and humans have suggested that the aberrant expression of Aid induced by Bcr-Abl kinase activity enhances genetic instability by augmenting the frequency of amplifications, deletions, and aberrant somatic hypermutation, which reportedly contribute to genomic instability and malignant transformation of BCR-ABL1+ B-ALL (13–16). Although the development of tyrosine kinase inhibitors (TKIs) has revolutionized therapy, resistance to TKIs leads to short-lasting responses for TKIs in patients with relapsed ALL (17,18). Consequently, new molecular mechanisms for controlling Bcr-Abl kinase need to be developed to cure BCR-ABL1+ B-ALL.
During the B-cell lineage commitment at the pro-B stage, the successful rearrangement of immunoglobulin heavy chain variable (V), diversity (D), and joining (J) gene segments generates the μ heavy chain (μHC) proteins. The μHC pairs with non-polymorphic surrogate light-chain components, λ5 and VpreB, to form the pre-B cell receptor (pre-BCR). The pre-BCR is an autonomously active receptor whose signals drive pre-B cell proliferation and differentiation into immature B cells (19,20). The initial step of pre-BCR signaling is the activation of the spleen tyrosine kinase (Syk), which has a crucial role in the activation of downstream pathways involved in the proliferation and differentiation of pre-B cells. Syk stimulates pre-B cell proliferation by activating PI3K and its downstream mediator Akt, which promotes phosphorylation and nuclear exclusion forkhead box protein O (Foxo) transcription factors, namely Foxo1, Foxo3a, and Foxo4. Thus, the expression levels of cell cycle arrest- and apoptosis induction-related, downstream target genes of Foxos, such as BIM, TRAIL, CDKN1B, and GADD45α, are decreased (21,22). As explained below, the proliferation process is also inhibited by extensive feedback of pre-BCR signaling components. Syk mediates differentiation by activating the adaptor protein Blnk, which could activate phosphatase and tensin homolog (Pten) or SH2-containing inositol phosphatase (Ship) to decrease Akt activity, resulting in decreased phosphorylation and increased stabilization of Foxo proteins. In addition, Blnk recruits Bruton's tyrosine kinase (Btk), phospholipase C-γ2 (Plcγ2), and growth factor receptor-bound protein 2 (Grb2) to initiate calcium-dependent signaling pathways, which is the key event in the signaling pathway of the development and maturation of B cells (23). Finally, the proliferation phase is terminated, and the differentiation process begins (24,25). Therefore, pre-BCR signaling activates seemingly opposing cellular programs, such as proliferation and differentiation. A large body of evidence has demonstrated that Bcr-Abl functions as a constitutively active kinase to initiate the downstream survival signaling in BCR-ABL1+ B-ALL blasts by mimicking pre-BCR pathways (26). Bcr-Abl activates the PI3K/Akt pathway to induce Foxo phosphorylation, leading to the nuclear exclusion of Foxos to stimulate abnormal cell growth (27–30). Although there are trillions of potential target cells, each containing hundreds of susceptible oncogenes, cancer occurs less than once in a lifetime. There are a variety of innate tumor inhibition mechanisms in mammalian cells. Once the proliferation is aberrant, these mechanisms will trigger apoptosis or senescence to prevent uncontrolled cell division (31). The differentiation signaling components Blnk and Btk have a synergistic effect in pre-B cell tumor suppression, because the incidence of pre-B cell leukemia in Blnk/Btk double-defect mice is significantly enhanced than that in Blnk single-deficient mice (32–34). However, the functional roles of differentiation signaling components have not been previously characterized in BCR-ABL1+ B-ALL.
The aim of the present study was to explore the tumor suppressive mechanism of differentiation signaling molecules in BCR-ABL1-transformed pro-B cells. The functional roles and the participation of differentiation signaling components Blnk and Foxo1 in BCR-ABL1-transformed pro-B cells were identified, which may provide a rationale for future BCR-ABL1+ B-ALL therapy.
Materials and methods
Cell lines
The Rag1 mutant mouse pro-B (D345) cell line was established by infection of bone marrow cells from BCL-2 transgenic mice with the v-Abl retrovirus, which was was kindly provided by Dr David Schatz (Yale University, New Haven, USA) and stored in our laboratory (35). The D345 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), non-essential amino acids and 1% penicillin-streptomycin (all from Hyclone; Cytiva) and β-mercaptoethanol (50 µM) at 37°C with 5% CO2. The 293T cells were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM (Hyclone; Cytiva) supplemented with 10% FBS, non-essential amino acids and penicillin-streptomycin (1%) at 37°C with 5% CO2.
Retroviral production and transduction
MSCV vector co-expressing human BCR-ABL1 (p210) and hCD4 (MSCV-BCR-ABL1-IRES-hCD4) and MSCV vector expressing hCD4 have been previously described (36). 293T cells were transfected with MSCV vectors and PKAT2 package vector using X-tremeGENE HP DNA transfection reagent (Roche Diagnostics) at 37°C. After 48 h of transfection, viral supernatants were collected, filtered, and stored at −80°C.
BCR-ABL1-transformed pro-B cells and the control cells were obtained by transduction of D345 cells with pMSCV-BCR-ABL1-hCD4 or pMSCV-EV-hCD4 viral supernatants with 1 µg/ml polybrene; then the six-well dishes were spun at 1,000 × g for 1.5 h at room temperature. The medium was changed after 4 h and incubated at 37°C. After 72 h, the cells were incubated with APC-conjugated anti-human CD4 antibody (cat. no. 561840; BD Biosciences) at a 1:10 dilution for 20 min at 4°C, and sorted by BD FACSAria II (BD Biosciences). Flow cytometric data were analyzed with Kaluza Analysis Software 2.1 (Beckman Coulter, Inc.).
Cell growth and viability
A Cell Counting Kit-8 (CCK-8) assay was used to detect cell growth according to the manufacturer's instructions. The cells were suspended with FBS-deficient RPMI-1640 medium supplemented with non-essential amino acids, penicillin-streptomycin, and β-mercaptoethanol (50 µM). Six replicates of 2×104 cells were seeded in a 96-well plate with 100 µl/well. Cells were cultured for 24, 48 and 72 h, then incubated with 10 µl CCK-8 solution (product no. FXP132-500; Beijing 4A Biotech Co., Ltd.) for 2 h. The absorbance at 450 nm was measured by a microplate reader.
The CCK-8 assay was also used to measure cell viability. Cells (2×105) were seeded in a 96-well plate with 100 µl/well in six replicates. Cells were treated with vehicle (PBS) or imatinib (0, 3, 6 and 9 µM) for 24 h. Subsequently, the cells were incubated with 10 µl CCK-8 solution for 2 h. The absorbance values at 450 nm were measured by a microplate reader.
Generation of Aid-, Blnk- and Foxo1-knockout cell lines
The pL-CRISPR.EFS.PAC was a gift from Dr Junjie Zhang of the University of Southern California (Los Angeles, USA). The AID, BLNK and FOXO1 single guide RNA (sgRNA) were designed at the CRISPR design website from Zhang laboratory (http://crispr.mit.edu/) and sequenced (Sunny Biotech Co., Ltd.), sgRNA that does not target the genome was used as a control, which was acquired from the Mouse GeCKOv2 Library (37,38). The gRNA sequences are listed in Table SI. The 293T cells were then independently transfected with the pCas9-AID, pCas9-BLNK and pCas9-FOXO1, pCas9-non-targeting guides along with ΔR9 and pVSVG helper plasmids using the X-tremeGENE HP DNA transfection reagent (Roche Diagnostics) at 37°C. The viral supernatants were collected 48 h after transfection.
BCR-ABL1-transformed pro-B cells were each infected with control, Aid-, Blnk-, or Foxo1-knockout viral supernatants in the presence of 1 µg/ml polybrene. Then the six-well dishes were spun at 1,000 × g for 1.5 h at room temperature. The medium was changed after 4 h and incubated at 37°C. After 72 h, cells were selected with puromycin (0.6 µg/ml) to enrich the transduced cells.
Apoptosis and BrdU proliferation assays
Cells (1×106) were treated with imatinib (0, 3, 6 and 9 µM), and after 24 h, the cells were collected and washed twice with cold PBS, and incubated for 20 min at room temperature in 1:20 diluted PE-conjugated Annexin V and anti-7-aminoactinomycin D (7-AAD) (cat. no. 559763; BD Biosciences), according to the manufacturer's instructions. Then, the cells were analyzed by flow cytometer (Beckman Coulter, Inc.). The Aid KO and Aid WT BCR-ABL1-transformed pro-B cells were starved in FBS-free medium for 24 h, and measured for apoptosis by flow cytometry. Flow cytometric data were analyzed with Kaluza Analysis Software 2.1 (Beckman Coulter, Inc.).
Proliferation assays were performed using the BrdU kit (cat. no. 552598; BD Biosciences) according to the manufacturer's instructions. Briefly, a population of 1×106 cells were cultured in 6-well plates and labeled with BrdU labeling reagent for 45 min before harvesting. The cells were centrifuged at 300 × g for 5 min at 4°C, then fixed and permeabilized with Cytofix/Cytoperm buffer and Cytoperm permeabilization buffer plus. Next, cells were treated with DNase for 1 h at 37°C and further incubated with APC-conjugated anti-BrdU for 20 min at room temperature. BrdU incorporation was measured using a flow cytometer (Beckman Coulter, Inc.). Flow cytometric data were analyzed with Kaluza Analysis Software 2.1 (Beckman Coulter, Inc.).
RNA extraction and reverse transcription-quantitative (RT-q)PCR
The total RNA of cell pellets was isolated by TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Before cDNA synthesis, 1 µg of total RNA was digested with 1 U/µl RNA-free recombination DNAse I. The cDNA was then transcribed with the PrimeScript™ RT reagent Kit (TaKaRa Bio, Inc.) according to the manufacturer's instructions. Quantitative PCR experiments were performed in triplicate with SYBR-Green dye (TaKaRa Bio, Inc.) on an Mx3000P qPCR system (Agilent Technologies, Inc.). All data were analyzed using the 2-ΔΔCq (relative quantification) method (39). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 10 min; 40 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec; and a final step at 95°C for 1 min, 55°C for 30 sec and 95°C for 30 sec. Gene expression was normalized to GAPDH. The primer sequences used for quantitative PCR are listed in Table SII. The gene expression levels of B-cell differentiation components were normalized to the GAPDH level in the corresponding cell line and represented as a heatmap using GraphPad Prism 8 (GraphPad Software, Inc.).
Western blotting
The cell pellets were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.4, 1% Triton X-100, 0.5% NaDoc, 10% glycerol, and 2.5% sodium deoxycholate) with protease and phosphatase inhibitors. Protein concentration was determined using a BCA kit (cat. no. P0010; Beyotime Institute of Biotechnology) and Multiskan™ FC Microplate Reader (Thermo Fisher Scientific Inc.), and analyzed using Skanit software version 3.1 (Thermo Fisher Scientific Inc.). A total of 60 µg of proteins were loaded onto 12% sodium dodecyl sulfide-polyacrylamide gel electrophoresis (SDS-PAGE) for electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; cat. no. IPVH00010; 0.45 µm; EMD Millipore). Membranes were blocked in 5% milk solution for 2 h at room temperature, and then incubated overnight at 4°C with indicated primary monoclonal antibodies with their respective dilutions (Table SIII). The membranes were then incubated with goat anti-rabbit IgG (H+L)-HRP (1:5,000; cat. no. 31466; Thermo Fisher Scientific Inc.), goat anti-rat IgG (H+L)-HRP (1:5,000; cat. no. 31470; Thermo Fisher Scientific Inc.) and goat anti-mouse IgG (H+L)-HRP (1:5,000; cat. no. 31431; Thermo Fisher Scientific Inc.) for 1 h at room temperature, and blots were visualized with Western Lighting Pro ECL (Fusion FX5; Vilber).
Immunofluorescence
Cells (1×104) were affixed to frosted X slides and fixed with 4% paraformaldehyde solution for 15 min at room temperature, followed by permeabilization with 0.2% Triton X-100 for 10 min at room temperature. The slides were blocked with 5% BSA and 10% horse serum in PBS at room temperature for 1 h and were incubated with anti-γH2AX (1:200; product no. 9718; Cell Signaling Technology, Inc.) at 4°C overnight. After three rinses with PBS, the cells were incubated with anti-rabbit IgG (H+L) Alexa Fluor® 555 Conjugate (1:500; product no. 4413; Cell Signaling Technology, Inc.) for 1 h. Then, the cells were washed twice and stained with 1 µg/ml DAPI for 10 min at room temperature. Images were acquired with a confocal microscope (Leica TCS SP8 STED 3X; Leica Microsystems, Inc.) at a magnification of ×40 and processed with ImageJ v.1.8.0 software (National Institutes of Health).
Mutation analysis of the ABL1 gene
Genomic DNA was extracted from 5×106 cells using the DNA Extraction Kit (cat. no. D824A; TaKaRa Bio, Inc.) according to the manufacturer's instructions. The ABL1 kinase portion of the BCR-ABL1 gene was amplified with two rounds of PCR. In the first round, the BCR-ABL1 fragments were amplified using BCR- (exon 13) and ABL1- (exon 9) specific primers to prevent co-amplification of normal ABL1. Primers for the second round of amplification focused on the ABL1 kinase domain (exons 3-8). The genomic fragments were amplified using high fidelity DNA polymerase KOD-Plus-Neo (code no. KOD-401; Toyobo Life Science). PCR products were purified with the Gel Extraction Kit (SKU no. D2500-01; Omega Bio-Tek, Inc.) and cloned into the pMD® 18-T vector (cat. no. 6011; TaKaRa Bio, Inc.). Clones were sequenced commercially (Sunny Biotech Co., Ltd.). PCR primers used for amplification are listed in Table SIV.
Statistical analysis
Unpaired t-test was performed for comparison of two groups and one-way ANOVA followed by Bonferroni's post hoc tests were performed for multiple comparisons using the SPSS 18.0 statistical software package (SPSS, Inc.). Results were presented as the mean ± SEM. P<0.05 was considered to indicate a statistically significant difference.
Results
Bcr-Abl kinase activity promotes proliferation of pro-B cells via the PI3K/Akt pathway
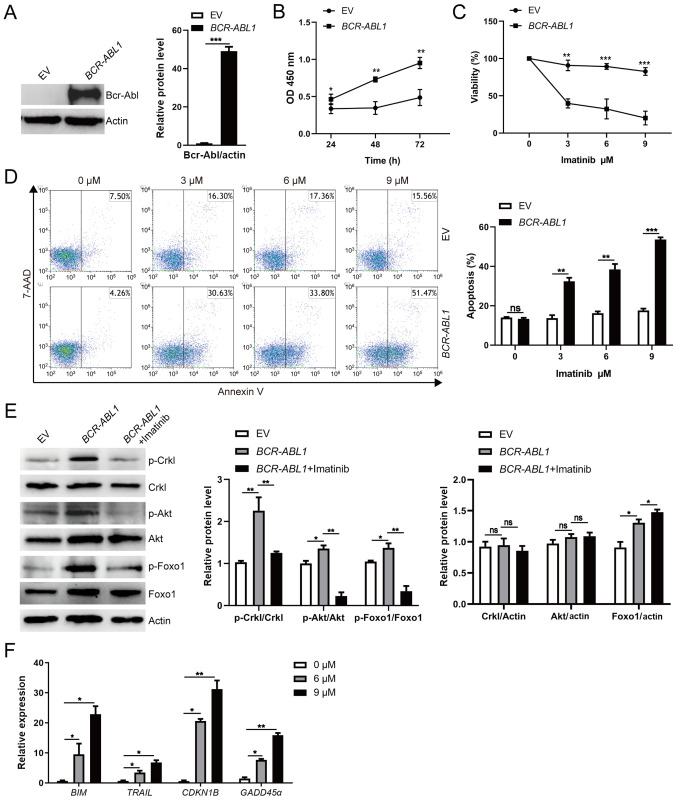
To demonstrate that Bcr-Abl kinase was sufficient to induce pro-B cell transformation, the retroviral vector pMSCV-BCR-ABL1-IRES-hCD4 and empty vector pMSCV–IRES-hCD4 were individually introduced into the D345 cell line. The D345 cell line harbors the Rag1 catalytic mutant (D708A), which retains the native ability to interact with Rag2 and bind to DNA but lacks catalytic activity, thus failing to induce the rearrangement of immunoglobulin genes, ultimately leading to arrested B-cell development at the pro-B stage (35). After flow cytometric sorting, over 90% of infected cells were positive for hCD4 expression (Fig. S1A and B). A large amount of Bcr-Abl expression was detected in pro-B cells infected with retroviral vectors carrying BCR-ABL1 (Fig. 1A). Faster growth of BCR-ABL1-transformed pro-B cells than the control cells in the absence of nutrition for 72 h was observed (Fig. 1B). After treatment with the tyrosine kinase inhibitor, imatinib, at the indicated concentrations for 24 h, the cell viability of BCR-ABL1-transformed pro-B cells significantly decreased compared to that of the control cells (Fig. 1C). The percentage of apoptotic cells was quantified by 7-AAD and Annexin V staining, and BCR-ABL1-transformed pro-B cells exhibited higher apoptotic cell populations than the control cells (Fig. 1D). Moreover, phosphorylated Crkl (p-Crkl) is the main target of Bcr-Abl tyrosine kinase and serves as the connector for downstream effector molecules (40). The p-Crkl levels as indicators of Bcr-Abl kinase activity were therefore determined, and it was revealed that p-Crkl was upregulated following BCR-ABL1 overexpression, while its levels were significantly reduced after imatinib treatment (Fig. 1E). The data revealed that Bcr-Abl kinase activity was required to promote pro-B cell growth in vitro.
Figure 1.
Enhanced pro-B cell survival is mediated by BCR-ABL1 overexpression. The pro-B cells were stably transfected with BCR-ABL1 or EV control. (A) Bcr-Abl expression was detected by western blotting. The protein levels were quantified by the ImageJ program and normalized to actin. (B) Cells were seeded and grown in a 96-well plate with an FBS-deficient culture medium; the absorbance of the CCK-8 solution was determined at 450 nm. (C) Cells were treated with imatinib (0, 3, 6 and 9 µM) for 24 h, and the surviving cells were analyzed by CCK-8 assay. (D) Cells were treated with imatinib (0, 3, 6 and 9 µM) for 24 h and harvested to analyze apoptosis by flow cytometry after Annexin V and 7-AAD staining. The cell population of apoptosis is indicated (left); the histogram reveals the percentages of apoptotic cells (right). (E) BCR-ABL1-transformed pro-B cells were treated or not with imatinib (6 µM) for 24 h. Western blotting analysis for phospho-specific antibodies against Foxo1 (S256), Akt (S473), Crkl (Y207), blots were next stripped and reprobed with antibodies against total-Foxo1, total-Akt, total Crkl. Phosphorylated protein levels were quantified by the ImageJ program and normalized to total form antibodies. The total protein levels were quantified by the ImageJ program and normalized to actin. (F) The mRNA levels of BIM, TRAIL, CDKN1B, and GADD45α were measured by RT-qPCR in BCR-ABL1-transformed pro-B cells treated with imatinib (0, 6 and 9 µM) for 24 h. GAPDH was used for normalization of cDNA amounts. Data are representative of at least three independent experiments. Error bars represent the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001. EV, empty vector; CCK-8, Cell Counting Kit-8; Foxo1, forkhead box protein O1; RT-qPCR, reverse transcription-quantitative PCR; p-phosphorylated; ns, not significant.
Considering the possibility that Bcr-Abl kinase enhances cell survival by activating the PI3K/Akt pathway to induce Foxo1 phosphorylation in BCR-ABL1-transformed pro-B cells (29), western blotting was performed to examine the phosphorylation of Akt and Foxo1 in BCR-ABL1-transformed pro-B cells. The data indicated that the levels of p-Akt and p-Foxo1 were increased in BCR-ABL1-transformed pro-B cells compared with that in control cells but were significantly reduced in the presence of 6 µM imatinib (Fig. 1E). Furthermore, the expression of Foxo1 target genes BIM, TRAIL, CDKN1B, and GADD45α, which are critical apoptosis and cell-cycle mediators (30), were upregulated after imatinib treatment in BCR-ABL1-transformed pro-B cells (Fig. 1F). Collectively, these results are consistent with previous studies revealing that Bcr-Abl kinase stimulates the PI3K/Akt pathway to inactivate Foxo1, leading to aberrant proliferation of pro-B cells (8,41,42).
Bcr-Abl kinase activity upregulates Aid expression causing genomic instability in pro-B cells
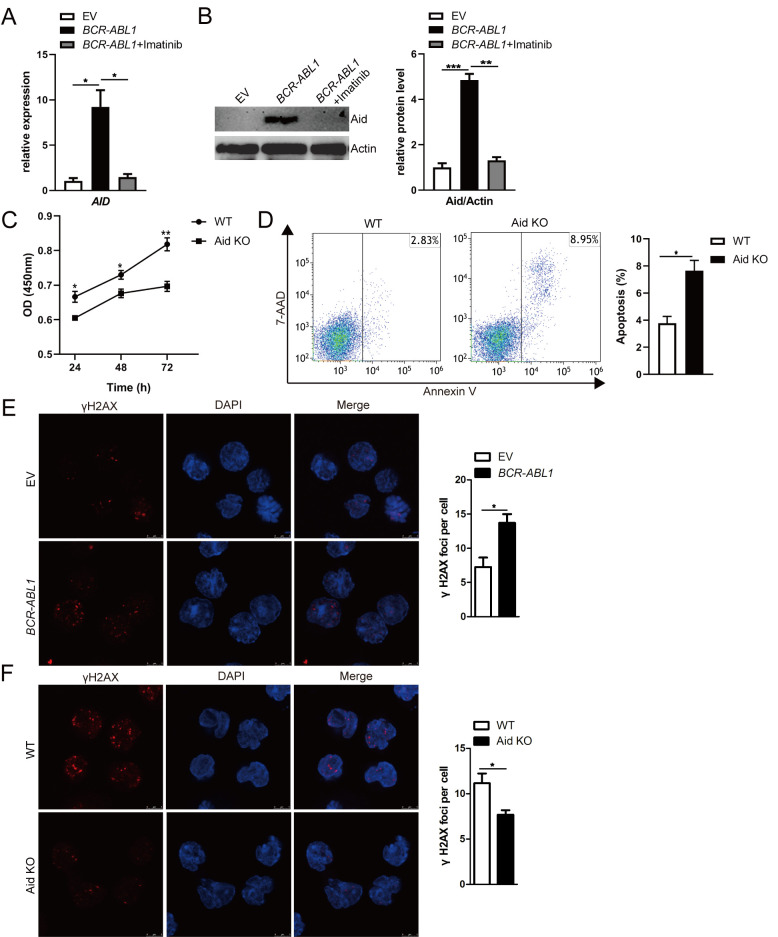
Previous studies have reported that aberrant Aid expression depends on Bcr-Abl kinase activity in BCR-ABL1+ B-ALL (14,43). Consistent with these findings, the mRNA and protein expression levels of AID gene were induced in BCR-ABL1-transformed pro-B cells, while imatinib treatment suppressed Aid expression (Fig. 2A and B). When Aid was deleted using CRISPR/Cas9 gene-editing, the protein level of Aid was significantly reduced in Aid-knockout (KO) BCR-ABL1-transformed pro-B cells compared to BCR-ABL1-transformed pro-B cells infected with non-targeting control viral supernatants (hereinafter named WT cells) (Fig. S2). Cell growth and apoptosis assays were used to determine the role of Aid in the cell survival of BCR-ABL1-transformed pro-B cells. By using the CCK-8 assay, significant inhibition of cell growth was revealed in Aid KO BCR-ABL1-transformed pro-B cells in the absence of nutrition for 72 h (Fig. 2C). The proportion of apoptotic cells was significantly increased in Aid KO cells compared to WT cells (Fig. 2D). These results revealed that Aid can enhance BCR-ABL1-transformed pro-B cell survival.
Figure 2.
Aid induces genomic instability dependent on Bcr-Abl kinase activity in pro-B cells. (A and B) AID mRNA and Aid protein expression were detected by (A) RT-qPCR and (B) western blotting; BCR-ABL1-transformed pro-B cells were treated with or without imatinib (6 µM) for 24 h. GAPDH was used to normalize cDNA amounts, and actin was selected as the loading control for western blotting; the protein levels were quantified by the ImageJ program and normalized to actin. (C) Cells were cultured in a 96-well plate with FBS-deficient culture medium, and the absorbance of the CCK-8 solution was determined at 450 nm. (D) Cells were starved for 24 h, and cell apoptosis was detected by flow cytometry. The cell population of apoptosis is indicated (left); the histogram reveals the percentages of apoptotic cells (right). (E and F) Immunofluorescence of indicated cells stained with γH2AX antibodies (red) and counterstained with DAPI (blue). Bars, 5 µm. The representative images of γH2AX foci of these cells are presented (left), and the histogram revealed the number of γH2AX foci/cell (right). Data are representative of three independent experiments. Error bars represent the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001. Aid, activation-induced cytidine deaminase; CKK-8, Cell Counting Kit-8; RT-qPCR, reverse transcription-quantitative PCR; EV, empty vector; WT, wild-type; KO, knockout.
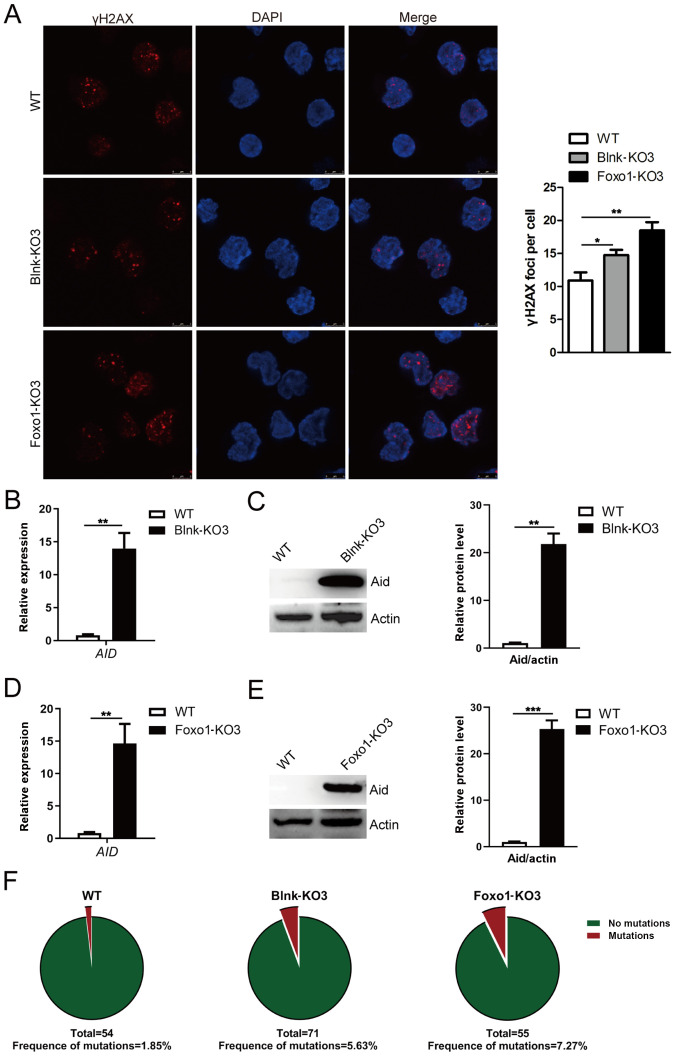
The Rag mutant cell line was utilized to directly examine the contribution of dysregulated Aid on genomic instability. The γH2AX foci, a reliable marker of DNA double-strand breaks (DSB), was assessed through immunofluorescence to characterize the genomic instability (44). As anticipated, increased γH2AX foci per cell were observed in BCR-ABL1-transformed pro-B cells compared with control cells (Fig. 2E). However, the γH2AX foci per cell were less numerous in Aid-KO BCR-ABL1-transformed pro-B cells than WT cells (Fig. 2F), suggesting that Bcr-Abl kinase activity-mediated aberrant Aid expression contributed to genomic instability in BCR-ABL1-transformed pro-B cells. It was concluded that Bcr-Abl kinase activity initiated malignant transformation characterized by pro-B cell proliferation and genomic instability.
B-cell differentiation components Blnk and Foxo1 suppress the survival of BCR-ABL1-transformed pro-B cells
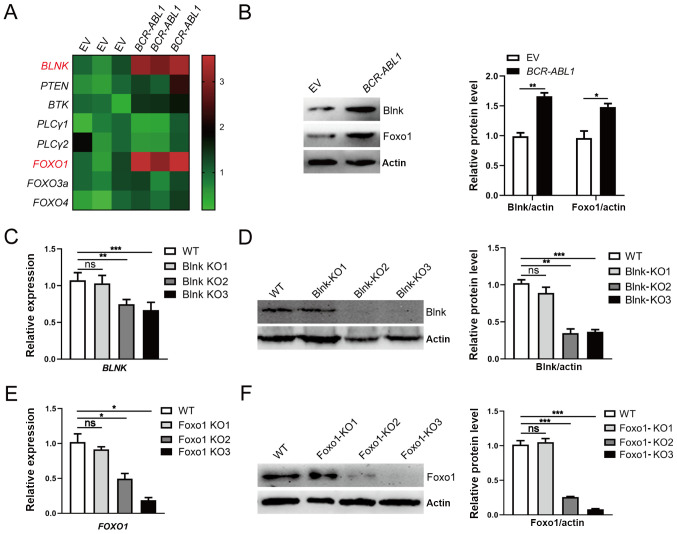
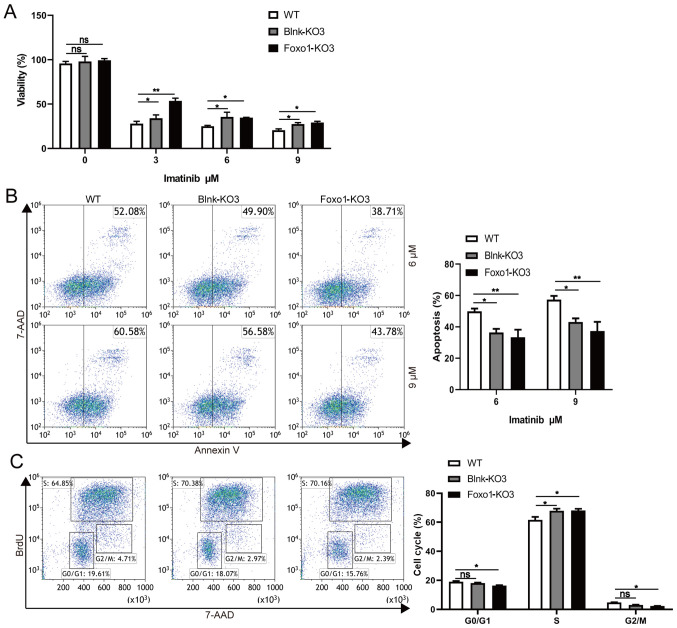
To investigate B-cell differentiation function of components in BCR-ABL1-transformed pro-B cells, the expression of eight key components associated with the pre-BCR differentiation pathway was analyzed. The results revealed that the mRNA and protein expression levels of BLNK and FOXO1 were upregulated in BCR-ABL1-transformed pro-B cells compared to control cells (Figs. 1E and 3A and B). To ascertain the exact role of Blnk and Foxo1 in BCR-ABL1-transformed pro-B cells, the Blnk- or Foxo1-KO BCR-ABL1-transformed pro-B cell lines were generated using CRISPR/Cas9-based gene-editing technology. The Blnk KO3 cells and the Foxo1 KO3 cells exhibited the greatest knockout efficiency (Fig. 3C-3F). After being treated with imatinib, the depletion of Blnk or Foxo1 in BCR-ABL1-transformed pro-B cells revealed the higher levels of viable cells and lower levels of apoptotic cells, indicating that either Blnk or Foxo1 deletion improved the anti-apoptosis ability (Fig. 4A and B). In addition, the cell growth ability of Blnk-KO3 or Foxo1-KO3 BCR-ABL1-transformed pro-B cells was assessed by the BrdU incorporation assay and the results revealed that depletion of Blnk or Foxo1 induced a significant increase in the S phase and a reduction in the G0/G1 phase (Fig. 4C). Collectively, these observations indicated the inhibitory effects of Blnk and Foxo1 on cell growth in BCR-ABL1-transformed pro-B cells.
Figure 3.
Upregulation of Blnk and Foxo1 responding to oncogenic stress in BCR-ABL1-transformed pro-B cells. (A) Heatmap of the transcription levels of B-cell differentiation components. The gene expression levels were measured by RT-qPCR, and the relative expression of each gene was normalized to the GAPDH level and presented as a heatmap. (B) Western blotting analysis for Blnk and Foxo1 expression. The protein levels were quantified by the ImageJ program and normalized to actin. (C and D) Blnk-knockout BCR-ABL1-transformed pro-B cells was generated by CRISPR/Cas9-mediated deletion using three guide RNAs (Blnk KO1-KO3). The depleting efficiency was confirmed by (C) RT-qPCR and (D) western blotting. Blnk KO3 exhibited the best efficiency for Blnk knockout. GAPDH was used to normalize cDNA, and the protein levels were quantified by the ImageJ program and normalized to actin. (E and F) Foxo1-knockout BCR-ABL1-transformed pro-B cells were generated by CRISPR/Cas9-mediated deletion using three guide RNAs (Foxo1 KO1-KO3). The depleting efficiency was confirmed by (E) RT-qPCR and (F) western blotting. Foxo1-KO3 cells exhibited the best depletion efficiency. GAPDH was used to normalize cDNA, and the protein levels were quantified by the ImageJ program and normalized to actin. Data are representative of three independent experiments. Error bars represent the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001. Blnk, B-cell linker; Foxo1, forkhead box protein O1; RT-qPCR, reverse transcription-quantitative PCR; KO, knockout; EV, empty vector; WT, wild-type; ns, not significant.
Figure 4.
Blnk or Foxo1 deletion promotes the cell growth in BCR-ABL1-transformed pro-B cells. (A) WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells were treated with imatinib (0, 3, 6 and 9 µM) for 24 h, respectively. After treatment, cell viability was detected by CCK-8 assay. (B) Flow cytometric analysis of apoptosis cells in WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells treated with imatinib (6 and 9 µM) for 24 h. The cell population of apoptosis is indicated (left); the histogram reveals the percentages of apoptotic cells (right). (C) Flow cytometric analysis of the cell cycle of WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells. The cell population of G0/G1, S, G2/M phase is indicated (left); the histogram reveals the percentages of each phase (right). Data are representative of at least three independent experiments. Error bars represent the mean ± SEM. *P<0.05, **P<0.01. Blnk, B-cell linker; Foxo1, forkhead box protein O1; CCK-8, Cell Counting Kit-8; KO, knockout; WT, wild-type; ns, not significant.
Blnk and Foxo1 reduce the genomic instability of BCR-ABL1-transformed pro-B cells
The effects of Blnk or Foxo1 deletion on genomic instability were then further determined in BCR-ABL1-transformed pro-B cells. Increased levels of γH2AX were detected in Blnk- or Foxo1-deficient BCR-ABL1-transformed pro-B cells (Fig. 5A). Furthermore, higher AID transcript levels were accompanied by a corresponding change of protein in Blnk- or Foxo1-deficient BCR-ABL1-transformed pro-B cells (Fig. 5B-E). To analyze the somatic mutations of non-Ig genes dependent on Aid, genomic DNA was purified from WT, Blnk-KO3, and Foxo1-KO3 BCR-ABL1-transformed pro-B cells. An 852-bp segment located in the ABL1 kinase domain (exons 3-8), a known target of Aid, was amplified, sequenced, and analyzed for point mutations (Fig. S3A-C). The data revealed that the ABL1 region exhibited higher mutation frequencies in Blnk-KO3 cells (6.607×10−5 mutations per bp) and Foxo1-KO3 cells (8.533×10−5 mutations per bp) than that in WT counterparts (2.171×10−5 mutations per bp) (Fig. 5F; Table SV). Thus, the results indicated that Blnk or Foxo1 negatively regulated the genomic instability in BCR-ABL1-transformed pro-B cells.
Figure 5.
Blnk or Foxo1 deletion leads to increased genomic instability in BCR-ABL1-transformed pro-B cells. (A) Immunofluorescence of WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells stained with γH2AX antibodies (red) and counterstained with DAPI (blue). Bars, 5 µm. The representative images of γH2AX foci of these cells are presented (left), and the histogram reveals the number of γH2AX foci/cell (right). The (B) mRNA and (C) protein levels of AID gene in WT and Blnk-KO3 BCR-ABL1-transformed pro-B cells. The (D) mRNA and (E) protein levels of AID in WT and Foxo1-KO3 BCR-ABL1-transformed pro-B cells. GAPDH was used for normalization of cDNA; the protein levels were normalized to actin. (F) WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells were subjected to amplification and sequence analysis for ABL1 kinase domain mutation; the frequencies of mutation are presented at the bottom for each sample. Data are representative of at least three independent experiments. Error bars represent the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001. Blnk, B-cell linker; Foxo1, forkhead box protein O1; Aid, activation-induced cytidine deaminase; WT, wild-type; KO, knockout.
Blnk and Foxo1 participate in regulation of Bcr-Abl kinase in BCR-ABL1-transformed pro-B cells
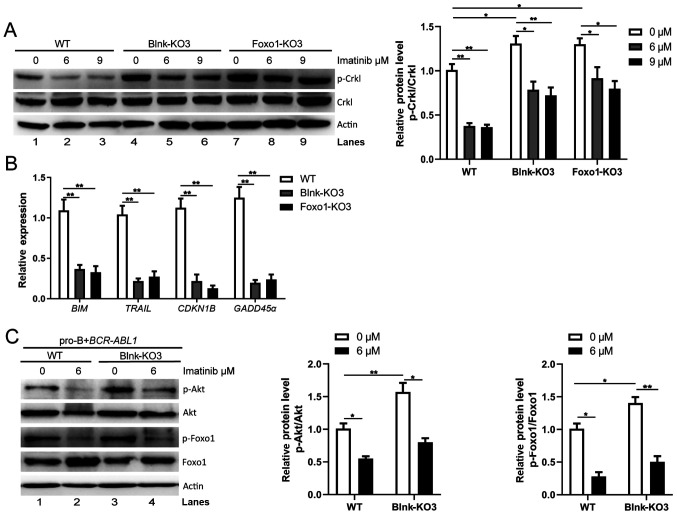
As Blnk and Foxo1 decreased cell growth ability and genomic instability in BCR-ABL1-transformed pro-B cells, it was hypothesized that Blnk and Foxo1 regulated Bcr-Abl kinase during the pro-B cell stage. p-Crkl in Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells were detected. Notably, following Blnk or Foxo1 deletion, the level of p-Crkl was significantly increased (Fig. 6A, lanes 1, 4, and 7). Notably, imatinib treatment attenuated p-Crkl in WT, Blnk-KO3, and Foxo1-KO3 BCR-ABL1-transformed pro-B cells (Fig. 6A, lanes 2, 3, 5, 6, 8 and 9), indicating that either Blnk- or Foxo1-deficient BCR-ABL1-transformed pro-B cells still responded to imatinib treatment. Thus, based on these results, it was argued that Blnk and Foxo1 served as tumor suppressors by inhibiting the Bcr-Abl kinase activity, hindering tumor initiation and progression in BCR-ABL1+ B-ALL.
Figure 6.
Blnk and Foxo1 limit the activity of Bcr-Abl kinase in BCR-ABL1-transformed pro-B cells. (A) Western blotting analysis of p-Crkl was performed in WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells treated with imatinib (0, 6 and 9 µM) for 24 h, respectively. The p-Crkl levels were quantified by the ImageJ program and normalized to total Crkl. Actin was used as a loading control. (B) The transcription of BIM, TRAIL, CDKN1B, and GADD45α in WT, Blnk-KO3 and Foxo1-KO3 BCR-ABL1-transformed pro-B cells. Gene mRNA fold expression values were normalized to the GAPDH. (C) Effects of deletion of Blnk on p-Foxo1 and p-Akt were measured with phospho-specific antibodies in WT and Blnk-KO3 BCR-ABL1-transformed pro-B cells treated with imatinib (0 and 6 µM). Blots were next stripped and reprobed with antibodies against total-Foxo1, total-Akt. Phosphorylated protein levels were quantified by the ImageJ program and normalized to total form antibodies. Actin served as a loading control. Data are representative of three independent experiments. Error bars represent the mean ± SEM. *P<0.05 and **P<0.01. Blnk, B-cell linker; Foxo1, forkhead box protein O1; WT, wild-type; KO, knockout; p-phosphorylated.
To elucidate the interaction of Blnk and Foxo1 in BCR-ABL1-transformed pro-B cells, gene expression changes of Foxo1 target genes were assayed following Blnk or Foxo1 knockout. The results revealed that the expression levels of Foxo1 target genes BIM, TRAIL, CDKN1B and GADD45α were lower in Blnk- or Foxo1-deficient BCR-ABL1-transformed cells than in the WT cells (Fig. 6B). During B-cell development, Blnk inhibits Akt activity and activates Foxo proteins to initiate differentiation (22). Consistent with these findings, the present results revealed higher levels of p-Foxo1 and p-Akt (Fig. 6C, lanes 1 and 3) in Blnk-KO3 BCR-ABL1-transformed pro-B cells compared to the WT counterparts, and imatinib caused a decrease in p-Foxo1 and p-Akt levels (Fig. 6C, lanes 2 and 4), suggesting that Blnk restrains Akt activity-mediated reduction of Foxo1 phosphorylation in BCR-ABL1-transformed pro-B cells. The present findings indicated that, in BCR-ABL1-transformed pro-B cells, Blnk reduced Foxo1 phosphorylation and maintained Foxo1 activity via PI3K/Akt pathway inactivation, which ultimately was involved in regulation of the Bcr-Abl kinase.
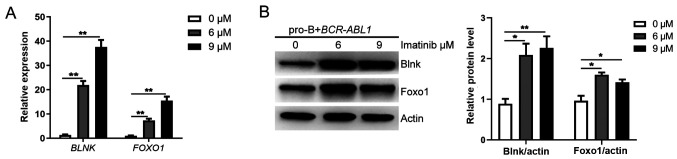
Although overactive oncogenes induce apoptosis or senescence, the oncogenes could also suppress these ‘commits suicide’ processes to allow aberrant proliferation (45). It is worth mentioning that the mRNA and protein expression levels of BLNK and FOXO1 were significantly increased in the BCR-ABL1-transformed pro-B cells treated with imatinib (Figs. 1E and 7A and B). These results indicated the inhibitory effect of Bcr-Abl kinase on the expression of Blnk and Foxo1 in BCR-ABL1-transformed pro-B cells. Therefore, it was inferred that pro-B cells have an intrinsic antitumor ability, mediated by Blnk and Foxo1 upregulation in response to oncogenic stress. In contrast, Bcr-Abl kinase inhibited the expression of Blnk and Foxo1, thus affecting their tumor suppressor function and fueling malignant transformation of pro-B cells, marking the first step toward tumorigenesis.
Figure 7.
Bcr-Abl kinase activity reversely inhibits Blnk and Foxo1 expression in BCR-ABL1-transformed pro-B cells. The (A) mRNA and (B) protein levels of BLNK and FOXO1 gene in BCR-ABL1-transformed, imatinib-treated (0, 6 and 9 µM for 24 h) pro-B cells. Gene mRNA fold expression values have been normalized to the GAPDH; the protein levels were quantified by the ImageJ program and normalized to actin. Data are representative of three independent experiments. Error bars represent the mean ± SEM. *P<0.05 and **P<0.01. Blnk, B-cell linker; Foxo1, forkhead box proteinO1.
Discussion
BCR-ABL1+ B-ALL defines the most common and prognostically unfavorable subtype of ALL. Although recent studies indicated that TKIs combined with conventional chemotherapy could improve clinical outcomes, the overall prognosis of BCR-ABL1+ B-ALL patients remains poor due to high rates of resistance to therapy (46,47). Therefore, novel, more effective therapy is required to improve the outcome of BCR-ABL1+ B-ALL. In the present study, BCR-ABL1-transformed pro-B cells characterized by cell growth advantage and genomic instability caused by Bcr-Abl kinase activity were established. It was demonstrated that the expression of Blnk and Foxo1 were enhanced in response to oncogenic stress of BCR-ABL1. Blnk and Foxo1 reduced Bcr-Abl kinase activity, thus weakening the aberrant proliferation and preventing genomic instability in BCR-ABL1-transformed pro-B cells. The present findings established the antitumor role of Blnk and Foxo1 in Bcr-Abl kinase regulation in BCR-ABL1-transformed pro-B cells.
BCR-ABL1+ B-ALL initiates with great efficiency in B-cell progenitors, including pro-B cells, but not in more mature pre-B cells (48). The D345 pro-B cell line was utilized to simulate the initiation stage of BCR-ABL1+ B-ALL. Consistent with previous studies, the present results demonstrated that Bcr-Abl kinase activity promoted cell proliferation by activating the PI3K/Akt signaling pathway to facilitate the phosphorylation of Foxo1, which was reversed by imatinib treatment (49,50). In the experimental setting, the Rag1 mutant (D708A) of the pro-B cell line lacked catalytic activity, which excludes the role of Rag recombinase and focuses on Aid contribution to genomic instability. Aid induced global genetic instability by hypermutation on the tumor suppressor and DNA repair genes, leading to poor clinical outcomes of patients (51–53). Previous studies have revealed that Bcr-Abl kinase activity upregulates Aid through transcriptional inhibition of ID2 or the absence of miR155 (14,43). Similar to this data, the present results verified that Bcr-Abl kinase induced aberrant Aid expression to mediate genomic instability in BCR-ABL1-transformed pro-B cells, while the mechanism of Bcr-Abl signaling pathway-regulation of Aid expression in BCR-ABL1-transformed pro-B cells warrants further study.
The present data established a tight association between Bcr-Abl and the B-cell differentiation components Blnk and Foxo1 in BCR-ABL1-transformed pro-B cells. Aberrant activation of oncogenic signaling are necessary to produce pre-malignant tumors. Concurrently, oncogenic signaling triggers cell apoptosis or senescence through the Arf/p53/p21, the p16/pRb and the DDR pathways (54–57). Previously published results have highlighted the mechanism that BCR-ABL1+ B-ALL initiation in the B-cell lineage is developmentally regulated; the oncogene-induced stress leads to higher expression of the tumor suppressor gene ARF in pre-B cells rather than pro-B cells, and Arf induces apoptosis and suppresses the occurrence of ALL at pre-B cell stage (48). However, the studies left important gaps regarding whether there are tumor suppressors that regulate B-ALL initiation at the pro-B cell stage. Studying the specific process in response to oncogenic stress at the pro-B cell stage, in the present study it was determined that the expression levels of the B-cell differentiation components Blnk and Foxo1 were increased after BCR-ABL1 oncogene expression. In fact, Blnk and Foxo1 are considered initiators of the differentiation program of the pre-BCR signaling pathway and critical tumor suppressors (20). One interpretation for the enhanced expression of Blnk and Foxo1 in BCR-ABL1-transformed pro-B cells is that Blnk and Foxo1 antagonize the function of Bcr-Abl kinase activity. Consistent with this argument, Blnk- or Foxo1-deficient BCR-ABL1-transformed pro-B cells revealed enhanced Bcr-Abl kinase activity, accompanied by marked cell survival and increased genomic instability mediated by enhanced Aid expression. It has been reported that the aberrant expression of Aid initiating SHM on Ig and non-Ig genes is driven by carcinogenic Bcr-Abl kinase and is related to the poor prognosis of BCR-ABL1+ leukemia (43) Continuous upregulation of BCR-ABL1 not only induces the survival advantage of leukemic cells, but also provides a target for Aid (58,59). Numerous studies have suggested that Aid promotes the mutations of BCR-ABL1 to cause imatinib-resistance and leads to the rapid progression of CML-blast crisis progression (13,60,61). We question whether Aid may lead to imatinib-resistance in BCR-ABL1+ B-ALL. Sequencing of the BCR-ABL1 gene in Blnk-KO3 cells and Foxo1-KO3 cells revealed increased kinase domain mutation frequencies while it failed to cause drug resistance, which may be explained by leukemic cell dependency shift from Bcr-Abl signaling toward the SRC kinase signaling (62,63). Furthermore, a previous study by Plas and Thompson revealed that, Foxo1 is a major downstream effect factor of the PI3K/Akt pathway. The ultilization of Akt inhibitor LY294002 confirmed that the protein levels of Foxo1 were regulated by proteasome-dependent Akt activation (64). Akt-mediated Foxo1 phosphorylation promoting the transport of phosphorylated Foxo1 from the nucleus to the cytoplasm, and then phosphorylated Foxo1 are rapidly degraded (4,5,65). This is a key step for cell proliferation. However, Blnk can change the fate of cells from proliferation to differentiation (32,66). In previous studies, the relationship of Blnk/PI3K-Akt/Foxo1 was clarified by Blnk ectopic expression; Blnk counteracted the function of the PI3K/Akt pathway and enabled Foxo1 to activate target genes in the nucleus (23,25). According to the present data, the expression levels of Foxo1 target genes were lower in Blnk-deficient BCR-ABL1-transformed cells than in WT cells, and the levels of p-Foxo1 and p-Akt were increased in Blnk-KO3 BCR-ABL1-transformed pro-B cells, indicating that Blnk maintained Foxo1 activity by inhibiting the PI3K/Akt pathway in BCR-ABL1-transformed pro-B cells. We will further explore the specific interaction of Blnk/PI3K-Akt/Foxo1 in subsequent studies. Based on these observations, it was concluded that Blnk and Foxo1 exerted an antitumor role by inhibiting Bcr-Abl kinase in BCR-ABL1-transformed pro-B cells. However, oncogenes may bypass and/or inhibit these mechanisms, further fueling tumor cell survival (45,67). The present results indicated that Bcr-Abl kinase activity inhibition results in stronger upregulation of the tumor suppressor genes Blnk and Foxo1, demonstrating that Bcr-Abl kinase reversely inhibited Blnk and Foxo1 expression in pro-B cells. The present data indicated a network of interaction between tumor suppressors and Bcr-Abl, which requires further study on the more detailed regulatory mechanism.
In conclusion, the present study provided evidence of the pivotal function of Blnk and Foxo1 for Bcr-Abl kinase regulation in BCR-ABL1-transformed pro-B cells. The results provided novel insights into the roles of Blnk and Foxo1 tumor suppressors in BCR-ABL1-transformed pro-B cells, which will aid in developing new therapeutic strategies for BCR-ABL1+ B-ALL.
Supplementary Material
Acknowledgments
The authors would like to thank Dr David G. Schatz (Yale University, New Haven, USA) for providing D345 pro-B and BD pre-B v-abl cell lines, and we thank Dr Junjie Zhang (University of Southern California, Los Angeles, USA) for providing pL-CRISPR.EFS.PAC plasmids. The authors also thank the Core Facilities Sharing Platform of Xi'an Jiaotong University for providing the confocal microscope (Leica TCS SP8 STED 3X; Leica Microsystems, Inc.).
Glossary
Abbreviations
- B-ALL
B-acute lymphoblastic leukemia
- BCR-ABL1+ B-ALL
BCR-ABL1-positive B-cell acute lymphoblastic leukemia
- TKIs
tyrosine kinase inhibitors
- pre-BCR
pre-B cell receptor
- RT-qPCR
reverse transcription-quantitative PCR
- EV
empty vector
- CCK-8
Cell Counting Kit-8
- p-
phosphorylated
- Aid
activation-induced cytidine deaminase
- Blnk
B-cell linker
- Foxo1
forkhead box protein O1
- WT
wild-type
- KO
knockout
- ns
not significant
Funding
This work was supported by grants (grant nos. 81670157 and 81801581) from the National Natural Scientific Foundation of China (to YJ and YD) and by a grant (grant no. 2016JZ030) from the Natural Scientific Foundation of Shaanxi (to YJ).
Availability of data and materials
The datasets analyzed during the present study are available from the corresponding author upon reasonable request.
Author's contributions
PZ and YJ conceived and designed the experiments. PZ and YW performed the majority of experiments and wrote the manuscript. MQ and DL performed the retroviral production and transfection. WOO was involved in manuscript review and DNA extraction. MY and ZL performed the CCK-8 assay. CL and YM revised the work critically for important intellectual content. YD and YJ were major contributors in funding acquisition and assisted in the data analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.El Fakih R, Jabbour E, Ravandi F, Hassanein M, Anjum F, Ahmed S, Kantarjian H. Current paradigms in the management of Philadelphia chromosome positive acute lymphoblastic leukemia in adults. Am J Hematol. 2018;93:286–295. doi: 10.1002/ajh.24926. [DOI] [PubMed] [Google Scholar]
- 2.Lee HJ, Thompson JE, Wang ES, Wetzler M. Philadelphia chromosome-positive acute lymphoblastic leukemia: current treatment and future perspectives. Cancer. 2011;117:1583–1594. doi: 10.1002/cncr.25690. [DOI] [PubMed] [Google Scholar]
- 3.Vinhas R, Lourenço A, Santos S, Lemos M, Ribeiro P, de Sousa AB, Baptista PV, Fernandes AR. A novel BCR-ABL1 mutation in a patient with Philadelphia chromosome-positive B-cell acute lymphoblastic leukemia. OncoTargets Ther. 2018;11:8589–8598. doi: 10.2147/OTT.S177019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köhrer S, Havranek O, Seyfried F, Hurtz C, Coffey GP, Kim E, Ten HE, Jäger U, Vanura K, O'Brien S. Pre-BCR signaling in precursor B-cell acute lymphoblastic leukemia regulates PI3K/AKT, FOXO1, and MYC, and can be targeted by SYK inhibition. Leukemia. 2016;30:1246–1254. doi: 10.1038/leu.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neshat MS, Raitano AB, Wang HG, Reed JC, Sawyers CL. The survival function of the Bcr-Abl oncogene is mediated by Bad-dependent and -independent pathways: Roles for phosphatidylinositol 3-kinase and Raf. Mol Cell Biol. 2000;20:1179–1186. doi: 10.1128/MCB.20.4.1179-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roumiantsev S, Aos IED, Varticovski L, Ilaria RL, Etten RAV. The Src homology 2 domain of Bcr/Abl is required for efficient induction of chronic myeloid leukemia–like disease in mice but not for lymphoid leukemogenesis or activation of phosphatidylinositol 3-kinase. Blood. 2001;97:4–13. doi: 10.1182/blood.V97.1.4. [DOI] [PubMed] [Google Scholar]
- 7.Sheng Z, Ma L, Sun JE, Zhu LJ, Green MR. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011;118:2840–2848. doi: 10.1182/blood-2010-12-322537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, Mccubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 9.Iacobucci I, Lonetti A, Messa F, Ferrari A, Cilloni D, Soverini S, Paoloni F, Arruga F, Ottaviani E, Chiaretti S, et al. Different isoforms of the B-cell mutator activation-induced cytidine deaminase are aberrantly expressed in BCR-ABL1-positive acute lymphoblastic leukemia patients. Leukemia. 2010;24:66–73. doi: 10.1038/leu.2009.197. [DOI] [PubMed] [Google Scholar]
- 10.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon SM, Trageser D, Hasselfeld B, Henke N, Mooster J, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong Y, Liu F, Wu C, Li S, Zhao X, Zhang P, Jiao J, Yu X, Ji Y, Zhang M. Illegitimate RAG-mediated recombination events are involved in IKZF1 Δ3-6 deletion in BCR-ABL1 lymphoblastic leukaemia. Clin Exp Immunol. 2016;185:320–331. doi: 10.1111/cei.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruber TA, Mi SC, Sposto R, Müschen M. Activation-induced cytidine deaminase accelerates clonal evolution in BCR-ABL1-driven B cell lineage acute lymphoblastic leukemia. Cancer Res. 2010;70:7411–7420. doi: 10.1158/0008-5472.CAN-10-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm L, Duy C, Iacobucci I, Kuchen S, Levetzow GV, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messina M, Chiaretti S, Iacobucci I, Tavolaro S, Lonetti A, Santangelo S, Elia L, Papayannidis C, Paoloni F, Vitale A, et al. AICDA expression in BCR/ABL1-positive acute lymphoblastic leukaemia is associated with a peculiar gene expression profile. Br J Haematol. 2015;152:727–732. doi: 10.1111/j.1365-2141.2010.08449.x. [DOI] [PubMed] [Google Scholar]
- 16.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, Mcbride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fielding AK. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia in adults: a broader range of options, improved outcomes, and more therapeutic dilemmas. Am Soc Clin Oncol Educ Book. 2015;35:e352–9. doi: 10.14694/EdBook_AM.2015.35.e352. [DOI] [PubMed] [Google Scholar]
- 18.Xing H, Yang X, Liu T, Lin J, Chen X, Gong Y. The study of resistant mechanisms and reversal in an imatinib resistant Ph+ acute lymphoblastic leukemia cell line. Leuk Res. 2012;36:509–513. doi: 10.1016/j.leukres.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Donahue AC, Fruman DA. Proliferation and survival of activated B cells requires sustained antigen receptor engagement and phosphoinositide 3-kinase activation. J Immunol. 2003;170:5851–5860. doi: 10.4049/jimmunol.170.12.5851. [DOI] [PubMed] [Google Scholar]
- 20.Übelhart R, Werner M, Jumaa H. Assembly and function of the precursor B-cell receptor. Curr Top Microbiol Immunol. 2016;393:3–25. doi: 10.1007/82_2015_475. [DOI] [PubMed] [Google Scholar]
- 21.Burgering B. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 22.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 23.Koretzky GA, Abtahian F, Silverman MA. SLP76 and SLP65: Complex regulation of signalling in lymphocytes and beyond. Nat Rev Immunol. 2006;6:67–78. doi: 10.1038/nri1750. [DOI] [PubMed] [Google Scholar]
- 24.Garg M, Wahid M, Khan F. Regulation of peripheral and central immunity: Understanding the role of Src homology 2 domain-containing tyrosine phosphatases, SHP-1 & SHP-2. Immunobiology. 2020;225:151847. doi: 10.1016/j.imbio.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldhahn N, Klein F, Mooster JL, Hadweh P, Sprangers M, Wartenberg M, Bekhite MM, Hofmann WK, Herzog S, Jumaa H, et al. Mimicry of a constitutively active pre–B cell receptor in acute lymphoblastic leukemia cells. J Exp Med. 2005;201:1837–1852. doi: 10.1084/jem.20042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey FB, England K, Cotter TG. Bcr-Abl regulates osteopontin transcription via Ras, PI-3K, aPKC, Raf-1, and MEK. J Leukoc Biol. 2005;78:289–300. doi: 10.1189/jlb.1104655. [DOI] [PubMed] [Google Scholar]
- 28.Kim E, Koehrer S, Wang Z, O'Brien S, Wierda WG, Thomas DA, Estrov Z, Kantarjian HM, Lannutti B, Davis RE. The PI3K delta inhibitor idelalisib interferes with pre-B cell receptor signaling in acute lymphoblastic leukemia (ALL): a new therapeutic concept. Blood. 2013;122:2632. doi: 10.1182/blood.V122.21.2632.2632. [DOI] [Google Scholar]
- 29.Pellicano F, Scott MT, Helgason GV, Hopcroft LE, Allan EK, Aspinall-O'Dea M, Copland M, Pierce A, Huntly BJ, Whetton AD, et al. The antiproliferative activity of kinase inhibitors in chronic myeloid leukemia cells is mediated by FOXO transcription factors. Stem Cells. 2014;32:2324–2337. doi: 10.1002/stem.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szydlowski M, Kiliszek P, Sewastianik T, Jablonska E, Bialopiotrowicz E, Gorniak P, Polak A, Markowicz S, Nowak E, Grygorowicz MA, et al. FOXO1 activation is an effector of SYK and AKT inhibition in tonic BCR signal-dependent diffuse large B-cell lymphomas. Blood. 2015;127:739–748. doi: 10.1182/blood-2015-06-654111. [DOI] [PubMed] [Google Scholar]
- 31.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 32.Flemming A, Brummer T, Reth M, Jumaa H. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol. 2003;4:38–43. doi: 10.1038/ni862. [DOI] [PubMed] [Google Scholar]
- 33.Hendriks RW, Kersseboom R. Involvement of SLP-65 and Btk in tumor suppression and malignant transformation of pre-B cells. Semin Immunol. 2006;18:67–76. doi: 10.1016/j.smim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Kersseboom R, Middendorp S, Dingjan GM, Dahlenborg K, Reth M, Jumaa H, Hendriks RW. Bruton's tyrosine kinase cooperates with the B cell linker protein SLP-65 as a tumor suppressor in Pre-B cells. J Exp Med. 2003;198:91–98. doi: 10.1084/jem.20030615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Peng C, Hu Y, Li H, Sheng Z, Chen Y, Sullivan C, Cerny J, Hutchinson L, Higgins A, et al. The Blk pathway functions as a tumor suppressor in chronic myeloid leukemia stem cells. Nat Genet. 2012;44:861–871. doi: 10.1038/ng.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.de Jong R, ten Hoeve J, Heisterkamp N, Groffen J. ten HJ, Heisterkamp N and Groffen J: Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene. 1997;14:507–513. doi: 10.1038/sj.onc.1200885. [DOI] [PubMed] [Google Scholar]
- 41.Huang R, Liu H, Chen Y, He Y, Kang Q, Tu S, He Y, Zhou X, Wang L, Yang J, et al. EPS8 regulates proliferation, apoptosis and chemosensitivity in BCR-ABL positive cells via the BCR-ABL/PI3K/AKT/mTOR pathway. Oncol Rep. 2018;39:119–128. doi: 10.3892/or.2017.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Chen Y, Chen X, Zhao W, Zhong Y, Wang R, Jin M, Qiu Y, Kong D. In Vitro Antileukemia Activity of ZSTK474 on K562 and Multidrug Resistant K562/A02 Cells. Int J Biol Sci. 2016;12:631–638. doi: 10.7150/ijbs.14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giebel B. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J Exp Med. 2007;204:1157–1166. doi: 10.1084/jem.20062662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valdiglesias V, Giunta S, Fenech M, Neri M, Bonassi S. γH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat Res. 2013;753:24–40. doi: 10.1016/j.mrrev.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Larsson LG. Oncogene- and tumor suppressor gene-mediated suppression of cellular senescence. Semin Cancer Biol. 2011;21:367–376. doi: 10.1016/j.semcancer.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Bassan R, Rohatiner AZ, Lerede T, Di BE, Rambaldi A, Pogliani E, Rossi G, Fabris P, Morandi S, Casula P, et al. Role of early anthracycline dose-intensity according to expression of Philadelphia chromosome/BCR-ABL rearrangements in B-precursor adult acute lymphoblastic leukemia. Hematol J Off J Eur Haematol Assoc. 2000;1:226–234. doi: 10.1038/sj.thj.6200032. [DOI] [PubMed] [Google Scholar]
- 47.Daenen S. Imatinib in Philadelphia chromosome positive acute lymphoblastic leukemia (ALL) Blood. 2003;102:97–100. [Google Scholar]
- 48.Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Immature B-cell progenitors survive oncogenic stress and efficiently initiate Ph+ B-acute lymphoblastic leukemia. Blood. 2010;116:2522–2530. doi: 10.1182/blood-2010-01-264093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gishizky ML. Molecular mechanisms of Bcr-Abl-induced oncogenesis. Cytokines Mol Ther. 1996;2:251–261. [PubMed] [Google Scholar]
- 50.Skorski T, Kanakaraj P, Nieborowska-Skorska M, Ratajczak MZ, Wen SC, Zon G, Gewirtz AM, Perussia B, Calabretta B. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 1995;86:726–736. doi: 10.1182/blood.V86.2.726.bloodjournal862726. [DOI] [PubMed] [Google Scholar]
- 51.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 52.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 54.Collado M, Serrano M. Senescence in tumours: Evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 56.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 57.Haigis KM, Sweet-Cordero A. New insights into oncogenic stress. Nat Genet. 2011;43:177–178. doi: 10.1038/ng0311-177. [DOI] [PubMed] [Google Scholar]
- 58.Sharma N, Magistroni V, Piazza R, Citterio S, Mezzatesta C, Khandelwal P, Pirola A, Gambacorti-Passerini C. BCR/ABL1 and BCR are under the transcriptional control of the MYC oncogene. Mol Cancer. 2015;14:132. doi: 10.1186/s12943-015-0407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clapper E, Wang S, Raninga PV, Di TG, Tonissen KF. Cross-talk between Bcr-abl and the thioredoxin system in chronic myeloid leukaemia: implications for CML treatment. Antioxidants. 2020;9:9. doi: 10.3390/antiox9030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 61.McCarron SL, Maher K, Kelly J, Ryan MF, Langabeer SE. Rapid evolution to blast crisis associated with a Q252H ABL1 kinase domain mutation in e19a2 BCR-ABL1 chronic myeloid leukaemia. Case Rep Hematol. 2013;2013:490740. doi: 10.1155/2013/490740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blake RA, Broome MA, Liu X, Wu J, Gishizky M, Sun L, Courtneidge SA. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol. 2000;20:9018–9027. doi: 10.1128/MCB.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 65.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 66.Meixlsperger S, Köhler F, Wossning T, Reppel M, Müschen M, Jumaa H. Conventional light chains inhibit the autonomous signaling capacity of the B cell receptor. Immunity. 2007;26:323–333. doi: 10.1016/j.immuni.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Schmitt CA. Cellular senescence and cancer treatment. Biochim Biophys Acta. 2007;1775:5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the present study are available from the corresponding author upon reasonable request.