Summary
The isolation of human antibodies with naturally paired heavy and light chains is crucial for understanding the human antibody immune response. Here, we present a protocol for antibody cloning from the sorted single human memory B cells recognizing hepatitis B virus (HBV) S antigen (HBsAg). A two-fluorescent-dye labeling strategy against HBsAg allows for an improved sorting specificity, while non-relevant protein staining allows for the exclusion of non-specific B cells. This protocol could also be widely adapted for other antigens.
For complete details on the use and execution of this protocol, please refer to Wang et al. (2020).
Graphical Abstract
Highlights
-
•
Human memory B cells recognizing HBsAg were single-cell sorted into 96-well plates
-
•
A two-fluorescent-dye labeling strategy was used to improve the sorting specificity
-
•
Three nested PCRs were performed using reverse-transcribed cDNA of sorted single cells
-
•
Cloning of naturally paired antibody is critical for studying the human antibody response
The isolation of human antibodies with naturally paired heavy and light chains is crucial for understanding the human antibody immune response. Here, we present a protocol for antibody cloning from the sorted single human memory B cells recognizing hepatitis B virus (HBV) S antigen (HBsAg). A two-fluorescent-dye labeling strategy against HBsAg allows for an improved sorting specificity, while non-relevant protein staining allows for the exclusion of non-specific B cells. This protocol could also be widely adapted for other antigens.
Before You Begin
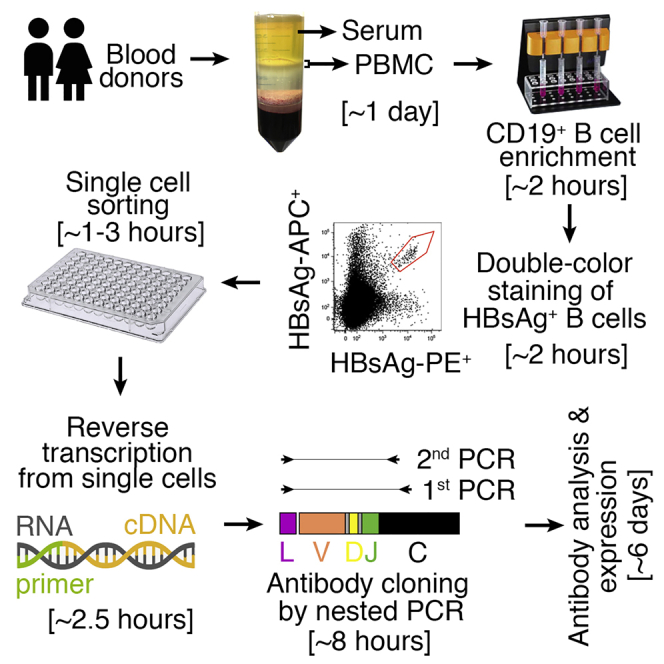
The main steps of this protocol include CD19+ B cell enrichment from the preserved human peripheral blood mononuclear cells (PBMCs), B cell labeling, single-cell flow cytometry sorting, reverse transcription and nested polymerase chain reaction (PCR) from single cells for antibody cloning, and antibody analysis to identify the antibodies with functional and naturally paired heavy and light chains.
Before starting the protocol, please ensure that the human PBMCs have been separated from peripheral blood, aliquoted, and cryopreserved. Researchers should proceed for sorting right after the CD19+ B cell enrichment and labeling; therefore it is important to make a reservation for sorting service.
Processing and Storage of PBMCs
Timing: ~1 day
-
1.
Blood draw.
Note: Approximately 400 mL (or less) blood is collected at clinics or blood centers. The whole blood in a standard blood bag containing anticoagulants (heparin) should be rapidly and reliably transferred to the laboratory for processing.
Note: From healthy human donor, the yield of PBMCs per mL blood ranges between 0.5–3 × 106 cells. The percentage of CD19+ B cells among PBMCs is approximately 5%–10%, and 10%–20% of these B cells are IgG+. Since the percentage of HBsAg+ cells among CD19+ CD20+ IgG+ B cell population is very low, around 0.01%–0.07% (Wang et al., 2020), we suggest to draw at least 200 mL of blood to collect sufficient PBMCs.
-
2.
Processing of PBMCs.
Note: Researchers should follow the standard principles and steps during the processing procedures of PBMCs. Different preparation techniques of PBMCs could be chosen, including the method by using cell separation tube with Frit barrier (CSTFB) or Ficoll gradient method.
-
3.
Aliquoting and storage.
Note: Aliquot 1 mL freezing medium (1–2 × 107 cells/mL) per cryovial (Cat#5000-1020, Nalgene). Immediately transfer the cryovials to −80°C freezer for controlled-rate freezing. One day later, transfer all cryovials to liquid nitrogen for long-term storage.
Biotinylation and Fluorophore-Labeling of Bait Proteins
Timing: ~1 day
-
4.Biotinylation of HBV antigen HBsAg and ovalbumin.
-
a.Refer to EZ-Link Micro Sulfo-NHS-LC-Biotinylation Kit (Cat#21935, Thermo Fisher Scientific) or EZ-Link Micro NHS-PEG4-Biotinylation Kit (Cat#21955, Thermo Fisher Scientific) User Guide for detailed instructions.
-
b.50–200 μg of commercially available HBsAg (0.6 mg/mL) (Cat#HBS-875, ProSpec) and ovalbumin (1 mg/mL) (Cat#S7951, Sigma-Aldrich) is incubated with biotin stock solution (prepared in water) at room temperature (25°C) for 30–60 min.
-
c.Buffer exchange and excess biotin removal. Apply the HBsAg-biotin mixture sample directly onto the center of the resin bed of equilibrated Zeba Spin Desalting column (included in these kits). Centrifuge the column at 1,000 × g for 2 min and collect the flow-through solution as the biotinylated HBsAg sample.
-
a.
Note: Use water as solvent to prepare the biotin stock solution.
Note: After biotinylation, both HBsAg and ovalbumin do not lose stability when stored at 4°C.
-
5.Fluorophore-labeling of biotinylated bait proteins.
-
a.Add streptavidin-fluorophore into 3 μg of biotinylated proteins (in PBS). Use streptavidin-fluorophore at 1:10 of total volume and incubate at 4°C for 1 h in the dark.
-
a.
Note: Label HBsAg by streptavidin-phycoerythrin (strep-PE) (Cat#12-4317-87, eBioscience) and streptavidin-allophycocyanin (strep-APC) (Cat#554067, BD Biosciences) respectively, while label ovalbumin by streptavidin-Alexa Fluor 488 (Cat#S32354, Thermo Fisher Scientific). Other fluorophores are also suitable depending on the sorting instrument.
-
b.Keep these mixtures at 4°C in dark and add them directly to enriched B cells later on.
-
b.
Buffer Preparation
Timing: ~1 h
-
6.
Prepare FACS buffer: 500 mL PBS (Cat#10010023, Thermo Fisher Scientific) with 10–15 mL fetal bovine serum (FBS) (final concentration 2%–3%) (Cat#SH3007103, Thermo Fisher Scientific).
-
7.
Prepare MACS buffer: 500 mL PBS with 2.5 g bovine serum albumin (BSA) (final concentration 0.5%) (Cat#A0281, Sigma-Aldrich) and 2 mL of 0.5 M ethylenediaminetetraacetic acid (EDTA) (final concentration 2 mM) (Cat#15575020, Thermo Fisher Scientific).
-
8.
Filter the buffer after preparation using a 0.22 μm filter (Cat#431118, CORNING) and keep buffer cold (2°C–8°C).
-
9.
Warm a bottle of RPMI 1640 medium (Cat#11875093, Thermo Fisher Scientific) to 37°C before experiment.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-Cy7 anti-human CD20 (Clone L27) | BD Biosciences | Cat#335811; RRID: AB_399985 |
| Bv421 anti-human CD19 (Clone HIB19) | BD Biosciences | Cat#562440; RRID: AB_11153299 |
| Bv421 anti-human IgG (Clone G18-145) | BD Biosciences | Cat#562581; RRID: AB_2737665 |
| PE anti-human CD27 (Clone M-T271) | BD Biosciences | Cat#555441; RRID: AB_395834 |
| APC anti-human IgG (Clone G18-145) | BD Pharmingen | Cat#550931; RRID: AB_398478 |
| Alexa Fluor 488 anti-human CD19 (Clone HIB19) | BD Biosciences | Cat#557697; RRID: AB_396806 |
| Biological Samples | ||
| Human Blood or Human Peripheral Blood Mononuclear Cells (PBMCs) | (Wang et al., 2020) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| HBsAg adr CHO | ProSpec | Cat#HBS-875 |
| Ovalbumin (257–264) Chicken | Sigma-Aldrich | Cat#S7951 |
| Streptavidin-allophycocyanin (strep-APC) | BD Biosciences | Cat#554067; RRID: AB_10050396 |
| Streptavidin-phycoerythrin (strep-PE) | eBioscience | Cat#12-4317-87 |
| Streptavidin-Alexa Fluor 488 | Thermo Fisher Scientific | Cat#S32354 |
| PBS, pH 7.4 | Thermo Fisher Scientific | Cat#10010023 |
| PBS (10×), pH 7.2 | Thermo Fisher Scientific | Cat#70013032 |
| HyClone™ Fetal Bovine Serum | Fisher Scientific | Cat#SH3007103 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A0281 |
| UltraPure™ 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat#15575020 |
| RPMI 1640 Medium | Thermo Fisher Scientific | Cat#11875093 |
| Human Fc Block | BD Biosciences | Cat#564220 |
| RNAsin Plus RNase Inhibitor | Promega | Cat#N2615 |
| Nuclease-Free Water | QIAGEN | Cat#129114 |
| DTT 0.1 M Solution | Thermo Fisher Scientific | Cat# 707265ML |
| IGEPAL® CA-630 | Sigma-Aldrich | Cat#I8896 |
| dATP Solution (100 mM) | Thermo Fisher Scientific | Cat#R0141 |
| dTTP Solution (100 mM) | Thermo Fisher Scientific | Cat#R1191 |
| dCTP Solution (100 mM) | Thermo Fisher Scientific | Cat#R0151 |
| dGTP Solution (100 mM) | Thermo Fisher Scientific | Cat#R0161 |
| UltraPure™ Sucrose | Thermo Fisher Scientific | Cat#15503022 |
| Cresol Red | Sigma-Aldrich | Cat#114472 |
| Critical Commercial Assays | ||
| EZ-Link™ Micro Sulfo-NHS-LC-Biotinylation Kit | Thermo Fisher Scientific | Cat#21935 |
| EZ-Link™ Micro NHS-PEG4-Biotinylation Kit | Thermo Fisher Scientific | Cat#21955 |
| CD19 MicroBeads, Human | Miltenyi Biotech | Cat#130-097-055 |
| QuadroMACS™ Separator and Starting Kits | Miltenyi Biotech | Cat#130-091-051 |
| LS Magnetic Columns | Miltenyi Biotech | Cat#130-042-401 |
| Superscript III Reverse Transcriptase | Thermo Fisher Scientific | Cat#18080044 |
| HotStarTaq DNA Polymerase | QIAGEN | Cat#203209 |
| Oligonucleotides | ||
| F1-HC (5′-ACAGGTGCCCACTCCCAGG TGCAG) |
(Wang et al., 2020) | N/A |
| F2-HC (5′-AAGGTGTCCAGTGTGARGT GCAG) |
(Wang et al., 2020) | N/A |
| F3-HC (5′-CCCAGATGGGTCCTGTCCCAG GTGCAG) |
(Wang et al., 2020) | N/A |
| F4-HC (5′-CAAGGAGTCTGTTCCGAGGT GCAG) |
(Wang et al., 2020) | N/A |
| R1-HC (5′-GGAAGGTGTGCACGCCGCT GGTC) |
(Wang et al., 2020) | N/A |
| R2-HC (5′-GTTCGGGGAAGTAGTCCTT GAC) |
(Wang et al., 2020) | N/A |
| F1-Kappa (5′-ATGAGGSTCCCYGCTCA GCTGCTGG) |
(Wang et al., 2020) | N/A |
| F2-Kappa (5′-CTCTTCCTCCTGCTACTC TGGCTCCCAG) |
(Wang et al., 2020) | N/A |
| F3-Kappa (5′-ATTTCTCTGTTGCTCTGG ATCTCTG) |
(Wang et al., 2020) | N/A |
| F4-Kappa (5′-ATGACCCAGWCTCCAB YCWCCCTG) |
(Wang et al., 2020) | N/A |
| R1-Kappa (5′-GTTTCTCGTAGTCTGC TTTGCTCA) |
(Wang et al., 2020) | N/A |
| R2-Kappa (5′-GTGCTGTCCTTGCT GTCCTGCT) |
(Wang et al., 2020) | N/A |
| F1-Lambda (5′-GGTCCTGGGCCCAGTCT GTGCTG) |
(Wang et al., 2020) | N/A |
| F2-Lambda (5′-GGTCCTGGGCCCAGTCT GCCCTG) |
(Wang et al., 2020) | N/A |
| F3-Lambda (5′-GCTCTGTGACCTCCTAT GAGCTG) |
(Wang et al., 2020) | N/A |
| F4-Lambda (5′-GGTCTCTCTCSCAGCYTG TGCTG) |
(Wang et al., 2020) | N/A |
| F5-Lambda (5′-GTTCTTGGGCCAATTTTA TGCTG) |
(Wang et al., 2020) | N/A |
| F6-Lambda (5′-GGTCCAATTCYCAGGCT GTGGTG) |
(Wang et al., 2020) | N/A |
| F7-Lambda (5′-GAGTGGATTCTCAGACT GTGGTG) |
(Wang et al., 2020) | N/A |
| R1-Lambda (5′-CACCAGTGTGGCCTTGT TGGCTTG) |
(Wang et al., 2020) | N/A |
| R2-Lambda (5′-CTCCTCACTCGAGG GYGGGAACAGAGTG) |
(Wang et al., 2020) | N/A |
| Random Primers | Thermo Fisher Scientific | Cat#48190011 |
| Recombinant DNA | ||
| IGγ1 Expression Vector | (von Boehmer et al., 2016) | N/A |
| IGκ Expression Vector | (von Boehmer et al., 2016) | N/A |
| IGλ Expression Vector | (von Boehmer et al., 2016) | N/A |
| Software and Algorithms | ||
| IgBlast | (Ye et al., 2013) | http://www.ncbi.nlm.nih.gov/igblast/ |
| IMGT/V-QUEST | (Brochet et al., 2008) | http://www.imgt.org/IMGT_vquest/vquest |
| Other | ||
| General Long-Term Storage Cryogenic Tubes | Nalgene | Cat#5000-1020 |
| SimpliAmp™ Thermal Cycler | Thermo Fisher Scientific | Cat#A24811 |
| 500 mL Bottle Top Vacuum Filter, 0.22 μm Pore | CORNING | Cat#431118 |
| Pipet-Lite Multi Pipette L12-20XLS+ | RAININ | Cat#17013808 |
Step-By-Step Method Details
Purification of CD19+ B Cells from PBMCs
Timing: ~2 h
This step will provide a detailed procedure for enrichment of CD19+ B cell from human PBMCs.
Note: Use a vial of PBMCs from naïve donor or an extra vial from selected donors with elite serum neutralizing activity to process alongside for flow cytometry compensation and for background determination during sorting.
Note: All the procedures should be performed following institutional biohazard guidelines for human sample preparation.
Note: Refer to CD19-MicroBeads (Cat#130-097-055, Miltenyi Biotech) User Guide for detailed instructions.
-
1.
Take the cryovials of PBMCs from liquid nitrogen and quickly thaw them in the 37°C warm water until little ice left. Transfer all PBMCs into a 50 mL falcon tube filled with 40 mL warmed RPMI 1640 medium and mix well.
-
2.
Pre-cool the centrifuge to 4°C. Spin cells 300 × g for 5 min at 4°C.
-
3.
Discard the supernatant into bleach and resuspend the cell pellet with pre-chilled MACS buffer (100 μL of buffer per 1 × 107 total cells).
-
4.
Add 20 μL of CD19 MicroBeads (Cat#130-097-055, Miltenyi Biotech) per 1 × 107 total cells.
Note: For example, if five cryovials (2 × 107 cells per cryovial) are thawed, resuspend the cell pellet with 1 mL pre-chilled MACS buffer and further add 200 μL of CD19 MicroBeads for incubation.
-
5.
Mix well and incubate with rotation for 20 min at 4°C.
-
6.
During the incubation, prepare the QuadroMACS Separator (Cat#130-091-051, Miltenyi Biotech) for LS columns (Cat#130-042-401, Miltenyi Biotech), and place LS column in the magnetic field of a QuadroMACS Separator.
Note: One LS column is sufficient for positive selection for 1 × 108 labeled cells from 2 × 109 total cells.
-
7.
Rinse the LS columns with 3 mL MACS buffer.
-
8.
When the incubation is over, fill the tubes with chilled MACS buffer. Spin cells 300 × g for 5 min at 4°C and discard the supernatant. Resuspend the washed cell pellet in 2 mL cold MACS buffer.
-
9.
Proceed to magnetic separation and apply cell suspension onto the rinsed LS columns.
-
10.
Collect the CD19- cells that pass through into a 15 mL falcon tube.
Note: This is unlabeled CD19- cell fraction. Discard these cells or use them for other purposes.
-
11.
Perform washing steps by adding 3 mL MACS buffer three times.
Note: Wait until the column reservoir is empty and then add the fresh buffer to wash.
-
12.
Remove the LS column from the MACS Separator and place it on a 15 mL falcon collection tube.
-
13.
Pipette 5 mL MACS buffer onto the column. Immediate take the plunger and flush out the magnetically labeled cells by pushing the plunger (supplied with the LS column) firmly and gently.
Note: If there are two LS column with the same sample of PBMCs, repeat the same with the second LS column and flush out the CD19+ cells into the same collection tube.
-
14.
Fill the collection tube with chilled FACS buffer. Spin cells 300 × g for 5 min at 4°C.
-
15.
There will be a small CD19+ cell pellet in the collection tube. Discard the supernatant and resuspend the washed cell pellet in cold FACS buffer (use 300–500 μL for original 1 × 108 total cells).
Optional: The number of the enriched CD19+ B cells in this step could be determined.
Note: The purity of enriched B cells, assessed by flow cytometry using CD19 or CD20 staining, reaches 96%–98%.
Alternatives: For the enrichment of CD19+ cells, alternative magnetic separation beads, such as Dynabeads magnetic beads, could be used.
Labeling of HBsAg+ B Cells and Single-Cell Sorting
Timing: ~3–6 h
This step will provide a detailed procedure for fluorescent staining of the enriched CD19+ cells. The two-color (HBsAg-PE+ and HBsAg-APC+) with a dump channel (ovalbumin-Alexa Fluor 488+) staining strategy increases the specificity of the sorted single cells.
-
16.
Add Human Fc Block (Cat#564220, BD Biosciences) into the suspended cells at 1:50 of total volume and incubate at 4°C for 20 min.
Note: Human Fc Block is used to block unwanted binding of antibodies to human Fc receptor-expressing cells and to decrease staining background. Human Fc Block (Cat#564220, BD Biosciences) is widely used due to its high specificity, but alternative products or homemade reagents could also be used.
-
17.
Wash the cells with chilled FACS buffer. Spin cells 300 × g for 5 min at 4°C.
-
18.
Discard the supernatant and resuspend the washed cell pellet in cold FACS buffer (use 300 μL for original 1 × 108 total cells). Use a small proportion of cells for flow cytometry compensation, while the rest for the real sample staining.
-
19.Generation of single stained controls and flow cytometry compensation.
-
a.Split the cells used for compensation into six aliquots (eppendorf tubes or six wells in a 96-well plate).
-
b.Generate single stained controls by adding PE-Cy7 anti-human CD20 (Cat#335811, BD Biosciences), PE anti-human CD27 (Cat#555441, BD Biosciences), APC anti-human IgG (Cat#550931, BD Pharmingen), Alexa Fluor 488 anti-human CD19 (Cat#557697, BD Biosciences) and Bv421 anti-human CD19 (Cat#562440, BD Biosciences) antibodies, respectively, to five of the aliquoted samples. All antibodies are added in a 1:10–1:20 volume dilution. The sixth aliquot is a non-staining control.
-
c.Incubate at 4°C for 30 min in the dark.
-
d.Wash twice with cold FACS buffer by centrifugation at 300 × g for 5 min at 4°C.
-
e.Resuspend the cells in 200 μL FACS buffer.
-
f.Perform fluorescence-activated cell sorting (FACS) compensation using flow cytometer (BD FACSAria™ II Cell Sorter, BD Biosciences) before single-cell sorting.
-
a.
Note: Prepare the samples for compensation in parallel with the following staining.
-
20.Generation of fully stained samples.
-
a.Add the pre-incubated strep-PE-HBsAg, strep-APC-HBsAg, and strep-Alexa Fluor 488-ovalbumin (dump channel) simultaneously to the enriched CD19+ cell fraction.
-
a.
CRITICAL: For every original 108 total cells, add 3 μg of each fluorescently labeled HBsAg and 3 μg ovalbumin for staining. Lower amount of proteins may lead to inefficient staining.
-
b.Incubate at 4°C in the dark for 30 min.
-
c.Add PE-Cy7 anti-human CD20 (Cat#335811, BD Biosciences) and Bv421 anti-human IgG (Cat#562581) directly to the CD19+ cell fraction without washing.
-
b.
Note: Anti-human IgG staining is optional.
-
d.Incubate at 4°C for another 20 min in the dark.
-
e.After incubation, wash twice with cold FACS buffer by centrifugation at 300 × g for 5 min at 4°C.
-
f.Resuspend each CD19+ cell sample in 1 mL FACS buffer.
-
d.
Optional: Viability dye could be used in the dump channel to gate out dead cells and debris.
-
21.
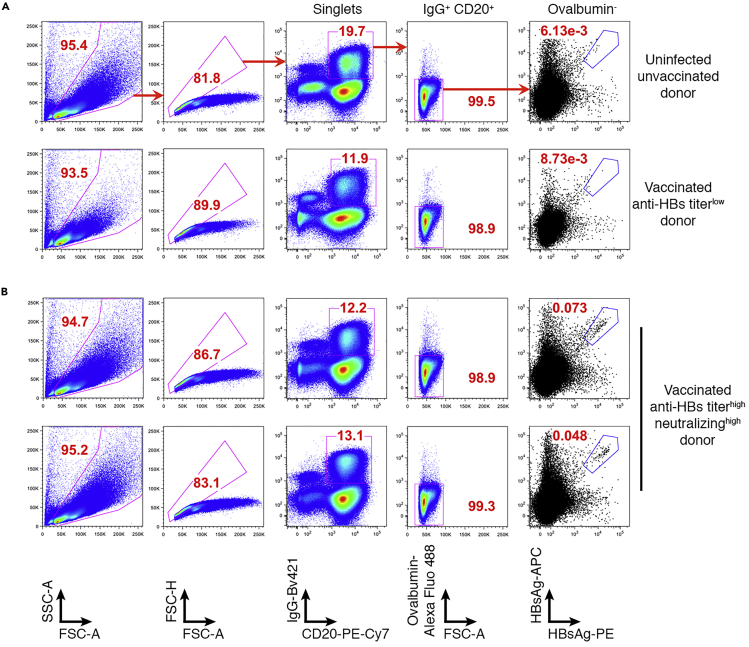
Run the control samples (CD19+ cells enriched from a naïve donor or a vaccinated donor with low level of serum anti-HBs titer) to confirm proper compensation on the sorter and set the gate for sorting (CD20-PE-Cy7+, IgG-Bv421+, HBsAg-PE+, HBsAg-APC+, ovalbumin-Alexa Fluor 488−) (Figure 1A).
Figure 1.
Gating Strategy and Representative Data for Flow Cytometry Analysis
(A) Samples with a low level of anti-HBs titers. Each panel from left to right corresponds to a sequential gating strategy. Frequencies of HBsAg-specific memory B cells in PBMCs are shown in the right most panel. FSC, forward scatter; SSC, side scatter (-A, area; -H, height; -W, width).
(B) Samples with a high level of anti-HBs titers and a high level of serum neutralizing activities. The percentages of IgG+ memory B cells that bind to HBsAg antigen (HBsAg-PE+ and HBsAg-APC+) are significantly increased.
-
22.Prepare the lysis buffer for single-cell sorting.
-
a.Recipe for the lysis buffer (for ten 96-well plates in theory).
-
a.
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| 10× PBS (Cat#70013032, Thermo Fisher Scientific) | 0.5× | 200 |
| 0.1 M DTT (Cat#707265ML, Thermo Fisher Scientific) | 10 mM | 400 |
| RNAsin Plus RNase inhibitor (40 U/μL) (Cat#N2615, Promega) |
3 U/μL | 300 |
| Nuclease-Free Water (Cat#129114, QIAGEN) | n/a | 3,100 |
| Total | n/a | 4,000 |
-
b.Add 4 μL into each well of 96-well plates using multichannel pipette (Cat#17013808, RAININ). Avoid bubbles during adding.
-
b.
CRITICAL: Keep the plates on ice before sorting.
CRITICAL: Perform this step in the DNA-free hood to avoid RNase contamination.
-
23.
Perform the calibration procedure of the sorting instrument for 96-well plate sorting prior to the single-cell sorting.
-
24.
Perform single-cell FACS using flow cytometer (BD FACSAria™ II Cell Sorter, BD Biosciences). Sort CD20-PE-Cy7+ IgG-Bv421+ HBsAg-PE+ HBsAg-APC+ ovalbumin-Alexa Fluor 488− single cells into each well of the 96-well plates (Figure 1B).
Note: Two-color flow cytometry of HBsAg-PE and HBsAg-APC yield diagonal patterns, eliminating B cells with non-specific binding to PE or APC.
-
25.
After sorting, seal the 96-well plates immediately by using aluminum foil.
-
26.
Put the 96-well plates on dry ice immediately and store them at −80°C.
Pause Point: RNA in the sorted single cells could be stable at −80°C for at least 2 months.
Reverse Transcription from Sorted Single Cells
Timing: ~2.5 h
This step will provide a detailed procedure for reverse transcription of the sorted HBsAg-binding single B cells.
CRITICAL: To avoid DNA/RNA and RNase contamination is crucial for this step. Perform this step in the DNA-free hood, and decontaminate the hood prior to use.
-
27.
Take plates out from −80°C and thaw on ice for 5–10 min.
-
28.
Centrifuge at 300 × g for 5 min at 4°C.
-
29.Prepare reverse transcription mixture (RT mix-I) under sterile conditions and on ice.
-
a.Recipe for RT mix-I (for one 96-well plate).
-
a.
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| Random Primers (0.3 μg/μL) (Cat#48190011, Thermo Fisher Scientific) |
18.3 ng/μL | 47 |
| 10% IGEPAL® CA-630 in Water (Cat#I8896, Sigma-Aldrich) | 0.61% | 47 |
| RNAsin Plus RNase Inhibitor (40 U/μL) (Cat#N2615, Promega) |
0.78 U/μL | 15 |
| Nuclease-Free Water (Cat#129114, QIAGEN) | n/a | 661 |
| Total | n/a | 770 |
-
b.Add 7 μL into each well of 96-well plates using multichannel pipette. Avoid bubbles during adding. Now there are 11 μL in each well.
-
c.Pipette the liquid to rinse the sides of wells and then mix 10 times using a multichannel pipette
-
b.
CRITICAL: Keep the plates on ice.
-
30.
Cover plates with sterile aluminum sealing foil and centrifuge at 300 × g for 1 min at 4°C.
-
31.
Incubate plates for 5 min at 65°C in the preheated thermocycler (Cat#A24811, Thermo Fisher Scientific).
-
32.
After incubation, chill the plate on ice for 2–5 min.
-
33.Prepare reverse transcription mixture (RT mix-II) under sterile conditions and on ice.
-
a.Recipe for RT mix-II (for one 96-well plate).
-
a.
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| SuperScript™ III Reverse Transcriptase (200 U/μL) (Cat#18080044, Thermo Fisher Scientific) |
7.14 ng/μL | 27.5 |
| RNAsin Plus RNase Inhibitor (40 U/μL) (Cat#N2615, Promega) |
0.57 U/μL | 11 |
| 0.1 M DTT (supplied with SuperScript™ III Reverse Transcriptase) | 14.29 mM | 110 |
| dNTP (25 mM) (see Note blow) | 1.79 mM | 55 |
| 5× First-Strand Buffer (supplied with SuperScript™ III Reverse Transcriptase) | n/a | 330 |
| Nuclease-Free Water (Cat#129114, QIAGEN) | n/a | 236.5 |
| Total | n/a | 770 |
Note: Prepare dNTP (25 mM) by mixing stock of dATP (Cat#R0141, Thermo Fisher Scientific), dTTP (Cat#R1191, Thermo Fisher Scientific), dCTP (Cat#R0151, Thermo Fisher Scientific), and dGTP solutions (Cat#R0161, Thermo Fisher Scientific) at 1:1:1:1 ratio. Aliquot and store them at −20°C.
-
b.Add 7 μL into each well of 96-well plates using multichannel pipette. Avoid bubbles during adding. Now there are 18 μL in each well.
-
c.Mix 10 times using a multichannel pipette.
-
b.
-
34.
Cover plates with sterile aluminum sealing foil and quick centrifuge at 4°C.
-
35.
Place plates into PCR thermocycler (Cat#A24811, Thermo Fisher Scientific) and run the program as below.
| Thermocycler Conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Annealing | 25°C | 10 min | 1 |
| Extension | 50°C | 60 min | 1 |
| Inactivation | 70°C | 15 min | 1 |
| Hold | 4°C | Forever | |
-
36.
Remove the plates from the PCR thermocycler and quick centrifuge at 4°C.
Pause Point: Seal the plates by wrapping the sides with parafilm membranes and store the plates at −80°C. The transcribed cDNA could be stable at −80°C for at least several months.
Note: Researchers could also proceed to perform the PCR amplification immediately.
Antibody Cloning – 1st PCR
Timing: ~4 h
This step will provide a detailed procedure for 1st round of nested PCR reactions.
-
37.
Take the plates out from −80°C and thaw on ice for 10 min.
-
38.
Add 10 μL of pre-chilled nuclease-free water (Cat#129114, QIAGEN) and mix 10 times using a multichannel pipette. Centrifuge at 300 × g for 1 min at 4°C. Now there are 28 μL in each well. Keep the plates on ice for the first round of nested PCR reactions.
-
39.Prepare primers for PCR reactions.
-
a.Dilute all primers to 50 μM each.
-
b.To prepare forward primer mixture, mix forward primers at 1:1:1 ratio for immunoglobulin heavy chain, Kappa light chain, and Lambda light chain respectively.
-
c.Use forward primer mixtures and individual reverse primers (1st PCR) for the first round of nested PCR reactions.
-
a.
Note: Forward primers mixture for heavy chain: F1-HC (5′-ACAGGTGCCCACTCCCAGGTGCAG) + F2-HC (5′-AAGGTGTCCAGTGTGARGTGCAG) + F3-HC (5′-CCCAGATGGGTCCTGTCCCAGGTGCAG) + F4-HC (5′-CAAGGAGTCTGTTCCGAGGTGCAG). Reverse primer for heavy chain (1st PCR): R1-HC (5′-GGAAGGTGTGCACGCCGCTGGTC).
Note: Forward primers mixture for kappa light chain: F1-Kappa (5′-ATGAGGSTCCCYGCTCAGCTGCTGG) + F2-Kappa (5′-CTCTTCCTCCTGCTACTCTGGCTCCCAG) + F3-Kappa (5′-ATTTCTCTGTTGCTCTGGATCTCTG) + F4-Kappa (5′-ATGACCCAGWCTCCABYCWCCCTG). Reverse primer for kappa light chain (1st PCR): R1-Kappa (5′-GTTTCTCGTAGTCTGCTTTGCTCA)
Note: Forward primers mixture for lambda light chain: F1-Lambda (5′-GGTCCTGGGCCCAGTCTGTGCTG) + F2-Lambda (5′-GGTCCTGGGCCCAGTCTGCCCTG) + F3-Lambda (5′-GCTCTGTGACCTCCTATGAGCTG) + F4-Lambda (5′-GGTCTCTCTCSCAGCYTGTGCTG) + F5-Lambda (5′-GTTCTTGGGCCAATTTTATGCTG) + F6-Lambda (5′-GGTCCAATTCYCAGGCTGTGGTG) + F7-Lambda (5′-GAGTGGATTCTCAGACTGTGGTG). Reverse primer for lambda light chain (1st PCR): R1-Lambda (5′-CACCAGTGTGGCCTTGTTGGCTTG).
-
40.Prepare mixture for the first round of nested PCR reactions (1st PCR mix) under sterile conditions and on ice.
-
a.Recipe for 1st PCR mix (for one 96-well plate).
-
a.
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| HotStarTaq DNA Polymerase (5 U/μL) (Cat#203209, QIAGEN) |
0.05 U/μL | 42 |
| Forward Primer Mixture (50 μM) (see Note above) |
0.29 μM | 23 |
| Reverse Primer (50 μM) (see Note above) |
0.19 μM | 15 |
| dNTP (25 mM) (see Note blow) | 0.31 mM | 48 |
| 10× PCR Buffer (supplied with HotStarTaq DNA Polymerase) | n/a | 420 |
| Nuclease-Free Water (Cat#129114, QIAGEN) | n/a | 3,352 |
| Total | n/a | 3,900 |
Note: Prepare dNTP (25 mM) by mixing stock of dATP (Cat#R0141, Thermo Fisher Scientific), dTTP (Cat#R1191, Thermo Fisher Scientific), dCTP (Cat#R0151, Thermo Fisher Scientific), and dGTP solutions (Cat#R0161, Thermo Fisher Scientific) at 1:1:1:1 ratio. Aliquot and store them at −20°C.
-
b.Add 38 μL into each well of 96-well plates using multichannel pipette. Avoid bubbles during adding.
-
c.Prepare PCR plates for heavy chain, kappa light chain, and lambda light chain respectively.
-
b.
-
41.
Transfer 4 μL diluted cDNA from each well of cDNA plates to the corresponding wells in the prepared 1st PCR plates. Mix 10 times using a multichannel pipette.
Note: Transfer three times respectively: 4 μL diluted cDNA to the heavy chain-1st PCR plate, 4 μL diluted cDNA to the kappa light chain-1st PCR plate, and 4 μL diluted cDNA to the lambda light chain-1st PCR plate.
Note: After use, put the cDNA plates at 4°C for short-term storage and put them back to −80°C for long-term storage.
-
42.
Cover the prepared 1st PCR plates with sterile aluminum sealing foil and centrifuge at 300 × g for 1 min at 4°C.
-
43.
Place the 1st PCR plates into PCR thermocyclers and run the program as below.
| PCR Cycling Conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 15 min | 1 |
| Denaturation | 94°C | 30 s | 50 cycles |
| Annealing | 46°C | 30 s | |
| Extension | 72°C | 55 s | |
| Final extension | 72°C | 10 min | 1 |
| Hold | 4°C | Forever | |
-
44.
Remove the 1st PCR plates from the PCR thermocyclers and quick centrifuge at 4°C.
Pause Point: Researchers could also proceed to perform the 2nd round of nested PCR amplification immediately or store the 1st PCR plates at −20°C for long-term storage.
Antibody Cloning – 2nd PCR
Timing: ~4 h
This step will provide a detailed procedure for 2nd round of nested PCR reactions.
-
45.
Take the 1st PCR plates. If they are frozen, thaw on ice for 10 min.
-
46.Prepare 40% sucrose loading buffer for the 2nd PCR reaction.
-
a.Dissolve 20 g of sucrose (Cat#15503022, Thermo Fisher Scientific) in 50 mL nuclease-free water (Cat#129114, QIAGEN) in a 50 mL falcon tube.
-
b.Add cresol red (Cat#114472, Sigma-Aldrich) to quick dissolve and a red color is produced.
-
a.
Note: Add sufficient (but not excessive) cresol red as a dye for agarose gel electrophoresis later after the 2nd PCR reactions.
-
47.
Prepare primers for PCR reactions.
Note: Forward primers mixtures for heavy chain, kappa light chain, and lambda light chain are the same as the ones for the 1st PCR reactions.
Note: Reverse primers for the 2nd PCR reaction are different. Reverse primer for heavy chain (2nd PCR): R2-HC (5′-GTTCGGGGAAGTAGTCCTTGAC). Reverse primer for kappa light chain (2nd PCR): R2-Kappa (5′-GTGCTGTCCTTGCTGTCCTGCT). Reverse primer for lambda light chain (2nd PCR): R2-Lambda (5′-CTCCTCACTCGAGGGYGGGAACAGAGTG).
-
48.Prepare mixture for the second round of nested PCR reactions (2nd PCR mix) under sterile conditions and on ice.
-
a.Recipe for 2nd PCR mix (for one 96-well plate).
-
a.
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| HotStarTaq DNA Polymerase (5 U/μL) (Cat#203209, QIAGEN) |
0.05 U/μL | 42 |
| Forward Primer Mixture (50 μM) (see Note above) |
0.29 μM | 23 |
| Reverse Primer (50 μM) (see Note above) |
0.19 μM | 15 |
| dNTP (25 mM) (see Note blow) | 0.31 mM | 48 |
| 10× PCR Buffer (supplied with HotStarTaq DNA Polymerase) | ~1.1× | 420 |
| 40% Sucrose Loading Buffer (see above) | 8.2% | 800 |
| Nuclease-Free Water (Cat#129114, QIAGEN) | n/a | 2,552 |
| Total | n/a | 3,900 |
Note: Prepare dNTP (25 mM) by mixing stock of dATP (Cat#R0141, Thermo Fisher Scientific), dTTP (Cat#R1191, Thermo Fisher Scientific), dCTP (Cat#R0151, Thermo Fisher Scientific), and dGTP solutions (Cat#R0161, Thermo Fisher Scientific) at 1:1:1:1 ratio. Aliquot and store them at −20°C.
Note: The 2nd PCR mix is pink or light pink due to the presence of cresol red dye.
-
b.Add 38 μL into each well of 96-well plates using multichannel pipette. Avoid bubbles during adding.
-
c.Prepare the 2nd PCR plates for heavy chain, kappa light chain, and lambda light chain respectively.
-
b.
-
49.
Transfer 4 μL from each well of the 1st PCR plates to the corresponding wells in the prepared 2nd PCR plates. Mix 10 times using a multichannel pipette.
Note: Transfer three times respectively: 4 μL of 1st PCR product to the heavy chain-2nd PCR plate, 4 μL of 1st PCR product to the kappa light chain-2nd PCR plate, and 4 μL of 1st PCR product to the lambda light chain-2nd PCR plate.
Note: After use, put the 1st PCR plates at 4°C for short-term storage and freeze them at −20°C for long-term storage.
-
50.
Cover the prepared 2nd PCR plates with sterile aluminum sealing foil and centrifuge at 300 × g for 1 min at 4°C.
-
51.
Place the 2nd PCR plates into PCR thermocyclers and run the program as below.
| PCR Cycling Conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 15 min | 1 |
| Denaturation | 94°C | 30 s | 50 cycles |
| Annealing | 57°C | 30 s | |
| Extension | 72°C | 50 s | |
| Final extension | 72°C | 10 min | 1 |
| Hold | 4°C | Forever | |
-
52.
Remove the 2nd PCR plates from the PCR thermocyclers and quick centrifuge at 4°C.
Pause Point: Researchers could put the 2nd PCR plates at 4°C for short-term storage and −20°C for long-term storage.
Antibody Sequencing and Analysis
Timing: ~6 days
This step will provide a detailed procedure for the sequencing and analysis of the amplified variable regions.
-
53.
Take 2nd PCR plates. If they are frozen, thaw on ice for 10 min.
-
54.
Directly load 5 μL of 2nd PCR product from each well onto 2% agarose gel with ethidium bromide.
Note: The presence of cresol red requires no other loading dye.
Alternatives: Many popular ethidium bromide alternatives, such as SYBR Safe (Thermo Fisher Scientific) or Gel Red (Biotium), could be used instead.
Note: After use, cover the remaining 2nd PCR plates with sterile aluminum sealing foil and put them back to 4°C for short-term storage.
-
55.
Run the gel.
Note: The expected size of PCR product is around 500 base pairs for heavy and light chains (Figure 2).
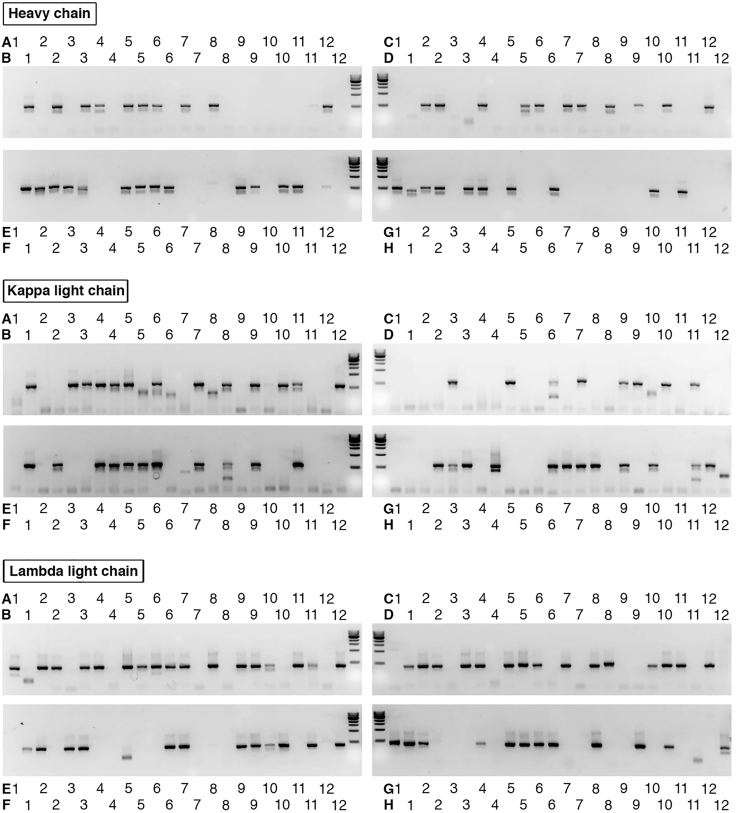
Figure 2.
Representative Figures of Gel Electrophoresis
Each band of immunoglobulin heavy, kappa, and lambda light chains is amplified from the sorted single B cells by two rounds of nested PCR reactions.
Note: If a multichannel pipette is used for sample loading, the sample order is as shown in Figure 2.
-
56.
Cut the gel bands or purify the PCR products for sequencing. Sequence the PCR product using the reverse primers for the 2nd PCR reactions.
Note: Sequencing primers: R2-HC (5′-GTTCGGGGAAGTAGTCCTTGAC) for heavy chain; R2-Kappa (5′-GTGCTGTCCTTGCTGTCCTGCT) for kappa light chain; and R2-Lambda (5′-CTCCTCACTCGAGGGYGGGAACAGAGTG) for lambda light chain.
-
57.
Use IMGT/V-QUEST (http://www.imgt.org/IMGT_vquest/vquest) (Brochet et al., 2008) or IGBLAST (https://www.ncbi.nlm.nih.gov/igblast/) (Ye et al., 2013) tools to analyze all the sequencing results of heavy and kappa/lambda light chains.
Note: The productivity of both amplified heavy and light chains will be determined, and the rearranged V-, D-, and J-genes will be assigned.
-
58.
Identify the antibodies with functional and paired heavy and light chain.
-
59.
Determine the antibody clonality. The antibodies with the same V and J allele assignments, the same CDR3 length, and ≥80% CDR3 identity are from an expanded clone.
-
60.
Choose the antibodies from clones for vector construction and antibody expression as previously described (von Boehmer et al., 2016).
Expected Outcomes
After the purification of CD19+ B cells from PBMCs, a small CD19+ cell pellet will be seen at the bottom of the collection tube.
The two-color-labeled HBsAg-binding B cells are single-cell sorted into individual wells of 96-well plates. Representative results highlight the gating strategy and expected results (Figure 1). Cells were first gated based on forward scatter area (FSC-A) and side scatter area (SSC-A), while the aggregated cells and debris are eliminated by FSC-A/H and SSC-A/W, respectively (Figure 1). IgG-Bv421+ CD20-PE-Cy7+ and ovalbumin-Alexa Fluor 488- cells are further selected. Double positive cells (HBsAg-PE+ and HBsAg-APC+) are selected based on the staining in negative controls (naïve donor and vaccinated donor but without anti-HBs antibody) (Figure 1).
After reverse transcription and nested PCR from single cells, the amplified products are examined by gel electrophoresis (Figure 2). The bands for heavy and kappa/lambda light chains (around 500 base pairs) are amplified from each individual well of 96-well plates (Figure 2).
Finally, the amplified V(D)J fragments are sequenced and analyzed to identify the antibodies with functional and naturally paired heavy and light chains. The details of how to analyze antibody sequences are not described in the current protocol, but details can be found in previous publications (Brochet et al., 2008; von Boehmer et al., 2016; Ye et al., 2013).
Limitations
Forward primers used during nested PCR are designed based on the leader sequence of V genes; therefore, no mutation on the V region would be introduced due to primer mismatch. However, the forward primer mixture, especially for immunoglobulin heavy chains, could not cover and amplify all V genes. Therefore, some of the wells with single B cells have no heavy chain V(D)J fragment amplified (Figure 2, Heavy chain).
Troubleshooting
Described below are some potential problems and recommendations for troubleshooting.
Problem 1
The percentage of all IgG+ memory B cells that bind to both PE- and APC-tagged HBsAg antigen proteins (HBsAg-PE+ and HBsAg-APC+) is too low (step 24).
Potential Solution
This donor might have low level of serum anti-HBs antibody and low level of HBsAg-binding memory B cells. The potential solution is to change to a different donor. More importantly, to decrease the background staining, viability dye could be used in the dump channel to gate out dead cells and debris.
Problem 2
All the wells have bands amplified after 2nd PCR reaction and their sizes on the 2% agarose gel are similar (step 55).
Potential Solution
This is likely due to contamination. Contamination by constructed antibody vectors in any reagents during sorting, reverse transcription, or nested PCR could lead to the amplification of bands in all wells. Potential solution is to exclude the possibly contaminated reagents, to thoroughly clean the working bench and DNA-free hood, and to restart the experiments.
Problem 3
There are very few or no bands amplified after the 2nd PCR reaction (step 55).
Potential Solution
If there is no band amplified, make sure that the single cells are successfully sorted into individual wells. If there are very few bands amplified, make sure that the sorted single B cells and the reverse-transcribed products are well stored in −80°C and avoid repeated freeze and thaw cycles.
Problem 4
The Sanger sequencing quality is too low for the amplified bands (step 56).
Potential Solution
This might due to a mixture of more than one amplified bands by PCR. Researchers could clone the bands into vector for sequencing.
Problem 5
Both kappa and lambda light chains are amplified from the same single cell (step 58).
Potential Solution
Analysis of sequencing results could reveal that one of the light chains is not functional due to the presence of stop codon or the shift of reading frame. It is also possible that both light chains are functional.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Qiao Wang (wangqiao@fudan.edu.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate/analyze new datasets/code.
Acknowledgments
This work was supported by National Natural Science Foundation of China 31872730 (to Q.W.) and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning TP2017010 (to Q.W.). The project was also supported in part by NIH grants 2U19AI111825-06 (to M.C.N.) and K08DK090576 (to Y.P.J.). Support was also provided by the Robertson Therapeutic Development Fund (to Q.W. and M.C.N.). M.C.N. is an HHMI Investigator.
Author Contributions
Y.Z., Z.L., and Z.W. wrote the protocol. Q.Z. and T.S. assisted with the protocol development. C.T.M. critically read and revised this protocol. M.C.N., Y.P.J., and Q.W. conceived the protocol.
Declaration of Interests
The authors declare no competing interests.
Contributor Information
Michel C. Nussenzweig, Email: nussen@mail.rockefeller.edu.
Ype P. de Jong, Email: ydj2001@med.cornell.edu.
Qiao Wang, Email: wangqiao@fudan.edu.cn.
References
- Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue) doi: 10.1093/nar/gkn316. W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer L., Liu C., Ackerman S., Gitlin A.D., Wang Q., Gazumyan A., Nussenzweig M.C. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- Wang Q., Michailidis E., Yu Y., Wang Z., Hurley A.M., Oren D.A., Mayer C.T., Gazumyan A., Liu Z., Zhou Y. A combination of human broadly neutralizing antibodies against hepatitis B virus HBsAg with distinct epitopes suppresses escape mutations. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41(Web Server issue):W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze new datasets/code.