Summary
Quiescent muscle stem cells, also called satellite cells (SCs), are essential for muscle regeneration. However, quiescent SCs are quickly activated during fluorescence-activated cell sorting (FACS) isolation. Here, we present an optimized protocol to isolate quiescent muscle stem cells from fixative-perfused mice and generate high-quality cDNA libraries for RNA-sequencing analysis. Fixation preserves the signatures of quiescent muscle stem cells in vivo. Isolated SCs can be used for downstream analysis such as immunofluorescence, RNA sequencing, and mass spectrometry.
For complete information on the use and execution of this protocol, please refer to Yue et al. (2020).
Graphical Abstract

Highlights
-
•
An optimized protocol for quiescent muscle stem cell isolation
-
•
Use of fixative perfusion to preserve the gene expression signature in vivo
-
•
Adaptable for various downstream analyses (e.g., transcriptome, proteome)
-
•
Fixative perfusion extendable to the isolation of other cell types from tissues in vivo
Quiescent muscle stem cells, also called satellite cells (SCs), are essential for muscle regeneration. However, quiescent SCs are quickly activated during fluorescence-activated cell sorting (FACS) isolation. Here, we present an optimized protocol to isolate quiescent muscle stem cells from fixative-perfused mice and generate high-quality cDNA libraries for RNA-sequencing analysis. Fixation preserves the signatures of quiescent muscle stem cells in vivo. Isolated cells can be used for downstream analysis such as immunofluorescence, RNA sequencing, and mass spectrometry.
Before You Begin
Preparation of Fix Sort Reagents
Timing: 3–4 h, can be made prior to theday of theprocedure
-
1.
1× PBS, 500 mL. (Stored at 4°C, enough for 8 mice. No need to freshly prepare each time.)
-
2.
2 M glycine, 500 mL. Refer to Materials and Equipment for the buffer recipe. (Stored at 4°C, enough for 16 mice. No need to freshly prepare each time.)
-
3.
Wash Medium, 500 mL. Refer to Materials and Equipment for the buffer recipe. (Stored at 4°C, enough for 2–3 mice. If only performing cell sorting for 1 mouse, ∼250 mL Wash Medium is sufficient.)
-
4.
Prepare Collagenase II for the first digestion: For FACS preparation of unfixed muscles (i.e., from a PBS-perfused mouse), weigh 10,000 U Collagenase II per mouse. For FACS preparation of fixed muscles (i.e., from a PFA-perfused mouse), weigh 20,000 U Collagenase II per mouse. Put the Collagenase II powder into a 50 mL conical tube. Wrap the tube with foil. Stored at 4 °C.
-
5.
Prepare Collagenase II aliquots for the second digestion: 3,000 U/mL in Wash Medium, 30 mL (enough for 30 mice). 1 mL for each aliquot (Stored at −20°C).
-
6.
Prepare Dispase II aliquots for the second digestion: 30 U/mL in Wash Medium, 30 mL (enough for 30 mice). 1 mL for each aliquot (Stored at −20°C).
-
7.
Prepare PFA aliquots: 32% PFA, 20 mL (enough for 40 mice). 500 μL for each aliquot (Stored at −80°C).
-
8.Fix sort tools (per mouse):
-
a.250 mL glass beaker ×3;
-
b.No. 11 blade ×1
-
c.No. 20 blade ×1
-
d.a pair of blunt-end forceps (i.e., Narrow Pattern Forceps, 11002-13, FST)
-
e.a pair of pointy-end forceps (i.e., Dumont #5 Forceps, 11251-30, FST)
-
f.a pair of dissection scissors (i.e., Fine Scissors-ToughCut, 14058-11, FST)
-
g.10 cm glass petri dish ×1
-
h.10 mL syringe ×3
-
i.25-gauge needle ×3
-
j.30 mL syringe ×1
-
k.50 mL conical tube ×4
-
l.40 μm cell strainer ×2
-
m.5 mL round-bottom polypropylene tube ×2
-
n.Insulin syringe 30G ×1 (Optional. For intraperitoneal injection of Avertin.)
-
a.
Note: This protocol describes the use of a transgenic mouse line, Tg:Pax7nGFP, expressing green fluorescent protein (GFP) under the control of the Pax7 promoter and marking quiescent muscle stem cells in resting skeletal muscles (Sambasivan et al., 2009). Other reporter lines can also be used.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse Pax7 (Optional) | Developmental Studies Hybridoma Bank |
RRID: AB_528428 |
| Anti-mouse Myod1, Clone 5.8A (Optional) | Dako (Now part of Agilent) | Catalog #: M3512; RRID: AB_2148874 |
| Anti-Dek (Optional) | ProteinTech Group | Catalog #: 16448-1-AP; RRID: AB_2092097 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphate buffered saline | Sigma-Aldrich | Catalog #: P3813 |
| 32% Paraformaldehyde Aqueous Solution, EM Grade | Electron Microscopy Sciences | Catalog #: 15714 |
| Glycine, for electrophoresis, ≥99% | Sigma-Aldrich | Catalog #: G8898 |
| Nutrient mixture F-10 Ham | Sigma-Aldrich | Catalog #: N6635 |
| Horse serum | Invitrogen | Catalog #: 16050114 |
| Penicillin Streptomycin (10,000 U/mL) | Thermo Scientific | Catalog #: 15140122 |
| Collagenase, Type 2 | Worthington Biochemical | Catalog #: LS004177 |
| Dispase II, powder | Thermo Fisher Scientific | Catalog #: 17105041 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | Catalog #: D1306; RRID: AB_2629482 |
| 2,2,2-Tribromoethanol, 99% (Avertin, optional) | Acros Organics | Catalog #: 421430100 |
| ECM Gel from Engelbreth-Holm-Swarm murine sarcoma, liquid, cell culture tested | Sigma-Aldrich | Catalog #: E1270-10ML |
| Critical Commercial Assays | ||
| miRNeasy FFPE Kit | Qiagen | Catalog #: 217504 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tg:Pax7nGFP | A kind gift from Shahragim Tajbakhsh (Institut Pasteur) | N/A |
| Software and Algorithms | ||
| ZEN software (blue edition) | Carl Zeiss | N/A |
| Other | ||
| Syringe filter, minisart high flow, 0.22 μm, PES membrane, sterile, 28 mm | Sartorius | Catalog #: 1209Z85 |
| 40 μm Cell strainer | SPL Life Sciences | Catalog #: 93040 |
| Filter Upper Cup, PES 0.22 μm, 500 mL, 75 mm | JET biofil | Catalog #: FPE214150 |
| Eppendorf Centrifuge 5804R | Eppendorf | Catalog #: 5804R |
| Eppendorf Centrifuge 5427R | Eppendorf | Catalog #: 5427R |
| Shaking water bath | Memmert | Catalog #: WNB 22 |
| 10 mL Syringe | Terumo | Catalog #: 130113L |
| Disposable needle 25G × 5/8 inch Mekim #13534 | Terumo | Catalog #: NN-2516R |
| Round-Bottom Polypropylene Tube, 5 mL | Falcon | Catalog #: 352063 |
| Size calibration beads (Optional) | Thermo Scientific | Catalog #: F13838 |
| RNase AWAY™ Surface Decontaminant (Optional) | Thermo Scientific | Catalog #: 7002PK |
| Qubit RNA HS Assay Kit (Optional) | Thermo Scientific | Catalog #: Q32852 |
| Hemocytometer | Gizmo Supply Co | Catalog #: B-CNT-SLDE-V2 |
Materials and Equipment
0.5% Paraformaldehyde (PFA)
| Solution | Volume |
|---|---|
| 32% PFA | 468.75 μL |
| Cold 1× PBS | 29.5 mL |
| Total | 30 mL |
CRITICAL:
PFA is water-soluble and should always be used with adequate ventilation, preferably in a fume hood. Eyes and skin exposure should be avoided. Follow the safety data sheet when handling PFA.
2 M Glycine
| Reagent | Amount |
|---|---|
| Glycine | 75 g |
| 1× PBS | 500 mL |
| Total | 500 mL |
Note: Glycine takes a few hours to dissolve. Use a stir bar to help dissolve the glycine. Filter with a 0.22 μm filter (Catalog #: FPE214150).
Wash Medium
| Solution | Volume (mL) |
|---|---|
| Ham’s F10 | 445 |
| Horse Serum (HS) | 50 |
| Penicillin and Streptomycin (P/S) | 5 |
| Total | 500 |
Step-By-Step Method Details
Mouse Perfusion
Timing: ∼20 min per mouse
Mouse perfusion is critical and the most difficult part of this protocol. Successful perfusion allows PFA solution to go through the circulation system from the left ventricle to the right atrium of the heart. For beginners, it takes some practices to be experienced in the mouse perfusion. In general, it is helpful to perform the perfusion in a place where you could position your hands comfortably and with sufficient lighting.
-
1.
Place a 250 mL beaker with cold 1× PBS on ice, 30 mL 1× PBS per mouse. Label it as “PBS.”
-
2.
Prepare 0.5% PFA solution freshly, 30 mL per mouse (refer to Materials and Equipment for the buffer recipe). Place a 250 mL beaker with cold 0.5% PFA on ice. Label it as “PFA.”
-
3.
Place a 250 mL beaker with cold 2 M glycine on ice, 30 mL per mouse. Label it as “Glycine.”
-
4.
Prepare the perfusion syringe by adapting a 25-gauge needle to a 10 mL syringe. Prepare a total of three syringes and label them as “PBS,” “PFA,” and “Glycine,” respectively.
CRITICAL: To better control the incision when you pierce the heart using a needle and inject the solution, it is recommended to use a piece of tape to wrap below the pointy end of the needle (See Figure 1). If the incision is too deep, the needle will pierce through the left ventricle to the left atrium, and the solution will pass through the pulmonary circulation (See TroubleshootingProblem 1 for details) and result in a failure of fixation. If the incision is too shallow, the PFA solution may not enter the left ventricle, which also leads to a failure of fixation.
Figure 1.
Example of a Wrapped Needle
Illustration (A) and image (B) of how the needle is wrapped by the tape to control the depth of the incision.
-
5.
Anesthetize the mouse. We administrate ∼400 μL 1.2% Avertin solution for a 20 g mouse (2,2,2-Tribromoethanol 250 mg/kg body weight of the mouse) through intraperitoneal injection. Alternative anesthetization methods can be used.
-
6.
After the mouse is anesthetized, lay the mouse on the dissection board in a supine position. Stabilize its feet with tapes/needles. Open the chest and expose the heart for perfusion (See Methods Video S1).
-
a.Use the forceps to pick up the skin around the bottom of the sternum (the highest point of the mouse chest when lying down). Make an incision using a pair of scissors.
-
b.Use the forceps to lift the bottom of the sternum and cut along the diaphragm using the scissors. Try to hold the forceps high and keep the scissor tip up to avoid damaging any organ.
-
c.Lift the rib cage and expose the heart.
-
d.Make an incision on the right atrium (in a deep red color) using the scissors (i.e., Fine Scissors-ToughCut, 14058-11, FST) so the blood can bleed out (Figure 2).
-
a.
Figure 2.
Schematic Illustration of the Incision Sites of the Heart
Video of the Chest Opening, Right Atrium Incision, and Perfusion. Related to Step 6
-
7.
Use blunt-end forceps to gently hold the heart, then pierce the heart near the bottom of the left ventricle using the perfusion syringe (Figure 2). Perfuse 10 mL cold PBS using a 10 mL syringe with a 25G needle to wash away the blood. While you are piercing into the heart and pushing the solution in, the pressure would spontaneously draw the needle into the left ventricle (See Video S1).
CRITICAL: After the needle is already pierced into the left ventricle, keep the tip of the needle slightly tilted up at ∼60° above the horizontal line (See Video S1). Then you can stabilize your syringe with the blunt-end forceps during perfusion, making sure the syringe does not pierce into the left atrium during perfusion.
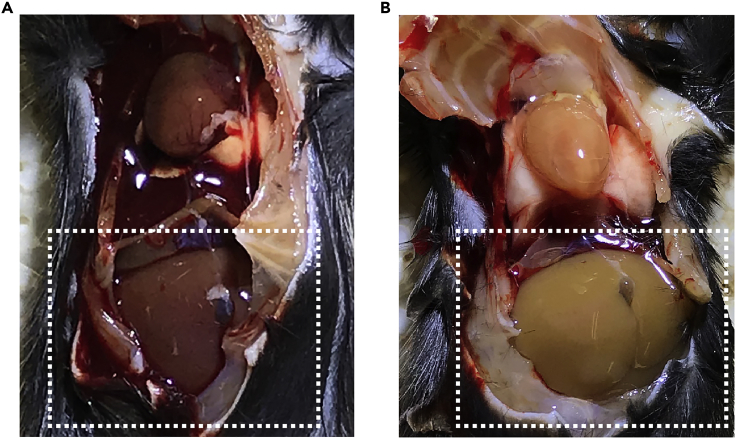
Note: A sign of good perfusion is that the solution comes out from the right atrium. Usually, the liver will become pale within a few seconds (Figure 3).
Figure 3.
Image of Liver Before and After Perfusion
Image of the mouse liver (A) before and (B) after perfusion. The dotted white square shows the liver.
-
8.
Repeat step 7 twice, so perfuse a total of 30 mL cold PBS three times (10 mL PBS each time).
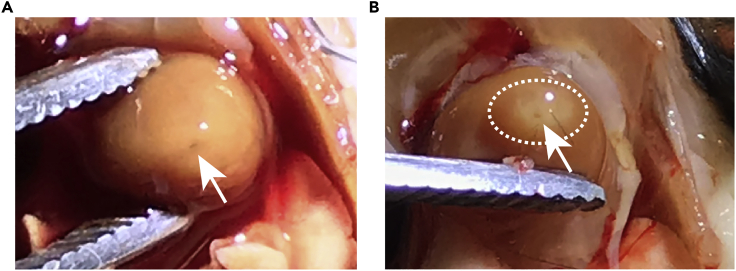
Note: When exposing the heart under the dissection light, you should be able to see the incision site of the needle. Try to inject into the same site when you repeat the perfusion step. Under good lighting conditions, you would see the incision site already after PBS perfusion (See Figure 4A). Thus, it is critical to perform perfusion under sufficient light.
Figure 4.
Image of the Incision Site by the Needle after PBS or PFA Perfusion
Image of the incision site by the needle (A) after PBS perfusion and (B) after PFA perfusion. Arrows point to the incision site. The dotted white circle indicates the tissue around the incision site after PFA perfusion, which becomes white.
-
9.
Start a 5 min timer. Perfuse 10 mL cold 0.5% PFA using a 10 mL syringe with a 25-gauge needle 3 times, so the total perfusion volume is 30 mL. The speed of the perfusion is around 10 mL solution in 1 min by hand perfusion (See Video S1). Wait until the timer rings so the total fixation time is 5 min.
Note: Right after the first PFA perfusion, you will notice the tissue around the incision site becomes rigid and white (See Figure 4B). From this point on, it should be easy to see the incision site and inject the solution into the same site again.
-
10.
Perfuse 10 mL cold 2 M glycine using a 10 mL syringe with a 25-gauge needle 3 times, so the total perfusion volume is 30 mL.
Note: After perfusion, the tail of the mouse will become rigid. Usually, a light fixative (0.5% PFA) would not make the muscles very rigid, so you should not feel a huge difference when touching the muscles after perfusion.
FACS Preparation + Sorting
Timing: FACS preparation, 3–4 h for 1–4 mice; cell sorting, 1 h for 1 mouse if a high-speed sorter is used (i.e., BD influx)
FACS preparation involves a series of enzyme digestions and washing steps to obtain single-cell suspension from the tissue. The FACS preparation for unfixed SCs was previously published in Liu et al., 2015. We further optimized it for a higher yield. The FACS preparation protocol for unfixed SCs and fixed SCs are very similar except for step 17 (A total of 10,000 U Collagenase II is used for unfixed SC isolation while a total of 20,000 U Collagenase II is used for fixed SC isolation).
-
11.
Pour ∼10 mL cold Wash Medium into a 10 cm glass petri dish.
-
12.
Use a No.11 blade and scissors to dissect the hindlimb muscles out. Put them into the petri dish.
-
13.
Take the muscle pieces out. Mince them with a No.20 blade on the cover of the glass petri dish.
Note: Fixed muscles are slightly more rigid compared with the unfixed muscles. You may notice the resistance when you mince the muscles using the blade.
-
14.
Pour ∼2 mL first enzyme digestion solution to the minced muscle slurry. Use scissors to further cut it.
-
15.
Pour the muscle slurry back to the first enzyme digestion solution in a 50 mL conical tube.
-
16.
Wrap the cap of the 50 mL tube with parafilm to prevent leakage.
-
17.
First enzyme digestion: Put the tube into the 37°C shaking water bath at medium-high speed for 90 min.
Note: The Collagenase II used in the first enzyme digestion for fixed cells is 2,000 U/mL × 10 mL, which doubles the amount compared with the enzyme used for unfixed cells (1,000 U/mL × 10 mL).
Optional: Pre-cool the centrifuge 5408R to 4°C.
-
18.
After the first enzyme digestion, take the tube out and fill it up with cold Wash Medium (50 mL in total). Flip the tube upside down for a few times to mix.
-
19.
Pour 25 mL of the slurry to another tube. Fill up both tubes with cold Wash Medium (50 mL in total for each tube).
Note: Splitting the slurry into two tubes helps to reduce the complexity of the solution for debris clearing.
-
20.
Centrifuge at 500 × g, 4°C, for 10 min using the centrifuge 5408R.
Note: During the centrifugation, warm the Collagenase II and Dispase II aliquots on a 37°C heat block.
-
21.
Take the tubes out carefully. Use the aspirator to aspirate away the Wash Medium till ∼12.5 mL solution remains.
Note: Place a 1 mL pipette tip in front of the aspirator to avoid aspirating too fast. Do not disrupt the pellet.
-
22.
Add 1 mL Collagenase II (3,000 U/mL) and 1 mL Dispase II (30 U/mL) to one tube. Mix it by pipetting up and down 5 times using a 25 mL serological pipette.
Note: Dispase II may have precipitates after long term storage, but it does not affect the second digestion. You can spin the precipitates down and use only the supernatant. Either way works in our lab.
-
23.
Transfer the solution with enzymes to another tube. Mix it by pipetting up and down 5 times using the same 25 mL serological pipette.
-
24.
Add cold Wash Medium to 30 mL (i.e., the concentration of the second digestion enzyme is 100 U/mL for Collagenase and 1 U/mL for Dispase II).
-
25.
Second enzyme digestion: Put the tube into the 37°C shaking water bath at medium-high speed for 30 min. Wrap the cap of the 50 mL tube with parafilm to prevent leakage.
-
26.
After the second enzyme digestion, mix the cell solution using a 30 mL syringe adapted to a 20-gauge needle ten times.
Note: It is important to syringe the cell solution ten times to obtain high yield. Any undigested small muscle pieces that clog the needle should be removed.
-
27.
Filter the cell solution with a 40 μm filter adapted to a new 50 mL conical tube.
-
28.
Add cold Wash Medium to 50 mL.
-
29.
Centrifuge at 500 × g, 4°C, for 10 min using the centrifuge 5408R.
-
30.
Take the tubes out carefully. Use the aspirator to aspirate away the Wash Medium until ∼2–3 mL solution remains.
Note: Place a 1 mL pipette tip in front of the aspirator to avoid aspirating too fast. Furthermore, place a 200 μL pipette tip in front of the aspirator when the solution goes down to ∼15 mL, and do not disrupt the pellet. Aspirate until the solution reaches 2–3 mL.
-
31.
Use a 1 mL pipette to resuspend the cell solution. Filter the cell solution with a 40 μm filter adapted to a new 50 mL conical tube.
-
32.
Transfer the single-cell suspension to a 5 mL round-bottom polypropylene tube as the loading tube.
Note: If using size calibration beads for size reference, dilute individual size beads at around 1:100 in PBS respectively in each loading tube (i.e., add 3 μL beads to 297 μL PBS). Sonicate the beads before usage to avoid aggregates. Load individual size beads and record ∼1,000–2,000 events for each beads.
-
33.
Sort cells using a cell sorter.
Note: For Pax7nGFP mice, SCs are marked by GFP signals. Gate for singlets and then the GFP+ population (See sorting scheme in Figure 5). Our lab usually uses a BD Influx cell sorter for cell sorting. The sorting time is ∼1 h per mouse worth of single-cell suspension.
Figure 5.
FACS Gating Plots and Size Analysis of pSCs and fSCs
FACS plot and population hierarchy of fSCs and pSCs. Each gate shows the enrichment corresponding to the mononuclear cell population amount of the total cell population in the previous gate. The forward scatter (FSC) and side scatter (SSC) analysis plots of the size calibration beads are used as size references. The FSC and SSC analysis of fSC and pSC populations are revealed in pseudocolor plots. The sort plots have previously been published in Yue et al. (2020).
-
34.
Collect the sorted cells in a 5 mL round-bottom polypropylene tube with 1 mL cold Wash Medium. Sorting does not need to be at 4°C.
RNA Isolation from Fixed Cells [Optional]
Timing: ∼1.5 h
The RNA isolation protocol was adapted from the manufacturer’s protocol with modifications (Thomsen et al., 2016). 100K fSCs are tested and sufficient for RNA isolation and later library generation step. To handle RNA, all pipette tips, microcentrifuge tubes, and reagents should be RNase-free. All the reagents used in the RNA isolation step are included in the miRNeasy FFPE Kit.
-
35.
Transfer the sorted cells to a 2 mL round-bottom microcentrifuge tube.
-
36.
Centrifuge the cell at 15,000 × g, 4°C for 3 min using a pre-chilled centrifuge 5427R or equivalent.
-
37.
Pipette away the supernatant. For 100K fSCs input, you will observe a small white pellet near the bottom of the tube. Do not disturb the pellet.
Note: If desire, the procedure can be stopped here. Snap freeze the cell pellet using liquid N2. Store the cell pellet at −80°C.
-
38.
Add 100 μL buffer PKD. Mix by pipetting up and down.
-
39.
Add 10 μL proteinase K. Mix by pipetting up and down.
-
40.
Incubate at 56°C for 1 h.
-
41.
Cool down the tubes on ice. Then centrifuge at 20,000 × g, 20°C–25°C (room temperature in lab, RT) for 20 min.
-
42.
Transfer the supernatant to a 2 mL microcentrifuge tube.
-
43.
Add 10 μL DNase Booster Buffer and 10 μL DNase I stock solution. Mix by pipetting up and down.
-
44.
Incubate at RT for 15 min.
-
45.
Add 320 μL buffer RBC. Mix by pipetting up and down.
-
46.
Add 1120 μL ethanol (100%). Mix well by pipetting up and down 5 times.
-
47.
Transfer 700 μL sample to the RNeasy MinElute spin column.
-
48.
Centrifuge at 15,000 × g, 30 s at RT.
-
49.
Discard the flow-through.
-
50.
Repeat step 48–49 until all sample has passed through the column.
-
51.
Add 500 μL buffer RPE to the column. Centrifuge at 15,000 × g, 30 s at RT.
-
52.
Discard the flow-through.
-
53.
Add 500 μL buffer RPE to the column. Centrifuge at 15,000 × g, 30 s at RT.
-
54.
Discard the flow-through.
-
55.
Place the column into a new 2 mL collection tube, open the lid of the spin column, and centrifuge at 15,000 × g for 5 min.
-
56.
Place the column into a new 1.5 mL collection tube. Add 14 μL RNase-free water to the membrane of the column. Wait for 5 min. Close the lip during elution.
-
57.
Centrifuge at 15,000 × g for 1 min.
-
58.
Discard the column. Keep the collection tube.
-
59.
Take 1 μL RNA for quantification using Qubit RNA HS Assay. Alternative RNA quantification method that is suitable for low input can be used.
Note: Store the RNA at −80°C. The isolated RNA is now compatible for traditional cDNA generation methods such as the SMART-seq 2 protocol.
Expected Outcomes
Sort Plot, Sort Yield, and Purity
The sort plots of satellite cells from PFA-perfused mice (fSCs) and PBS-perfused mice (pSCs), are similar (Figure 5). We usually get ∼100–300K fSCs from one Pax7nGFP mouse. Strictly following the protocol and not omitting any step (i.e., step 26) help to increase the yield.
To assess the purity, sorted cells can be assessed by Pax7 immunostaining (Figure 6). Other quiescent satellite cell markers can also be used.
Figure 6.
Immunostaining of Sorted Cells
25K sorted cells were plated down in one well of the 8-well chamber slide for 1 h for cells to sink. The chamber slides were pre-coated with ECM (1:250 in cold F10) for 30 min. After 1 h, cells were fixed and stained with Pax7. Nuclei were stained by DAPI. Scale bar, 50 μm.
Fixation Quality Control
In the first few trials, before you are confident with the perfusion technique, there are a few fixation quality control analyses to ensure the sorted fSCs are fixed.
To ensure that the fixation works, sorted cells can be plated down and cultured for a few days. Unfixed cells (i.e., pSC) would be activated and spread out during the culture whereas the cell morphology of fixed cells (i.e., fSCs) should not change (Figure 7).
Figure 7.
Bright Field Images of Sorted Cells during 3-Day Culture
100K sorted cells were plated down in one well of the 8-well chamber slide and cultured in wash medium (F10 + 10% HS + P/S) for 3 days. The chamber slides were pre-coated with ECM (1:250 in cold F10) for 30 min. Bright field images were taken every 24 h.
To ensure that the fixation works, cultured cells can also be stained with activation marker proteins. For example, unfixed cells (i.e., pSC) would be activated and express activation-related proteins such as MyoD and Dek whereas fixed cells (i.e., fSCs) would be negative for activation markers even after 3 day culture (Figure 8). Other activation markers can also be used.
Figure 8.
Immunostaining of Sorted Cells 3 days after Culture
100K cells from Pax7nGFP mice were plated down in one well of the 8-well chamber slide and cultured in wash medium (F10 + 10% HS + P/S) for 3 days. The chamber slides were pre-coated with ECM (1:250 in cold F10) for 30 min. 3 days after culture, cells were fixed and stained with GFP, MyoD, and Dek. Nuclei were stained by DAPI. Scale bar, 50 μm.
The cDNA size distribution of cDNA from unfixed control (i.e.pSCs) and fSCs are similar (Figure 9). The gene body coverage of RNA-seq library from unfixed control (i.e.pSCs) and fSCs are similar (Figure 10).
Figure 9.
Bioanalyzer Analyzer Traces of cDNA from pSCs and fSCs
Two biological replicates are shown. These data have previously been published in Yue et al. (2020).
Figure 10.
Gene Body Coverage Analysis of RNA-Seq Libraries from pSCs and fSCs
Two biological replicates are shown. These data have previously been published in Yue et al. (2020).
Limitations
The perfusion technique is key to a successful cell fixation. It takes practice to be familiar with the perfusion protocol and get consistent results. A typical sort yield is ∼100–300K fSCs from one Pax7nGFP mouse.
The VCAM1 staining failed to work on our hands using fSCs. Thus, we are unable to use the wildtype mouse to isolate fSCs using our usual lin-ve VCAM1+ve sorting scheme (CD31-ve, CD45-ve, Sca-1-ve, VCAM1+ve) (Liu et al., 2015). We have not further characterized other quiescent satellite cell markers such as CD34. For other cell surface markers, a test trial should be done to see whether the staining works on fixed cells.
Troubleshooting
Problem 1
Cells are not fixed. Isolated fSCs activate after culture.
Potential Solution
-
1.
Make sure that the 0.5% PFA is freshly prepared and mixed well.
-
2.
Make sure that the solutions have perfused into the heart from the left ventricle and come out from the right atrium. As the tissue around the incision becomes rigid during PFA perfusion, the incision site will become bigger. Also, there can be solution leakage from the injection site of the left ventricle rather than pushing through the circulation. A sign of good perfusion is that the liver turns pale (Figure 3).
-
3.
Make sure there is no solution coming out from the mouth/nose. Make sure that the lung is not swollen. These all indicate that the needle has poked to the left atrium so the solution has gone through the pulmonary circulation instead of the systemic circulation. If you see any sign of pulmonary circulation before or during PFA perfusion, the fixation is usually failed.
Problem 2
Low yield in sorted fSCs number, e.g., 10K cells only.
Potential Solution
-
1.
Make sure that the 0.5% PFA is freshly prepared and mixed well. High concentration of PFA (e.g., 2% PFA) would result in a low yield.
-
2.
Make sure you are familiar with the regular sorting protocol without fixation. A typical yield of a fresh sort is around 600–800K quiescent SCs from one Pax7nGFP mouse. Fixation is expected to decrease cell yield to around 100K–300K cells from one Pax7nGFP mouse. The cell count should be examined by hemocytometer and not rely on the count from the cell sorter.
-
3.
During the FACS preparation, carefully aspirate the wash medium away after each centrifugation step. Do not disturb the cell pellet. Because satellite cells are small, they are usually on the surface of the cell pellet and can be easily aspirated away. After the last centrifuge (step 30), try to leave some medium so you minimize the chance to disturb the cell pellet.
Problem 3
Fragmented cDNA from fSCs.
Potential Solution
-
1.
RNA isolated from fSCs without proper reverse crosslinking will result in low-quality RNA that hinders the subsequent cDNA generation. The quality of these RNA could not be analyzed by the RNA fragment analyzer analysis. Using the miRNAeasy FFPE isolation kit is critical as it involves a de-crosslinking step to allow satisfied isolated RNA for the following cDNA library generation step. From our experience, using a regular RNA isolation kit (i.e., Nucleospin XS), the generated cDNA will be fragmented (∼400 bp) from the RNA isolated from fSCs and the sequencing result will have poor gene coverage.
-
2.
While handling a low amount of RNA, try to keep an RNase-free working environment to avoid RNA degradation. Degraded RNA will also generate fragmented cDNA. It is a good habit to set up an RNA-work bench. The bench and pipettes should be cleaned and wrapped by RNase AWAY™ Surface Decontaminant (Optional) before the RNA isolation work. Frequently clean your gloves with RNase AWAY™ Surface Decontaminant if necessary.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Tom Cheung (tcheung@ust.hk).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate/analyze any datasets/code.
Acknowledgments
We thank Cheung lab members for discussions. We thank S. Tajbakhsh for providing Pax7-nGFP mice. This work was supported by research grants from the Hong Kong Research Grant Council (GRF660313, C6015-14G, C6003-14G, C6009-15G, AoE/M-604/16, T13-607/12R, T13-605/18-W), the National Key R&D Program of China (2018YFE0203600), the Guangdong Provincial Key S&T Program (2018B030336001), Lee Hysan Foundation (LHF17SC01), Hong Kong Epigenome Project (Lo Ka Chung Charitable Foundation), and the Croucher Innovation Award (CIA14SC04) from Croucher Foundation. This study was supported in part by the Innovation and Technology Commission (ITCPD/17-9). L.Y. is a recipient of the Hong Kong PhD Fellowship Scheme. T.H.C. is the S.H. Ho Associate Professor of Life Science at HKUST.
Author Contributions
Conceptualization, L.Y. and T.H.C.; Investigation, L.Y.; Writing – Original Draft, L.Y.; Writing – Review & Editing, L.Y. and T.H.C.; Funding Acquisition, T.H.C.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100128.
Contributor Information
Lu Yue, Email: lyue@ust.hk.
Tom H. Cheung, Email: tcheung@ust.hk.
References
- Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R.G., Kelly R., Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Thomsen E.R., Mich J.K., Yao Z., Hodge R.D., Doyle A.M., Jang S., Shehata S.I., Nelson A.M., Shapovalova N.V., Levi B.P. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods. 2016;13:87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L., Wan R., Luan S., Zeng W., Cheung T.H. Dek modulates global intron retention during muscle stem cells quiescence exit. Dev. Cell. 2020;53:661–676.e6. doi: 10.1016/j.devcel.2020.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video of the Chest Opening, Right Atrium Incision, and Perfusion. Related to Step 6
Data Availability Statement
This study did not generate/analyze any datasets/code.