Abstract
Maternal prenatal exposures, including bisphenol A (BPA), are associated with offspring’s risk of disease later in life. Alterations in DNA methylation may be a mechanism through which altered prenatal conditions (e.g. maternal exposure to environmental toxicants) elicit this disease risk. In the Michigan Mother and Infant Pairs Cohort, maternal first-trimester urinary BPA, bisphenol F, and bisphenol S concentrations were tested for association with DNA methylation patterns in infant umbilical cord blood leukocytes (N = 69). We used the Illumina Infinium MethylationEPIC BeadChip to quantitatively evaluate DNA methylation across the epigenome; 822 020 probes passed pre-processing and quality checks. Single-site DNA methylation and bisphenol models were adjusted for infant sex, estimated cell-type proportions (determined using cell-type estimation algorithm), and batch as covariates. Thirty-eight CpG sites [false discovery rate (FDR) <0.05] were significantly associated with maternal BPA exposure. Increasing BPA concentrations were associated with lower DNA methylation at 87% of significant sites. BPA exposure associated DNA methylation sites were enriched for 38 pathways significant at FDR <0.05. The pathway or gene-set with the greatest odds of enrichment for differential methylation (FDR <0.05) was type I interferon receptor binding. This study provides a novel understanding of fetal response to maternal bisphenol exposure through epigenetic change.
Keywords: environmental epigenomics, prenatal exposure, DNA methylation
Introduction
Bisphenol A (BPA), a chemical commonly used in receipts, plastics, and food packaging, is considered to be a ‘ubiquitous exposure’, principally because of its wide-spread usage and high rate (over 95%) of detection in human urine [1]. Exposure to BPA and two of its commonly used replacement analogues, bisphenol F (BPF), and bisphenol S (BPS), are readily detectable in the US populations [1–4]. BPF and BPS are now increasingly utilized in place of BPA particularly as a result of consumer and scientific-based advocacy efforts. This pressure effectively elicited the US Food and Drug Administration’s ban of BPA in infant-related plastics and products [5]. However, significantly less is known about BPF and BPS, with new evidence suggesting that these replacement chemicals with close structural similarities to BPA may have comparable or increased levels of potency as endocrine disruptors and may also negatively impact the reproductive system [5–7]. In the USA, BPF and BPS were only recently added to the list of chemicals measured in the National Health and Nutrition Examination Survey (NHANES), appearing for the first time from 2013 to 2014 [4].
While the proportion of adults and children with detectable levels of these bisphenols is concerning, the exposure patterns experienced by pregnant and lactating mothers introduce an additional layer of consideration. Specifically, when the potential impact on fetal development and lifetime health trajectory are evaluated. Pregnant women in the USA and internationally are typically exposed to or have biological concentrations of urinary bisphenols at similar levels to non-pregnant women [8–11]. Furthermore, BPA, BPF, and BPS have the potential to cross the placenta at differing rates and with inter-individual variation [12–14].
Environmental research establishes the framework of time around conception, gestation, and birth as one of the most developmentally susceptible times of life. This aligns with the Developmental Origins of Health and Disease hypothesis, which recognizes the connection between maternal exposure during pregnancy and the risks posed to her offspring’s health and later-life disease [15]. An increasing number of studies have investigated the impact of prenatal exposure to bisphenols on phenotypic outcomes in infants and children. Maternal and prenatal exposure to bisphenols in humans is associated with pregnancy duration and birth weight [16–19], increased risk of preeclampsia [20], early childhood behavior [21–23], childhood body mass index (BMI) [24], and peripubertal metabolic homeostasis [25, 26]. Studies in mice have demonstrated that prenatal or early-life exposure to bisphenols is associated with altered brain development and behavior [27] as well as disruptions in metabolic homeostasis [28–31], glucose metabolism [32, 33], neuroendocrine function [34, 35], and immune function [36, 37]. Despite these developments in understanding of the association between prenatal bisphenol exposure and phenotypic outcomes in offspring, less is known of the possible mechanism through which bisphenols elicit these outcomes.
Recent work indicates that environmentally induced disease etiology may be mediated by changes in the epigenetic profile [38–40]. For the purposes of this investigation, we define the epigenome as consisting of chemical modifications (e.g. DNA methylation and histone modification) that are mitotically heritable and regulate gene expression but are not the result of a change in the DNA sequence [15]. Currently, very few studies exist that evaluate prenatal bisphenol exposure and its consequent longitudinal impact on the fetal and later-life epigenome [41, 42]. Most studies were completed in mice; with evidence suggesting that prenatal exposure to bisphenols is associated with changes in DNA methylation in genes regulating hepatic function [43], metabolism [44, 45], neuronal [46] and inflammatory pathways [47], and other regulatory epigenetic machinery [48]. Four human studies have evaluated the epigenetic impact of prenatal exposure to bisphenols [49–52]. From these collective investigations comes significant insight into elements of the association between prenatal BPA exposure and DNA methylation in offspring growth and neurological function in addition to its sexually dimorphic nature. However, these studies are not uniform across their approach in three key elements: (i) the time point and sample type in which bisphenol exposure was measured (e.g. urinalysis during pregnancy or cord blood), (ii) the type of DNA methylation profiling (e.g. in candidate genes or epigenome-wide), and (iii) the timepoint at which DNA methylation was analyzed in offspring. With advances in exposure science and DNA methylation technology, it is critical to evaluate exposure to multiple bisphenols from the first trimester, a time during which the epigenome is highly susceptible to reprogramming; and measure outcomes at birth, utilizing methods that generate data at all genes.
This study aimed to test the association between maternal exposure to the bisphenol BPA or its substitute chemicals, BPF and BPS, and cord blood leukocyte DNA methylation at >800 000 loci in a longitudinal pregnancy cohort. This study is of the few to evaluate prenatal exposure to bisphenols during the first trimester and its epigenome-wide association with DNA methylation infant cord blood. Importantly, we are the first to use this method to also investigate the replacement phenols BPF and BPS.
Methods
Study Population
The samples used in this study were derived from the Michigan Mother−Infant Pairs pregnancy cohort (MMIP), which initiated in 2011. Briefly, women providing informed, written consent were enrolled during their first prenatal visit to the University of Michigan Women’s Hospital clinic. At this visit, maternal first-trimester blood and urine were collected. Women also completed a questionnaire that gathered socio-demographic factors, health behaviors, food consumption and personal care product use, among other measures. Exclusion criteria included: age <18 years, prior infertility treatment, pregnancy with multiple fetuses, and pregnancy <8 weeks or >14 weeks gestation. Women were provided study materials between weeks 34 and 38 of gestation for blood and urine collection upon admission into labor. Maternal blood and urine were collected when admitted and umbilical cord blood samples were collected at delivery. At the time of writing, 331 mothers have enrolled in MMIP, and 200 have been followed-up through labor and delivery. For the analysis described here, a subset of MMIP families enrolled between 2011 and 2017 with first-trimester exposure assessment of three urinary bisphenols and DNA methylation analysis via the Infinium EPIC were included (n = 69). The University of Michigan Medical School Institutional Review Board approved all study procedures (HUM00017941).
Epigenome-Wide DNA Methylation Analysis of Infant Umbilical Cord Blood
Infant cord blood samples (N = 69) were collected into PaxGene Blood DNA tubes (PreAnalytix) with the use of butterfly needles at the time of birth and stored at −80°C until processing. Total DNA was extracted with the PaxGene Blood DNA kit. DNA quality and concentration were assessed via Qubit at the University of Michigan Advanced Genomics Core. DNA was bisulfite converted using the EZ-96 DNA Methylation Kit (Zymo), wherein ∼500 ng of input DNA was used. The kit utilized sodium bisulfite to convert un-methylated cytosines to uracil and ultimately thymine, while methylated cytosines were protected [53].
Following bisulfite treatment, DNA methylation at >850 000 CpG sites was evaluated using the Illumina Infinium MethylationEPIC BeadChip (EPIC) at the University of Michigan Advanced Genomics Core according to standard protocols. Cord blood samples were run on three separate days, and these experimental batches are considered in statistical models.
Processing and Quality Control of Infinium MethylationEPIC Data
Arrays were assessed for quality of samples and probes using a standard pipeline. Briefly, the pipeline utilized the minfi package [54] (R Project for Statistical Computing) to read in raw data image files. Quality control of samples was assessed by comparing estimated sex (from methylation values on the X and Y chromosomes) with known infant sex, detection P-values of probes, and intensity signals.
Probes with poor detection (positions that failed detection in more than 10% of samples N = 1475), cross-reactive probes, and probes that target polymorphic CpG sites in the Illumina HumanMethylation arrays were dropped [55]. The Functional Normalization [56] R package was used to correct for background and perform dye-bias normalization.
Using estimateCellCounts, the relative proportion of B cells, CD4, CD8T, granulocytes, monocytes, neutrophils, and nucleated red blood cells (nRBCs) were estimated for each cord blood sample using an established algorithm based on DNA methylation profiles of sorted major cord blood cell types [57]. estimateCellCounts is a cell proportion estimation algorithm that estimates the relative proportions of cell types within a given sample based on DNA methylation signatures of each cell type.
These preprocessing steps resulted in 822 020 retained probes from N = 69 cord blood samples that passed all quality control measures. Finally, M-values, defined as the log2 ratio of intensities of methylated probe versus unmethylated probes, were generated for each sample at these CpG sites and were used in downstream statistical analyses unless otherwise noted.
Maternal Bisphenol Measurement
Bisphenols (BPA, BPF, and BPS) were measured in spot urine samples collected from mothers during their first-trimester visit (between 8 and 14 weeks) for this subset of MMIP participants (n = 69). Samples were collected into polypropylene urine collection containers, aliquoted into glass vials, and frozen at −80°C until analysis. Total urinary BPA, BPF, and BPS were measured at NSF International (Ann Arbor, MI) using isotope dilution-liquid chromatography-tandem mass spectrometry (ID-LC-MS/MS), as reported previously [58]. Specific gravity was measured using a handheld digital refractometer (Atago Co., Ltd., Tokyo, Japan) at the time of sample analysis. Urinary bisphenol values below the limit of detection (LOD, 0.2 ng/ml) were replaced with LOD/√2 (0.141 ng/ml).
Statistical Analysis
All statistical analyses were performed in R version 3.6.0 (Platform: x86_64-apple-darwin15.6.0 (64-bit) & Running under: macOS Mojave 10.14.6). We first performed univariate analyses on all exposure biomarkers and potential covariates of interest. We then assessed relationships between exposures and covariates to identify potential confounders via chi-square tests, t-tests, and Spearman correlations. First-trimester urinary BPA was modeled as a continuous variable, and BPF and BPS were modeled as categorical (above or below the LOD) (Supplementary Tables S1−S3). Singular value decomposition (SVD) analysis was performed with the ChAMP package [59]. The correlation between principal components of the methylation data with biological and technical covariates was determined using linear regression (continuous variables) or Kruskal−Wallis (categorical variables). We did not identify potential confounders (i.e. covariates associated with both BPA and DNA methylation) to include in the model. However, due to their significant (P < 1 × 10−5) association with the DNA methylation data in the SVD analysis, infant sex, B cells, nRBCs, and sample-plate (batch) were selected as covariates to adjust for in final models. We also performed a sensitivity analysis on the sites significantly associated with BPA exposure to determine the effect of including gestational age and birth weight.
Single-Site Association Analysis
Linear regression was used to identify differentially methylated CpG sites (using M-values) by each maternal urinary bisphenol exposure, adjusting for covariates described above (infant sex, B cells, nRBCs, sample plate). An empirical Bayes method in the limma [60] R package was then used to shrink probe-wise variances toward a pooled estimate and calculate a moderated t-statistic. M-values were selected for statistical analysis given their advantages which include meeting the assumption of homoscedasticity and superior performance in detection rate and true positive rate, especially for highly methylated and unmethylated sites [61, 62]. P-value correction by the Benjamini−Hochberg false discovery rate (FDR) method was used [63], and a 5% FDR (i.e. q < 0.05) were considered significant.
Sensitivity analyses were performed. One maternal urinary BPA sample was identified as a statistical outlier [±2 standard deviations (SD) from the mean]. The outlier was removed, and the single-site analyses were rerun. The direction and significance of the sites identified as significant in the initial model were compared with the results from the model without the outlier. Additional analyses included examining scatterplots of the relationship between BPA and methylation at each significant site.
In order to test whether the bisphenol exposures may be influencing the same genes, we calculated the Pearson correlation between the effect estimates of all CpG sites from models for each bisphenol.
Lastly, we compared results of previously published epigenome-wide studies focused on BPA exposure with our results [49, 50]. Pearson correlation was run between the effect estimates for sites reported by Miura et al. [49] as significant at P ≤ 0.0001 for all infants and the corresponding results in our BPA model. Results from Junge et al. and Alavian-Ghavanini et al. were compared with our results for replication of the direction of the effect of BPA.
Differentially Methylated Regions
We utilized dmrcate [64] to test for differentially methylated regions (DMRs) by maternal first-trimester urinary phenols exposure. A DMR had to consist of at least two consecutive probes. Probes that were two nucleotides or closer to a single-nucleotide polymorphism that had minor allele frequency >0.05 were filtered out first. The model was adjusted for cell type (Bcell and nRBC), infant sex, and batch. GenomicRanges [65] was used to graph an annotated representation of the DMRs. GenomicRanges requires the use of beta values (e.g. proportion of DNA methylation at CpG sites), and data are displayed as averaged across quartiles of BPA. Quartile cutoffs are as follows: Q1 [<LOD, 0.348], Q2 [0.349, 0.897], Q3 [0.898,1.90], Q4 [1.91, 6.76] in nanograms per milliliters BPA. DMRcate analysis was also repeated without the BPA outlying subject.
Pathway Analysis
LRpath [66] was utilized to perform gene-set enrichment across all probes annotated to genes (using Entrez Gene IDs) using concepts (also known as gene-sets) from KEGG and GO (Biological Process, Molecular Function, and Cellular Component). LRpath uses raw P-values, fold changes, and Entrez gene IDs for each probe mapping to a known gene from the single-site linear model for the association between each bisphenol and DNA methylation. LRPath utilizes logistic regression in determining gene-set membership status (dependent variable) by the statistical significance of genes’ differential methylation (independent variable, raw P-value). Concepts from both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were selected from LRPath’s internal annotation database of gene-sets (concepts) as those onto which our data should be mapped, and only gene-sets with a minimum of 10 and a maximum of 250 genes were used; a directional test was included based on the direction of association between BPA and DNA methylation at each site. LRPath tests the odds that the genes in a concept have higher significance values (e.g. lower P-values from the differential methylation analysis) than expected at random, and FDR of 5% was considered a statistically significant enriched gene-set.
Results
Study Population Characteristics
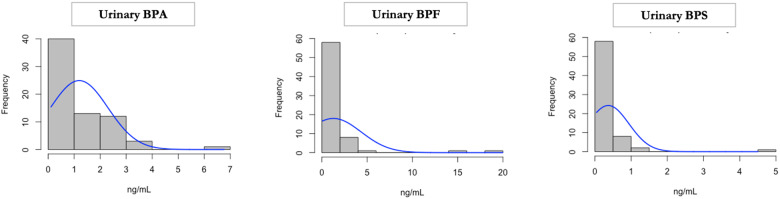
Table 1 contains the demographic data of the maternal−infant pairs included in this study. The mean maternal age was 32, and on average, the number of weeks to delivery was 39.5 weeks. After adjusting for SG, mean maternal, first-trimester urinary BPA concentration was 1.19 ng/ml (range < LOD: 6.78 ng/ml) (Fig. 1). The highest maternal BPA exposure was determined to be an outlier (e.g. greater than two SD away from the mean). However, her exposure levels were biologically plausible, given its fitting within the distribution of measured samples in the most recent NHANES report from 2014 to 2015. Therefore, this sample was retained. Fifty-nine of 69 (85.5%) maternal samples had urinary BPA levels above the LOD. Mean maternal first-trimester urinary BPA concentration was 1.27 ng/ml (range < LOD: 19.97 ng/ml) (Fig. 1). Thirty-nine of 69 (56.5%) maternal samples had urinary BPF levels above the LOD, and two outliers were detected. Mean maternal first-trimester urinary BPS concentration was 0.37 ng/ml (range < LOD: 4.50 ng/ml) (Fig. 1). Forty of 69 (57.9%) maternal samples had urinary BPS levels above the LOD.
Table 1:
Descriptive statistics [median (25th, 75th percentiles) or n (%)] for N = 69 mother−infant pairs in the MMIP cohort
| Maternal age (years) | 32 (30, 34) |
| Number of days to delivery (days) | 277 (273, 282) |
| Maternal race/ethnicity | |
| White | 66 (95%) |
| African American | 1 (1.45%) |
| Asian | 1 (1.45%) |
| Other or mixed race | 1 (1.45%) |
| B-cell proportion | 0.0890 (0.0625, 0.111) |
| Monocyte proportion | 0.0916 (0.0767, 0.105) |
| nRBC proportion | 0.0696 (0.0481, 0.113) |
| CD4+ cell proportion | 0.151 (0.114, 0.189) |
| CD8+ cell proportion | 0.124 (0.100, 0.148) |
| Natural killer cell proportion | 0.00563 (0.0, 0.0270) |
| Maternal urinary BPA (ng/ml) | 0.898 (0.349, 1.91) |
| Maternal urinary BPF (ng/ml) | 0.298 (0.177, 0.820) |
| Maternal urinary BPS (ng/ml) | 0.226 (0.145, 0.365) |
| Infant sex | |
| Female | 37 (53.6%) |
| Male | 32 (46.3%) |
| Infant birth weight (g) | 3500 (3270, 3820) |
Limit of detection (LOD) <0.2 ng/ml. Urinary bisphenol measures adjusted for specific-gravity.
Figure 1:
Maternal urinary bisphenol measures. BPA, BPF, and BPS were measured in urine collected from MMIP women during their first-trimester visit. Bisphenols were adjusted for specific-gravity (SG), and they are represented as nanograms per milliliters
When we assessed relationships between maternal first-trimester urinary BPA exposure and covariates of interest, including maternal characteristics and estimated cord blood cell-type proportions (Supplementary Table S1) using Spearman correlations or t-tests, none were statistically significant. Similarly, t-tests and chi-square tests for covariates of interest with maternal first-trimester urinary BPF and BPS (modeled as categorical variables) were not statistically significant except for pre-pregnancy BMI by BPF (detected vs. <LOD) (Supplementary Tables S2 and S3).
Single-Site DNA Methylation
Single-site association analysis revealed maternal first-trimester urinary BPA exposure was associated with 38 differentially methylated sites (DMS) in infant cord blood at q < 0.05. The genomic inflation factor (λ) for the analysis was 0.823. Increasing BPA concentrations were associated with lower DNA methylation at 87% of significant sites (Table 2). The five most significantly DMS (q < 0.003) were within the genes SLC2A1-AS1, KIF21B, CRYL1, HSPBAP1, and FN1. For interpretability, Table 2 shows effect estimates from a model of the beta-values. To more clearly demonstrate percent difference in methylation at each site, M-values were replaced with beta values in the single-site analysis. For example, for every 1 ng/ml increase in BPA, DNA methylation at SLC2A1-AS1 decreased by 10%, while DNA methylation at KIF21B increased by 2.7%.
Table 2:
Differentially methylated CpG sites associated with maternal first-trimester urinary BPA exposure
| Locus | Gene name | Relation to CpG island | Effect estimate using beta-values | q-value |
|---|---|---|---|---|
| chr1: 43437674 | SLC2A1-AS1 | Open sea | −0.10 | 0.00069 |
| chr1: 14591868 | Open sea | −0.060 | 0.00154 | |
| chr1: 200992656 | KIF21B | Island | 0.027 | 0.00154 |
| chr19: 36661673 | Open sea | 0.031 | 0.00155 | |
| chr13: 21049223 | CRYL1 | Open sea | −0.063 | 0.00166 |
| chr18: 33160855 | North Shore | −0.0071 | 0.00239 | |
| chr3: 122512541 | HSPBAP1 | Island | 0.028 | 0.00239 |
| chr8: 10622805 | Open sea | −0.027 | 0.00290 | |
| chr2: 216237359 | FN1 | Open sea | −0.057 | 0.00290 |
| chr16: 51184562 | SALL1 | Island | −0.00096 | 0.00290 |
| chr20: 10199434 | SNAP25 | North Shore | −0.0011 | 0.00290 |
| chr15: 85660361 | PDE8A | Open sea | −0.060 | 0.00290 |
| chr8: 72756155 | MSC | Island | −0.00077 | 0.00333 |
| chr12: 121698404 | CAMKK2 | Open sea | −0.058 | 0.00333 |
| chr19: 3180815 | South Shore | −0.00096 | 0.00333 | |
| chr2: 85822726 | RNF181 | Island | −0.0038 | 0.00370 |
| chr2: 239039182 | ESPNL | North Shore | −0.045 | 0.00402 |
| chr8: 33342681 | MAK16 | Island | −0.00086 | 0.00402 |
| chr7: 142536625 | Open sea | −0.049 | 0.00402 | |
| chr11: 26595206 | MUC15 | Open sea | −0.060 | 0.00487 |
| chr2: 71017846 | FIGLA | Island | −0.017 | 0.00712 |
| chr7: 1068244 | C7orf50 | Island | 0.013 | 0.00712 |
| chr5: 106879524 | EFNA5 | Open sea | −0.030 | 0.00753 |
| chr2: 172957268 | North Shore | −0.015 | 0.00913 | |
| chr12: 11324011 | SMIM10L1 | Island | −0.00060 | 0.00963 |
| chr3: 46752152 | TMIE | Open sea | −0.037 | 0.01499 |
| chr16: 8735575 | METTL22 | Open sea | −0.028 | 0.01543 |
| chr12: 62653559 | USP15 | North Shore | −0.0021 | 0.01769 |
| chr3: 56502021 | ERC2 | Island | −0.012 | 0.01797 |
| chr1: 111098247 | Island | −0.00062 | 0.01906 | |
| chr12: 123380878 | VPS37B | Island | −0.0083 | 0.02749 |
| chr19: 2462065 | Island | −0.00094 | 0.02749 | |
| chr10: 636076 | DIP2C | Open sea | −0.027 | 0.02775 |
| chr14: 62210927 | HIF1A | Open sea | −0.017 | 0.03846 |
| chr4: 154400013 | KIAA0922 | Open sea | −0.031 | 0.03846 |
| chr4: 154349775 | Open sea | −0.015 | 0.03846 | |
| chr12: 1058965 | RAD52 | Island | 0.011 | 0.03990 |
| chr8: 11059042 | XKR6 | Island | −0.00049 | 0.04156 |
Note: Results shown are for CpG sites associated with maternal urinary first-trimester BPA exposure below false discovery rate (FDR) significance of q < 0.05. Model was adjusted for infant sex, nRBCs, Bcells, and sample plate (batch). Effect estimate is the unit change with each 1 ng/ml increase in BPA from the model of M-values (logit-transformed beta values). Beta is the effect estimate when modeling the proportion of methylation (beta value) at the same CpG site instead and represents the increase in proportion methylated per each nanograms per milliliters increase in first-trimester BPA. The beta estimate is included for interpretation purposes; significance values are generated from the M-value analysis.
Supplementary Table S4 provides results for the 38 DMS associated with prenatal BPA exposure when the model is run without the BPA outlying subject. When the outlier was removed, only two sites remained significantly associated with BPA at P ≤ 0.0001, λ = 0.948 (in SLC2A1-AS1 and RAD52). The remaining CpG sites may be false positives or may only be perturbed at higher levels of exposure; this should be tested further in future studies. Supplementary Table S5 displays the sensitivity analysis, run utilizing the same model as BPA, but including gestational age and birth weight, and indicated that the 38 CpG sites were as or more significantly associated with BPA.
BPF exposure, dichotomized as below or above the LOD, was not associated with DMS at FDR of q < 0.05, but was associated with 19 DMS at P ≤ 0.0001, λ = 0.788. BPS exposure, also dichotomized as below or above the LOD, was not associated with DMS at the FDR of q < 0.05 but was associated with one DMS at VPS53 at P ≤ 0.0001, λ = 0.674 (Supplementary Files S2 and S3). The effect estimates from the BPA, BPF, and BPS models were significantly correlated. BPA and BPF (cor = 0.194), BPA and BPS (cor = 0.116), and BPF and BPS (cor = 0.179) were each significantly, positively correlated at P < 2.2e−16.
Differentially Methylated Regions
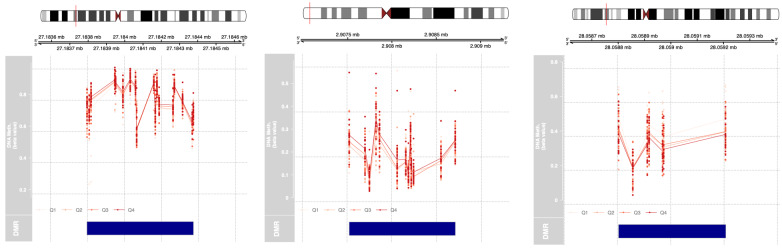
Three DMRs were detected in association with maternal first-trimester urinary BPA exposure, wherein each region possessed at least seven CpG sites. These genes were HOXA-AS3, PRSS22, and ZSCAN12P1. Two of the three regions (HOXA-AS3 and PRSS22) displayed an increase in DNA methylation with increasing BPA (Table 3). Figure 2 includes the 18 HOXA-AS3 CpG sites contained within the DMR. Similarly, for PRSS22, across its 13 CpG sites higher maternal exposure first-trimester BPA exposure was associated with increased percent DNA methylation, and this association remained after exclusion of the BPA outlier (P = 0.00000752). Alternatively, in the seven CpG sites of ZSCAN12P1, higher maternal first-trimester urinary BPA exposure was associated with lower percent methylation.
Table 3:
Differentially methylated regions in association with maternal first-trimester urinary BPA exposure
| Chromosome | Gene Name | Start (bp) | End (bp) | Number of CpG Sites | P-valuea | Max Beta Change per nanograms per milliliters BPA increaseb |
|---|---|---|---|---|---|---|
| 7 | HOXA-AS3 | 27183794 | 27184375 | 18 | 1.79E−14 | 0.0190 |
| 16 | PRSS22 | 2907517 | 2908715 | 13 | 6.83E−18 | 0.0376 |
| 6 | ZSCAN12P1 | 28058802 | 28059208 | 7 | 2.25E−13 | −0.0388 |
Significance considered at q < 0.05.
Minimum FDR P-value for the region.
For interpretability, changes across the DMR are reported as proportion methylated (beta), though models used logit-transformed beta values (M-values).
Figure 2:
Differentially methylated regions (DMRs) associated with first-trimester urinary BPA exposure. Three DMRS in cord blood leukocytes in (a) HOXA-AS3, (b) PRSS22, and (c) ZSCAN12P1 were identified via DMRcate that were associated with first-trimester BPA levels (modeled as a continuous variable and adjusted for infant sex, batch, and estimated nRBCs and B cells). Here, proportion of DNA methylation (beta values) at CpG sites within the DMR are displayed, averaged across quartiles of BPA. Quartile cutoffs are as follows: Q1 [<LOD, 0.348], Q2 [0.349, 0.897], Q3 [0.898, 1.90], and Q4 [1.91, 6.76] in nanograms per milliliters BPA.
Pathway Analysis
BPA exposure associated DNA methylation sites were enriched for 38 pathways significant at FDR <0.05. Higher BPA exposure was associated with increased methylation for all enriched pathways (Table 4). The pathway or concept with the greatest odds of enrichment for differential methylation was type I interferon (IFN) receptor binding; pathways related to type I IFN activity appeared four additional times. Other highly enriched pathways included JAK/STAT signaling and response; G-protein coupled receptor (GPCR) signaling, and immune response (Table 4). In general, the enriched pathways were associated with the nervous system, immune response, and neuroinflammation.
Table 4:
Gene-sets enriched for differentially methylated genes in cord blood leukocytes by maternal first-trimester urinary bisphenol exposures using LRPath.
| Pathway ID | Pathway name | Database with concept | No. of genes in concept | q-value | Direction |
|---|---|---|---|---|---|
| Panel A: pathways associated with maternal first-trimester urinary BPA exposure | |||||
| GO: 0005132 | Type I interferon receptor binding | GOMF | 13 | 7.82E−07 | Up |
| GO: 0000786 | Nucleosome | GOCC | 86 | 1.53E−04 | Up |
| GO: 0044815 | DNA packaging complex | GOCC | 92 | 1.53E−04 | Up |
| GO: 0005549 | Odorant binding | GOMF | 81 | 3.07E−04 | Up |
| GO: 0033139 | Regulation of peptidyl-serine phosphorylation of STAT protein | GOBP | 18 | 0.00206 | Up |
| GO: 0033141 | Positive regulation of peptidyl-serine phosphorylation of STAT protein | GOBP | 17 | 0.00206 | Up |
| GO: 0002323 | Natural killer cell activation involved in immune response | GOBP | 25 | 0.00258 | Up |
| GO: 0042501 | Serine phosphorylation of STAT protein | GOBP | 22 | 0.00258 | Up |
| GO: 1900424 | Regulation of defense response to bacterium | GOBP | 11 | 0.00258 | Up |
| GO: 0002922 | Positive regulation of humoral immune response | GOBP | 15 | 0.00359 | Up |
| GO: 0001055 | RNA polymerase II activity | GOMF | 10 | 0.00570 | Up |
| GO: 0007259 | JAK-STAT cascade | GOBP | 155 | 0.00584 | Up |
| GO: 0042100 | B-cell proliferation | GOBP | 81 | 0.00584 | Up |
| GO: 0043330 | Response to exogenous dsRNA | GOBP | 40 | 0.00584 | Up |
| GO: 0097696 | STAT cascade | GOBP | 155 | 0.00584 | Up |
| GO: 0006959 | Humoral immune response | GOBP | 157 | 0.00631 | Up |
| GO: 0042742 | Defense response to bacterium | GOBP | 205 | 0.00837 | Up |
| GO: 0071880 | Adenylate cyclase-activating adrenergic receptor signaling pathway | GOBP | 18 | 0.00917 | Up |
| hsa04623 | Cytosolic DNA-sensing pathway | KEGG | 51 | 0.00982 | Up |
| GO: 0016290 | Palmitoyl-CoA hydrolase activity | GOMF | 11 | 0.0105 | Up |
| GO: 0007189 | Adenylate cyclase-activating G-protein coupled receptor signaling pathway | GOBP | 82 | 0.0164 | Up |
| GO: 0007192 | Adenylate cyclase-activating serotonin receptor signaling pathway | GOBP | 11 | 0.0164 | Up |
| GO: 0071875 | Adrenergic receptor signaling pathway | GOBP | 25 | 0.0184 | Up |
| GO: 0032993 | Protein-DNA complex | GOCC | 154 | 0.0209 | Up |
| hsa04630 | Jak-STAT signaling pathway | KEGG | 145 | 0.0213 | Up |
| hsa04140 | Regulation of autophagy | KEGG | 30 | 0.0213 | Up |
| GO: 0033617 | Mitochondrial respiratory chain complex IV assembly | GOBP | 13 | 0.0337 | Up |
| GO: 0034340 | Response to type I interferon | GOBP | 79 | 0.0337 | Up |
| GO: 0060337 | Type I interferon signaling pathway | GOBP | 75 | 0.0337 | Up |
| GO: 0071357 | Cellular response to type I interferon | GOBP | 75 | 0.0337 | Up |
| GO: 0097034 | Mitochondrial respiratory chain complex IV biogenesis | GOBP | 13 | 0.0337 | Up |
| hsa05320 | Autoimmune thyroid disease | KEGG | 44 | 0.0341 | Up |
| GO: 0050830 | Defense response to Gram-positive bacterium | GOBP | 65 | 0.0381 | Up |
| hsa05322 | Systemic lupus erythematosus | KEGG | 118 | 0.0403 | Up |
| GO: 0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | GOBP | 183 | 0.0457 | Up |
| GO: 0002286 | T-cell activation involved in immune response | GOBP | 84 | 0.0460 | Up |
| GO: 0019731 | Antibacterial humoral response | GOBP | 35 | 0.0460 | Up |
| GO: 0005665 | DNA-directed RNA polymerase II, core complex | GOCC | 17 | 0.0497 | Up |
| Panel B: pathway associated with maternal first-trimester urinary BPF exposure | |||||
| hsa05322 | Systemic lupus erythematosus | KEGG | 118 | 0.0296 | Up |
Note: Significance was considered at FDR q < 0.05.
‘Concept’ represents gene-sets.
GO, Gene Ontology; MF, molecular function; BP, biological process; CC, cellular component; KEGG databases, Kyoto Encyclopedia of Genes and Genomes databases.
Results from the BPF exposure were enriched for smaller P-values in one pathway: systemic lupus erythematosus (SLE) (q = 0.0295). Higher BPF exposure was associated with increased methylation in genes of this pathway (Table 4). BPS exposure associated DNA methylation sites were not enriched for pathways at FDR <0.05.
Comparison of Results with Previously Published BPA Studies
Forty-two of the 45 probes reported in Miura et al. as significantly associated with BPA exposure at P < 0.0001 were included in our dataset. Pearson’s correlation between effect estimates at these 42 sites revealed a slightly positive correlation (cor = 0.106) that was not significant (P = 0.503) (Supplementary Table S5). The direction of the effect of BPA exposure on DNA methylation in MEST and RAB408 in our results did not correspond to that detected by Junge et al. However, Junge et al. modeled BPA as high versus low BPA exposure. The direction of the effect of BPA exposure on DNA methylation in GRIN2B also did not correspond to that reported by Alavian-Ghavanini et al. Again, this group chose to model BPA as an ordered categorical variable and as forth quartile versus 1st quartile and reported odds ratios (Supplementary Table S6).
Discussion
Utilizing the Illumina Infinium MethylationEPIC BeadChip (EPIC) array to quantify DNA methylation in infant cord blood leukocytes at over 800 000 CpG sites, this pilot study of 69 samples identified that maternal prenatal BPA exposure was associated with DNA methylation at 38 CpG sites while BPF and BPS in this same subset were not associated with specific CpG sites. Upon exclusion of one maternal−child pair on the basis of outlying maternal BPA exposure level, three CpG sites remained significant. There is increasing evidence of BPA exposure enhancing autoimmunity in humans [67–69]; this pilot study provides additional support of this association.
The preconception period and early pregnancy is a sensitive developmental time period for both physiological development and epigenetic reprogramming. During embryonic development, primordial germ cells and preimplantation embryos undergo two waves of methylation reprogramming [70, 71]. During the first wave, the paternal genome is actively demethylated and the maternal genome is passively demethylated, followed by reprogramming and remethylation of somatic embryonic stem cells and primordial germ cells in accordance with infant sex [72]. The interface of this essential reprogramming event with potential environmental or maternal exposures leaves the fetal epigenome extremely vulnerable to insult or alteration [73]. It is therefore possible that exposures experienced during this time period may alter DNA methylation in somatic embryonic stem cells and primordial germ cells; changes which could be propagated to subsequent cells and possibly influence development and disease later in life [73].
Considering very early in development is the most susceptible and a critical period for epigenetic effects [42], the focus of this investigation centered on maternal exposure to bisphenols during the first trimester. Our single-site analysis revealed 38 individual CpG sites in infant cord blood leukocytes that were differentially methylated in relation to early maternal BPA exposure (Table 2) and 3 DMRs in the genes HOXA-AS3, PRSS22, and ZSCAN12P1 (Table 3). However, the association of prenatal BPA exposure to DNA methylation in most of these genes was diminished when sensitivity analyses that excluded one outlying subject were performed. Associations with BPA and DNA methylation at CpG sites in SLC2A1-AS1 and RAD52 and the DMR in PRSS22 remained (P < 0.001) after outlier exclusion emphasizing the need to study these further in other birth cohorts with phenol exposures.
Using raw P-values from all model results, differentially methylated genes associated with BPA were enriched in gene-sets related to the nervous system, immune response, and neuroinflammation. These included JAK/STAT signaling and response [44, 47]; GPCRs, which play an important role in the nervous system [74–76]; and lastly, the IFN1 receptor and immune function pathways. Of particular interest as related to IFN1 receptor binding is its role in the severity and manifestation of SLE [77]. It has been demonstrated that BPA can stimulate estrogen-receptor alpha (ERalpha) and IFN signaling in myeloid cells and immune pathways resulting in activation of innate immune sensors [78], and increasing evidence supports the B-cell receptor pathway and IFN signaling in SLE pathogenesis [79]. Despite not reaching significance in the single-site models, urinary BPF exposure was also associated with the SLE gene-set during enrichment analysis.
Currently published literature of prenatal exposure to BPA and its epigenetic impact present similar findings of genes and gene-sets related to neurological function and inflammation. Junge et al. detected hypomethylation at two CpG sites in infant cord blood in response to maternal prenatal exposure to BPA: cg17580798 in the MEST promoter region and cg23117250 in an intronic region of RAB408 [50]. These sites were not significantly associated with prenatal BPA exposure in our study; however, MEST expression in mesenchymal tissue and mesenchymal stem cells (MSCs) and its functional significance to adipogenesis, particularly in the context of BPA exposure, is relevant to the role of HOXA-AS3, one of the DMRs detected in this study. HOXA-AS3 has a distinct role as an epigenetic switch in the lineage specification of MSCs as either promoting the adipogenic or osteogenic induction of MSCs [80]. Although we did not detect differential DNA methylation in the same genes, there is concordance between our results and those reported by Junge et al. in the potential for prenatal BPA exposure to impact genes related to MSCs, adipogenesis, and perhaps long-term body weight. Montrose et al. investigated the impact of maternal first-trimester urinary BPA exposure on DNA methylation in candidate genes in the same MMIP cohort [52]. Urinary BPA exposure was associated with a decrease in DNA methylation in IGF2 and PPARA in female infants; highlighting both the sexually dimorphic response of exposure to bisphenols and its association with disruption of genes related to growth and, adipogenesis, and metabolism. Alavian-Ghavanini et al. [51], a priori selected GRIN2B, a gene involved in neural function, and assessed associations between prenatal BPA exposure and DNA methylation at this gene in buccal DNA of 7-year old children. While GRIN2B was not associated with prenatal BPA in this study, GRIN2B and two genes associated with BPA in the present study, SLC2A1 and HIF1A, are related to one another via overlapping biological pathways [51, 81, 82]. Miura et al. [49] utilized a Japanese cohort for whom they measured BPA concentrations in cord blood and evaluated cross-sectional epigenome-wide associations with cord blood DNA methylation. A principle element of the study involved sex-stratified analyses, which detected significant differences in the response of male and female infants. They detected 28 DMS (q < 0.05) in male infants and 16 DMS in female infants [49]. While the same genes were not significant in the present study, there was concordance between the genes Miura et al., detected and the results presented in this study as related to gene families. For example, PRSS is a gene family for which we detected a DMR (PRSS22), and CpG sites within SLC and KIAA were associated with BPA in the Japanese cohort.
The discovery that neither BPF nor BPS maternal exposures were significantly associated with differential DNA methylation in the infant cord blood in this study was not surprising given the small sample size of the study, the necessity to model these exposures as categorical, and given that roughly half of mothers had undetectable levels of these bisphenols in their urine. Despite this, it was and is important to include BPF and BPS in the investigation of maternal exposure to bisphenols. BPF and BPS were first included in NHANES in 2013 − 14, and Lehmler et al. found that exposures to BPA, BPF, and BPS among adults and children could be considered near-ubiquitous [4]. We recommend assessment of other bisphenols in epigenetic studies in the future, because as the use of BPA substitutes in consumer products and manufacturing increases, it is pertinent to not only evaluate population exposure, but also to determine the impact of that exposure. Furthermore, we consider it valuable to simultaneously assess multiple bisphenols in human exposure studies so as to classify and categorize the similarities and differences of these toxicants.
Limitations and Future Directions
The MMIP cohort used in this study is based out of the University of Michigan Hospital in Ann Arbor, Michigan, and the majority of participants enrolled into the study were non-Hispanic White. This may limit the generalizability of the results. The final number of mother−infant pairs included in this study was determined by the availability of samples with data (e.g. maternal first-trimester urine with exposure assessment and infant umbilical cord blood). This limited our statistical power to detect DMS by all bisphenols, and broader biological pathways in association with maternal bisphenol exposure. However, in line with our recommendations for the inclusion of these bisphenols in exposure studies, the non-significant results that we detected for BPF and BPS still allow us to observe trends of exposure over time and a baseline to which we can compare future studies.
We also acknowledge that the small sample size may lead to spurious effects from statistical outliers. Thus, we report results with and without one BPA outlier. Since we cannot determine in this study whether individuals with higher exposure levels would display similar associations with BPA, we recommend future studies of prenatal BPA exposure and the offspring epigenome be performed in populations with a wide range of exposure to better understand how families with increased toxicant burdens may be impacted.
Additionally, we recognize the potential limitations of using infant cord blood as a surrogate tissue for evaluating the impact of prenatal exposures. While we control for cell-type heterogeneity with the use of a cord blood-specific cell-type reference panel, cord blood is still principally made up of immune cells. This may explain, in part, why some of the single sites that we detected were associated with immune function. However, we consider it a distinct strength of this study that we chose first-trimester maternal exposure assessment, particularly because we expect changes induced early in pregnancy to propagate across all germ layers and tissues of the developing fetus.
This pilot study examined the association between maternal first-trimester urinary bisphenol exposure and DNA methylation in infant cord blood. Maternal BPA exposure was associated with differential methylation at 38 single-sites in genes related to pathways of neurological function, inflammation, and in particular SLE. Given, however, that many of these associations lose statistical significance with the exclusion of a biological outlier, these associations must be interpreted with caution. With mounting evidence of the consequences associated with exposure to endocrine disrupting chemicals comes the sincere need to evaluate a variety of exposures across many populations. BPA and its replacement chemicals, BPF and BPS, remain heavily utilized in manufacturing, and exposure to these chemicals is considered ubiquitous. This study adds to the body of evidence about prenatal exposure to bisphenols and its association with differential DNA methylation in infants. These data begin to elucidate the correlation between these chemicals and ultimately provide additional tools that may be integrated in risk assessment and mitigation in individuals or populations with higher bisphenol exposure levels. Future studies should target a larger study population or meta-analysis, including participants from diverse backgrounds and a wide range of prenatal bisphenol exposures and accompanying RNA-seq data to determine the functional significance of differential DNA methylation. Furthermore, an important next step would be to assess the impact of prenatal bisphenol exposure on DNA methylation profiles in sorted CD4T or CD8T cells for determining the connection between BPA and autoimmune disease.
Supplementary Data
Supplementary data are available at EnvEpig online.
Funding
This work was funded by the National Institute of Environmental Health Sciences (NIEHS) Children’s Health Exposure Analysis Resource (CHEAR) program (grant no. U2CES026553) in addition to the following grants from the National Institutes of Health: grant nos R01ES017500, P01ES022844, P30ES017885, and 1UG3 OD023285-01. Grant RD83543601 from the US Environmental Protection Agency (US EPA) also supported this work. Contents are solely the responsibility of the grantees and do not necessarily represent the official views of the National Institutes of Health or the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
Conflict of interest statement. None declared.
Data Availability
Data will be available through a public repository. DOI will be provided before publication.
Supplementary Material
References
- 1. Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL.. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect 2008;116:39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thayer KA, Taylor KW, Garantziotis S, Schurman SH, Kissling GE, Hunt D, Herbert B, Church R, Jankowich R, Churchwell MI, Scheri RC, Birnbaum LS, Bucher JR.. Bisphenol a, bisphenol s, and 4-hydro xyphenyl 4-isopro oxyphenyl sulfone (bpsip) in urine and blood of cashiers. Environ Health Perspect 2016;124:437–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL.. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ Health Perspect 2005;113:391–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lehmler HJ, Liu B, Gadogbe M, Bao W.. Exposure to bisphenol A, bisphenol F, bisphenol S, in U. S. adults and children: the National Health and Nutrition Examination Survey 2013-2014. ACS Omega 2018;3:6523–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siracusa JS, Yin L, Measel E, Liang S, Yu X.. Effects of bisphenol A and its analogs on reproductive health: a mini review. Reprod Toxicol 2018;79:96–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rochester JR, Bolden AL.. Bisphenol S and F: a systematic review and comparison of the hormonal activity of bisphenol a substitutes. Environ Health Perspect 2015;123:643–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eladak S, Grisin T, Moison D, Guerquin M-J, N'Tumba-Byn T, Pozzi-Gaudin S, Benachi A, Livera G, Rouiller-Fabre V, Habert R.. A new chapter in the bisphenol a story: bisphenol S and bisphenol F are not safe alternatives to this compound. Fertil Steril 2015;103:11–21 [DOI] [PubMed] [Google Scholar]
- 8. Callan AC, Hinwood AL, Heffernan A, Eaglesham G, Mueller J, Odland JØ.. Urinary bisphenol A concentrations in pregnant women. Int J Hyg Environ Health 2013;216:641–4 [DOI] [PubMed] [Google Scholar]
- 9. Woodruff TJ, Zota AR, Schwartz JM.. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ Health Perspect 2011;119:878–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerona RR, Pan J, Zota AR. et al. Direct measurement of Bisphenol A (BPA), BPA glucuronide and BPA sulfate in a diverse and low-income population of pregnant women reveals high exposure, with potential implications for previous exposure estimates: a cross-sectional study. Environ Heal A Glob Access Sci Source 2016;15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arbuckle TE, Marro L, Davis K, Fisher M, Ayotte P, Bélanger P, Dumas P, LeBlanc A, Bérubé R, Gaudreau É, Provencher G, Faustman EM, Vigoren E, Ettinger AS, Dellarco M, MacPherson S, Fraser WD.. Exposure to free and conjugated forms of bisphenol A and triclosan among pregnant women in the MIREC cohort. Environ Health Perspect 2015;123:277–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J, Choi K, Park J, Moon H-B, Choi G, Lee JJ, Suh E, Kim H-J, Eun S-H, Kim G-H, Cho GJ, Kim SK, Kim S, Kim SY, Kim S, Eom S, Choi S, Kim YD, Kim S.. Bisphenol A distribution in serum, urine, placenta, breast milk, and umbilical cord serum in a birth panel of mother–neonate pairs. Sci Total Environ 2018;626:1494–501 [DOI] [PubMed] [Google Scholar]
- 13. Grandin FC, Lacroix MZ, Gayrard V, Gauderat G, Mila H, Toutain P-L, Picard-Hagen N.. Bisphenol S instead of bisphenol A: toxicokinetic investigations in the ovine materno-feto-placental unit. Environ Int 2018;120:584–92 [DOI] [PubMed] [Google Scholar]
- 14. Grandin FC, Lacroix MZ, Gayrard V, Viguié C, Mila H, de Place A, Vayssière C, Morin M, Corbett J, Gayrard C, Gely CA, Toutain P-L, Picard-Hagen N.. Is bisphenol S a safer alternative to bisphenol A in terms of potential fetal exposure? Placental transfer across the perfused human placenta. Chemosphere 2019;221:471–8 [DOI] [PubMed] [Google Scholar]
- 15. Dolinoy DC, Weidman JR, Jirtle RL.. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol 2007;23:297–307 [DOI] [PubMed] [Google Scholar]
- 16. Wan Y, Huo W, Xu S. et al. Relationship between maternal exposure to bisphenol S and pregnancy duration. Environ Pollut 2018;238:717–24 [DOI] [PubMed] [Google Scholar]
- 17. Weinberger B, Vetrano AM, Archer FE, Marcella SW, Buckley B, Wartenberg D, Robson MG, Klim J, Azhar S, Cavin S, Wang L, Rich DQ.. Effects of maternal exposure to phthalates and bisphenol A during pregnancy on gestational age. J Matern Neonatal Med 2014;27:323–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferguson KK, Meeker JD, Cantonwine DE, Chen YH, Mukherjee B, McElrath TF.. Urinary phthalate metabolite and bisphenol A associations with ultrasound and delivery indices of fetal growth. Environ Int 2016;94:531–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veiga-Lopez A, Kannan K, Liao C, Ye W, Domino SE, Padmanabhan V.. Gender-specific effects on gestational length and birth weight by early pregnancy BPA exposure. J Clin Endocrinol Metab 2015;100:E1394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cantonwine DE, Meeker JD, Ferguson KK, Mukherjee B, Hauser R, McElrath TF.. Urinary concentrations of bisphenol A and phthalate metabolites measured during pregnancy and risk of preeclampsia. Environ Health Perspect 2016;124:1651–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans SF, Kobrosly RW, Barrett ES, Thurston SW, Calafat AM, Weiss B, Stahlhut R, Yolton K, Swan SH.. Prenatal bisphenol A exposure and maternally reported behavior in boys and girls. Neurotoxicology 2014;45:91–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braun JM, Yolton K, Dietrich KN, Hornung R, Ye X, Calafat AM, Lanphear BP.. Prenatal bisphenol A exposure and early childhood behavior. Environ Health Perspect 2009;117:1945–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perera F, Vishnevetsky J, Herbstman JB, Calafat AM, Xiong W, Rauh V, Wang S.. Prenatal bisphenol a exposure and child behavior in an inner-city cohort. Environ Health Perspect 2012;120:1190–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harley KG, Schall RA, Chevrier J, Tyler K, Aguirre H, Bradman A, Holland NT, Lustig RH, Calafat AM, Eskenazi B.. Prenatal and postnatal bisphenol A exposure and body mass index in childhood in the CHAMACOS cohort. Environ Health Perspect 2013;121:514–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watkins DJ, Peterson KE, Ferguson KK, Mercado-García A, Tamayo y Ortiz M, Cantoral A, Meeker JD, Téllez-Rojo MM.. Relating phthalate and BPA exposure to metabolism in peripubescence: the role of exposure timing, sex, and puberty. J Clin Endocrinol Metab 2016;101:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ashley-Martin J, Dodds L, Arbuckle TE. et al. A birth cohort study to investigate the association between prenatal phthalate and bisphenol A exposures and fetal markers of metabolic dysfunction. Environ Heal 2014;13:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jašarević E, Williams SA, Vandas GM, Ellersieck MR, Liao C, Kannan K, Roberts RM, Geary DC, Rosenfeld CS.. Sex and dose-dependent effects of developmental exposure to bisphenol A on anxiety and spatial learning in deer mice (Peromyscus maniculatus bairdii) offspring. Horm Behav 2013;63:180–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Esterik JCJ, Dollé MET, Lamoree MH, van Leeuwen SPJ, Hamers T, Legler J, van der Ven LTM.. Programming of metabolic effects in C57BL/6JxFVB mice by exposure to bisphenol A during gestation and lactation. Toxicology 2014;321:40–52 [DOI] [PubMed] [Google Scholar]
- 29. Anderson OS, Peterson KE, Sanchez BN, Zhang Z, Mancuso P, Dolinoy DC.. Perinatal bisphenol a exposure promotes hyperactivity, lean body composition, and hormonal responses across the murine life course. FASEB J 2013;27:1784–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, Yu P, Qian W, Li Y, Zhao J, Huan F, Wang J, Xiao H.. Perinatal bisphenol A exposure and adult glucose homeostasis: identifying critical windows of exposure. PLoS One 2013;8:e64143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alonso-Magdalena P, Vieira E, Soriano S, Menes L, Burks D, Quesada I, Nadal A.. Bisphenol a exposure during pregnancy disrupts glucose homeostasis in mothers and adult male offspring. Environ Health Perspect 2010;118:1243–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. García-Arévalo M, Alonso-Magdalena P, Servitja J-M, Boronat-Belda T, Merino B, Villar-Pazos S, Medina-Gómez G, Novials A, Quesada I, Nadal A.. Maternal exposure to bisphenol-A during pregnancy increases pancreatic β-cell growth during early life in male mice offspring. Endocrinology 2016;157:4158–71 [DOI] [PubMed] [Google Scholar]
- 33. Li J, Wang Y, Fang F. et al. Bisphenol A disrupts glucose transport and neurophysiological role of IR/IRS/AKT/GSK3β axis in the brain of male mice. Environ Toxicol Pharmacol 2016;43:7–12 [DOI] [PubMed] [Google Scholar]
- 34. Franssen D, Gérard A, Hennuy B, Donneau AF, Bourguignon JP, Parent AS.. Delayed neuroendocrine sexual maturation in female rats after a very low dose of bisphenol a through altered gabaergic neurotransmission and opposing effects of a high dose. Endocrinology 2016;157:1740–50 [DOI] [PubMed] [Google Scholar]
- 35. Witchey SK, Fuchs J, Patisaul HB.. Perinatal bisphenol A (BPA) exposure alters brain oxytocin receptor (OTR) expression in a sex- and region- specific manner: a CLARITY-BPA consortium follow-up study. Neurotoxicology 2019;74:139–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weinhouse C, Anderson OS, Bergin IL, Vandenbergh DJ, Gyekis JP, Dingman MA, Yang J, Dolinoy DC.. Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ Health Perspect 2014;122:485–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fischer C, Mamillapalli R, Goetz LG, Jorgenson E, Ilagan Y, Taylor HS.. Bisphenol A (BPA) exposure in utero leads to immunoregulatory cytokine dysregulation in the mouse mammary gland: a potential mechanism programming breast cancer risk. Horm Canc 2016;7:241–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cardenas A, Lutz SM, Everson TM, Perron P, Bouchard L, Hivert MF.. Mediation by placental DNA methylation of the association of prenatal maternal smoking and birth weight. Am J Epidemiol 2019;188:1878–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Witt SH, Frank J, Gilles M, Lang M, Treutlein J, Streit F, Wolf IAC, Peus V, Scharnholz B, Send TS, Heilmann-Heimbach S, Sivalingam S, Dukal H, Strohmaier J, Sütterlin M, Arloth J, Laucht M, Nöthen MM, Deuschle M, Rietschel M.. Impact on birth weight of maternal smoking throughout pregnancy mediated by DNA methylation. BMC Genomics 2018;19:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ladd-Acosta C, Fallin MD.. Invited commentary: is DNA methylation an actionable mediator of prenatal exposure effects on child health? Am J Epidemiol 2019;188:1887–9 [DOI] [PubMed] [Google Scholar]
- 41. Kundakovic M, Champagne FA.. Epigenetic perspective on the developmental effects of bisphenol A. Brain Behav Immun 2011;25:1084–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCabe C, Anderson OS, Montrose L, Neier K, Dolinoy DC.. Sexually dimorphic effects of early-life exposures to endocrine disruptors: sex-specific epigenetic reprogramming as a potential mechanism. Curr Envir Health Rpt 2017;4:426–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strakovsky RS, Wang H, Engeseth NJ, Flaws JA, Helferich WG, Pan Y-X, Lezmi S.. Developmental bisphenol A (BPA) exposure leads to sex-specific modification of hepatic gene expression and epigenome at birth that may exacerbate high-fat diet-induced hepatic steatosis. Toxicol Appl Pharmacol 2015;284:101–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anderson OS, Kim JH, Peterson KE, Sanchez BN, Sant KE, Sartor MA, Weinhouse C, Dolinoy DC.. Novel epigenetic biomarkers mediating bisphenol A exposure and metabolic phenotypes in female mice. Endocrinology 2017;158:en.2016–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, Kannan K, Weinhouse C, Rozek LS, Dolinoy DC.. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen 2012;53:334–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kundakovic M, Gudsnuk K, Franks B, Madrid J, Miller RL, Perera FP, Champagne FA.. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc Natl Acad Sci U S A 2013;110:9956–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weinhouse C, Sartor MA, Faulk C, Anderson OS, Sant KE, Harris C, Dolinoy DC.. Epigenome-wide DNA methylation analysis implicates neuronal and inflammatory signaling pathways in adult murine hepatic tumorigenesis following perinatal exposure to bisphenol A. Environ Mol Mutagen 2016;57:435–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Senyildiz M, Karaman EF, Bas SS, Pirincci PA, Ozden S.. Effects of BPA on global DNA methylation and global histone 3 lysine modifications in SH-SY5Y cells: an epigenetic mechanism linking the regulation of chromatin modifying genes. Toxicol Vitr 2017;44:313–21 [DOI] [PubMed] [Google Scholar]
- 49. Miura R, Araki A, Minatoya M, Miyake K, Chen M-L, Kobayashi S, Miyashita C, Yamamoto J, Matsumura T, Ishizuka M, Kubota T, Kishi R.. An epigenome-wide analysis of cord blood DNA methylation reveals sex-specific effect of exposure to bisphenol A. Sci Rep 2019;9:12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Junge KM, Leppert B, Jahreis S, Wissenbach DK, Feltens R, Grützmann K, Thürmann L, Bauer T, Ishaque N, Schick M, Bewerunge-Hudler M, Röder S, Bauer M, Schulz A, Borte M, Landgraf K, Körner A, Kiess W, von Bergen M, Stangl GI, Trump S, Eils R, Polte T, Lehmann I.. MEST mediates the impact of prenatal bisphenol A exposure on long-term body weight development. Clin Epigenet 2018;10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alavian-Ghavanini A, Lin P-I, Lind PM, Risén Rimfors S, Halin Lejonklou M, Dunder L, Tang M, Lindh C, Bornehag C-G, Rüegg J.. Prenatal bisphenol A exposure is linked to epigenetic changes in glutamate receptor subunit gene Grin2b in female rats and humans. Sci Rep 2018;8:11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Montrose L, Padmanabhan V, Goodrich JM, Domino SE, Treadwell MC, Meeker JD, Watkins DJ, Dolinoy DC.. Maternal levels of endocrine disrupting chemicals in the first trimester of pregnancy are associated with infant cord blood DNA methylation. Epigenetics 2018;13:301–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grunau C, Barks A, Dolinoy DC. et al. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 2001;29:65e–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA.. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL.. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data 2016;9:22–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD.. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 2014;15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, L. McKenney S, Witter F, Walston J, Feinberg AP, Fallin MD.. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 2016;11:354–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goodrich JM, Ingle ME, Domino SE, Treadwell MC, Dolinoy DC, Burant C, Meeker JD, Padmanabhan V.. First trimester maternal exposures to endocrine disrupting chemicals and metals and fetal size in the Michigan Mother–Infant Pairs study. J Dev Orig Health Dis 2019;10:447–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE.. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017;33:3982–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smyth GK. limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, and Dudoit S (eds). Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer-Verlag, 2005, 397–420. [Google Scholar]
- 61. Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, Lin SM.. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xie C, Leung YK, Chen A, Long DX, Hoyo C, Ho SM.. Differential methylation values in differential methylation analysis. Bioinformatics 2019;35:1094–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hochberg Y, Benjamini Y.. More powerful procedures for multiple significance testing. Statist Med 1990;9:811–8 [DOI] [PubMed] [Google Scholar]
- 64. Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, Clark SJ, Molloy PL.. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ.. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim JH, Karnovsky A, Mahavisno V, Weymouth T, Pande M, Dolinoy DC, Rozek LS, Sartor MA.. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics 2012;13:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Alhomaidan HT, Rasheed N, Almatrafi S, Al-Rashdi FH, Rasheed Z.. Bisphenol A modified DNA: A possible immunogenic stimulus for anti-DNA autoantibodies in systemic lupus erythematosus. Autoimmunity 2019;52:272–80 [DOI] [PubMed] [Google Scholar]
- 68. Chailurkit L, Aekplakorn W, Ongphiphadhanakul B.. The association of serum bisphenol A with thyroid autoimmunity. Int J Environ Res Public Health 2016;13:1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sriphrapradang C, Chailurkit L, Aekplakorn W, Ongphiphadhanakul B.. Association between bisphenol A and abnormal free thyroxine level in men. Endocrine 2013;44:441–7 [DOI] [PubMed] [Google Scholar]
- 70. Reik W, Dean W, Walter J.. Epigenetic reprogramming in mammalian development. Science 2001;293:1089–93 [DOI] [PubMed] [Google Scholar]
- 71. Messerschmidt DM, Knowles BB, Solter D.. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 2014;28:812–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stein RA, Lee Davis D.. Nutrition and epigenetics In: Encyclopedia of Lifestyle Medicine & Health. Thousand Oaks: SAGE Publications, 2012, 404 [Google Scholar]
- 73. Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol 2015;218:71–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Arambula SE, Belcher SM, Planchart A, Turner SD, Patisaul HB.. Impact of low dose oral exposure to bisphenol a (BPA) on the neonatal rat hypothalamic and hippocampal transcriptome: a clarity-bpa Consortium study. Endocrinology 2016;157:3856–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Martínez R, Esteve-Codina A, Herrero-Nogareda L, Ortiz-Villanueva E, Barata C, Tauler R, Raldúa D, Piña B, Navarro-Martín L.. Dose-dependent transcriptomic responses of zebrafish eleutheroembryos to bisphenol A. Environ Pollut 2018;243:988–97 [DOI] [PubMed] [Google Scholar]
- 76. Jadhav R, Santucci-Pereira J, Wang Y, Liu J, Nguyen T, Wang J, Jenkins S, Russo J, Huang T, Jin V, Lamartiniere C.. DNA methylation targets influenced by bisphenol A and/or genistein are associated with survival outcomes in breast cancer patients. Genes (Basel) 2017;8:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol 2014;192:5459–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Panchanathan R, Liu H, Leung Y-K, Ho S, Choubey D.. Bisphenol A (BPA) stimulates the interferon signaling and activates the inflammasome activity in myeloid cells. Mol Cell Endocrinol 2015;415:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jarvinen TM, Hellquist A, Zucchelli M, Koskenmies S, Panelius J, Hasan T, Julkunen H, D'Amato M, Kere J.. Replication of GWAS-identified systemic lupus erythematosus susceptibility genes affirms B-cell receptor pathway signalling and strengthens the role of IRF5 in disease susceptibility in a Northern European population. Rheumatology 2012;51:87–92 [DOI] [PubMed] [Google Scholar]
- 80. Zhu X-X, Yan Y-W, Chen D, Ai C-Z, Lu X, Xu S-S, Jiang S, Zhong G-S, Chen D-B, Jiang Y-Z.. Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2016;7:63561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi M-B, Harpole D, Lancaster JM, Berchuck A, Olson JA, Marks JR, Dressman HK, West M, Nevins JR.. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353–7 [DOI] [PubMed] [Google Scholar]
- 82. Wu G, Feng X, Stein L.. A human functional protein interaction network and its application to cancer data analysis. Genome Biol 2010;11:R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available through a public repository. DOI will be provided before publication.