Summary
This protocol offers a detailed procedure for the in vitro differentiation of human pluripotent stem cells (hPSCs) to multipotent hematopoietic progenitors that arise from SOX17+ hemogenic endothelium, mimicking intra-embryonic, HOXA-positive, aorta-gonad mesonephros (AGM) hematopoiesis. The generated endothelium displays transcriptional similarities to cells sorted from human 5-week AGM, and CD45+CD34+RUNX1C+ progenitors share an accessible chromatin profile with adult hematopoietic stem cells and multipotent progenitors. Therefore, this protocol is suitable for the mechanistic study of human multipotent progenitor development and for modeling childhood leukemias.
For complete details on the use and execution of this protocol, please refer to Nafria et al. (2020).
Graphical Abstract

For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.xpro.2020.100130.
Highlights
-
•
Protocol generating definitive hematopoiesis from human pluripotent stem cells
-
•
Mimics intra-embryonic hematopoiesis from HOXA+ SOX17+ hemogenic endothelium
-
•
Generates blood progenitors resembling adult HSCs and multipotent progenitors
-
•
Enables the study of human hematopoiesis under unperturbed and leukemic conditions
This protocol offers a detailed procedure for the in vitro differentiation of human pluripotent stem cells (hPSCs) to multipotent hematopoietic progenitors that arise from SOX17+ hemogenic endothelium, mimicking intra-embryonic, HOXA-positive, aorta-gonad mesonephros (AGM) hematopoiesis. The generated endothelium displays transcriptional similarities to cells sorted from human 5-week AGM, and CD45+CD34+RUNX1C+ progenitors share an accessible chromatin profile with adult hematopoietic stem cells and multipotent progenitors. Therefore, this protocol is suitable for the mechanistic study of human multipotent progenitor development and for modeling childhood leukemias.
Before You Begin
Preparation of Stock Solutions and Aliquots
Timing: up to 2 days
Prepare all stock and media solutions, following the guidelines and storage indications listed in the Materials and Equipment section. Refer to Key Resources Table for a complete list of reagents and materials.
Cell Culture of Human Pluripotent Stem Cells
Timing: 1–2 weeks
In this protocol, human pluripotent stem cells (hPSCs) – including human embryonic stem cells (hESCs) and human Induced pluripotent stem cells (hiPSCs) – are co-cultured with mitotically inactivated mouse embryonic fibroblasts (feeder cells). Cells are grown under antibiotic-free conditions using a KnockOutTM Serum Replacement (KOSR)-containing medium. This version of the differentiation protocol does not work reliably for hPSC cultures grown in feeder-free conditions and using albumin-free defined media.
-
1.
Pre-coat flasks with feeder medium for at least 8 h before plating any cells. It is useful to prepare a stock of coated flasks in advance and store them in the incubator at 37°C. Pre-coated flasks are used to directly plate feeder cells or to passage hPSCs prior to the start of differentiation. Pre-coating the flasks enhances the adhesion and survival of feeder cells.
-
2.
Plate feeder cells at a density of 0.5 x106 cells per T25 pre-coated flask and 1–1.5 x106 cells per T75 pre-coated flask. Feeder cells rapidly adhere to the flask and can be used to seed hPSCs within 30 min of plating.
-
3.
Culture hPSCs on feeder cells and in hPSC medium under 5% O2 in a 5% CO2 incubator at 37°C. Cells are cultured under hypoxic conditions, since a low oxygen concentration mimicking physiological levels increases lifespan, slows growth rate and reduces differentiation and stress responses, hence limiting DNA damage, genetic instability, and telomere shortening.
-
4.Passage hPSC cells 1:3–1:4 every 2 or 3 days (once the cells appear 90%–100% confluent) onto a new flask with feeder cells, by performing a 3-min incubation with sufficient volume of TrypLETM Select dissociation Enzyme to cover the flask surface (usually 0.5 mL per T25 flask and 1.5 mL per T75 flask). For passaging hiPSCs, viability is improved by performing a 3-min incubation with EDTA solution, using sufficient volume to cover the flask surface. This preferentially dissociates the culture into small clumps rather than to single cells. Pellet the harvested cells by centrifugation, resuspend the cells in culture medium and proceed to plating:
-
a.Remove the feeder medium from the flask with feeder cells, once feeder cells have adhered to the surface.
-
b.Wash once with PBS.
-
c.Plate 1/4 - 1/3 of the harvested hPSCs onto the flask of the same surface area containing feeder cells. Culture the cells in 5 mL of hPSC medium per T25 flask and in 15 mL medium per T75 flask.
-
d.Perform a complete medium change every day.
-
a.
CRITICAL: Make sure cell lines have been passaged for a week before starting any differentiation.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-human CD38 | BD Pharmingen | Cat # 555462; RRID: AB_398599 |
| APC anti-human CD90 | BD Pharmingen | Cat # 559869; RRID: AB_398677 |
| BV-421 anti-human CD34 | BioLegend | Cat # 343609; RRID: AB_11147951 |
| BV-421 anti-human CD43 | BD Horizon | Cat # 562916; RRID: AB_2737890 |
| BV-421 anti-human CD45 | BioLegend | Cat # 304032; RRID: AB_2561357 |
| BV-421 anti-human CD90 | BioLegend | Cat # 328121; RRID: AB_10933261 |
| PeCy7 anti-human CD16 | BioLegend | Cat # 302015; RRID: AB_314215 |
| PeCy7 anti-human CD34 | BioLegend | Cat # 343516; RRID: AB_1877251 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| α-MTG (α-Monothioglycerol) | Merck | Cat # M6145 |
| 2-Mercaptoethanol | Thermo Fisher Scientific | Cat # 21985-023 |
| AA2P (Ascorbic acid 2-phosphate) | Merck | Cat # A8960 |
| Accutase® solution | Merk | Cat # A6964 |
| Activin A | R&D Systems | Cat # 338-AC |
| BMP4 (Bone morphogenetic protein 4) | R&D Systems | Cat # 314-BP |
| Bovostar acid-stripped Bovine Serum Albumin | Bovogen, Australia (12 Williams Ave, Vic 3033) | Cat # BSAS 0.1 |
| CHIR99021 | Tocris Biosciences | Cat # 4423 |
| Collagenase Type 4 | Worthington | Cat # CLS-4 |
| Distilled water | Life Technologies | Cat # 15230-162 |
| DMEM/F12 (DMEM Nutrient Mixture F-12 1× + L-glutamine Na bicarbonate) | Thermo Fisher Scientific | Cat # 11320-033 |
| EDTA (Ethylenediaminetetraacetic acid), 0.5 M, UltraPure, pH 8.0 | Thermo Fisher Scientific | Cat # 15575020 |
| EtOH (Ethanol), 100%, AnalaR/Molecular biology grade | Merck | Cat # E7023 |
| FBS (Fetal Bovine Serum), Gibco™, qualified, heat inactivated, EU-approved, South America Origin | Thermo Fisher Scientific | Cat # 11550356 |
| FGF2 (Fibroblast Growth Factor) | PeproTech | Cat # 100-18B |
| FLT3 (FMS-like tyrosine kinase 3 receptor) ligand | PeproTech | Cat # 300-19 |
| GMAX (GlutaMAX™) 100× | Thermo Fisher Scientific | Cat # 35050-061 |
| Ham’s F12 | Thermo Fisher Scientific | Cat # 11765-062 |
| IGF2 (Insulin-like growth factor 2) | PeproTech | Cat # 100-12 |
| IL-3 (Interleukin 3) | PeproTech | Cat # 200-03 |
| IL-6 (Interleukin 6) | PeproTech | Cat # 200-06 |
| IMDM (Iscove's Modified Dulbecco's Media) with no phenol red | Themo Fisher Scientific | Cat # 21056-023 |
| ITS-E (Insulin-Transferrin-Selenium-E) | In Vitria | Cat # 777ITS092 |
| KOSR (KnockOutTM Serum Replacement) | Thermo Fisher Scientific | Cat # 10828028 |
| L-AA (L-Ascorbic acid) | Merck | Cat # A4403 |
| Linoleic acid | Merck | Cat # L2376 |
| Linolenic acid | Merck | Cat # L1376 |
| Matrigel® Growth Factor Reduced phenol red- free | In Vitro Technologies | Cat # FAL356231 |
| NaCl (Sodium chloride) | Merk | Cat # S5886 |
| NEAA (Non-essential amino acids) 100× | Thermo Fisher Scientific | Cat # 11140-050 |
| OsrHSA (Recombinant human serum albumin derived from Oryza sativa rice grain) | ScienCell Research Labs, Carlsbad, CA | Cat # OsrHSA |
| PBS (Phosphate Buffered Saline), no calcium, no magnesium | Thermo Fisher Scientific | Cat # 14190144 |
| Pen/Strep (Penicillin/Streptomycin) | Thermo Fisher Scientific | Cat # 15140122 |
| PFHMII (Protein Free Hybridoma Medium II) | Thermo Fisher Scientific | Cat # 12040077 |
| Polysorbate 80 (Tween 80) | Merck | Cat # P1754 |
| PVA (Polyvinyl alcohol) | Merck | Cat # P8136 |
| SB431542 | Sapphire Bioscience | Cat # 13031 |
| Soybean Oil / lecithin | Merck | Cat # S7381 |
| SCF (Stem cell factor) | PeproTech | Cat # 300-07 |
| SyntheChol | Merck | Cat # S5442 |
| TPO (Thrombopoietin) | PeproTech | Cat # 300-18 |
| TrypLETM Select dissociation Enzyme | Thermo Fisher Scientific | Cat # 12563011 |
| VEGF (Vascular endothelial growth factor) | PeproTech | Cat # 100-20 |
| Experimental Models: Cell Lines | ||
| SOX17mCHERRY/wRUNX1CGFP/w hESC H9 line | Ng et al., 2016 | N/A |
| Inducible RUNX1-ETO SOX17mCHERRY/wRUNX1CGFP/w hESC H9 line | Nafria et al., 2020 | N/A |
| Other | ||
| 18G needle × 1.5 inches, Terumo® AGANITM | Medisave | Cat # AN-1838R |
| 21G needle × 2 inches, Terumo® AGANITM | Medisave | Cat # AN-2150R |
| 23G needle × 1.25 inches, Terumo® AGANITM | Medisave | Cat # AN-2332R |
| 25G needle × 1 inch, Terumo® AGANITM | Medisave | Cat # AN-2525R |
| 5 mL round-bottom tubes for flow cytometry, Polypropylene, FalconTM REF 352002 | Thermo Fisher Scientific | Cat# 10519901 |
| 50 μm strainer filter, Sysmex CellTricksTM | Wolflabs | Cat # 04-004-2327 |
| 70 μm sterile strainer cap, Greiner Bio-One EASYstrainer | Kisker Biotech | Cat # 542070 |
| Low attachment round-bottomed 96-well plates (Sterile) | Costar | Cat # COR3788 |
| Sterile reagent reservoir for multichannel pipettes | StarLab | Cat # S4026-5806 |
| T25: Corning® cell culture flasks, surface area 25 cm2, canted neck, cap (vented) | Merck | Cat # 430639 |
| T75: Corning® cell culture flasks, surface area 75 cm2, canted neck, cap (vented) | Merck | Cat # 430641U |
| Tissue culture treated 6-well plates | Merck | Cat # CLS3516 |
| Vacuum Filtration system, Corning® 500 mL, 0.22 μm Pore 33.2 cm2 PES Membrane, Sterile | Merck | Cat # 431097 |
| Variable oxygen control CO2 incubator: Forma™ Series 3 Water Jacketed | Thermo Fisher Scientific | Cat # 4140TS |
Materials and Equipment
Equipment and Basic Materials
For this protocol it will be preferable to have access to an incubator in which the O2 concentration can be controlled, because hPSC cultures are better maintained in an undifferentiated state at ~5% O2. Differentiating hPSC cultures are maintained under normoxic condition in a standard CO2 incubator. Other essential materials include:
-
•
Sterile round-bottom low attachment 96-well plates, for the setup of the spin embryoid bodies.
-
•
Standard tissue culture treated flasks and 6-well plates.
-
•
Multichannel pipette of 8 or 12 rows (20–200 μL). It is important these pipettes are well calibrated and work smoothly.
-
•
Filter cup units and a suction system.
Frozen Stock Solutions
Working stock solutions can be stored at −20°C as single use aliquots containing the corresponding volume to be used in differentiation media, thus avoiding freeze-thaw cycles.
Note: Aliquot volumes described in this protocol are sufficient to prepare a final volume of 200 mL of media.
α-Monothioglycerol (α-MTG)
| Reagent | Final Concentration | Volume |
|---|---|---|
| α-MTG (11.3 M stock) | 150 mM | 130 μL of stock |
| IMDM or Ham’s F12 | n/a | 10 mL |
| Total | n/a | 10 mL |
Note: Mix well by pipetting, as α-MTG is viscous and denser than the media, and aliquot into volumes of 620 μL.
Ascorbic Acid 2-phosphate (AA2P)
| Reagent | Final Concentration | Mass/Volume |
|---|---|---|
| AA2P | 10 mg/mL | 500 mg |
| Distilled water | n/a | 50 mL |
| Total | n/a | 50 mL |
Note: Shake well and let sit for 30 min at room temperature (20°C–25°C) to dissolve. Do not heat. Aliquot into 1 mL volumes.
L-Ascorbic acid (L-AA)
| Reagent | Final Concentration | Mass/Volume |
|---|---|---|
| L-AA | 10 mg/mL | 500 mg |
| Distilled water | n/a | 50 mL |
| Total | n/a | 50 mL |
Note: Shake well and let sit for 30 min at room temperature (20°C–25°C) to dissolve. Do not heat. Aliquot into 1 mL volumes.
Linoleic and Linolenic Acids and Soybean Oil Mix
| Reagent | Final Concentration | Mass/Volume |
|---|---|---|
| Linoleic acid | 1 mg/mL | 10 mg (12 μL of stock) |
| Linolenic acid | 1 mg/mL | 10 mg (12 μL of stock) |
| Soybean Oil | 20 mg/mL | 200 mg (200 μL of stock) |
| EtOH 100% | n/a | 10 mL |
| Total | n/a | 10 mL |
-
•
Prepare a mixture of 500 μL of pure linolenic acid and 500 μL pure linoleic acid oils. Store at −20°C for future use.
-
•
Add 25 μL Linoleic/Linolenic oil pre-mix and 200 μL pure Soybean oil into a tube with 10 mL pure ethanol (EtOH).
-
•
Disperse the oil droplets by using a vortex and invert the tube several times, until the oil is fully dissolved.
Note: The soybean oil is very dense and may sink to the bottom of the tube. Make sure it is fully dissolved.
-
•
Aliquot into 1 mL volumes and store at −20°C. Both the Linoleic/Linolenic oil pre-mix and the Linoleic/Linolenic/Soybean oil pre-mix can be subjected to several freeze-thaw cycles. Aliquots are also stable at 4°C for several months.
Other Stock Solutions
Albumin and other described stock solutions in this section can be stored at 4°C for several months.
EDTA Dissociation Solution
| Reagent Stocks | Final Concentration | Volume |
|---|---|---|
| PBS (Ca2+ Mg2+ free) | n/a | 496.5 mL |
| EDTA (in dH2O ultrapure, 0.5 M) | 0.5 mM | 0.5 mL |
| NaCl (5 M) | 30 mM | 3 mL |
| Total | n/a | 500 mL |
-
•
Filter sterilize and store at 4°C.
10% Bovostar Bovine Serum Albumin
| Reagent Stocks | Final Concentration | Mass/Volume |
|---|---|---|
| Bovostar Bovine Serum Albumin | 10% | 10 g |
| Distilled water | n/a | 98 mL |
| NaCl (5 M) | 0.1 M | 2 mL of stock |
| Polysorbate 80 (10 mg/mL) | 3.2 μg/mL | 32 μL of stock |
| Total | n/a | 100 mL |
-
•
Add 10 g Bovostar and 98 mL sterile distilled water into a sterile bottle and place at 4°C to dissolve.
Note: It might take up to 2 days to fully dissolve.
-
•
Add the NaCl and polysorbate 80 (Tween 80) and swirl the bottle to mix.
-
•
Filter sterilize using a vacuum filtration system.
10% OsrHSA Albumin (Recombinant Human Serum Albumin Derived from Oryza sativa Rice Grain)
| Reagent Stock | Final Concentration | Mass/Volume |
|---|---|---|
| OsrHSA | 10% | 10 g |
| Distilled water | n/a | 98 mL |
| NaCl (5 M) | 0.1 M | 2 mL of stock |
| Polysorbate 80 (10 mg/mL) | 3.2 μg/mL | 32 μL of stock |
| Total | n/a | 100 mL |
-
•
Transfer the 10 g OsrHSA into a sterile bottle. Rinse the container with 50 mL distilled water and leave it to dissolve for 30 min to collect the residual albumin remaining in the container.
-
•
Transfer the 50 mL into the bottle and add 48 mL distilled water, to give a final volume of 98 mL.
Note: Do not shake the bottle, let it sit overnight (12–18 h) at 4°C to dissolve.
-
•
Add the NaCl and polysorbate 80 (Tween 80) and swirl the bottle to mix.
-
•
Filter sterilize using a filtration cup unit.
10 % Polyvinyl alcohol (PVA)
| Reagent | Final Concentration | Mass/Volume |
|---|---|---|
| PVA powder | 10% | 10 g |
| Distilled water | n/a | 100 mL |
| Total | n/a | 100 mL |
-
•
Place 100 mL distilled water into a sterile glass container.
Note: Use a flask with a broad base, such an Erlenmeyer flask.
-
•
Gradually sprinkle the PVA directly over the water surface, taking care not to touch the neck of the flask.
CRITICAL: Add the PVA slowly, allowing the white powder to become translucent before adding more.
-
•
Place the flask at 4°C overnight (12–18 h), allowing the PVA to fully dissolve.
Note: If any PVA lumps are still present after 24 h, break them down by vigorously shaking the flask and place again at 4°C to allow the contents to settle.
-
•
Heat the PVA solution in a microwave, bringing the solution to boil and simmering for 1 min.
-
•
Allow to cool to room temperature (20°C–25°C) before storing at 4°C.
Basal Media Solutions with 0.06% PVA
| Reagent | Final Concentration | Volume |
|---|---|---|
| IMDM (phenol red free) | n/a | 497 mL |
| 10% PVA stock | 0.06% | 3 mL of stock |
| Total | n/a | 500 mL |
| Reagent | Final Concentration | Volume |
|---|---|---|
| Ham’s F12 | n/a | 497 mL |
| 10% PVA stock | 0.06% | 3 mL of stock |
| Total | n/a | 500 mL |
-
•
Add 3 mL of the 10% PVA stock solution into each bottle of basal medium.
-
•
Cap the bottle and shake vigorously for 1 min, until very frothy.
-
•
Let the bottles sit inside the tissue culture hood for a couple of hours, or until the froth subsides, before storing at 4°C.
Matrigel Solution
| Reagent | Final Concentration | Volume |
|---|---|---|
| IMDM (phenol red free) | n/a | 49.5 mL |
| Matrigel® Growth Factor Reduced, phenol red free |
1:200 dilution of stock | 250 μL of stock |
| Penicillin-Streptomycin (5,000 U/mL) (Pen/Strep) | 1:200 of stock | 250 μL of stock |
| Total | n/a | 50 mL |
Note: Prepare this from a freshly thawed aliquot of frozen concentrated Matrigel® stock. It can be stored in the refrigerator at 4°C for 1–2 weeks. Keep the thawed stock Matrigel® and the Matrigel solution on ice to prevent premature gelling.
Cell Culture Media Solutions
All media solutions need to be filter-sterilized and can be stored at −20°C if not immediately required.
Feeder Medium
| Reagent | Final Concentration | Volume |
|---|---|---|
| DMEM/F12 | 1× | 439 mL |
| Fetal Bovine Serum (FBS) | 10% | 50 mL |
| Non-Essential Amino Acids (NEAA) 100X | 1× | 5 mL of stock |
| GlutaMAX™ (GMAX) 200 mM | 2 mM | 5 mL of stock |
| 2-Mercaptoethanol 55 mM | 0.11 mM | 1 mL of stock |
| Total | n/a | 500 mL |
hESC Medium
| Reagent | Final Concentration | Volume |
|---|---|---|
| DMEM/F12 | 1× | 194.5 mL |
| KOSR | 20% | 50 mL |
| NEAA 100× | 1× | 2.5 mL of stock |
| GMAX 200 mM | 2 mM | 2.5 mL of stock |
| 2- Mercaptoethanol 55 mM | 0.11 mM | 500 μL of stock |
| FGF2 100 ng/μL | 10 ng/mL | 25 μL of stock |
| Total | N/A | 250 mL |
Note: For hiPSC medium, use 50 ng/mL FGF2 (125 μL for 250 mL final volume of medium).
CRITICAL: Do not heat inactivate KOSR, as a serum substitute it does not contain complement. Store the KOSR in frozen aliquots of 50 mL to avoid more than two freeze-thaw cycles.
STAPEL Differentiation Medium
STAPEL medium (Ng et al., 2008, 2016) is the base medium solution used throughout the differentiation protocol. Growth factors are added fresh to the media at different stages. The composition shown below includes a couple of minor modifications to the originally published APEL medium.
| Reagent | Stock Concentration | Final Concentration | Volume |
|---|---|---|---|
| IMDM/0.06% PVA | 1× | ~ 0.05% PVA | 90 mL |
| Ham’s F12/0.06% PVA | 1× | 90 mL | |
| OsrHSA | 10% | 0.5% | 5 mL |
| Bovostar | 10% | 0.5% | 5 mL |
| Protein Free Hybridoma Medium II (PFHMII) | 1× | 5% | 10 mL |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-E) | 100× | 1× | 2 mL |
| GMAX | 200 mM | 2 mM | 2 mL |
| AA2P | 10 mg/mL | 50 μg/mL | 1 mL |
| L-AA | 10 mg/mL | 50 μg/mL | 1 mL |
| α-MTG | 150 mM | 450 μM | 600 μL |
| Linoleic/Linolenic/Soybean oil pre-mix | L-L acids: 1 mg/mL Soybean oil: 20 mg/mL |
125 ng/mL | 50 μL |
| SyntheChol | 16 mg/mL | 4 μg/mL | 50 μL |
| TOTAL | N/A | N/A | 200 mL |
Note: IMDM/PVA and Ham’s F12/PVA are added first to the filtration cup and slightly in excess (3 mL extra of each media, included in the Volume in the table above). The volume calculated accounts for a loss of ~5 mL during filtration due to the volume of liquid required to wet the membrane.
Alternative sources of albumin: Different sources of albumin might be used in place of the ones listed in this protocol (OsrHSA and Bovostar). This will depend on the local availability and may be cell line dependent. We have found that some forms of albumin are associated with cellular toxicity, so this reagent needs to be tested.
Growth Factor Combinations for STAPEL Differentiation Medium
Reconstitute growth factors at a concentration of 100 ng/μL and small molecule inhibitors – the activin inhibitor SB431542 (SB) and the WNT agonist CHIR99021 (CHIR) – at 10 mM, following the manufacturers’ guidelines. Aliquot in small volumes to reduce freeze-thaw cycles.
CRITICAL: Store aliquoted growth factor stocks at −20°C and do not keep working stocks at 4°C for longer than a week. Growth factors are best added fresh to the media, but solutions can be kept at 4°C for a week or can be frozen for future use without apparent loss of activity.
Note: The indicated volumes of media with added growth factors are sufficient for a differentiation of ten 96-well plates using the central 60 wells of each plate.
Stage 1 STAPEL: BVSAF + 0.5 CHIR
| Growth Factor | Final Concentration | Volume |
|---|---|---|
| STAPEL medium | n/a | 50 mL |
| CHIR99021 10 mM | 0.5 μM | 2 μL |
| BMP4 100 ng/μL | 20 ng/mL | 10 μL |
| VEGF 100 ng/μL | 25 ng/mL | 12.5 μL |
| SCF 100 ng/μL | 25 ng/mL | 12.5 μL |
| Activin A 100 ng/μL | 7.5 ng/mL | 3.75 μL |
| FGF2 100 ng/μL | 10 ng/mL | 5 μL |
| Total | n/a | 50 mL |
Note: A volume of 80 μL STAPEL is added per well, thus 4.8 mL/plate (using the inner 60 wells only) is required. A volume of 50 mL is sufficient for ten 96-well plates.
STAPEL SB/CHIR
| Small Molecule Inhibitor | Final Concentration | Volume per Plate | Total Volume |
|---|---|---|---|
| STAPEL medium | n/a | 1.2 mL | 13 mL |
| SB431542 (SB) 10 mM | 3.8 μM | 2.28 μL | 24.7 μL |
| CHIR99021 (CHIR) 10 mM | 3 μM | 1.8 μL | 19.5 μL |
| Total | n/a | n/a | 13 mL |
Note: The volume per plate allows for 20 μL of STAPEL SB/CHIR medium added to 80 μL STAPEL BVSAF already present in the well, making a final volume of 100 μL per well of a 96-well plate (using 60 wells/plate only). Therefore, STAPEL SB/CHIR medium is prepared at a higher concentration to achieve the appropriate final concentration in each well. A volume of 13 mL STAPEL SB/CHIR will be enough (in slight excess) for ten 96-well plates.
Stage 2 STAPEL: BVSIF
| Growth factor | Final Concentration | Volume |
|---|---|---|
| STAPEL media | n/a | 65 mL |
| BMP4 100 ng/μL | 20 ng/mL | 13 μL |
| VEGF 100 ng/μL | 50 ng/mL | 32.5 μL |
| SCF 100 ng/μL | 50 ng/mL | 32.5 μL |
| IGF2 100 ng/μL | 10 ng/mL | 6.5 μL |
| FGF2 100 ng/μL | 10 ng/mL | 6.5 μL |
| Total | n/a | 65 mL |
Note: A volume of 100 μL STAPEL BVSIF is added per well, thus 6 mL/plate of 60 wells is required. A volume of 65 mL is sufficient (in excess) for ten 96-well plates.
Stage 3
| Growth Factor | Final Concentration | Volume |
|---|---|---|
| STAPEL media | n/a | 100 mL |
| BMP4 100 ng/μL | 10 ng/mL | 10 μL |
| SCF 100 ng/μL | 100 ng/mL | 100 μL |
| FLT3 100 ng/μL | 100 ng/mL | 100 μL |
| TPO 100 ng/μL | 50 ng/mL | 50 μL |
| VEGF 100 ng/μL | 50 ng/mL | 50 μL |
| IL-6 100 ng/μL | 25 ng/mL | 25 μL |
| IL-3 100 ng/μL | 25 ng/mL | 25 μL |
| IGF2 100 ng/μL | 10 ng/mL | 10 μL |
| FGF2 100 ng/μL | 10 ng/mL | 10 μL |
| Pen/Strep (optional) | 1:200 | 500 μL |
| Total | n/a | 100 mL |
Note: This growth factor mix will allow proper development of stromal layers and vasculature prior to the emergence of blood progenitors. However, other combinations and concentrations might be suitable, depending on cell lines and experimental questions. A volume of 100 mL is prepared initially, as this medium will be used to refresh the adherent cultures in 6-well plates by performing half medium changes of 2 mL every two days.
Stage 3 Specific
| Growth Factor | Final Concentration (ng/mL) | Volume |
|---|---|---|
| STAPEL media | n/a | 100 mL |
| SCF 100 ng/μL | 100 ng/mL | 100 μL |
| FLT3 100 ng/μL | 100 ng/mL | 100 μL |
| TPO 100 ng/μL | 50 ng/mL | 50 μL |
| IL-6 100 ng/μL | 25 ng/mL | 25 μL |
| IL-3 100 ng/μL | 25 ng/mL | 25 μL |
| Pen/Strep (optional) | 1:200 | 500 μL |
| Total | n/a | 100 mL |
Alternatives: At this step, growth factors used will depend on the cell line and the purpose of the experiments. These growth factors will favor stem cell and myeloid differentiation.
Step-By-Step Method Details
This protocol mainly consists of three major Stages, which are defined by the combination of growth factors used in the mediaum. Stage 1 (days 0–4) involves the setup of the spin Embryoid Bodies (EBs), after a feeder-depletion step. A critical step of this protocol is addition of SB and CHIR at day 1.7, which patterns the mesoderm toward the definitive program. During Stage 2 (days 4–8), SB and CHIR are removed from the cultures and the hematopoietic mesoderm within the EBs develops to endothelium. Stage 3 marks the start of the adherent culture, which results in the growth of stroma and hemogenic endothelium and the subsequent generation of hematopoietic progenitor cells from day 14 onwards. Harvesting of hematopoietic progenitors can be carried out at different time points from day 16 up until day 28, depending on the cell fractions of interest. A comprehensive overview of the protocol is shown as the graphical abstract.
Depletion of Feeder Cells from hPSC Cultures
Timing: 30 min (+ at least 2 h of incubation)
The concentration of mouse feeder cells needs to be reduced in the hPSC cultures, which have been cultured on feeders in T75 flasks, before starting the differentiation protocol. Passaging hPSCs onto feeder medium-coated flasks is performed at least 2 h before the differentiation start point. This step also ensures that all the hPSCs used for the differentiation are growing and healthy, as dying cells will not survive the dissociation. This helps to synchronize differentiation kinetics of the cells.
-
1.
Harvest hPSCs using TrypLETM Select for hESCs and EDTA based dissociation medium for hiPSCs, adding a sufficient volume of the reagent to cover the flask surface (usually 0.5 mL per T25 flask and 1.5 mL per T75 flask).
-
2.
Before plating the cells, remove feeder medium from the pre-coated T75 flask (or flask of the same surface area, pre-coated with feeder medium for at least 8 h), rinse with PBS, and add 15 mL of hESC or hiPSC media.
-
3.
Pellet cells and re-plate at a 1:1 ratio.
-
4.
Incubate in low O2 conditions. Allow at least 2 h for the cells to re-attach.
Note: Cells from a confluent (95%–100%) T75 flask are used to set up ten 96-well plates with 60 EBs/plate, which corresponds to approximately 5,000–6,000 input cells/well. Scale the number of 96-well plates depending on the size of the starting cell culture flask. Using this guide avoids the need for cell counting, which increases the speed of the differentiation setup. We find that this yields a higher proportion of viable cells per EB (Troubleshooting 1).
Stage 1 (Day 0): Setup of the Spin Embryoid Bodies (EBs)
Timing: 1 h
This step describes the generation of EBs using a spin-based method. Briefly, undifferentiated hPSCs (following feeder depletion) are aggregated by centrifugation onto round-bottom, low attachment 96-well plates, resulting in the formation of one EB per well (Figure 1).
-
5.
Using a multichannel pipette, add 70 μL sterile distilled water to the outer 30 wells of ten 96-well plates. This step reduces evaporation of medium from the inner 60 wells that will contain the EBs.
-
6.
Prepare 50 mL Stage 1 differentiation media with BVSAF cytokines.
-
7.
Harvest cells from the T75 flask (95%–100% confluent) using Accutase® solution, by 3-min incubation at 37°C.
CRITICAL: The use of Accutase® instead of TrypLETM Select reduces cell death during the harvesting step, and therefore yields better EB formation and overall differentiation.
-
8.
Transfer the cell pellet into the 50 mL Stage 1 differentiation media. Rinse out the tube with differentiation medium to collect all the cells.
-
9.
Mix gently and transfer to a sterile reagent reservoir.
-
10.
Using a multichannel pipette, aliquot 80 μL per well of the cell-medium mixture, using only the inner 60 wells of the ten low attachment 96-well plates previously prepared.
-
11.
Aggregate the cells at the bottom of each well by centrifugation (2 min at 4°C at 400 × g). After centrifugation, the cell aggregate should be visible at the bottom of each well.
-
12.
Place the plates into a 5% CO2 incubator under normoxic conditions at 37°C.
Note: Note when the plates are placed to the incubator as the differentiation initiation time. This is required for subsequent steps.
-
13.
On day 1 of differentiation, evaluate EB formation under the microscope: ideally, the single cells have aggregated and condensed within the wells to form spheres with an “edge” or “rind,” with little associated cell death at their periphery (Figure 2).
Figure 1.
Generation of Intra-embryonic Hematopoietic Progenitors from hPSCs Using a Spin-EB Method
The timeline of the protocol is shown at the top of the figure. In order to set up the spin-EB culture, single-cell dissociated hPSCs are plated in non-adherent round-bottom 96-well plates and are aggregated at the bottom of the wells by centrifugation, yielding 60 EBs/plate. EBs are cultured in Stage 1 medium for 4 days. At day 1.7, EBs are treated with an Activin inhibitor (SB) and WNT-agonist (CHIR) to pattern the mesoderm toward the intra-embryonic, HOXA-positive hematopoietic program, also inhibiting the generation of extra-embryonic blood progenitors. At day 4, medium containing SB/CHIR is removed using a multichannel pipette, placed in an angle to not disturb the non-adherent EBs present at the bottom of the wells. EBs are subsequently cultured in Stage 2 growth factors that promote endothelial development until day 8, when EBs are transferred to adherent wells in Stage 3 media to allow the outgrowth of stromal and endothelial cells. EB cultures generate CD34+ RUNC1C+ blood progenitors that arise from cell clusters located within arterial structures of SOX17+ hemogenic endothelium. Hematopoietic progenitors detach from the endothelium and continue maturing in suspension. To avoid removal of blood progenitors in suspension, half medium changes are performed from day 14, by tilting the pipette and removing only the upper 2 mL of medium and then adding 2 mL of fresh medium. Blood progenitors and EBs can be harvested at different stages of the differentiation and subjected to a range of analyses, including gene and protein expression, chromatin accessibility and immunoprecipitation, and growth and differentiation assays.
Figure 2.
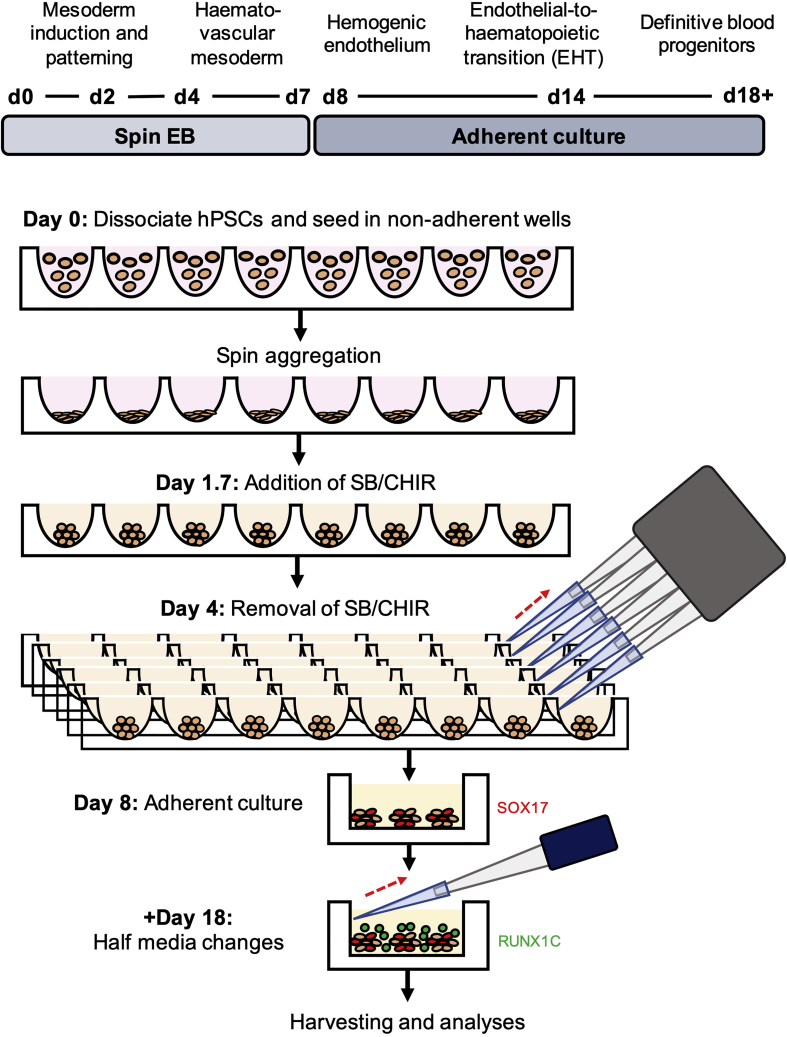
Non-adherent EBs at Day 1 of Differentiation
EBs are formed 1 day after the spin-EB setup and appear as dense spheres with a clearly visible border around them. EBs should present with very little associated cell death, observed as debris surrounding the EB. Scale bars represent 100 μm.
Stage 1 (Day 1.7): Addition of SB/CHIR
Timing: 30 min
This step describes the addition of an Activin inhibitor (SB431542) (Kennedy et al., 2012) and WNT agonist (CHIR99021) (Sturgeon et al., 2014), which, together, pattern the mesoderm toward the definitive hematopoietic program (Ng et al., 2016). These two small molecules are added approximately 6 h before reaching the 48 h time point from the spin EB plate setup (approximately day 1.7 of differentiation).
-
14.
Prepare a mixture of STAPEL medium with added SB and CHIR to make the following final concentrations per well: SB431542 at 3.5 μM and CHIR99021 at 3 μM (see materials and methods for calculations).
-
15.
Place this mixture in a sterile reagent reservoir.
-
16.
Using a multichannel pipette, add 20 μL of the STAPEL + SB/CHIR mixture into each well of the spin-EBs plates, giving a final volume of 100 μL/well.
-
17.
Place the plates back into the incubator.
CRITICAL: The timing of SB/CHIR may vary slightly with the rate of differentiation of the cell line used. Adding it too early may inhibit mesoderm formation, too late may not adequately suppress extra-embryonic, yolk sac-type hematopoiesis. With the cell lines used in this manuscript, the optimal time of SB/CHIR addition was approximately 42 h after the onset of differentiation. This protocol works for both unmodified and genetically modified hPSCs, but the timing of SB/CHIR might need to be optimized within a 4–6 h window around this time of addition.
Stage 2 (Day 4): Removal of SB/CHIR
Timing: 1 h
At day 4 of the differentiation, the inhibitors are removed from the EB plates and replaced with media containing a mix of growth factors to support development of the hematopoietic mesoderm to endothelium.
-
18.
Check under the microscope that EBs appear round with defined sharp edges (Figure 3).
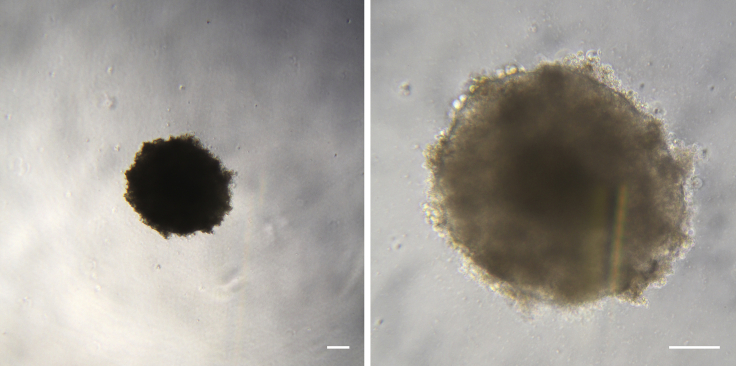
Figure 3.
Non-adherent EBs at Day 4 of Differentiation
At day 4, after a 2-day treatment with SB431542 and CHIR9902, EBs appear homogeneous, they have increased in size and display a very clearly defined edge. Scale bars represent 100 μm.
-
19.
Prepare Stage 2 differentiation mixture, consisting of STAPEL media with BVSIF cytokines. Approximately, 6 mL of medium will be needed per plate.
-
20.
Using a multichannel pipette, remove as much Stage 1 medium as possible (70–80 μL) from each well, without disturbing the EBs. For that, tilt the tips so the tips do not touch the bottom of the well and pipette up media slowly (Figure 1, day 4).
Note: Place the removed medium in a sterile reservoir, do not discard immediately. Change pipette tips if doing more than ten plates and/or different cell lines, to avoid contamination.
-
21.
Holding the plates against the light, check that no EBs are missing. Any removed EBs might be in the medium discarded in the sterile reservoir and can be placed back to their respective wells.
-
22.
Place the Stage 2 medium into a clean sterile reagent reservoir and, using a multichannel pipette, add 100 μL medium per well.
-
23.
Place the plates back into the incubator and leave until day 7 of differentiation.
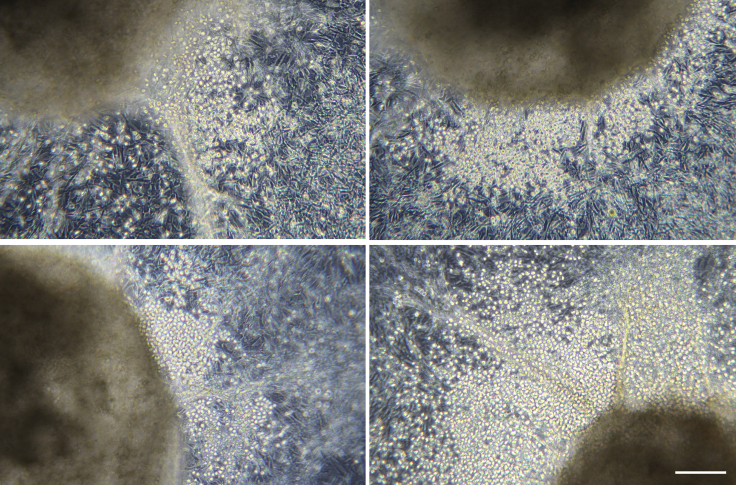
Stage 2 (Day 7): Flow Cytometry Checkpoint
Timing: 1 h
At day 7 of differentiation, mesoderm patterning and differentiation kinetics are evaluated by flow cytometric analysis, before starting the adherent phase of cell culture. This step is important to ensure that the CD34+ endothelium has formed and that the extra-embryonic program has been successfully inhibited.
-
24.
Check under the microscope that EBs display a sharp edge without notable amount of cell growth around them. They may display a small number of protruding cells, as the EB tries to attach to the bottom of the well (Figure 4).
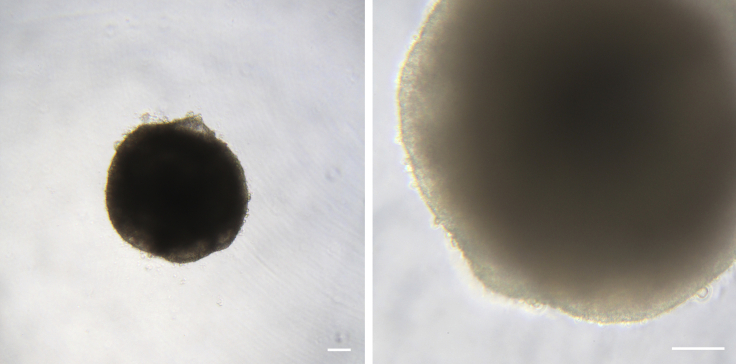
Figure 4.
Example of Non-adherent EBs at Day 7 of Differentiation
At day 7, EBs should appear round and dense and the edge should be very well defined. Some EBs might display a small number of protruding cells, as the EB tries to attach to the surface of the well. Scale bars represent 100 μm.
-
25.Check cell surface marker expression using flow cytometry in a sample of the EBs:
-
a.Collect 10–20 EBs in a tube and let them settle by gravity. This usually takes approximately 2 min. Alternatively, pellet the EBs by centrifugation at 400 × g for 2 min.
-
b.Carefully remove the supernatant and incubate the EBs in 1 mL TrypLETM Select for 30 min in a water bath at 37°C.
-
c.Dissociate the EBs by passing them through a 23G needle 10 times, or until no clumps remain in the suspension. The EBs should break up readily if the incubation period with TrypLETM Select has been sufficient.
-
d.Filter the cell suspensions through a 50 μm strainer filter for sample preparations into a 5-mL round-bottom tube used for flow cytometry.
-
e.Wash the cells with FACS buffer (phosphate buffered saline (PBS) with 2% FBS) and pellet by centrifugation.
-
f.Remove the supernatant and resuspend the cell pellet in 60 μL FACS buffer. Transfer 10 μL into a fresh tube to use as an unstained or isotype control and stain 50 μL with the following antibodies and dilutions: CD90-APC (1:100), CD34-PeCy7 (1:100), CD43-BV (1:50).
-
g.Incubate for 15 min at 4°C in a covered ice bucket or in the fridge.
-
h.To remove unbound antibodies, wash the tubes twice by adding 2 mL of FACS buffer and pelleting by centrifugation.
-
i.Resuspend in 300 μL PBS prior to analysis in the flow cytometer.
-
a.
Expected outcome: At this time point, almost all the cells (>90%) should express CD90, around 20%–40% of the cells should express CD34 and very few of the cells (<3%–5%) should display CD43. The presence of cells that express the earliest blood cell marker, CD43, suggests that the extra-embryonic hematopoietic program has not been adequately suppressed (Troubleshooting 2).
-
26.
On day 7, coat tissue culture treated 6-well plates with 2 mL of Matrigel Solution and place into a 5% CO2 incubator under normoxic conditions at 37°C overnight (12–18 h) or for a minimum of 60 min. The EBs attach better when this step is done on the day prior to plating. At least three 6-well plates will be required: approximately 20–30 EBs will be plated per well next day, depending on experimental design; EBs are plated at a higher density if they are to be harvested at earlier time points.
Note: Tissue culture 10 cm plates might also be suitable (with 70–90 EBs/plate) and appear to yield an increased number of progenitors as compared to differentiations using 6-well plates. However, larger plates do not allow as many experimental variables to be tested, they require greater volumes of medium, they are more difficult to handle and there is a higher risk of contamination.
Stage 3 (Days 8 and 9): Setup of the Adherent Culture
Timing: 1 h on day 8 and 30 min on day 9
At day 8, the EBs are transferred onto tissue culture treated plates to establish the adherent stage of the differentiation culture. Next day, once the EBs have attached, the medium is replaced by Stage 3 medium.
-
27.At day 8, transfer the EBs from the 96-well plates into the Matrigel pre-coated plates using a multichannel pipette:
-
a.Aspirate the Matrigel solution from all wells.
-
b.Set the multichannel pipette at 100 μL and use only four pipette tips, as they will fit a single well of a 6-well plate.
-
c.Transfer 20–30 EBs per well, depending at what time they are to be harvested for analysis; for harvesting at earlier time points (days 18–20) plate around 30 EBs/well. Do not change the medium the EBs are currently in.
-
a.
-
28.
Carefully place plates in the incubator, to allow the EBs to attach overnight (12–18 h). Do not move the plates during the first 24 h, as it might disturb the adhering EBs.
Note: A proportionally larger number of 70–90 EBs are plated when using 10-cm plates.
CRITICAL: There appears to be an optimum density of EBs at this stage that promotes efficient hematopoietic development. Some trial and error may be required to determine this, but we have found that EBs from cell lines that will proliferate readily and rapidly produce blood progenitors, might prefer to be plated at a lower density.
-
29.
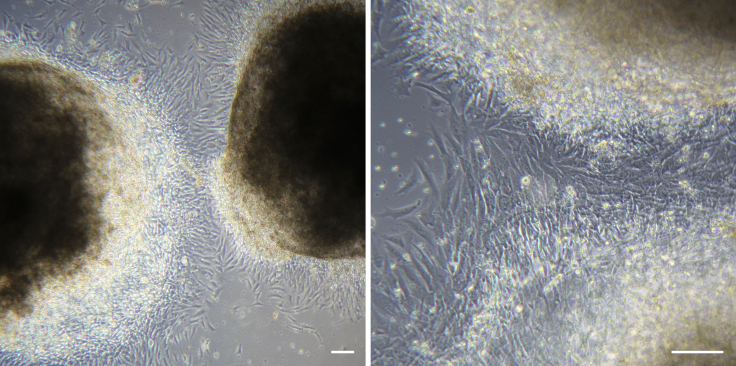
Next day (day 9), check under brightfield microscopy that EBs have successfully attached to the surface of the well and that growth of adherent endothelial and stromal cells around the EB is commencing (Figure 5).
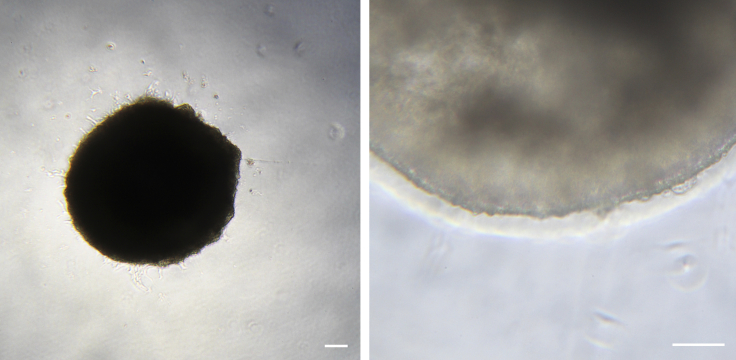
Figure 5.
EBs after Transferring to Adherent Culture
At day 9, EBs are attached to the surface of the tissue culture wells and endothelial and stromal cell outgrowth have commenced. Scale bars represent 100 μm.
-
30.
Gently remove the medium without disturbing the EBs and add 2 mL of Stage 3 medium (see materials section). Use 2 mL per well for 6-well plates or 10 mL for 10-cm plates.
-
31.
At day 11, top up the wells with 1 mL of Stage 3 medium (top up with 3–5 mL if using 10-cm plates).
CRITICAL: Be very gentle when changing the medium at day 9, as EBs might not be firmly attached to the well. To add the medium, place the pipette on the wall of the dish to avoid directing the medium stream onto the EBs. Leave any EBs that might detach in the well, as they may re-adhere over the following days.
Stage 3 (Days 13/15): The Onset of Hematopoietic Progenitor Formation
Timing: 15 min every 2 days
This step describes a full medium change to one that more specifically promotes the proliferation and differentiation of the desired hematopoietic lineage. This medium is used once blood progenitor cells start to appear in culture, usually around days 13–15.
-
32.
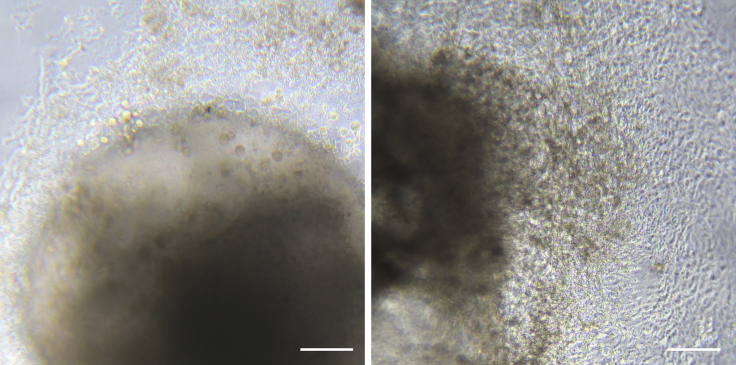
Under brightfield microscopy, check that blood progenitors are being generated from endothelium around the edges of the adhered EBs, but they remain attached and are not yet floating in suspension (Figure 6).
Figure 6.
Emergence of Hematopoietic Progenitors in Culture
Around day 14, the endothelial and stromal cells fully cover the well surface and the first hematopoietic progenitors start to emerge from the EBs. Images show EBs in adherent culture at day 14 of differentiation. Stromal and endothelial cells appear as flat elongated cells and immature progenitors appear as bright aggregates of small round cells, which are not yet in suspension. Scale bar represents 100 μm.
-
33.
Aspirate Stage 3 medium from the wells and add 2 mL per well of Stage 3 Specific medium.
Note: Add 10 mL of Stage 3 Specific medium when using 10-cm plates.
-
34.
Place plates back into the incubator.
-
35.
Two days later, top up the wells with 2–3 mL of fresh Stage 3 Specific medium.
-
36.
From then on, perform half medium changes every 2 days, as each well of a 6-well plate can hold a total volume of 4–5 mL. Carefully remove 2 mL from the surface of the medium, tilting the plates and aspirating from the edge, as most progenitors will remain in suspension close to the bottom of the well (Figure 1, +day 18). Add the 2 mL of fresh medium slowly, without disturbing the cultures.
Note: When using 10-cm plates, top up with 5 mL of fresh medium every 3–4 days, as they can hold up to 25 mL of medium. By using this method, cultures on 10-cm plates would not require half medium changes for harvestings performed up to days 20–22, therefore minimizing the loss of blood progenitors as compared to when performing half medium changes on 6-well plates. However, the risk of contamination is higher than when using smaller wells.
Alternatives: If there is concern over losing hematopoietic cells during the medium change, 2 mL medium from the culture can be transferred to a 10 mL conical polystyrene tube, centrifuged at 400 × g for 3 min and the supernatant can be discarded. Resuspend the pelleted hematopoietic cells with 2 mL of fresh medium and re-add this to the culture well.
Harvesting of Hematopoietic Cultures
Timing: 2 h
This step describes the harvest of the different cell fractions for subsequent analysis of the adherent hematopoietic cultures. It is important to first consider what cell fraction(s) is/are of interest for your experimental design and analyses. The majority of CD34+ blood progenitors will be in suspension in the cultures. However, many progenitor cells, often immature in phenotype, remain attached to the endothelium or are trapped within the EBs. The day of harvest may vary depending on the cells of interest. The first immature hematopoietic stem and progenitor cells are found immediately after the endothelial to hematopoietic transition, generally from days 13–15 of differentiation, and mostly remain adherent to the endothelium or within the EBs. These progenitors detach from the endothelium and appear in suspension in the medium from around days 16–20 and will progressively differentiate thereafter. The highest number of hematopoietic progenitors in suspension usually coincides with days 18–22 of the differentiation, with presence of both immature progenitors and more differentiated lineages. Cell sorting (Fluorescence-activated or magnetic-activated) can be performed immediately after harvesting to collect specific cell populations or to enrich for more immature cell fractions.
-
37.For harvesting the progenitors in the supernatant fraction:
-
a.Collect supernatants containing “floating” blood cells in a 50 mL tube and place them on ice. Cells from multiple wells may be pooled at this stage.
-
b.Rinse the wells gently with 1 mL PBS – to collect the remaining suspension and loosely attached blood cells – and combine into the main tube.
-
c.Pellet the cells by centrifugation at 4°C.
-
d.Keep tubes on ice until the other cell fractions are harvested or use directly for downstream analyses.
-
a.
-
38.To collect the remaining adherent cells (hematopoietic cells, stromal cells and endothelium):
-
a.Add 500 μL of TrypLETM and incubate at 37°C for 5 min.
-
b.Detach the EBs only, using a 1 mL pipette; the EBs are quite sticky so they will raft together.
-
c.Transfer the EBs into a sterile tube containing 1 mL of 2 mg/mL collagenase Type 4 diluted in IMDM.
-
d.Incubate for 30–45 min in a 37°C water bath.
-
e.Meanwhile, detach the remaining stromal-endothelial layer by pipetting up and down or by using a cell scraper, pellet cells by centrifugation, and keep on ice.
-
f.After the collagenase incubation, dissociate the EBs by passing the suspension through a range of increasing needle gauges; from 18G, 21G, 23G and finally a 25G needle (Troubleshooting 3). The cells should dissociate readily if the incubation period with collagenase has been of sufficient duration.
-
g.Pellet the cells to remove residual collagenase reagent and resuspend in PBS.
-
a.
Note: The distinct suspension and adherent cell fractions can be pooled or analyzed separately.
-
39.
Filter the cell suspensions through a 70 μm sterile strainer cap sitting on a 50 mL tube to remove remaining cell aggregates.
-
40.
Collect any remaining cells in the tube by rinsing with PBS and pass through the 70 μm filter.
-
41.
Pellet the filtered suspension for subsequent analysis, such as clonogenic assays and flow cytometry analysis or sorting based on hematopoietic cell surface markers (examples of conjugated antibodies to evaluate hematopoietic outcome are listed in Key Resources Table and include conjugated human CD34, CD45, CD90, CD38, and CD16).
Expected Outcomes
This protocol describes the differentiation of hPSCs to multilineage definitive hematopoietic progenitors that transcriptionally resemble cells arising from the aorta-gonad-mesonephros (AGM) region at the 5th week of embryonic development (Ng et al., 2016). EBs of uniform size are generated via a spin-based method (Ng et al., 2005). During the first four days (Stage 1), EBs are cultured in medium supplemented with growth factors that induce and pattern mesoderm (Ng et al., 2005). In order to activate the definitive (intra-embryonic) program, suppression of extra-embryonic, yolk sac-type hematopoiesis and upregulation of expression of members of the HOXA cluster of homeobox genes are required (Ng et al., 2016). This is achieved by addition of an Activin inhibitor (SB431542) and WNT agonist (CHIR99021) from day 2 to 4 of differentiation. After day 8, EBs are grown in adherent culture conditions.
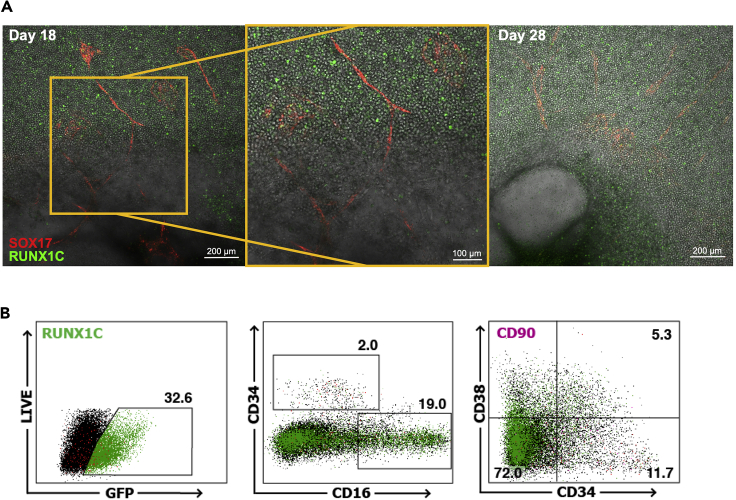
Vascular structures and hemogenic endothelium expressing the SOX17 marker will be generated within and around the EB from day 8 to 14 (Figure 7). The endothelial to hematopoietic transition occurs between day 10 and day 14 of differentiation, once the vascular structures have developed. From day 14 onward, SOX17− CD34+ RUNX1C−/+ hematopoietic progenitors are generated within the SOX17+ endothelium (Figure 8).
Figure 7.
Formation of SOX17+ Vascular Endothelium
(A) Confocal images of combined Z-stack layers of EB cultures at day 10 and day 14 of differentiation of the SOX17mCHERRY/wRUNX1CGFP/w hESC line (Ng et al., 2016). At day 10, EBs have developed SOX17+ endothelium and are surrounded by outgrowths of stromal and SOX17+ endothelial cells, which will cover the entire well surface by day 14. Fluorescence and brightfield channels are merged. Scale bar represents 200 μm. SOX17 (mCHERRY, red).
(B) Epifluorescence microscopy images of EB cultures at day 14, displaying a monolayer of SOX17+ endothelium (upper panels) and SOX17+ vessels (bottom panels) growing from the EB structures. The first formed hematopoietic progenitors (SOX17-) can be seen in cultures by day 14 (arrow). Panels show brightfield (left), epifluorescence (middle) and merged (right) fields. Scale bar represents 100 μm. SOX17 (mCHERRY, red).
Figure 8.
RUNX1C+ Hematopoietic Progenitors Emerge from SOX17+ Hemogenic Endothelium
Confocal images at day 17 of the adherent EB cultures. RUNX1C+ blood progenitors arise from hematopoietic cell clusters within vascular structures of SOX17+ hemogenic endothelium (arrows), mimicking the structures observed in the embryonic AGM. Bottom right image contains merged layers of a Z-stack capture. Scale bar represents 200 μm, 100 μm or 50 μm as indicated. SOX17 (mCHERRY, red) and RUNX1C (GFP, green).
Hematopoietic progenitors are present in suspension after detaching from the endothelium and are generated until day 28 to day 30 of differentiation (Figure 9A). Hematopoietic cells in suspension will continue to mature, yielding an accumulation of cells that have lost their CD90 and CD34 expression and gained expression of CD38 and of the myeloid markers such as CD33, CD14 or CD16 (Figure 9B). Depending on the growth factors used, this protocol can be biased toward myeloid or erythroid lineages (Ng et al., 2016). The derivation of T cells requires the culture of emerging hematopoietic cells under conditions of high NOTCH signaling, as described (Motazedian et al., 2020).
Figure 9.
Blood Progenitors Detach and Progressively Differentiate to Mature Lineages
(A) Confocal Z-stack images of EB cultures at day 18 and day 28 of differentiation. Blood progenitors (round cells) detach from the endothelium and accumulate in suspension in culture until later stages (day 28). Scale bars represent 200 μm or 100 μm, as indicated. SOX17 (mCHERRY, red) and RUNX1C (GFP, green).
(B) Flow cytometry analysis of the floating fraction of day 34 hematopoietic progenitors. Results are a representative of three biological replicates.
Cells derived from this protocol show similar transcriptional profiles and expression of cell surface markers and signaling molecules to cells sorted from human 5-week AGM (Ng et al., 2016). Moreover, CD45+CD34+RUNC1C+ progenitors generated using this protocol display an accessible chromatin pattern resembling that found in human adult HSC and multipotent hematopoietic progenitor cells (Nafria et al., 2020). For these reasons, this protocol is suitable for the mechanistic study of multipotent progenitor development and for the modeling of childhood leukemias. Differentiation of genetically engineered hPSCs – expressing RUNX1-ETO in an inducible manner – into definitive hematopoietic progenitors, using this protocol, has provided insight into the earliest reprograming events of the myeloid regulatory network after acquisition of this leukemic oncogene in a human setting (Nafria et al., 2020).
Limitations
CD34+ RUNX1C+ cells generated using this protocol show high clonogenic activity and are able to home to the bone marrow (Ng et al., 2016). However, similar to the “pre-HSCs” identified in the AGM region of the embryo, they do not show long-term repopulating capacity. For this reason, this protocol is constantly under review and improvement. We are aiming to attain a robust in vitro model that is able to generate long-term repopulating HSCs, which would be critical for generating cells for regenerative medicine as well as disease modeling.
Another limitation of this protocol is the requirement for hPSCs to be cultured on mitotically inactivated murine embryonic fibroblasts (feeder cells) in combination with KOSR-containing medium. Modified versions of this protocol, currently under evaluation, will enable reliable definitive differentiation using cultures grown on feeder-free conditions and in defined media.
Troubleshooting
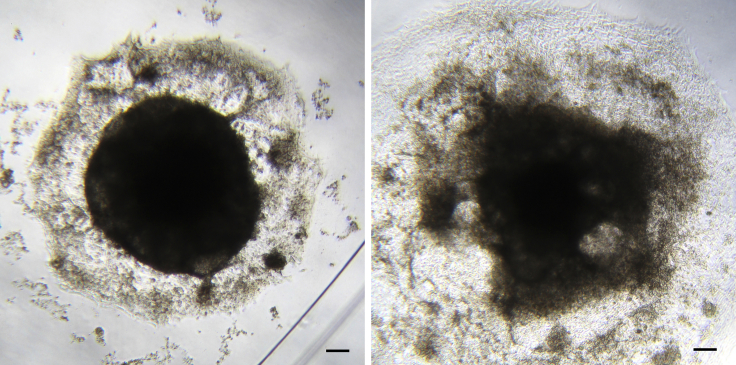
Problem 1
Excessive cell death during EBs formation and/or premature EB disintegration (step 18 and/or 24; Figure 10).
Figure 10.
Example of EBs Presenting Extensive Cell Death
Compromised EBs at day 4 (left) and day 7 (right) of differentiation, presenting large amounts of cell death and debris, displaying cavities and lighter areas and lacking the characteristic sharp edge of viable EBs. Scale bars represent 100 μm.
Potential Solution
The key to EB “happiness” is in the quality of starting hPSC population at the EB setup stage. Make sure that:
-
•
The hPSCs in culture are growing and are not over-confluent at the time of spin-EB setup (they should be 95%–100% confluent). These cells should also be undifferentiated, being EPCAM, CD9, and TRA-1-81 positive.
-
•
Cells are passaged 1:1 on feeder-free conditions on the day of spin-EB setup (allow at least 2 h for cells to re-attach). Evaluate the viability of the cells at this step, which should be 95%–100%, using a hemocytometer and trypan blue staining. This transient re-attachment step enriches for “good” cells, since compromised cells will not be able to re-attach, and removes residual feeder cells.
-
•
At the time of EB setup, harvest the hPSCs using Accutase® solution. The use of Accutase® results single-cell dissociation in a gentler manner than by using TrypLETM Select reagent, which reduces cell death.
-
•
To make the EB setup more efficient, avoid counting the cells and try to do this step as rapidly as possible.
Problem 2
Early formation of blood progenitors (step 18 and/or 24; Figure 11) or low/absent blood production in adherent culture (step 32).
Figure 11.
Example of EBs that Failed to Inhibit the Extra-embryonic Hematopoietic Program
Faulty mesoderm patterning toward the definitive program in EBs at day 4 (left image) and day 7 (right image) of differentiation. Defective inhibition of the extra-embryonic hematopoietic program (failed SB/CHIR treatment) results in formation of stroma (adherent cells) and blood progenitor cells (round bright cells) around the EBs during the non-adherent culture (during the first 8 days of differentiation). Scale bars represent 100 μm.
Potential Solution
Appearance of blood cells during the non-adherent EB culture, or low or absent blood production during the adherent culture might result as a faulty inhibition of the extra-embryonic hematopoietic program and hence with the failure of the intra-embryonic blood program. In order to overcome this problem, perform a flow cytometric analysis at day 7, include as many markers as necessary but it is essential that: most (95%–100%) of the cells must be CD90+, 20%–40% of the cells should be CD34+ and all cells should be CD43 and CD45 negative. Presence of CD43/45+ cells and of CD90- cells indicates failure of the SB/CHIR treatment.
-
•
SB/CHIR usually needs to be added at 42–44 h after the spin-EBs are set up. If there is evidence for excessive extra-embryonic hematopoietic cells emerging, try to start the SB/CHIR addition slightly earlier (between 36 and 42 h).
-
•
Make sure the multichannel pipette is properly calibrated and works smoothly, otherwise there is the risk that not all the wells in the plates will be treated equally.
-
•
Ensure that the hPSCs are not too high in passage number. We generally use cells for up to 20 passages after thaw. Older cells occasionally fail to differentiate properly.
Problem 3
Difficulties in dissociating the EBs (step 38f).
Potential Solution
EBs can be very hard and sticky depending on the cell line used and harvesting day, making them difficult to dissociate. To improve the single-cell dissociation:
-
•
Increase incubation time with Collagenase Type 4 in IMDM to 45–60 min (from 30 min) in a 37°C water bath.
-
•
Using a high gauge needle may shear the cells or cause the needle to block. Have the needles prepared in advance (18G, 21G, 23G, and 25G), to facilitate swapping between needles of different gauges in case the EBs are hard to break up. Dissociate the EBs starting with the 18G needle and do not swap to the next one until the EBs are able to pass through smoothly. Finally, keep dissociating them with the 25G needle until no clumps remain in the suspension.
Alternatives: If the above solution does not work for your cell line, EB dissociation can also be performed in the same way by using 1 mL of 2 mg/mL Collagenase Type 1 diluted in IMDM and a 15 to 30-min incubation in a 37°C water bath.
Resource Availability
Lead Contact
Further information can be provided by the Lead and Technical Contacts, Andrew Elefanty (andrew.elefanty@mcri.edu.au) and Elizabeth Ng (elizabeth.ng@mcri.edu.au).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate and/or analyze any datasets.
Acknowledgments
This protocol was developed by E.S.N. supported by funding from the National Health and Medical Research Council of Australia (GNT1068866, GNT1129861, GNT1138717), by the Australian Research Council Special Research Initiative in Stem Cells (Stem Cells Australia), the Children's Cancer Foundation, and by the Stafford Fox Medical Research Foundation. A.G.E. (GNT1117596) and E.G.S (GNT1079004) are Research Fellows of the National Health and Medical Research Council (Australia). Additional infrastructure funding to the Murdoch Children's Research Institute was provided by the Australian Government National Health and Medical Research Council Independent Research Institute Infrastructure Support Scheme and the Victorian Government's Operational Infrastructure Support Program. Published work using this protocol (Nafria et al., 2020) was funded by a U21 Birmingham/Melbourne joint studentship and a Wellcome Trust ISSF grant to the University of Birmingham held by M.N., and a development grant from the Birmingham Cancer Research UK Centre to C.B. M.N. was also awarded The Henry and Rachael Ackman Travelling Scholarship by the University of Melbourne Faculty of Medicine, Dentistry and Health Sciences.
Author Contributions
M.N. wrote the manuscript and performed experiments using this protocol. C.B., A.G.E., and E.G.S. supervised the experimental study. E.S.N. conceived and developed the protocol.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100130.
Contributor Information
Elizabeth Siewsun Ng, Email: elizabeth.ng@mcri.edu.au.
Andrew George Elefanty, Email: andrew.elefanty@mcri.edu.au.
Supplemental Information
References
- Kennedy M., Awong G., Sturgeon C.M., Ditadi A., LaMotte-Mohs R., Zúñiga-Pflücker J.C., Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Motazedian A., Bruveris F.F., Kumar S.V., Schiesser J.V., Chen T., Ng E.S., Chidgey A.P., Wells C.A., Elefanty A.G., Stanley E.G. Multipotent RAG1+ progenitors emerge directly from haemogenic endothelium in human pluripotent stem cell-derived haematopoietic organoids. Nat. Cell Biol. 2020;22:60–73. doi: 10.1038/s41556-019-0445-8. [DOI] [PubMed] [Google Scholar]
- Nafria M., Keane P., Ng E.S., Stanley E.G., Elefanty A.G., Bonifer C. Expression of RUNX1-ETO rapidly alters the chromatin landscape and growth of early human myeloid precursor cells. Cell Rep. 2020;31:107691. doi: 10.1016/j.celrep.2020.107691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng E.S., Davis R.P., Azzola L., Stanley E.G., Elefanty A.G. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–1603. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- Ng E.S., Davis R., Stanley E.G., Elefanty A.G. A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nat. Protoc. 2008;3:768–776. doi: 10.1038/nprot.2008.42. [DOI] [PubMed] [Google Scholar]
- Ng E.S., Azzola L., Bruveris F.F., Calvanese V., Phipson B., Vlahos K., Hirst C., Jokubaitis V.J., Yu Q.C., Maksimovic J. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 2016;34:1168–1179. doi: 10.1038/nbt.3702. [DOI] [PubMed] [Google Scholar]
- Sturgeon C.M., Ditadi A., Awong G., Kennedy M., Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014;32:554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate and/or analyze any datasets.