Abstract
Rab small GTPases regulate membrane traffic between distinct cellular compartments of all eukaryotes in a tempo‐spatially specific fashion. Rab small GTPases are also involved in the regulation of cytoskeleton and signalling. Membrane traffic and cytoskeletal regulation play pivotal role in the pathogenesis of Entamoeba histolytica, which is a protozoan parasite responsible for human amebiasis. E. histolytica is unique in that its genome encodes over 100 Rab proteins, containing multiple isotypes of conserved members (e.g., Rab7) and Entamoeba‐specific subgroups (e.g., RabA, B, and X). Among them, E. histolytica Rab7 is the most diversified group consisting of nine isotypes. While it was previously demonstrated that EhRab7A and EhRab7B are involved in lysosome and phagosome biogenesis, the individual roles of other Rab7 members and their coordination remain elusive. In this study, we characterised the third member of Rab7, Rab7D, to better understand the significance of the multiplicity of Rab7 isotypes in E. histolytica. Overexpression of EhRab7D caused reduction in phagocytosis of erythrocytes, trogocytosis (meaning nibbling or chewing of a portion) of live mammalian cells, and phagosome acidification and maturation. Conversely, transcriptional gene silencing of EhRab7D gene caused opposite phenotypes in phago/trogocytosis and phagosome maturation. Furthermore, EhRab7D gene silencing caused reduction in the attachment to and the motility on the collagen‐coated surface. Image analysis showed that EhRab7D was occasionally associated with lysosomes and prephagosomal vacuoles, but not with mature phagosomes and trogosomes. Finally, in silico prediction of structural organisation of EhRab7 isotypes identified unique amino acid changes on the effector binding surface of EhRab7D. Taken together, our data suggest that EhRab7D plays coordinated counteracting roles: a inhibitory role in phago/trogocytosis and lyso/phago/trogosome biogenesis, and an stimulatory role in adherence and motility, presumably via interaction with unique effectors. Finally, we propose the model in which three EhRab7 isotypes are sequentially involved in phago/trogocytosis.
Keywords: cytoskeleton, Entamoeba histolytica, lysosome, pathogenesis, phagocytosis, Rab7D, trogocytosis, vesicular traffic
1. INTRODUCTION
Entamoeba histolytica is the causative agent of human amebiasis, one of the 25 leading causes of global mortality from 1990 to 2010 (Lozano et al., 2012; UNICEF, 2019). Clinical manifestations include colitis, dysentery and extraintestinal (e.g., hepatic, pulmonary, and cerebral) abscesses (Shirley, Farr, Watanabe, & Moonah, 2018). E. histolytica exploits elaborate biological processes that contribute to its parasitism and pathogenesis, including motility, adherence, phago‐ and trogocytosis, cytotoxicity, defence against nutrient starvation, temperature changes, and host immune systems (Faust & Guillen, 2012; Marie & Petri Jr, 2014; Nakada‐Tsukui & Nozaki, 2016; Orozco, Guarneros, Martinez‐Palomo, & Sánchez, 1983; Ralston et al., 2014; Tavares et al., 2005; Tovy et al., 2011). All of these processes require coordinated regulation of signal transduction, cytoskeletal reorganization, and vesicular trafficking (Aguilar‐Rojas, Olivo‐Marin, & Guillen, 2016; Babuta, Bhattacharya, & Bhattacharya, 2020; Nozaki & Nakada‐Tsukui, 2006). Recent studies indicate in particular that trogo‐ and phagocytosis play a pivotal role in disease manifestations and host immune evasion (Miller, Suleiman, & Ralston, 2019; Nakada‐Tsukui & Nozaki, 2020; Ralston et al., 2014; Somlata, Nakada‐Tsukui, & Nozaki, 2017). It has been shown that both amebic trogo‐ and phagocytosis are energy dependent and require Gal/GalNAc lectin, protein and lipid kinases such as C2‐domain‐containing protein kinase (EhC2PK), atypical kinase (EhAK1), and phosphoinositide‐3‐kinase (PI3K), phosphatidylinositide‐binding proteins such as FYVE domain containing protein 4 (Nakada‐Tsukui, Okada, Mitra, & Nozaki, 2009) and SNXs (Watanabe, Nakada‐Tsukui, & Nozaki, 2020), and proteins involved in actin rearrangement such as calcium binding proteins, ArpA2/3, Rho/Rac, EhP3, and EhFP10 (Agarwal et al., 2019; Babuta et al., 2020; Babuta, Kumar, Gourinath, Bhattacharya, & Bhattacharya, 2018; Babuta, Mansuri, Bhattacharya, & Bhattacharya, 2015; Bharadwaj et al., 2018; Bharadwaj, Arya, Shahid Mansuri, Bhattacharya, & Bhattacharya, 2017; Gautam, Ali, Bhattacharya, & Gourinath, 2019; Nakada‐Tsukui et al., 2009; Powell et al., 2006; Ralston et al., 2014; Somlata, Bhattacharya, & Bhattacharya, 2011; Watanabe et al., 2020). EhAGC kinase 1 represents only protein that is known to be engaged with only trogocytosis, but not phagocytosis, whereas EhAGCK2 is involved in both processes, but on the different stages of target internalisation.
Rab small GTPases are the evolutionally conserved molecular switch that regulates directional and compartment‐specific membrane traffic. E. histolytica shows, despite its unicellularity, an exceptional expansion of Rab proteins encoded by its genome: 104 Rab genes that include 22 conserved Rab subfamilies such as Rab1, 2, 5, 7, 8, 11, and 21, and a number of unique lineage‐specific subfamilies such as RabA, B, and X (Nakada‐Tsukui, Saito‐Nakano, Husain, & Nozaki, 2010; Saito‐Nakano, Loftus, Hall, & Nozaki, 2005; Verma, Srivastava, & Datta, 2018), suggesting the functional diversification of Rabs in this parasite. Among evolutionarily conserved Rabs in E. histolytica, Rab7 represents the most divergent group. E. histolytica Rab7 subfamily is largely diversified and composed of 9 isotypes (Rab7A to Rab7I), designated alphabetically in a descending order of percentage identity to human Rab7 or yeast Ypt7 (Nakada‐Tsukui et al., 2010; Saito‐Nakano et al., 2005; Saito‐Nakano, Nakazawa, Shigeta, Takeuchi, & Nozaki, 2001). It was shown that six of nine Rab7 isotypes are expressed under axenic culture conditions (except for EhRab7C, EhRab7F, and EhRab7I, which are transcribed at the basal levels) (Saito‐Nakano, Mitra, Nakada‐Tsukui, Sato, & Nozaki, 2007). The presence of multiple Rab7 isoforms in E. histolytica likely indicates their functional specialisation and complexity of cellular trafficking they are involved in (e.g., lysosome biogenesis) as previously suggested (Mackiewicz & Wyroba, 2009). It was previously demonstrated that EhRab7A and EhRab7B are involved in lysosome and phagosome biogenesis (Nakada‐Tsukui, Saito‐Nakano, Ali, & Nozaki, 2005; Saito‐Nakano et al., 2007). EhRab7A is associated with retromer complex through its component Vps26 and is involved in the formation of the preparatory prephagosomal vacuole (PPV) in conjunction with EhRab5. EhRab7B is exclusively localised to the acidic compartment, lysosome, and contains lysosomal virulence‐related proteins such as amoebapore A and cysteine proteases. However, the individual roles of other Rab7 members remain elusive.
In this study, in order to better understand the significance of the multiplicity of Rab7 isotypes and a specific role of individual Rab7 isotypes in E. histolytica, we conducted functional characterisation of the third member of Rab7, EhRab7D. We chose this isotype because it was one of three Rab7 isotypes that were identified in phagosomes by two independent proteomic analysis of the purified phagosomes (Marion, Laurent, & Guillén, 2005; Okada et al., 2005). We took genetic approaches and created transformants in which EhRab7D was either overexpressed and transcriptionally gene silenced. Virulence‐associated cellular parameters were evaluated in these transformants, including adherence, motility, trogo‐ and phagocytosis, PPV formation, and acidification and degradation in phagosomes. Intracellular localization was also visualised by immunofluorescence imaging. Finally, we also conducted in silico analysis of all nine EhRab7 isotypes in comparison with mammalian counterpart. Our data suggest that EhRab7D plays apparently opposing (i.e., stimulatory or inhibitory) roles in vesicular traffic and adhesion/motility, respectively, during trogo‐ and phagocytosis of the host cells.
2. RESULTS
2.1. Overexpression of EhRab7D caused a reduction in phagocytosis and trogocytosis while EhRab7D gene silencing led to opposite phenotypes
The fact that EhRab7D was previously identified in purified phagosomes (Marion et al., 2005; Okada et al., 2005) prompted further characterisation of EhRab7D. To see if and to what extent EhRab7D is involved in phagocytosis and trogocytosis, we first examined phenotypic changes in internalisation of host cells caused by overexpression of HA‐tagged EhRab7D. Expression of HA‐EhRab7D in E. histolytica HM‐1 strain was verified by immunoblot analysis using antibody raised against recombinant EhRab7D and HA antibody (Figure 1a, Figure S1). The bands around 23 and 30 kDa corresponding to endogenous EhRab7D (23 kDa) and HA‐EhRab7D were observed by anti‐EhRab7D and anti‐HA antibody (Figure 1a).
FIGURE 1.
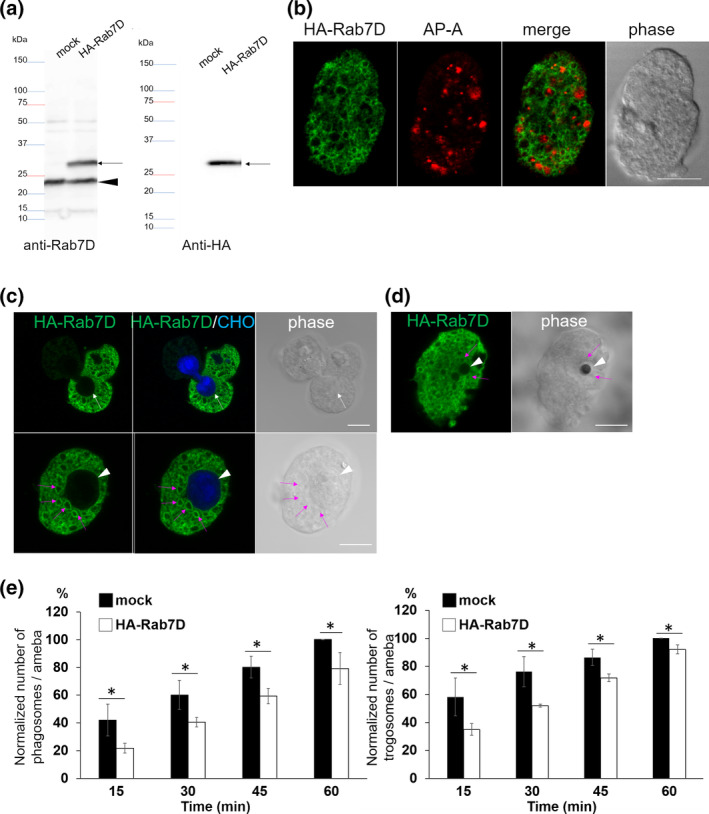
Expression and localization of HA‐EhRab7D and the effect of EhRab7D overexpression on phagocytosis and trogocytosis. (a) Immunoblot analysis of HA‐EhRab7D in E. histolytica transformants. Total cell lysates from mock‐transfected control (mock) and HA‐EhRab7D‐expressing transformant (HA‐Rab7D) were subjected to SDS‐PAGE and immunoblot analysis using the anti‐EhRab7D (left panel) and anti‐HA (right panel) antibodies. Arrows indicate HA‐EhRab7D. An arrowhead indicates intrinsic wild‐type EhRab7D. (b) Localization of HA‐EhRab7D in quiescent state. HA‐EhRab7D‐expressing transformant was fixed and reacted with anti‐HA (green) and anti‐AP‐A (red) antibodies. Bar, 10 μm. (c) Localization of HA‐EhRab7D during trogocytosis. HA‐EhRab7D expressing E. histolytica trophozoites were co‐cultured with CellTracker Blue‐stained live CHO cells for 15 min, fixed, and reacted with the anti‐HA antibody (green). A white arrow indicate trogocytic cup. An arrowhead indicate the trogosome. Magenta arrows indicate HA‐EhRab7D‐associated vesicles adjacent to the trogosome. Bar, 10 μm. (d) Localization of HA‐EhRab7D during phagocytosis of artificial beads. HA‐EhRab7D expressing E. histolytica trophozoites were incubated with human serum coated Dynabeads (2.8‐μm diameter) for 15 min, fixed, and reacted with the anti‐HA antibody (green). A white arrowhead and magenta arrows indicate the phagosome and HA‐EhRab7D‐associated vesicles, respectively. Bar, 10 μm. (e) The effect of HA‐ERab7D expression on phagocytosis and trogocytosis. Trophozoites of mock transfected and HA‐EhRab7D expressing strains were prestained with CellTracker Blue and co‐cultivated with live or heat‐killed CHO cells that had been prestained with CellTracker Orange to evaluate trogocytosis or phagocytosis, respectively. Microscopic images were captured on CQ1 every 15 min for 60 min and analysed to calculate the average numbers of CHO cell‐containing trogo‐/phagosomes per ameba. All data were normalised against the values obtained for control at 1 hr and are shown as percentages to these values. Data of one representative experiment was shown. Statistical significance was examined with Student's t test, *p < .05. n = 3
Immunofluorescence imaging revealed that HA‐Rab7D was mostly localised in the cytoplasm and occasionally on vesicular membranes in steady state (Figure 1b). HA‐Rab7D was also observed on the vacuoles that contain the lysosomal pore‐forming peptide, amebapore A (AP‐A), suggesting its partial localization to lysosomes, late endosomes, or non‐acidified prelysosomes. The localization of HA‐Rab7D detected by anti‐HA antibody was indistinguishable from that of endogenous EhRab7D (Figure S1). Next, we examined localization of HA‐Rab7D during trogocytosis (Figure 1c) and phagocytosis (Figure 1d). CellTracker Blue‐stained live Chinese hamster ovary (CHO) cells or serum coated Dynabeads were co‐cultured with HA‐EhRab7D expressing E. histolytica trophozoites for 15 min, then indirect immunofluorescence assay (IFA) was conducted to visualise HA‐EhRab7D with anti‐HA antibody. To our surprise, although HA‐EhRab7D signals were closely associated with trogosomes and phagosomes, HA‐EhRab7D was not precisely located to CHO cell‐containing trogosomes, the trogocytic cup, or bead‐containing phagosomes. It was unexpected because EhRab7D was previously reported to be associated with purified phagosomes (Marion et al., 2005; Okada et al., 2005).
As EhRab7D was observed closely associated with trogosomes and phagosomes (although the localization of EhRab7D on the phagosome/trogosome per se was not observed), we examined the effect of HA‐EhRab7D expression on phagocytosis and trogocytosis (Figure 1e). HA‐EhRab7D expressing and mock transfected E. histolytica strains (stained with CellTracker Blue) were co‐cultured with heat killed or live CHO cells (stained with CellTracker Orange), and internalisation of the prey was analysed by CQ1 confocal quantitative image cytometer. Internalisation of the prey was evaluated with three parameters, that is, the percentage of amebas containing CHO‐phagosomes/trogosomes, the number of CHO‐phagosomes/trogosomes per ameba, and the sum of the volume of CHO‐phagosomes/trogosomes per ameba. Data were normalised against those parameters obtained at 1 hr of coincubation and are shown as percentages of the values for the mock transformant. The average number of CHO‐phagosome/trogosome per ameba (Figure 1e), as well as other two parameters (data not shown) showed a consistent trend, showing that phagocytosis and trogocytosis were significantly inhibited by HA‐EhRab7D expression (p < .05).
2.2. Overexpression of EhRab7D also caused a reduction in erythrophagocytosis and the formation of prephagosomal vacuoles
We also observed a decrease in erythrophagocytosis by HA‐EhRab7D expressing cells compared to the mock control (Figure 2a, p = .02), which was similar to the phagocytosis/trogocytosis of CHO cells (Figure 1e). Expression of EhRab7D also inhibited acidification of phagosomes containing fluorescein isothiocyanate (FITC)‐labelled yeast (Figure 2b), suggesting involvement in phagosome maturation. It was previously shown that in the early phase of erythrophagocytosis (~10 min), an Entamoeba‐specific vesicular (vacuolar) compartment called prephagosomal vacuole (PPV), which is decorated with EhRab5 and EhRab7A (Saito‐Nakano, Yasuda, Nakada‐Tsukui, Leippe, & Nozaki, 2004), is formed; and in the later phase (at 30 min), Rab7A‐positive PPV fuses with erythrocyte‐containing phagosomes after EhRab5 is dissociated from PPV. PPVs are defined as EhRab7A‐associated vacuoles with the diameter of >3 μm (Saito‐Nakano et al., 2004) hereinafter. The distribution and dynamism of EhRab7A and EhRab7D was monitored during erythrophagocytosis. EhRab7A‐positive/EhRab7D‐negative PPVs were formed at 10 min (Figure 2c, red arrowheads). Subsequently, EhRab7A/EhRab7D‐double positive PPVs were observed at 30 min (Figure 2c, yellow arrowheads. EhRab7D‐positive PPVs were always colocalized with EhRab7A, and EhRab7A‐negative/EhRab7D‐positive PPVs were not observed. EhRab7D was seldom localised to erythrocyte‐containing phagosomes at 30 min (Figure 2c, erythrocytes were stained black). The PPVs were counted in HA‐EhRab7D expressing and mock transformants in a course of erythrophagocytosis. The number of EhRab7A‐positive PPVs decreased by approximately 70% in HA‐EhRab7D expressing cells at 10 min of erythrophagocytosis compared to mock control (EhRab7A‐positive PPVs per cell: 3.70 ± 0.63 in mock and 1.05 ± 0.2 in HA‐EhRab7D; p = .01) (Figure 2d, black and white bars; note that white bars are overlaid with grey bars). The number of EhRab7D‐positive PPVs increased in a course of erythrophagocytosis (EhRab7A/EhRab7D‐double positive PPVs per cell: 0.6 ± 0.1 at 10 min and 1.4 ± 0.3 at 30 min; p = .03) (Figure 2d, grey bars). These data indicate that EhRab7D is recruited to PPVs after EhRab7A during erythrophagocytosis and EhRab7D inhibits PPV formation.
FIGURE 2.
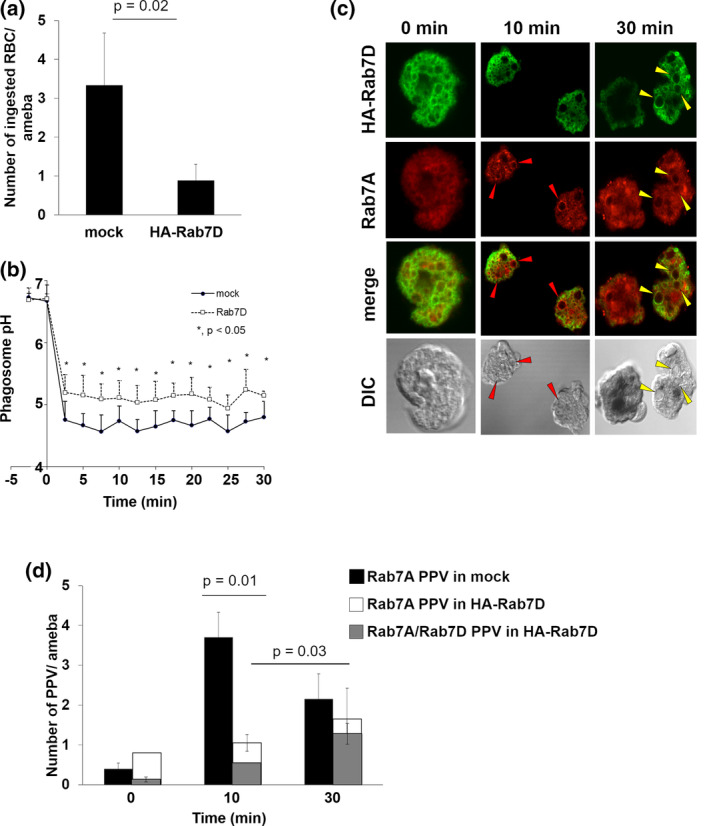
Effect of HA‐EhRab7D expression on erythrophagocytosis, phagosome acidification, and PPV formation. (a) Erythrophagocytosis by mock transfected and HA‐EhRab7D expressing strains. Trophozoites of mock transfected and HA‐EhRab7D expressing strains were incubated with hamster erythrocytes (RBCs) for 30 min. Cells were fixed and erythrocytes were visualised with diaminobenzidine. Number of RBCs per amoeba was counted. Statistical significance was inferred by t‐test, p = .02. (b) Kinetics of phagosome acidification in mock transfected (closed circles) and mRFP‐EhRab7D‐expressing (open squares) strains. Trophozoites were incubated with FITC‐conjugated yeasts at 1:10 ratio at 33°C to allow the amebae to ingest yeasts. mRFP‐EhRab7D‐expressing trophozoites that ingested only a single yeast were selected and phagosome pH was measured at 1 min intervals for 30 min. Data represent the average of 10 independent phagosomes. Statistical significance was examined with t‐test, *p < .05. (c) Localization of HA‐EhRab7D and EhRab7A during erythrophagocytosis. Trophozoites of HA‐EhRab7D‐expressing strain was incubated with RBCs for indicated time, fixed, reacted with anti‐EhRab7A or anti‐HA antibodies to visualise EhRab7A (red) or HA‐EhRab7D (green). Ingested RBCs were visualised by diaminobenzidine. Red arrowheads indicate EhRab7A‐associated vacuoles and yellow arrowheads indicate EhRab7A and HA‐EhRab7D double‐associated vacuoles. Merged images of EhRab7A and HA‐EhRab7D and differential interference contrast (DIC) images are also shown. (d) PPV formation during erythrophagocytosis in mock (black) and HA‐EhRab7D overexpressing (white and grey) transformants. Trophozoites of mock transfected and HA‐EhRab7D expressing strains were incubated with RBCs for indicated time, fixed, and reacted with anti‐EhRab7A (black and white), anti‐HA (grey), and diaminobenzidine as in (c). The numbers of EhRab7A‐associated vacuoles (PPV) and EhRab7A/HA‐RhRab7D‐double associated PPVs per ameba were calculated. Statistical significance was examined with t‐test, p = .01 and .03
2.3. Silencing of EhRab7D caused augmentation of trogocytosis and phagocytosis, but reduction in motility and adhesion
We next examined the effect of EhRab7D gene silencing on phagocytosis and trogocytosis. The silencing of the target gene was confirmed by RT‐PCR and immunoblot (Figure 3a). Non‐specific off‐target gene silencing of non‐targeted Rab genes was ruled out by RNA‐seq analysis (Table S1). EhRab7D gene transcript and encoded protein were totally abolished in the gene silenced strain (Figure 3a). EhRab7D gene silenced strain showed an enhancement in the formation of PPVs (EhRab7A‐positive, as defined as above) in the absence of erythrocytes (Figure 3b). The diameter of PPVs in EhRab7D gene silenced strain was larger than that of mock control strain (mock: 3.1 ± 0.3 μm, Rab7Dgs: 4.2 ± 0.7 μm, p = .02). In both strains, more than 85% of PPVs were not acidified, suggesting that lysosomes had not been fused with PPVs in the steady state. Phagocytosis and trogocytosis in EhRab7D gene silenced and mock transfected strains were quantified (Figure 3c). Both phagocytosis and trogocytosis were significantly increased in EhRab7D gene silenced strain. Together with the results of HA‐EhRab7D overexpression (Figure 1e), these data indicate that EhRab7D serves as negative regulator of phagocytosis and trogocytosis.
FIGURE 3.
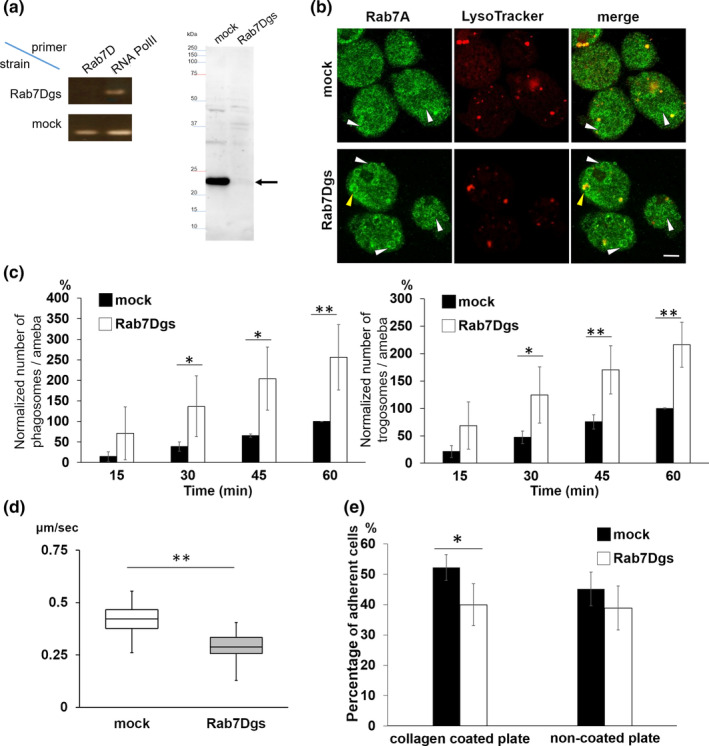
Establishment and phenotypes of EhRab7D gene silenced strain. (a) Confirmation of gene silencing by reverse transcriptase (RT)‐PCR (left panel) and immunoblot (right panel) analysis of mock transfected and EhRab7D gene silenced (gs) strain. Transcripts of EhRab7D and RNA polymerase II genes were amplified by RT‐PCR from cDNA isolated from the transformants and examined by agarose gel electrophoresis (left panel). Total lysate of the transformants were subjected to SDS‐PAGE and immunoblot analysis using the anti‐EhRab7D antibody (right panel). (b) Localization of EhRab7A and LysoTracker in EhRab7D gs and control strains. EhRab7D gs and mock transfected strains were stained with LysoTracker Red, fixed, and reacted with anti‐EhRab7A antibody (green). The diameter of the largest EhRab7A‐positive vacuole in each cell was measured in 50 trophozoites from three independent experiments. (c) The effect of gene silencing of EhRab7D gene on phagocytosis (left) and trogocytosis (right). Trophozoites of mock transfected and EhRab7D gene silenced strains prestained with CellTracker Blue were co‐cultivated with heat‐killed (left panel) or live (right panel) CHO cells prestained with CellTracker Orange to evaluate phagocytosis or trogocytosis, respectively. Also see the legend in Figure 1e. Statistical significance was examined with t‐test, *p < .05, **p < .01. n = 4. (d) Cell motility of mock transfected and EhRab7D gene silenced strain. The trophozoites of the indicated strains were stained with CellTracker Green were incubated with BI medium at 35.5°C on a collagen‐coated glass‐bottom dish. Time‐lapse images were collected every 30 s for 15 min on LSM780 and processed using Imaris software. Data form one representative experiment was shown. Statistical significance was examined with Dunnet test, **p < .05. n = 2. (e) Adhesion to collagen coated and non‐coated plastic plates of mock transfected and EhRab7D gene silenced strain. Trophozoites were prestained with CellTracker Green and incubated with BI medium on indicated plates at 35.5°C for 40 min. After incubation, the plates were gently rinsed with fresh warm BI medium, and the fluorescence intensity of each well before and after washing was measured, normalised against the value of the mock control at 60 min, and is shown in percentage. Statistical significance was examined with t‐test, *p < .05 and n = 4
Since the augmentation of phagocytosis and trogocytosis was observed as early as at 15 min, the observed phenotype may be attributable to activation of cytoskeletal rearrangement activity. We next examined motility of the trophozoites on the collagen coated glass surface and adhesion of the amebas to the collagen‐coated and non‐coated plastic plates (Figure 3d,e). Time‐lapse imaging and measurement of motility showed that the average velocity of motility of EhRab7D gene silenced strains was 0.29 ± 0.07 μm/s, which was significantly slower than that of control (0.43 ± 0.08 μm/s) (Figure 3d). Similarly, EhRab7D gene silenced strain showed significantly lower adhesion (12.3 ± 5.2%, p = .01) to a collagen‐coated plastic surface (Figure 3e). Adherence of EhRab7D gene silenced strain to non‐coated plastic surface was also reduced, although statistically insignificant.
2.4. In silico analysis of 9 isotypes suggests unique roles of EhRab7D in phago‐ and trogocytosis
To better understand structural and functional differences of EhRab7 isotypes, multiple sequence alignment of all nine EhRab7 isotypes with rat Rab7 was created (Figure 4). The nine EhRab7 isotypes are similar (about 40 to 50% identity) to one another. We found two unique features in EhRab7D. First, two unique amino acid substitutions (T/Q‐E/D/S to I/A‐P/T) in the inter‐switch region between Switch 1 and Switch 2 were identified. Second, an acidic amino acid stretch at aa.178–184 (“EDEEEPE”), was found in the natural disorder region at the carboxyl terminus (Figure 4). The inter‐switch region is near the binding site of the effector RILP for RnRab7 (Figure S2). Interestingly, the both features are also shared by EhRab7G, suggesting that EhRab7D and EhRab7G may share effector(s) and be involved in common pathways.
FIGURE 4.

Alignment of nine EhRab7 isotypes and rat Rab7 (RnRab7). Multiple amino Acid sequence alignment of all nine E. histolytica Rab7 isotypes with rat Rab7 (RnRab7, PDB ID: 1vg0B/UniProt: P09527) was constructed by using FAMSA with its default parameters. Switch 1 (SW1), switch 2 (SW2), and inter switch (IntSW) regions are depicted above the sequences. Two regions containing EhRab7D‐ and EhRab7G‐specific amino acid substitutions are coloured in yellow or green, and unique amino acids are highlighted with red squares. A region with yellow background corresponds to the interface between mammalian Rab7 effector RILP (see Supplementary Figure S2). Note the conserved motif of T/Q‐K/R‐E/D/S in all sequences except for EhRab7D and EhRab7G, in which the motif is replaced with I‐K‐P or A‐K‐T, respectively. The region with green background is located in an intrinsically disordered region, in which EhRab7D and EhRab7G contain a unique acidic amino acid stretch. “E” and “H” denote beta‐strand and alpha‐helix, respectively. “*” and “+” depict completely conserved and positively charged amino acids around SW2
3. DISCUSSION
3.1. A current model of EhRab7A, EhRab7B, and Rab7D in trogocytosis and phagocytosis
It has been well established that Rab7 GTPase has a broad range of roles in crucial cellular functions, including phagocytosis, endocytosis, autophagy, mitophagy, lipophagy, cytoskeletal dynamics, apoptosis, and tumour suppression (Guerra & Bucci, 2016). Based on the results of the current study and from literatures, we propose the following model for the role of three EhRab7 isotypes, EhRab7A, EhRab7B, and EhRab7D in E. histolytica during host cell internalisation (Figure 5, the number in a square bracket represents the step [or compartment] in the pathway in this figure). One of the hallmarks of the present study is the discovery that EhRab7D is involved in the formation of PPV, which is the unique preparatory compartment for phagocytosis, previously documented for the first time in this professional phagocytic eukaryote (Saito‐Nakano et al., 2004). PPV serves as a preparatory compartment for degradative functions to deliver digestive enzymes to phago‐ and trogosomes.
FIGURE 5.
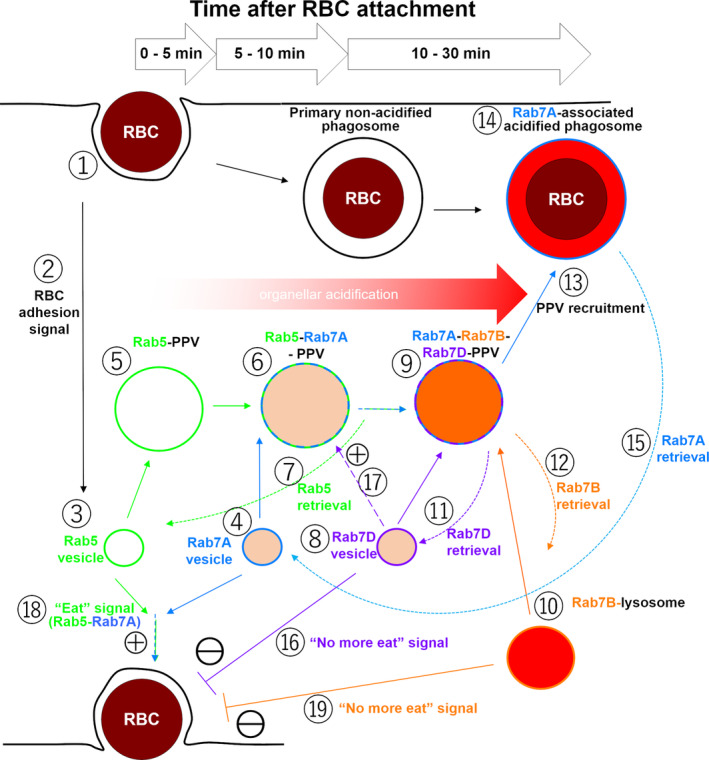
A proposed model of the roles of EhRab7A, EhRab7B, and EhRab7D during erythrophagocytosis of E. histolytica. Refer to Discussion
Interaction with the host cell (any nucleated cells or erythrocyte) [1, 2] induces the formation of PPV [5], and this process is apparently mediated by EhRab5 and EhRab7A [3, 4]. Under quiescent state (before erythrocyte interaction) EhRab5 and EhRab7A are localised to independent small vesicles [3, 4]. EhRab5 vesicles [3] fuse (homotypically or heteropypically with unknown vesicles) and form EhRab5‐PPVs [5]. Subsequently EhRab7A vesicles [4] fuse with EhRab5‐PPVs and form EhRab5/EhRab7A double‐positive PPVs [6] within 10‐min after initiation of phagocytosis (Saito‐Nakano et al., 2004). Under normal conditions, EhRab7A, but not EhRab5, partially overlaps with lysosomes (Nakada‐Tsukui et al., 2005; Saito‐Nakano et al., 2004), suggesting EhRab7A only partly localises to late endosomes and lysosomes [3, 4]. PPVs become EhRab7A single‐positive after release of EhRab5 [7]. EhRab7D, which is localised to a non‐acidified compartment under normal conditions [8], is then recruited to PPVs and colocalized with EhRab7A [9]. EhRab7B, in lysosomes [10], is also recruited to PPVs after EhRab5 is dissociated from PPVs [7] and EhRab7D gets associated with PPV [9]. Now PPVs are acidified and contain lysosomal proteins, such as amoebapore A and cysteine protease 2 (Saito‐Nakano et al., 2004, 2007) [9]. Contrary to EhRab5 and EhRab7D, EhRab7B is exclusively localised to lysosomes [10] and only partially co‐localised with EhRab7A in steady state (Saito‐Nakano et al., 2007). EhRab7B‐positive lysosomes are probably responsible for PPV acidification. Finally, EhRab7B and EhRab7D are dissociated from PPVs (with an unknown trigger) [11, 12], while PPVs are still decorated with EhRab7A, prior to subsequent fusion to the primary phago‐ or trogosome [13] to fully mature them [15]. One should not that exact timing and signal for the dissociation of EhRab5 from PPV [7], the recruitment to and dissociation from PPV of EhRab7A [4, 15], EhRab7B [10, 12], and EhRab7D [8, 11] remain to be clearly demonstrated.
3.2. Rab7D is a negative regulator of trogocytosis and phagocytosis
Another significance of the present study is the discovery of EhRab7D acting as a negative regulator of trogo‐ and phagocytosis. Overexpression of EhRab7D clearly inhibited phago and trogocytosis, while EhRab7D gene silencing enhanced them [16]. This is apparently linked with the fact that EhRab7D overexpression repressed PPV formation [17]. Similar EhRab mediated regulation of phagocyotosis and trogocytosis was demonstrated. Co‐expression of EhRab5 and EhRab7A caused enhancement of erythrophagocytosis, suggesting that either or both of EhRab5 and EhRab7A are involved in triggering of phagocytosis (Saito‐Nakano et al., 2004) [18]. EhRab7B overexpression also induced formation of EhRab7A‐positive PPVs without phagocytosis stimuli and caused a reduction in phagocytosis (Saito‐Nakano et al., 2007) [19]. EhRab7D is needed for maturation and acidification of PPV, as suggested by the evidence that gene‐silencing of EhRab7D caused enlargement of EhRab7A‐positive vacuoles (Figure 3b) and that EhRab7A‐positive enlarged PPVs in EhRab7D gene‐silenced cells were not acidified (Figure 3b). Thus, although EhRab7A, EhRab7B, and EhRab7D are involved in PPV biogenesis during phagocytosis, they seem to play counteracting roles on PPV formation and phago‐ and trogocytosis.
3.3. Effectors of Rab7 isotypes
It was previously demonstrated that EhRab7A binds to the retromer complex, via a unique carboxyl terminal extension in Vps26 (Nakada‐Tsukui et al., 2005). While effectors and binding proteins for EhRab7D remain elusive, the effector of EhRab7A was previously demonstrated (Nakada‐Tsukui et al., 2005). Retromer complex, the EhRab7A effector, is assumed to be involved in the forward transport of hydrolytic enzymes or their cargo receptors to trogosomes and phagosomes or in the transport in a reverse direction in E. histolytica (Nakada‐Tsukui et al., 2005; Picazarri et al., 2015; Watanabe et al., 2020). Retromer is composed of Vps26, Vps29, Vps35 and SNXs (although direct binding between Vps26/29/35 and SNXs has not yet been demonstrated). Interestingly, EhSNX1, but not EhSNX2, interacts with the Vps26/Vps29/Vps35 complex and thus can subsequently interacts with EhRab7A. Since EhSNX1 binds to Arp2/3 complex (Babuta et al., 2015, 2018, also see next section), EhRab7A may be also involved in the regulation of cytoskeleton through this pathway. Thus, EhRab7A may have dual roles in vesicular traffic via retromer recruitment and cytoskeletal regulation via Arp2/3 recruitment. On the other hand, effectors for EhRab7B and EhRab7D remain elusive.
Unique features shared by EhRab7D and EhRab7G are the a.a. substitutions in the inter‐switch region and the acidic amino acid stretch at the carboxyl terminus (Figure S2). This inter‐switch region was reported to be an interface between Rab7 and Rab interacting lysosomal protein (RILP) (Cantalupo, Alifano, Roberti, Bruni, & Bucci, 2001; Harrison, Bucci, Vieira, Schroer, & Grinstein, 2003; Jordens et al., 2001), which serves as a bridge between microtubule and Rab7‐associated compartment. It is believed that the perinuclear localization of late endosomes/lysosomes is achieved by the interaction between Rab7 and microtube network mediated by RILP. Although RILP is not apparently conserved in E. histolytica and late endosomes/lysosomes are not enriched at the perinuclear region, this unique inter‐switch region of EhRab7D and EhRab7G may be, by analogy, involved in the organelle‐to‐organelle interaction via a not‐yet‐identified effector that links EhRab7D positive compartment (e.g., PPV). Acidic amino acid stretch is also known as a binding domain of actin cytoskeleton regulators. e.g., the acidic amino acid stretch in Wiskott‐Aldrich Syndrome Protein (WSAP) family and related protein Scar were known to bind to actin brunching complex Arp2/3 complex (Pollitt & Insall, 2009; Rougerie, Miskolci, & Cox, 2013). These features may reflect unique effector specificity in EhRab7D. Further study is needed to identify EhRab7D effector.
3.4. EhRab7D Is involved in motility and adherence via Actin regulation
We have shown that gene silencing of EhRab7D caused reduction in motility and adhesion, while EhRab7D overexpression resulted in the opposite phenotypes. Thus, EhRab7D appears to positively regulate motility and adherence, most likely via regulation of actin cytoskeleton. Actin is the major skeletal components in E. histolytica; however, the regulatory mechanisms are not fully understood. E. histolytica conserves a panel of Rho and Rac small GTPases and components of actin nucleation/branching complex, Arp2/3. However, all components known to be involved in actin nucleation/branching such as the major actin nucleation initiator, WASP family proteins (Manich et al., 2018), Cdc42, mDia, and ROCK (Nobes & Hall, 1995; Rougerie et al., 2013; Tojkander, Gateva, & Lappalainen, 2012) are absent in the E. histolytica genome. Arp2/3 complex was previously implicated for actin nucleation and formation of branched F‐actin structure in E. histolytica and demonstrated to be associated with actin dots, adhesion plates, and macropinosomes (Manich et al., 2018). It was also reported that Arp2/3 complex is recruited to the phagocytic (Babuta et al., 2015) and trogocytic cup, where SNX1/retromer can interact with Arp2/3 (Watanabe et al., 2020). Thus, it is plausible that EhRab7A may interact with Arp2/3 via SNX1/retromer and regulates motility and adherence by actin regulation.
In general, several Rab7 effectors are known to link Rab7 with microtubules and actin filaments, for example, RILP, FYCO1, myosin II, and Rac1 (Borg, Bakke, & Progida, 2014; Cantalupo et al., 2001; Pankiv et al., 2010; Sun, Büki, Ettala, Vääräniemi, & Väänänen, 2005), or with intermediate filaments, for example, vimentin and peripherin (Margiotta, Progida, Bakke, & Bucci, 2017). However, none of them are conserved in E. histolytica except for Rac. RILP and FYCO1 are known to connect Rab7 with microtubules and myosin II, and Rac1 connects Rab7 with actin filaments (Borg et al., 2014; Cantalupo et al., 2001; Pankiv et al., 2010; Sun et al., 2005). Thus, Rab7 is surely involved in cell migration and adhesion via interaction with cytoskeletons in other organisms. Thus, it is conceivable that Entamoeba also utilise Rab7‐mediated cytoskeletal regulation with the analogous, but not yet identified mechanisms.
4. EXPERIMENTAL PROCEDURES
4.1. Cells and cultures
Trophozoites of E. histolytica clonal strain HM1:IMSS cl6 (cl‐6) and G3 were axenically cultured in 13x100 mm screw‐capped Pyrex glass tubes in BI‐S‐33 (BIS) medium at 35.5°C, as described (Diamond, Harlow, & Cunnick, 1978; Diamond, Mattern, & Bartgis, 1972). CHO cells were cultured in 25 cm2 tissue culture flask containing F12 medium (Thermo Fisher Scientific, MA, USA) with 10% heat‐inactivated fetal bovine serum (Sigma‐Aldrich, MO, USA).
4.2. Antibodies
The anti‐EhRab7A antibody was described previously (Nakada‐Tsukui et al., 2005). The anti‐HA monoclonal antibody (clone 16B12) was purchased from COVANCE (Berkeley, CA, USA). Antiserum was commercially raised against recombinant GST‐EhRab7D (see below for its production) in rabbits (Eurofins Genomics, Tokyo, Japan). HRP‐conjugated anti‐mouse IgG and anti‐rabbit IgG antibody and Alexa Fluor‐488 conjugated anti‐mouse IgG were purchased from Thermo Fisher Scientific.
4.3. Establishment of transgenic E. histolytica strains for overexpression of wildtype EhRab7D and EhRab7D gene silencing
To construct a plasmid to express EhRab7D fused with three tandem copies of the HA epitope at the N terminus, a 615 bp DNA fragment containing EhRab7D protein coding sequence was amplified by PCR from genomic DNA with primers containing BglII and XhoI sites and cloned into the BglII and XhoI sites of pEhEx (Nozaki et al., 1999). A DNA fragment encoding three tandem copies of the haemagglutinin (HA) peptide was inserted at the region corresponding to the amino terminus of EhRab7B gene, as previously described (Saito‐Nakano et al., 2004). To produce a plasmid to express in amebae EhRab7D fused with monomeric red fluorescence protein (mRFP) at the N terminus, a protein coding sequence of EhRab7D was amplified by PCR, and ligated into SmaI and XhoI sites of pKT‐MR (Saito‐Nakano et al., 2007). Each plasmid were transfected to E. histolytica cl‐6 strain by lipofection (Nozaki et al., 1999). For antisense small RNA‐mediated transcriptional gene silencing (Bracha, Nuchamowitz, Anbar, & Mirelman, 2006) of EhRab7D gene, a fragment containing the first 420 bp of EhRab7D coding sequence was amplified by PCR with appropriate primers containing StuI and SacI sites, and ligated to the compatible sites of the double digested pSAP2‐Gunma plasmid (Mi‐ichi, Makiuchi, Furukawa, Sato, & Nozaki, 2011). The plasmid was transfected to E. histolytica G3 strain as above (Bracha et al., 2006; Nakada‐Tsukui, Tsuboi, Furukawa, Yamada, & Nozaki, 2012) to establish EhRab7D gene silenced strain. Drug selection was conducted with 1 μg/ml Geneticin after 24 hr of transfection, and the drug concentration was gradually increased for 2 weeks until it reached 10 μg/ml. All the transformants were maintained with the BIS medium containing 10 μg/ml geneticin.
4.4. Immunoblot analysis
The lysates from amebas harvested in logarithmic growth phase cells was prepared by incubating the cells with lysis buffer (50 mM Tris–HCl, pH 7,5, 150 mM NaCl containing 1% Triton‐X 100 and 0.5 mg/ml E64 on ice for 20 min, followed by centrifugation at 15,000g for 5 min. The supernatant was recovered and protein amount was measured using Lowry method by Bio‐rad DC protein Assay (Bio‐rad, CA, USA). SDS‐PAGE and western blot analyses were performed as previously described (Sambrook & Russell, 2001) to check protein expression of EhRab7D. For immunoblot analysis, anti‐HA mouse monoclonal antibody 16B12 (at a dilution of 1/1000), anti‐EhRab7A and anti‐EhRab7D rabbit polyclonal antibodies (at a dilution of 1/500) were used as primary antibodies, and HRP‐conjugated anti‐mouse IgG or anti‐rabbit IgG antibody (1/10,000) were used as secondary antibodies, respectively. After washing, chemiluminescence was developed with Immnobilon™ Western Chemiluminescent HRP Substrate (Merc Millipore, MA, USA) and signal was detected and captured using LAS 4000 (GE Healthcare).
4.5. Reverse transcriptase polymerase chain reaction (RT PCR)
Reverse transcriptase PCR was performed to examine mRNA levels of EhRab7D in EhRab7D gene silenced strain. Total RNA was isolated from trophozoites growing in logarithmic phase using TRIZOL (Thermo Fisher Scientific) and purified by DNAase I treatment according to the manufacturers' protocols (Thermo Fisher Scientific). Superscript® III First‐Strand Synthesis System (Thermo Fisher Scientific) and Ex Taq® PCR system (Takara, Kyoto, Japan) were used to synthesise first‐strand cDNA from purified total RNA and to amplify DNA form the cDNA template, respectively. PCR conditions were: initial denaturation step at 98°C for 30 s, followed by 30 cycles of denaturation step 98°C for 10 s, annealing step 55°C for 30 s, and extension step 72°C for 1 min, 30 cycles; with a final extension step at 68°C for 5 min. Primers used in PCR are as follows: for EhRab7D, 5′‐GAACAAGCACGAGAATGGTGTAAA‐3′ (7D1, forward) and 5′‐GGACTCGAGTTAGCAACACCCTCCTTCTTT‐3′ (7D2, reverse); for RNA polymerase II, 5′‐ATTCCATTCTTAGAAACATCTGCCAAAAAT‐3′ (forward) and 5′‐GAAGAAGAAGGAAATGTGATTAACAACATC‐3′ (reverse) .
4.6. Indirect immunofluorescence assay (IFA)
Immunofluorescence assays were performed as previously described (Saito‐Nakano et al., 2004). Briefly, amebae were harvested in logarithmic growth phase and transferred to 8 mm round wells on a slide glass. After 15–30 min of incubation in an anaerobic chamber at 35.5°C, CHO cells that had been prestained by 10 μM CellTracker Blue (Thermo Fisher Scientific) for 60 min or paramagnetic beads of 2.8 μm diameter (Dynabeads, Invitrogen, California, USA) coated with human serum (Sigma‐Alderich) were added to the well and the mixture was incubated for 15 min. Alternatively, trophozoites were mixed with hamster erythrocytes and co‐incubated for 10 and 30 min. After incubation, cells were fixed with 3.7% paraformaldehyde, permeabilized with 0.2% saponin, and reacted with anti‐HA mouse monoclonal antibody (1:500) or anit‐EhRab7A rabbit antiserum (1:100). To visualise ingested erythrocytes, they were stained with 0.84 mM diaminobenzidine for 5 min after fixation (Saito‐Nakano et al., 2004). Next, the samples were reacted with Alexa Fluor‐488 (green) conjugated anti‐mouse IgG (1:1000) antibody. Images were then captured by using LSM780 confocal laser scanning‐microscope (Carl Zeiss, Oberkochen, Germany). For staining of acidic endosomes and lysosomes, trophozoites were incubated with LysoTracker Red DND‐99 (Thermo Fisher Scientific) (1:500) for 18 h at 35°C.
4.7. Measurement of phagosome pH
The pH values of the phagosomes were monitored using FITC‐conjugated yeasts by ratiometry as previously described (Mitra, Kobayashi, Saito‐Nakano, & Nozaki, 2006; Mitra, Yasuda, Kobayashi, Saito‐Nakano, & Nozaki, 2005) with some modifications. Trophozoites transfected with pKT‐RFP7D were first examined for mRFP fluorescence to select cells that expressed a high level of mRFP1‐EhRab7D. Kinetics of acidification of phagosomes was monitored using FITC‐labelled yeasts using pKT‐MR, pKT‐RFP7D expressing cells as described previously (Mitra et al., 2005, 2006).
4.8. Trogocytosis and phagocytosis assay using confocal quantitative image cytometer
E. histolytica trophozoites grown in logarithmic growth phase were stained by incubation in the medium containing 20 μM CellTracker Blue (Thermo Fisher Scientific) for 1 hr. CHO cells were incubated in the medium containing 10 μM CellTracker Orange (Thermo Fisher Scientific) for 40 min. Approximately 1 x 104 amebas that had been washed and resuspended in 100 μl of OPTI‐MEM medium containing 1 mg/ml ascorbic acid, 5 mg/ml L‐cysteine and 15% heat‐inactivated adult bovine serum (Sigma‐Aldrich) were added into a well on a 96‐well glass‐bottom plate (IWAKI). The plate was incubated in an anaerobic chamber using Anaerocult (Merc Millipore) for 40 min at 35.5°C incubator. Approximately 5 × 104 of either live or heat‐killed CHO cells were resuspended in 100 μl of the above mentioned medium and added to each well containing amebas. To minimise aeration, 70 μl of mineral oil was layered on top of the medium, then the plate was covered with a plastic seal. Heat killing of CHO cells were conducted by incubating CHO cells resuspended in PBS at 55°C for 15 min. Images of amebas and CHO cells were captured using a 20x objective lens from five fields per well with five planes of 2 μm interval in Z axis, every 15 min for 1 hr on confocal quantitative image cytometer CQ1 (Yokogawa Electric Corporation, Tokyo, Japan) while incubating the plate at 35.5°C. Obtained images were analysed by the CQ1 softwares according to the manufacturer's protocol. The percentages of the amebas that had CHO cell‐containing trogo−/phagosomes, the average numbers of CHO cell‐containing trogo−/phagosomes per ameba, and the volume of all CHO cell‐containing trogo−/phagosomes per ameba were calculated. All data were then normalised against the values obtained for control at 1 hr and shown as percentages to these values.
4.9. Motility assay
Amebae grown and harvested in logarithmic growth phase were labelled with 10 μM CellTracker Green (Thermo Fisher Scientific) in BI medium for 40 min at 35.5°C. After washing with PBS, approximately 2 × 105 cells resuspended in 1.5 mL BI medium were transferred to a 35 mm collagen‐coated glass‐bottom dish (MatTek) and incubated in an anaerobic chamber at 35.5°C for 30 min. After the medium was removed, approximately 200 μl of warm BI was added to the center of the dish and covered with a cover glass. Time lapse images were captured on an LSM780 confocal laser scanning‐microscope (Carl Zeiss) using a 20× objective in every 30 s for 15 min. The motility of amebae was calculated as the total length of movement divided by duration of measurement by Imaris X64 software.
4.10. Adhesion assay
Approximately 4 × 104 trophozoites, stained with CellTracker Green as above, were seeded to each well of a 96‐well non‐coated (SPL Life Sciences) or a 96‐well collagen‐coated plate (Corning biocoat collagen I cellware), and incubated in an anaerobic chamber for 40 min at 35.5°C. After incubation, the fluorescence signal by excitation at 492 nm and emission at 532 nm was measured using a SpectrMax ABS Plus Microplate Reader (Molecular Devices, Ca, USA). After the medium was carefully removed, new warm medium was added. The fluorescence signal of the cells attached to the plate was measured again. The percentage of adherent cells was calculated as the fluorescence signal after wash divided by that before wash.
4.11. Transcriptome analysis
Transcriptome analysis was conducted as follows. E. histolytica trophozoites of mock and EhRab7D gene silenced strains were harvested form logarithmic growth phase. Total RNA was purified using TRIZOL reagent (Thermo Fisher Scientific) according to manufacturer's instruction. Total RNA samples from three biological replicates were sent for RNA sequencing (Macrogen, Kyoto, Japan). RNA‐seq libraries were generated by using TruSeq stranded mRNA kit. The protocol included polyA plus RNA purification, RNA fragmentation, random hexamer primed cDNA synthesis, linker ligation, PCR amplification, and gel purification. The libraries were then subjected to 100 nt paired‐end sequencing using an Illumina HiSeq2500 (Illumina, CA, USA). Averaged 5,059,781,884 total bases and 50,096,850 total reads was obtained. Data analysis was conducted by Tohoku Kagaku. Briefly, adaptor sequences were removed then low‐quality reads were removed. Obtained high‐quality reads were mapped to E. histolytica template genome then Fragments Per Kilobase of exon per Million mapped fragments (FPKM) were calculated and used for expression value (Tohoku kagaku, Iwate, Japan). The full dataset is available in NCBI GEO with the accession number of GSE152875.
4.12. Bioinformatic analysis of EhRab7D
In silico study based on Pylypenko, Hammich, Yu, & Houdusse, 2018 was performed for prediction of sites that are responsible for protein binding of EhRab7D based on its sequence alignment. Sequence alignment of all nine EhRab7 isotypes were also included. Multiple sequence alignment of all nine EhRab7 isotypes was constructed by using FAMSA (Deorowicz, Debudaj‐Grabysz, & Gudyś, 2016) with its default parameters. EhRab7D was also aligned with mammalian Rab7 (Rat Rab7/RnRab7, PDB ID:1vg0B/UniProt: P09527) (Rak et al., 2004), which is about 47% identity with EhRab7D, using SSEARCH with MIQS (Yamada & Tomii, 2014) to elucidate the residue positions in the known structure.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Tomoyoshi Nozaki, Yumiko Saito‐Nakano and Kumiko Nakada‐Tsukui: Conceptualization. Tomoyoshi Nozaki, Yumiko Saito‐Nakano, Kumiko Nakada‐Tsukui, and Ratna Wahyuni: Methodology. Kentaro Tomii: Software. Tomoyoshi Nozaki, Yumiko Saito‐Nakano and Kumiko Nakada‐Tsukui: Validation. Tomoyoshi Nozaki, Yumiko Saito‐Nakano, Kumiko Nakada‐Tsukui and Ratna Wahyuni: Formal Analysis. Tomoyoshi Nozaki, Yumiko Saito‐Nakano, Kumiko Nakada‐Tsukui and Ratna Wahyuni: Investigation. Tomoyoshi Nozaki, Yumiko Saito‐Nakano and Kumiko Nakada‐Tsukui: Resources. Tomoyoshi Nozaki, Ratna Wahyuni and Kumiko Nakada‐Tsukui: Data Curation. Yumiko Saito‐Nakano, Kumiko Nakada‐Tsukui and Ratna Wahyuni: Writing – Original Draft Preparation. Tomoyoshi Nozaki, Yumiko Saito‐Nakano, Kumiko Nakada‐Tsukui and Ratna Wahyuni: Writing – Review & Editing. Yumiko Saito‐Nakano, Kumiko Nakada‐Tsukui and Ratna Wahyuni: Visualization. Tomoyoshi Nozaki: Supervision. Tomoyoshi Nozaki, Yumiko Saito‐Nakano and Kumiko Nakada‐Tsukui: Project Administration. Tomoyoshi Nozaki, Yumiko Saito‐Nakano and Kumiko Nakada‐Tsukui: Funding Acquisition.
ETHICS STATEMENT
Animal procedures were approved by the by the Institutional Animal Care and Use Committee (No. 205089) and conducted at the AAALAC‐accredited National Institute of Infectious Diseases, Japan.
Supporting information
Figure S1. Localization of intrinsic EhRab7D. A wildtype trophozoite of E. histolytica HM‐1:IMSS cl6 strain was prestained with LysoTracker Red (red) to visualise lysosomes, fixed, and reacted with anti‐EhRab7D antibody (green) (1:200) to visualise EhRab7D.
Figure S2. Structural determination of human Rab7a (HsRab7a) binding with its effector RILP. Crystal structure of HsRab7 with RILP (PDB ID: 1yhn) with indication of amino acids located at the interface. Corresponding amino acid of EhRab7D unique I and P at the I‐K‐P stretch (see Figure 4) was shown in red sphere model and indicated by white arrows.
Table S1. Expression level of EhRab7 isotypes in EhRab7D gene silenced strain. Expression levels (FPKM) of three independent experiments and fold‐change of averaged FPKM value of nine EhRab7 isotypes in mock and EhRab7D gene silenced strain are shown.
ACKNOWLEDGMENTS
We thank Bithwa Nath Mitra for help in pH determination of phagosomes, Eiko Nakasone and Yuko Umeki, NIID, for technical assistance, and all members of Nozaki lab for discussions. This research is funded by Grants‐in‐Aid for Scientific Research (B) and Challenging Research (Exploratory) (KAKENHI JP26293093, JP17K19416, and JP18H02650 to T.N.), Scientific Research (B) and (C) and Scientific Research on Innovative Areas (JP16K08766, JP16H01210, JP19H03463, and JP20H05353 to K.N‐T., JP19K07531 to Y.S‐N.) from Ministry of Education, Culture, Sports, Science and Technology (MEXT) or Japan Society for Promotion of Sciences (JSPS), Grant for research on emerging and re‐emerging infectious diseases from Japan Agency for Medical Research and Development (AMED, JP20fk0108138 to Y.S‐N. and T.N.; JP20fk0108139 to K.N‐T and Y.S‐N), Grant for Science and Technology Research Partnership for Sustainable Development (SATREPS) from AMED and Japan International Cooperation Agency (JICA) (T.N.), and a grant for The Naito Foundation to Y.S‐N.
Saito‐Nakano Y, Wahyuni R, Nakada‐Tsukui K, Tomii K, Nozaki T. Rab7D small GTPase is involved in phago‐, trogocytosis and cytoskeletal reorganization in the enteric protozoan Entamoeba histolytica . Cellular Microbiology. 2021;23:e13267 10.1111/cmi.13267
Funding information Japan Agency for Medical Research and Development, Grant/Award Numbers: JP20fk0108138, JP20fk0108139; Japan Society for the Promotion of Science, Grant/Award Numbers: JP16H01210, JP16K08766, JP17K19416, JP18H02650, JP19H03463, JP19K07531, JP20H05353, JP26293093; Naito Foundation; AMED and Japan International Cooperation Agency (Science and Technology Research Partnership for Sustainable Development)
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- Agarwal, S. , Anand, G. , Sharma, S. , Parimita Rath, P. , Gourinath, S. , & Bhattacharya, A. (2019). EhP3, a homolog of 14‐3‐3 family of protein participates in actin reorganization and phagocytosis in Entamoeba histolytica . PLoS Pathogens, 15(5), e1007789 10.1371/journal.ppat.1007789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar‐Rojas, A. , Olivo‐Marin, J. C. , & Guillen, N. (2016). The motility of Entamoeba histolytica: Finding ways to understand intestinal amoebiasis. Current Opinion in Microbiology, 34, 24–30. 10.1016/j.mib.2016.07.016 [DOI] [PubMed] [Google Scholar]
- Babuta, M. , Bhattacharya, S. , & Bhattacharya, A. (2020). Entamoeba histolytica and pathogenesis: A calcium connection. PLoS Pathogens, 16(5), e1008214 10.1371/journal.ppat.1008214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babuta, M. , Kumar, S. , Gourinath, S. , Bhattacharya, S. , & Bhattacharya, A. (2018). Calcium‐binding protein EhCaBP3 is recruited to the phagocytic complex of Entamoeba histolytica by interacting with Arp2/3 complex subunit 2. Cellular Microbiology, 20(12), e12942 10.1111/cmi.12942 [DOI] [PubMed] [Google Scholar]
- Babuta, M. , Mansuri, M. S. , Bhattacharya, S. , & Bhattacharya, A. (2015). The Entamoeba histolytica, Arp2/3 complex is recruited to phagocytic cups through an atypical kinase EhAK1. PLoS Pathogens, 11(12), e1005310 10.1371/journal.ppat.1005310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, R. , Arya, R. , Shahid Mansuri, M. , Bhattacharya, S. , & Bhattacharya, A. (2017). EhRho1 regulates plasma membrane blebbing through PI3 kinase in Entamoeba histolytica . Cellular Microbiology, 19(10), e12751. 10.1111/cmi.12751 [DOI] [PubMed] [Google Scholar]
- Bharadwaj, R. , Sharma, S. , Janhawi, Arya, R. , Bhattacharya, S. , & Bhattacharya, A. (2018). EhRho1 regulates phagocytosis by modulating Actin dynamics through EhFormin1 and EhProfilin1 in Entamoeba histolytica . Cellular Microbiology, 20(9), e12851 10.1111/cmi.12851 [DOI] [PubMed] [Google Scholar]
- Borg, M. , Bakke, O. , & Progida, C. (2014). A novel interaction between Rab7b and actomyosin reveals a dual role in intracellular transport and cell migration. Journal of Cell Science, 127(Pt 22), 4927–4939. 10.1242/jcs.155861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha, R. , Nuchamowitz, Y. , Anbar, M. , & Mirelman, D. (2006). Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica . PLoS Pathogens, 2(5), e48 10.1371/journal.ppat.0020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo, G. , Alifano, P. , Roberti, V. , Bruni, C. B. , & Bucci, C. (2001). Rab‐interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. The EMBO Journal, 20(4), 683–693. 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deorowicz, S. , Debudaj‐Grabysz, A. , & Gudyś, A. (2016). FAMSA: Fast and accurate multiple sequence alignment of huge protein families. Scientific Reports, 6, 33964 10.1038/srep33964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, L. S. , Harlow, D. R. , & Cunnick, C. C. (1978). A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba . Transactions of the Royal Society of Tropical Medicine and Hygiene, 72(4), 431–432. 10.1016/0035-9203(78)90144-x [DOI] [PubMed] [Google Scholar]
- Diamond, L. S. , Mattern, C. F. , & Bartgis, I. L. (1972). Viruses of Entamoeba histolytica. I. Identification of transmissible virus‐like agents. Journal of Virology, 9(2), 326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust, D. M. , & Guillen, N. (2012). Virulence and virulence factors in Entamoeba histolytica, the agent of human amoebiasis. Microbes and Infection, 14(15), 1428–1441. 10.1016/j.micinf.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Gautam, G. , Ali, M. S. , Bhattacharya, A. , & Gourinath, S. (2019). EhFP10: A FYVE family GEF interacts with myosin IB to regulate cytoskeletal dynamics during endocytosis in Entamoeba histolytica . PLoS Pathogens, 15(2), e1007573 10.1371/journal.ppat.1007573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra, F. , & Bucci, C. (2016). Multiple roles of the small GTPase Rab7. Cells, 5(3), 34 10.3390/cells5030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, R. E. , Bucci, C. , Vieira, O. V. , Schroer, T. A. , & Grinstein, S. (2003). Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: Role of Rab7 and RILP. Molecular and Cellular Biology, 23(18), 6494–6506. 10.1128/mcb.23.18.6494-6506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens, I. , Fernandez‐Borja, M. , Marsman, M. , Dusseljee, S. , Janssen, L. , Calafat, J. , … Neefjes, J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein‐dynactin motors. Current Biology, 11(21), 1680–1685. 10.1016/s0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Lozano, R. , Naghavi, M. , Foreman, K. , Lim, S. , Shibuya, K. , Aboyans, V. , … Memish, Z. A. (2012). Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet (London, England), 380(9859), 2095–2128. 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz, P. , & Wyroba, E. (2009). Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evolutionary Biology, 9, 101 10.1186/1471-2148-9-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manich, M. , Hernandez‐Cuevas, N. , Ospina‐Villa, J. D. , Syan, S. , Marchat, L. A. , Olivo‐Marin, J. C. , & Guillén, N. (2018). Morphodynamics of the actin‐rich cytoskeleton in Entamoeba histolytica . Frontiers in Cellular and Infection Microbiology, 8, 179 10.3389/fcimb.2018.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margiotta, A. , Progida, C. , Bakke, O. , & Bucci, C. (2017). Rab7a regulates cell migration through Rac1 and vimentin. Biochimica et Biophysica Acta (BBA) ‐ Molecular Cell Research, 1864(2), 367–381. 10.1016/j.bbamcr.2016.11.020 [DOI] [PubMed] [Google Scholar]
- Marie, C. , & Petri, W. A., Jr. (2014). Regulation of virulence of Entamoeba histolytica . Annual Review of Microbiology, 68, 493–520. 10.1146/annurev-micro-091313-103550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, S. , Laurent, C. , & Guillén, N. (2005). Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: A proteomic approach. Cellular Microbiology, 7(10), 1504–1518. 10.1111/j.1462-5822.2005.00573.x [DOI] [PubMed] [Google Scholar]
- Mi‐ichi, F. , Makiuchi, T. , Furukawa, A. , Sato, D. , & Nozaki, T. (2011). Sulfate activation in mitosomes plays an important role in the proliferation of Entamoeba histolytica . PLoS Neglected Tropical Diseases, 5(8), e1263 10.1371/journal.pntd.0001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, H. W. , Suleiman, R. L. , & Ralston, K. S. (2019). Trogocytosis by Entamoeba histolytica mediates acquisition and display of human cell membrane proteins and evasion of Lysis by human serum. mBio, 10(2), e00068 10.1128/mBio.00068-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, B. N. , Kobayashi, S. , Saito‐Nakano, Y. , & Nozaki, T. (2006). Entamoeba histolytica: Differences in phagosome acidification and degradation between attenuated and virulent strains. Experimental Parasitology, 114(1), 57–61. 10.1016/j.exppara.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Mitra, B. N. , Yasuda, T. , Kobayashi, S. , Saito‐Nakano, Y. , & Nozaki, T. (2005). Differences in morphology of phagosomes and kinetics of acidification and degradation in phagosomes between the pathogenic Entamoeba histolytica and the non‐pathogenic Entamoeba dispar . Cell Motility and the Cytoskeleton, 62(2), 84–99. 10.1002/cm.20087 [DOI] [PubMed] [Google Scholar]
- Nakada‐Tsukui, K. , & Nozaki, T. (2016). Immune response of Amebiasis and immune evasion by Entamoeba histolytica . Frontiers in Immunology, 7, 175 10.3389/fimmu.2016.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada‐Tsukui, K. , & Nozaki, T. (2020). The emerging role of trogocytosis in the evasion of cancers and parasitic protists from immune cells. Biotarget, 4(4). 10.21037/biotarget.2020.03.02 [DOI] [Google Scholar]
- Nakada‐Tsukui, K. , Okada, H. , Mitra, B. N. , & Nozaki, T. (2009). Phosphatidylinositol‐phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica . Cellular Microbiology, 11(10), 1471–1491. 10.1111/j.1462-5822.2009.01341.x [DOI] [PubMed] [Google Scholar]
- Nakada‐Tsukui, K. , Saito‐Nakano, Y. , Ali, V. , & Nozaki, T. (2005). A retromerlike complex is a novel Rab7 effector that is involved in the transport of the virulence factor cysteine protease in the enteric protozoan parasite Entamoeba histolytica . Molecular Biology of the Cell, 16(11), 5294–5303. 10.1091/mbc.e05-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada‐Tsukui, K. , Saito‐Nakano, Y. , Husain, A. , & Nozaki, T. (2010). Conservation and function of Rab small GTPases in Entamoeba: Annotation of E. invadens Rab and its use for the understanding of Entamoeba biology. Experimental Parasitology, 126(3), 337–347. 10.1016/j.exppara.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Nakada‐Tsukui, K. , Tsuboi, K. , Furukawa, A. , Yamada, Y. , & Nozaki, T. (2012). A novel class of cysteine protease receptors that mediate lysosomal transport. Cellular Microbiology, 14(8), 1299–1317. 10.1111/j.1462-5822.2012.01800.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes, C. D. , & Hall, A. (1995). Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with Actin stress fibers, lamellipodia, and filopodia. Cell, 81(1), 53–62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Nozaki, T. , Asai, T. , Sanchez, L. B. , Kobayashi, S. , Nakazawa, M. , & Takeuchi, T. (1999). Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba . The Journal of Biological Chemistry, 274(45), 32445–32452. 10.1074/jbc.274.45.32445 [DOI] [PubMed] [Google Scholar]
- Nozaki, T. , & Nakada‐Tsukui, K. (2006). Membrane trafficking as a virulence mechanism of the enteric protozoan parasite Entamoeba histolytica . Parasitology Research, 98(3), 179–183. 10.1007/s00436-005-0079-6 [DOI] [PubMed] [Google Scholar]
- Okada, M. , Huston, C. D. , Mann, B. J. , Petri, W. A., Jr. , Kita, K. , & Nozaki, T. (2005). Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica . Eukaryotic Cell, 4(4), 827–831. 10.1128/EC.4.4.827-831.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, E. , Guarneros, G. , Martinez‐Palomo, A. , & Sánchez, T. (1983). Entamoeba histolytica. Phagocytosis as a virulence factor. The Journal of Experimental Medicine, 158(5), 1511–1521. 10.1084/jem.158.5.1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv, S. , Alemu, E. A. , Brech, A. , Bruun, J. A. , Lamark, T. , Overvatn, A. , … Johansen, T. (2010). FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end‐directed vesicle transport. The Journal of Cell Biology, 188(2), 253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picazarri, K. , Nakada‐Tsukui, K. , Tsuboi, K. , Miyamoto, E. , Watanabe, N. , Kawakami, E. , & Nozaki, T. (2015). Atg8 is involved in endosomal and phagosomal acidification in the parasitic protist Entamoeba histolytica . Cellular Microbiology, 17(10), 1510–1522. 10.1111/cmi.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt, A. Y. , & Insall, R. H. (2009). WASP and SCAR/WAVE proteins: The drivers of actin assembly. Journal of Cell Science, 122(Pt 15), 2575–2578. 10.1242/jcs.023879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, R. R. , Welter, B. H. , Hwu, R. , Bowersox, B. , Attaway, C. , & Temesvari, L. A. (2006). Entamoeba histolytica: FYVE‐finger domains, phosphatidylinositol 3‐phosphate biosensors, associate with phagosomes but not fluid filled endosomes. Experimental Parasitology, 112(4), 221–231. 10.1016/j.exppara.2005.11.013 [DOI] [PubMed] [Google Scholar]
- Pylypenko, O. , Hammich, H. , Yu, I. M. , & Houdusse, A. (2018). Rab GTPases and their interacting protein partners: Structural insights into Rab functional diversity. Small GTPases, 9(1‐2), 22–48. 10.1080/21541248.2017.1336191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak, A. , Pylypenko, O. , Niculae, A. , Pyatkov, K. , Goody, R. S. , & Alexandrov, K. (2004). Structure of the Rab7:REP‐1 complex: Insights into the mechanism of Rab prenylation and choroideremia disease. Cell, 117(6), 749–760. 10.1016/j.cell.2004.05.017 [DOI] [PubMed] [Google Scholar]
- Ralston, K. S. , Solga, M. D. , Mackey‐Lawrence, N. M. , Somlata, Bhattacharya, A. , & Petri, W. A., Jr. (2014). Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature, 508(7497), 526–530. 10.1038/nature13242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougerie, P. , Miskolci, V. , & Cox, D. (2013). Generation of membrane structures during phagocytosis and chemotaxis of macrophages: Role and regulation of the actin cytoskeleton. Immunological Reviews, 256(1), 222–239. 10.1111/imr.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito‐Nakano, Y. , Loftus, B. J. , Hall, N. , & Nozaki, T. (2005). The diversity of Rab GTPases in Entamoeba histolytica . Experimental Parasitology, 110(3), 244–252. 10.1016/j.exppara.2005.02.021 [DOI] [PubMed] [Google Scholar]
- Saito‐Nakano, Y. , Mitra, B. N. , Nakada‐Tsukui, K. , Sato, D. , & Nozaki, T. (2007). Two Rab7 isotypes, EhRab7A and EhRab7B, play distinct roles in biogenesis of lysosomes and phagosomes in the enteric protozoan parasite Entamoeba histolytica . Cellular Microbiology, 9(7), 1796–1808. 10.1111/j.1462-5822.2007.00915.x [DOI] [PubMed] [Google Scholar]
- Saito‐Nakano, Y. , Nakazawa, M. , Shigeta, Y. , Takeuchi, T. , & Nozaki, T. (2001). Identification and characterization of genes encoding novel Rab proteins from Entamoeba histolytica . Molecular and Biochemical Parasitology, 116(2), 219–222. 10.1016/s0166-6851(01)00318-8 [DOI] [PubMed] [Google Scholar]
- Saito‐Nakano, Y. , Yasuda, T. , Nakada‐Tsukui, K. , Leippe, M. , & Nozaki, T. (2004). Rab5‐associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica . The Journal of Biological Chemistry, 279(47), 49497–49507. 10.1074/jbc.M403987200 [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , & Russell, D. W. (2001). Molecular cloning: A laboratory manual (3 volume set) (3rd ed. New York: ). Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shirley, D. T. , Farr, L. , Watanabe, K. , & Moonah, S. (2018). A review of the global burden, new diagnostics, and current therapeutics for amebiasis. Open Forum Infectious Diseases, 5(7), ofy161 10.1093/ofid/ofy161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlata, Bhattacharya, S. , & Bhattacharya, A. (2011). A C2 domain protein kinase initiates phagocytosis in the protozoan parasite Entamoeba histolytica . Nature Communications, 2, 230 10.1038/ncomms1199 [DOI] [PubMed] [Google Scholar]
- Somlata, Nakada‐Tsukui, K. , & Nozaki, T. (2017). AGC family kinase 1 participates in trogocytosis but not in phagocytosis in Entamoeba histolytica . Nature Communications, 8(1), 101 10.1038/s41467-017-00199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Büki, K. G. , Ettala, O. , Vääräniemi, J. P. , & Väänänen, H. K. (2005). Possible role of direct Rac1‐Rab7 interaction in ruffled border formation of osteoclasts. The Journal of Biological Chemistry, 280(37), 32356–32361. 10.1074/jbc.M414213200 [DOI] [PubMed] [Google Scholar]
- Tavares, P. , Rigothier, M. C. , Khun, H. , Roux, P. , Huerre, M. , & Guillén, N. (2005). Roles of cell adhesion and cytoskeleton activity in Entamoeba histolytica pathogenesis: A delicate balance. Infection and Immunity, 73(3), 1771–1778. 10.1128/IAI.73.3.1771-1778.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander, S. , Gateva, G. , & Lappalainen, P. (2012). Actin stress fibers – assembly, dynamics and biological roles. Journal of Cell Science, 125(Pt 8), 1855–1864. 10.1242/jcs.098087 [DOI] [PubMed] [Google Scholar]
- Tovy, A. , Hertz, R. , Siman‐Tov, R. , Syan, S. , Faust, D. , Guillen, N. , & Ankri, S. (2011). Glucose starvation boosts Entamoeba histolytica virulence. PLoS Neglected Tropical Diseases, 5(8), e1247 10.1371/journal.pntd.0001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNICEF . (2019). Estimates of child cause of death, Diarrhoea 2018 Retrieved from https://data.unicef.org/topic/child-health/diarrhoeal-disease/
- Verma, K. , Srivastava, V. K. , & Datta, S. (2018). Rab GTPases take centre stage in understanding Entamoeba histolytica biology. Small GTPases, 11, 1–14. (Advance online publication). 10.1080/21541248.2018.1528840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N. , Nakada‐Tsukui, K. , & Nozaki, T. (2020). Two isotypes of phosphatidylinositol 3‐phosphate‐binding sorting nexins play distinct roles in trogocytosis in Entamoeba histolytica . Cellular Microbiology, 22(3), e13144 10.1111/cmi.13144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. , & Tomii, K. (2014). Revisiting amino acid substitution matrices for identifying distantly related proteins. Bioinformatics (Oxford, England), 30(3), 317–325. 10.1093/bioinformatics/btt694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Localization of intrinsic EhRab7D. A wildtype trophozoite of E. histolytica HM‐1:IMSS cl6 strain was prestained with LysoTracker Red (red) to visualise lysosomes, fixed, and reacted with anti‐EhRab7D antibody (green) (1:200) to visualise EhRab7D.
Figure S2. Structural determination of human Rab7a (HsRab7a) binding with its effector RILP. Crystal structure of HsRab7 with RILP (PDB ID: 1yhn) with indication of amino acids located at the interface. Corresponding amino acid of EhRab7D unique I and P at the I‐K‐P stretch (see Figure 4) was shown in red sphere model and indicated by white arrows.
Table S1. Expression level of EhRab7 isotypes in EhRab7D gene silenced strain. Expression levels (FPKM) of three independent experiments and fold‐change of averaged FPKM value of nine EhRab7 isotypes in mock and EhRab7D gene silenced strain are shown.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


