Summary
Alk-Ph is a clickable APEX2 substrate developed for spatially restricted protein/RNA labeling in intact yeast cells. Alk-Ph is more water soluble and cell wall permeable than biotin-phenol substrate, allowing more efficient profiling of the subcellular proteome in microorganisms. We describe the protocol for Alk-Ph probe synthesis, APEX2 expression, and protein/RNA labeling in yeast and the workflow for quantitative proteomic experiments and data analysis. Using the yeast mitochondria as an example, we provide guidelines to achieve high-resolution mapping of subcellular yeast proteome and transcriptome.
For complete details on the use and execution of this protocol, please refer to Li et al. (2020).
Graphical Abstract

Highlights
-
•
Synthesis of clickable APEX probe Alk-Ph
-
•
Proximity-dependent proteomic profiling in yeast
-
•
Spatially restricted RNA labeling in yeast
Alk-Ph is a clickable APEX2 substrate developed for spatially restricted protein/RNA labeling in intact yeast cells. Alk-Ph is more water soluble and cell wall permeable than biotin-phenol substrate, allowing more efficient profiling of the subcellular proteome in microorganisms. We describe the protocol for Alk-Ph probe synthesis, APEX2 expression, and protein/RNA labeling in yeast and the workflow for quantitative proteomic experiments and data analysis. Using the yeast mitochondria as an example, we provide guidelines to achieve high-resolution mapping of subcellular yeast proteome and transcriptome.
Before You Begin
Traditional physical or biochemical tools to isolate specific subcellular organelles for proteomic studies have inherent limitations. They are often prone to contamination and could not easily access transient or weak protein-protein interaction. The peroxidase-mediated proximity labeling technique has been developed and widely used to determine the protein inventories of distinct subcellular compartments. As an engineered ascorbate peroxidase, APEX2 oxidizes biotin-phenol in the presence of H2O2 to generate short-lived phenoxyl free radical that covalently links to nearby electron-rich amino acid residues, such as tyrosine. Biotin-phenol based APEX labeling has been used to map the proteome at various subcellular compartments, including both membrane-bound organelles (e.g., mitochondria) (Hung et al., 2017; Hung et al., 2014; Rhee et al., 2013) and membrane-less organelles (e.g., stress granules) (Markmiller et al., 2018). APEX techniques have also been applied in vivo, including Drosophila (Chen et al., 2015) and C.elegans (Reinke et al., 2017).
Despite the wide use of APEX in mammalian cell culture, its application in microorganisms, such as yeast, has been limited due to poor cell wall permeability of biotin-phenol (Hwang and Espenshade, 2016; Singer-Krüger et al., 2020). Zymolase digestion (Hwang and Espenshade, 2016) or freeze-thaw cycles (Singer-Krüger et al., 2020) were used to compromise the cell wall to facilitate probe penetration. Recently, we designed and synthesized a clickable APEX substrate, Alkyne-Phenol (Alk-Ph), by replacing the biotin moiety to a small clickable alkynyl group (Li et al., 2020), which exhibited higher solubility and improved membrane permeability than biotin-phenol. APEX2-mediated proximity-dependent labeling with Alk-Ph in intact yeast cells offers exceptional specificity (>94%) and high coverage. Moreover, we have demonstrated that this approach can also be extended to proximity-dependent RNA labeling in yeast. This protocol will describe procedures including Alk-Ph probe synthesis, APEX2 expression and labeling in yeast, proteomics analysis, and RNA quantification.
Yeast Cell Culture Medium
Timing: 4 h
-
1.Prepare the following yeast culture media in 1-L glass bottles and sterilize by autoclaving at 120°C for 2 h:
-
a.Synthetic drop-out liquid medium without tryptophan (SD liquid media):
-
-6.7 g nitrogen base
-
-900 mL sterile water
-
-
-
b.Synthetic drop-out solid medium without tryptophan (SD solid media):
-
-6.7 g nitrogen base
-
-15 g agar
-
-900 mL sterile water
-
-
-
c.Synthetic galactose minimal medium without tryptophan (SG media):
-
-6.7 g nitrogen base
-
-900 mL sterile water
-
-
-
d.Yeast extract peptone dextrose medium (YPD medium):
-
-20 g peptone
-
-10 g yeast extract
-
-950 mL sterile water
-
-
-
a.
-
2.
In the meantime, dissolve 60 g glucose into 150 mL sterile water, 20 g galactose into 50 mL sterile water, and dissolve 5.76 g yeast synthetic drop-out medium supplements without tryptophan into 150 mL sterile water, under vortex. Filter these solutions through Minisart® Syringe Filter (Φ=0.22 μm) for sterilization.
-
3.After autoclave sterilization is completed, let the media stand at 25°C until it is just cool enough to handle.
-
a.Add 50 mL glucose solution and 50 mL yeast synthetic drop-out medium supplements solution to the SD liquid media.
-
b.Add another 50 mL glucose solution and 50 mL yeast synthetic drop-out medium supplements solution to the SD solid media.
-
c.Add 50 mL galactose solution and 50 mL yeast synthetic drop-out medium supplements solution to the SG media.
-
d.Add 50 mL glucose solution to the YPD medium.
-
e.Add 1 mL 100 mg/mL ampicillin and 1 mL 50 mg/mL kanamycin to each media.
-
a.
-
4.
Pour the SD solid media into sterile plate until it covers the bottom (approximately 20 mL).
-
5.
After 1 h, store the cold SD media, SG media, YPD medium and SD plates into 4°C refrigerator. The frozen plates could be stored at 4°C for 1 month.
Note: All the steps after sterilization should be operated under a sterile environment, such as a Clean Bench.
Yeast Competent Cells Preparation
Timing: 24 h
-
6.
Inoculate frozen wild-type W303 yeast cells in 10 mL YPD medium and grow at 30°C to mid-log phase.
-
7.
Pellet the cells at 500 × g for 4 min and discard the supernatant.
-
8.
Add 10 mL EZ 1 solution (from the EZ frozen transformation kit, Zymo Research) to wash the cell pellet. Re-pellet the cells at 500 × g for 4 min and discard the supernatant again.
-
9.
Resuspend the pellet in 1 mL EZ 2 solution (from the EZ frozen transformation kit, Zymo Research) to obtain the W303 competent cells.
Pause Point: At this point, the competent cells can be free to use for transformation or stored frozen at −80°C for future use.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Streptavidin-HRP | Pierce | Cat# 21124 |
| Flag-Tag Monoclonal Antibody (2C5) | Biodragon | Cat# B1001 |
| Anti: GFP antibody | Abcam | Cat# Ab290 RRID: AB_303395 |
| HRP-Goat Anti-Mouse IgG (H+L) | Ruiying | Cat# RS0001 |
| HRP-Goat Anti-Rabbit IgG (H+L) | Ruiying | Cat# RS0002 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phanta® Max Super-Fidelity DNA Polymerase | Vazyme | Cat# P505-d2 |
| Gibson Assembly Master Mix | NEB | Cat# E2611S |
| 2× Pfu MasterMix (Dye) | Cwbio | Cat# CW0686A |
| D-(+)-Glucose | Sigma | Cat# G6152-500G |
| Yeast Nitrogen Base without Amino Acids | BD | Cat# 291940 |
| Yeast Synthetic Drop-out Medium Supplements without uracil, leucine, and tryptophan | Sigma | Cat# Y1876 |
| D-(+)-Galactose | Amresco | Cat# 0637 |
| Peptone | Amresco | Cat# J636 |
| Yeast Extract | OXOID | Cat# LP0021 |
| 4-Pentynoic acid | Ark | Cat# AK-32594 |
| N-Hydroxysuccinimide | J&K | Cat# 117997 |
| 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride | J&K | Cat# 211112 |
| Tyramine | J&K | Cat# 953409 |
| Triethylamine | J&K | Cat# 432915 |
| DAPI | Thermo Fisher | Cat# D1306 |
| Hoechst 33342 | Bioworld | Cat# BD5013 |
| Hydrogen peroxide aqueous solution | Xilong | Cat# S6364 |
| Sodium azide | Amresco | Cat# 0639-250G |
| Tetramethylrhodamine methyl ester | AAT Bioquest | Cat# 22221 |
| Sodium ascorbate | Aladdin | Cat# S105024 |
| 6-Hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) | Sigma | Cat# 238813 |
| Protease Inhibitor Cocktail (100×) | Cwbio | Cat# CW2200S |
| Glass beads | Sigma | Cat# G8772 |
| Biotin-(PEG)3-Azide | Click Chemistry Tools | Cat# AZ104 |
| Cy5 Azide | Click Chemistry Tools | Cat# AZ118 |
| BTTAA | Click Chemistry Tools | Cat# 1236 |
| THPTA | Click Chemistry Tools | Cat# 1010 |
| Copper sulfate pentahydrate | Aladdin | Cat# C112401 |
| Pierce Streptavidin Agarose | Pierce | Cat# 20349 |
| Trypsin | Promega | Cat# V5111 |
| Dithiothreitol | Sigma | Cat# D9163 |
| Iodoacetamide | Sigma | Cat# I6125 |
| Triethylammonium bicarbonate buffer | Sigma | Cat# T7408 |
| Formaldehyde solution | Sigma | Cat# 252549 |
| Formaldehyde solution 13C, d2 | Sigma | Cat# 596388 |
| Formic acid | Aladdin | Cat# F112032 |
| Formic acid | Fisher Scientific | Cat# A117-50 |
| HLB extraction cartridges | Waters | Cat# 186000383 |
| Phosphate-Buffered Saline (10×) pH 7.4, RNase-free | Invitrogen | Cat# AM9624 |
| BeyoPure™ Ultrapure Water (DNase/RNase-Free, Sterile) | BeyoPure | Cat# ST876 |
| TRIzol™ Reagent | Life Technologies | Cat# 15596018 |
| DNase I (RNase-free) | NEB | Cat# M0303 |
| Dynabeads® MyOne™ Streptavidin C1 | Invitrogen | Cat# 65002 |
| UltraPure™ 1 M Tris-HCI Buffer, pH 7.5 | Invitrogen | Cat# 15567-027 |
| NaCl (5 M), RNase-free | Invitrogen | Cat# AM9759 |
| EDTA (0.5 M), pH 8.0, RNase-free | Invitrogen | Cat# AM9260G |
| TWEEN® 20 | Sigma | Cat# P1379 |
| Sodium hydroxide solution | Sigma | Cat# S2770-100ML |
| Bovine serum albumin, fraction V, heat shock isolation | Sangon Biotech | Cat# A500023-0100 |
| Yeast tRNA | Invitrogen | Cat# 15401-011 |
| Urea | Sigma | Cat# U5378 |
| SDS, 10% Solution, RNase-free | Invitrogen | Cat# AM9822 |
| Formamide | Sigma | Cat# F9037 |
| D-Biotin | Invitrogen | Cat# B20656 |
| PowerUp™ SYBR™ Green Master Mix | Applied Biosystems | Cat# A25778 |
| HPLC-grade acetonitrile | Fisher Scientific | Cat# A998-4 |
| HPLC-grade water | Fisher Scientific | Cat# W5-4 |
| Alkyne-Phenol probe | This paper | N/A |
| Critical Commercial Assays | ||
| DNA extraction kit | TIANGEN | Cat# DP118-02 |
| Frozen-EZ Yeast Transformation II Kit™ | zymo research | Cat# T2001 |
| BCA Protein Assay Kit | Pierce | Cat# 23227 |
| RNA Clean & Concentrator-100 Kit | Zymo | Cat# R1019 |
| ProtoScript® II First Strand cDNA Synthesis Kit | NEB | Cat# E6560 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: Strain W303 (MATa) | Laboratory of Prof. Tao Liu and Ping Wei | N/A |
| Oligonucleotides | ||
| Primer for pCTCON2-Su9-APEX2-eGFP fwd: CCCCGGATCGAATTCCCTACTTCA rev: CTTGTACAGCTCGTCCATGCCG |
This paper | N/A |
| Primer for pCTCON2-Su9-eGFP fwd: CCCCGGATCGAATTCCCTACTTCA rev: CTTGTACAGCTCGTCCATGCCG |
This paper | N/A |
| Primer for pCTCON2-NLS-APEX2-eGFP fwd: ATGCCACCAAAAAAAAAAAGAAA AGTTAAGGACAATAGCTCGACGATTG rev: GATCCGCTAGCACCAGAGCCTC |
This paper | N/A |
| RT-PCR primer for ACT1 gene fwd: GAAATGCAAACCGCTGCTCA rev: TACCGGCAGATTCCAAACCC |
This paper | N/A |
| RT-PCR primer for TDH3 gene fwd: TCACGGTAGATACGCTGGTG rev: CCAGCGTCAATGTGCTTTTG |
This paper | N/A |
| RT-PCR primer for MT-COX1 gene fwd: GTGGTTTAACTGGTGTTGCCT rev: GTGAAAATGTCCCACCACGTA |
This paper | N/A |
| RT-PCR primer for MT-ATP6 gene fwd: TGCTTAAAGGACAAATTGGAGGTAA rev: CCAGCAGGTACGAATAATGAGA |
This paper | N/A |
| RT-PCR primer for MT-ATP9 gene fwd: TTGCTATCGTATTCGCAGCTTTrev: AGCTTCTGATAAGGCGAAACC | This paper | N/A |
| RT-PCR primer for MT-21S rRNA gene fwd: AGCGAAATTCCTTGGCCTATAA rev: CCGTCTTGCTGAAGGTACATAG |
This paper | N/A |
| Recombinant DNA | ||
| pCTCON2 | Laboratory of Prof. Alice Ting | Addgene# 41843 |
| Software and Algorithms | ||
| pFind studio (Version 3.0.11) | Chi et al., 2018 | http://pfind.ict.ac.cn/software/pFind3/index.html |
| pQuant | Liu et al., 2014 | http://pfind.ict.ac.cn/software/pFind3/index.html |
Materials and Equipment
Recipes of several solution or buffer mentioned in this protocol are described and listed in tables below.
Alk-Ph Stock
Dissolve 21.7 mg Alk-Ph to 200 μL DMSO (Cf = 500 mM). The stock could be stored at −20°C for 1 year. Dilute the stock into PBS solution for APEX labeling (1:200, Cf = 2.5 mM).
Quencher Solution
Dissolve 65.0 mg sodium azide to 1 mL of water (Cf = 1 M) to obtain sodium azide stock. Store at 25°C.
Dissolve 125.1 mg Trolox to 1 mL DMSO (Cf = 500 mM) to obtain Trolox stock. Store at −20°C.
Dissolve 11.9 mg sodium ascorbate to 6 mL PBS buffer (Cf = 10 mM) when use.
Mix them as the following table.
| Reagent | Final Concentration (mM) | Volume (μL) |
|---|---|---|
| Sodium ascorbate (10 mM) | 10 | 6000 |
| Sodium azide stock (1 M) | 10 | 60 |
| Trolox stock (500 mM) | 5 | 60 |
| Total | n/a | 6,120 |
The quencher buffer should be used in 30 min after prepared.
CRITICAL: Sodium azide is on the Hazardous Substance List as it can react with metal to form heavy metal azides or react with water to form hydrazoic acid. Heating at 300°C or reacting with metal also would lead to explosion. Contact with sodium azide should be minimized. Metallic container and spatula should be avoided.
Click Reaction Cocktail
Dissolve 4.44 mg azide-(PEG)3-biotin to 1 mL DMSO (Cf = 10 mM). Aliquot the stock to 50 μL and store at −20°C.
Dissolve 1.25 mg copper sulfate pentahydrate to 100 μL of water and 4.3 mg BTTAA to 200 μL of water. Mix the CuSO4 solution, BTTAA solution and extra 100 μL of water to obtain 400 μL Cu-BTTAA stock (Cf,Cu = 12.5 mM, Cf,BTTAA = 25 mM). Store at −20°C.
Dissolve 5 mg sodium ascorbate to 1 mL PBS buffer (Cf = 25 mM) when use.
Mix them as the following table.
| Reagent | Final Concentration (mM) | Volume (μL) |
|---|---|---|
| Azide-(PEG)3-biotin stock (10 mM) | 0.3 | 9 |
| Cu-BTTAA stock | 1 mM for Cu, 2 mM for BTTAA | 24 |
| Sodium ascorbate (25 mM) | 7.5 | 90 |
| PBS buffer | n/a | 177 |
| Total | n/a | 300 |
Triethylammonium bicarbonate buffer
| Reagent | Final Concentration (mM) | Volume (mL) |
|---|---|---|
| 10× triethylammonium bicarbonate buffer (1 M) | 100 | 1 |
| ddH2O | n/a | 9 |
| Total | n/a | 10 |
4% (v/v) CH2O solution
| Reagent | Final Concentration (v/v) | Volume (μL) |
|---|---|---|
| 37% (v/v) CH2O | 4% | 10.8 |
| ddH2O | n/a | 89.2 |
| Total | n/a | 100 |
4% (v/v) 13CD2O solution
| Reagent | Final Concentration (v/v) | Volume (μL) |
|---|---|---|
| 20% (v/v) 13CD2O | 4% | 20 |
| ddH2O | n/a | 80 |
| Total | n/a | 100 |
Sodium Cyanoborohydride Solution
Dissolve 3.97 mg sodium cyanoborohydride to 100 μL deionized water and mix.
2× B&W buffer
| Reagent | Final Concentration (mM) | Volume (mL) |
|---|---|---|
| Tris-HCl (1 M, pH 7.5) | 10 | 0.4 |
| NaCl (5 M) | 2,000 | 16 |
| EDTA (0.5 M) | 1 | 0.08 |
| 10% (w/v) Tween-20 | 0.2% (w/v) | 0.8 |
| RNase-free water | n/a | 22.72 |
| Total | n/a | 40 |
1× B&W buffer was made by mixing equal volume of 2× B&W buffer and RNase-free water.
RNA Block Buffer
Dilute BSA and yeast RNA with 2× B&W buffer to make a solution of 1 mg/mL BSA and 1 mg/mL yeast tRNA in 1× B&W buffer.
Step-By-Step Method Details
Synthesis of Alkyne-Phenol Probe
Timing: 2 days
The following steps describe the 0.2 gram-level synthesis and characterization of alkyne-phenol probe (compound 2), see Figure 1.
Note: All the procedures should be operated in a fume hood.
-
1.Synthesis of compound 1
-
a.Weigh 0.245 g of 4-pentynoic acid (2.5 mmol), 0.288 g N-hydroxysuccinimide (2.5 mmol) and 0.613 g of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDCI) hydrochloride (3.2 mmol) in a 50 mL round-bottom flask.
-
b.Add 20 mL anhydrous dimethylformamide (DMF) to the flask. Stir the solution at 25°C for 16 h.
-
c.Remove the solvent using the rotary evaporator to obtain a yellow residue. Dilute the residue with 15 mL ethyl acetate and 5 mL water. Wash the combined organic layer with 5 mL dilute hydrochloric acid solution (pH ~ 4), water, and brine successively. Dry the organic layer with 2 g of anhydrous sodium sulfate for 10 min and filter to remove the sodium sulfate. Condense the organic layer using the rotary evaporator to obtain the crude product of compound 1.
-
d.Purify the crude product by silica gel chromatography with 1% methanol in dichloromethane (DCM) to obtain 0.478 g of pure compound 1 (yield = 98%).
-
a.
Pause Point: compound 1 could be stored at −20°C for at least 1 year.
-
2.Synthesis of compound 2
-
a.Weigh 0.24 g of compound 1 (1.23 mmol) and 0.342 g tyramine (2.5 mmol) in a 50 mL round-bottom flask.
-
b.Add 860 μL of trimethylamine (TEA, 6.2 mmol) and 15 mL anhydrous DMF to the flask. Stir the solution at 25°C for 16 h.
-
c.Remove the solvent using the rotary evaporator to obtain a brown residue. Extract the product by 3 × 10 mL ethyl acetate. Wash the combined organic layer with 5 mL dilute hydrochloric acid solution (pH ~ 4), water, and brine successively. Dry the organic layer with 2 g of anhydrous sodium sulfate for 10 min and filter to remove the sodium sulfate. Condense the organic layer using the rotary evaporator to obtain the crude product of compound 2.
-
d.Purify the crude product by silica gel chromatography with 2% methanol in DCM to obtain 0.216 g of pure compound 2 (yield = 81%).
-
e.Characterize compound 2 by 1H-NMR spectroscopy and electrospray ionization UPLC-MS. (See the Expected Outcome section).
-
a.
CRITICAL: Excess tyramine is necessary to drive conversion of compound 1 to compound 2 and to avoid the generate of N, O-disubstituted by-product.
Note: The Alk-Ph probe is available from the Lead Contact upon request. (See the Materials Availability section)
Figure 1.
Synthesis of Alkyne-Phenol Probe (Compound 2)
Expression of APEX2 in Yeast
Timing: 5 days
The following steps describe the expression of APEX2-fusion in transformed yeast cells.
-
3.
Mix 0.1–0.5 μg plasmid DNA (in less than 2 μL volume) with 20 μL of competent cells. Add 200 μL EZ 3 solution (from the EZ frozen transformation kit, Zymo Research) to the mixture.
-
4.
Incubate at 30°C for 1.5 h. Mix the transformation solution vigorously by vortexing three times at 15 min intervals during the incubation.
-
5.
Spread the above transformation mixture on a SD plate. Incubate the plate at 30°C for 3 days to obtain transformant colonies.
-
6.
Inoculate a single colony of transformants containing Su9-APEX2-eGFP plasmid from SD plates into 5 mL SD media. Grow the transformants at 30°C to mid-log phase for approximately 24 h.
-
7.
Measure the absorption of grown yeast cells in SD media at OD600. The absorbance was typically between 15 and 20. Dilute the cells to 100 mL SG media such that OD600 ~ 0.1. Grow the cells at 30°C for 8 h to OD600 ~ 1.0.
Note: The volume of diluted SG media is dependent to the following experiments. Generally, 5 mL SG media are enough for western blotting analysis, 60 mL SG media is sufficient for proteomic analysis, and 30 mL SG media is adequate for RNA labeling.
Pause Point: After step 5. The transformed plates could be stored at 4°C for up to 1 month.
APEX Labeling and Click Reaction
Timing: 10 h
The following steps describe the protocols of APEX2 labeling in living yeast cells and click reaction on protein level.
-
8.
Pellet APEX-expressing cells from 60 mL SG media at 4,000 × g for 5 min and discard the supernatant. Wash the pellet with 12 mL PBS twice.
-
9.
Resuspend the cell pellet in 6 mL PBS buffer containing 2.5 mM Alk-Ph probe (See Materials and Equipment section). Incubate at 25°C for 30 min.
Note: It is necessary to use 2.5 mM or higher concentration of Alk-Ph probe to achieve high labeling efficiency in yeast.
-
10.
In the meantime, prepare a quencher solution consisting 10 mM sodium azide, 10 mM sodium ascorbate and 5 mM Trolox in 6 mL PBS buffer (See Materials and Equipment section).
CRITICAL: The quencher solution should be freshly prepared each time and used within 30 min.
-
11.
Add 60 μL 100 mM H2O2 to the yeast suspension and mix thoroughly by a vortex.
-
12.
After 1 min, add prepared 6 mL quencher solution to stop APEX labeling and mix thoroughly by a vortex again.
-
13.
Pellet cells at 12,000 × g for 2 min and discard the supernatant. Wash the cells with 12 mL PBS twice.
-
14.
Resuspend the cell pellet in 600 μL PBS buffer containing 1% (v/v) protease inhibitor cocktail. Add an equal volume glass beads to the suspension. Vibrate the mixture at 2,000 rpm at 4°C for 1 min. Thereafter, cool the cells on ice for 1 min. Repeat the “shake-cool” cycle for ten times to lyse yeast cells (see Troubleshooting 1).
-
15.
Centrifuge at 12,000 × g and 4°C for 5 min and collect the supernatant. Add 6 mL cold methanol to the supernatant and store at −80°C for 3 h to precipitate yeast proteins and remove excess small molecules.
-
16.
Centrifuge at 12,000 g and 4°C for 2 min and discard the supernatant. Resolubilize the protein pellet in 600 μL 0.5% SDS aqueous solution.
Note: Heating at 95°C or pipetting are helpful to resolubilize the protein pellet in 0.5% SDS aqueous solution.
-
17.
Mix the protein solution with a click reaction cocktail containing 0.3 mM azide-(PEG)3-biotin reagent, 1 mM CuSO4, 2 mM BTTAA, and 7.5 mM sodium ascorbate in 300 μL PBS buffer (See Materials and Equipment section). Incubate the reaction mixture at 25°C for 1 h.
-
18.
Take 75 μL protein solution for western blotting analysis, add 8 mL cold methanol to the remaining protein sample and store at −80°C for 3 h to precipitate yeast proteins and remove excess click reagents.
CRITICAL: Cells in 60 mL SG media are only corresponding to one contrast. Please prepare more cells if need more contrasts, such as omit probe or omit H2O2.
Note: For proteomic experiments, azide-(PEG)3-biotin reagent is conjugated for enrichment and western blotting analysis. For characterization by in-gel fluorescence cells from 5 mL SG media are sufficient and conjugation should be performed instead with azide-Cy5.
Pause Point: After step 15, precipitated protein sample could be stored in methanol at −80°C for a few days, which also facilitates more thorough precipitation.
Streptavidin Blot and In-Gel Fluorescence Characterization of APEX2 Labeling
Timing: 8 h
The following steps describe the protocol of western blotting or in-gel fluorescence characterization for characterization of APEX2 labeling.
-
19.
Add 19 μL 5× protein loading buffer to the 75 μL protein sample mentioned in step 18 of APEX labeling and click reaction section and boil at 95°C for 10 min. Separate the labeled protein on a 12% SDS-PAGE gel.
-
20.Blotting analysis and in-gel fluorescence analysis. (See Troubleshooting 2 and 3)
-
a.Blotting analysis
-
i.Transfer the gels to nitrocellulose membrane.
-
ii.Block the membrane with 20 mL 3% BSA in TBST buffer (TBS buffer with 0.1% Tween20) at 4°C for 16 h or at 25°C for 1 h.
-
iii.For detecting biotin labeling signal, immerse the membrane with 0.25 μg/mL streptavidin-HRP at 25°C for 1 h.
-
iv.For detecting the expression of Flag or GFP, probe the blots with α-Flag (1:5,000) monoclonal antibody or rabbit α-GFP (1:5,000) as primary antibodies for 1 h, followed by HRP-conjugated goat α-mouse IgG (1:5,000) or α-rabbit IgG (1:5,000) as the secondary antibodies for 1 h.
-
v.Image the chemiluminescence of the blot on a Chemidoc imager.
-
i.
-
b.In-gel fluorescence analysis
-
i.Rinse the gel with destaining solution (60% v/v water, 30% v/v methanol, 10% v/v acetic acid) for 16 h.
-
ii.Image the Cy5 fluorescence signal on Typhoon FLA 9500 imager.
-
i.
-
a.
Enrichment of Biotinylated Proteins and Dimethylation Labeling
Timing: 24 h
The following steps describe the preparation of sample for proteomic analysis.
-
21.
Pellet the refrigerated sample mentioned in step 18 of APEX labeling and click reaction section at 12,000 × g for 2 min and discard the supernatant. Resolubilize the protein pellet in 800 μL 0.5% SDS aqueous solution.
-
22.
Measure the protein concentration with BCA protein assay before enrichment.
-
23.
Add 200 μL streptavidin agarose beads to the protein solution and incubate at 25°C for 2 h with gentle rotation.
Note: Based on our previous experiments, 100 μL streptavidin agarose beads are corresponding to 1 mg protein input approximately.
-
24.
Centrifuge the beads at 3,000 g for 2 min and discard the supernatant. Wash beads twice with1 mL 2% SDS in water, twice with 1 mL 8 M urea, and twice with 1 mL 2 M sodium chloride, successively.
-
25.
Centrifuge the beads at 3,000 g for 2 min and discard the supernatant. Resuspend the beads in 500 μL 6 M urea in PBS buffer. Add 25 μL 200 mM dithiothreitol and incubate at 60°C for 15 min to reduce the disulfide bond.
-
26.
Cool the beads to 25°C. Add 25 μL 400 mM iodoacetamide and incubate at 30°C in dark for 30 min to block free sulfydryl.
Note: Dithiothreitol is added to reduce the disulfide bond in native protein to free sulfydryl, which is subsequently alkylated with iodoacetamide. The alkylation is both uniform and irreversible, which facilitates downstream mass spectrometry data analysis.
-
27.
Wash the beads twice with 1 mL triethylammonium bicarbonate buffer (See Materials and Equipment section) to remove excess small molecules.
-
28.
Resuspend the beads in 200 μL triethylammonium bicarbonate buffer. Add 4 μg sequencing-grade trypsin (dissolved in 4 μL triethylammonium bicarbonate buffer) and incubate at 37°C for 16 h.
-
29.
Centrifuge at 15,000 × g for 10 min and discard the beads pellet. Add 12 μL 4% (v/v) CH2O or 12 μL 4% (v/v) 13CD2O on ice, respectively (See Materials and Equipment section). Add 12 μL 39.68 mg/mL NaBH3CN to the peptide solution and incubate at 25°C for 1 h.
-
30.
Add 48 μL 1% (v/v) ammonia solution and 24 μL formic acid into the solution and vibrate thoroughly. Mix the light and heavy isotopically labeled sample together.
-
31.
Desalt with HLB extraction cartridges.
-
32.Desalt peptide samples with 10-mg HLB SPE cartridge with gravity flow.
-
a.Condition each cartridge with 1 mL HPLC-grade acetonitrile.
-
b.Equilibrate each cartridge with 2 mL HPLC-grade water (1 mL each time).
-
c.Load ~100 μL of each sample onto each cartridge.
-
d.Wash each cartridge with 1 mL HPLC-grade water.
-
e.Elute peptides from each cartridge with HLB eluting solution.
-
f.Evaporate eluted peptide samples with vacuum centrifugation to dryness.
-
a.
CRITICAL: All the doses of used reagents are prepared for one contrast in the demethylation labeling experiment.
LC-MS/MS Analysis
Timing: 3–4 h
The next section describes the protocol and parameters for LC-MS/MS analysis.
-
33.
Resuspend the dried peptide samples with 12 μL LC-MS sampling buffer (0.1% formic acid, 5% acetonitrile).
-
34.
Remove undissolved substances by centrifugation at 20,000 × g for 10 min at 4°C.
-
35.
Precool LC auto-sampler to 4°C.
-
36.
For each sample, inject half of dissolved peptide solution into the LC-MS/MS at a flow rate 600 nL/min.
-
37.The followed LC-MS/MS parameters were used to analyze samples:
-
a.Pack 2-cm micro-capillary pre-column, with C18 (3-μm, 120-Å).
-
b.Pack 12-cm, 150-μm micro-capillary analytical column with C18 (1.9-μm, 120-Å) and equip it with a homemade electrospray emitter tip or a commercial electrospray tip.
-
c.Collect pre-column and analytic columns.
-
d.Set NanoLC mobile phase condition as followed: 0 min, 7% B (B = 80% acetonitrile/0.1% formic acid); 14 min, 10% B; 51 min, 20% B; 68 min, 30% B; 67–75 min, 95% B.
-
e.Set the flow rate 600 nL/min.
-
f.Mass spectrometer parameters are listed in Table 1.
-
a.
Table 1.
Parameters of Mass Spectrometer
| Parameters | Settings |
|---|---|
| Spray voltage | 2.1 kV |
| Heated capillary temperature | 320°C |
| Data recording method | Data-dependent, Top 20 |
| Resolution | 70,000 |
| AGC | 3.00E+06 |
| Max injection time | 20 ms |
| Isolation window | 1.0-m/z |
| Normalized collision energy | 28 |
Yeast RNA Labeling and Enrichment
Timing: 3 days
The following steps describe the protocols of RNA labeling in living yeast cells and subsequent real-time quantitative reverse transcription PCR (RT-qPCR) analysis. The procedures for APEX labeling and cell lysis for RNA identification is the same as described in APEX labeling and click reaction section (step 8–14). For RNA analysis, yeast cells from 30 mL SG media were used. After cell lysis, centrifuge the sample at 12,000 × g for 5 min and keep the supernatant.
-
38.
Extract yeast RNA from supernatant using TRIzol™ Reagent according to manufactural instructions. For every 200 μL supernatant, add 1 mL TRIzol. Measure the concentration of extracted RNA using NanoDrop™ One Spectrophotometers and check RNA purity according to A260/280.
Note: a ratio of ~2.0 is generally accepted as high purity for RNA.
-
39.
Treat RNA with DNase I according to manufactural instructions to remove residual DNA.
-
40.
For samples labeled with Alk-Ph and the corresponding control samples, RNA was bio-conjugated with 0.1 mM Biotin-(PEG)3-Azide, 2 mM THPTA, 0.5 mM Copper sulfate pentahydrate, and 5 mM sodium ascorbate at 25°C for 10 min.
-
41.
Purify all sample by RNA Clean & Concentrator – 100 according to manufacturer’s instructions.
-
42.
For 50 μg DNase I digested total RNA, enrich biotinylated RNA with 10 μL Dynabeads MyOne Streptavidin C1 beads (C1 beads) in 1.5-mL EP tube. Wash C1 beads by 1-mL 1 x B&W buffer (5 mM Tris-HCl, pH7.5, 1 M NaCl, 0.5 mM EDTA, 0.1% v/v Tween-20 in H2O. See Materials and Equipment section) three times, NaOH solution (0.1 M NaOH and 0.05 M NaCl in H2O) twice, and once in 0.1 M NaCl solution at 25°C.
Note: Perform washing by vortexing at 600–1,000 rpm for 1 min. Put tubes on magnetic stand for 2 min and discard the supernatant. For 10–20 μL C1 beads, use 200 μL wash buffer each time. The volume of buffer used for washing should be adjusted based on the amounts of beads.
-
43.
Block C1 beads with 1 mL Block buffer (See Materials and Equipment section) on a thermoshaker at 25°C for 2 h. Wash beads by 1 mL 1× B&W buffer three times.
-
44.
Dilute biotinylated RNA of each sample to the same concentration with H2O and mix RNA with equal volume of 2× B&W buffer. Add the mixture to blocked beads and rotate at 25°C for 45 min.
-
45.
Wash the RNA-loaded beads by 1 mL 1× B&W buffer three time, twice with 1× PBS supplemented with 4 M urea and 0.1% SDS, and twice with 1× PBS at 25°C.
-
46.
Incubate C1 beads with 50 μL Elution Buffer (95% formamide, 10 mM EDTA, 1.5 mM D-biotin) at 50°C for 5 min, and 90°C for 5 min. Put samples on magnetic stand and carefully pipette out the supernatant to a new 1.5-mL EP tube.
-
47.
Extract RNA by TRIzol from supernatant according to manufactural instructions. Dissolve RNA in 10–20 μL H2O.
-
48.
Reverse transcribe enriched RNA into cDNA by ProtoScript® II First Strand cDNA Synthesis Kit.
-
49.
Mix the cDNA product with PowerUp™ SYBR™ Green Master Mix and corresponding qPCR primers and quantify cDNA on StepOnePlus™ Real-Time PCR System.
CRITICAL: All experiments should be performed in RNase-free apparatus at AirClean 600 PCR WorkStation until RNA is reversibly transcribed to cDNA.
Expected Outcomes
Synthesis of Alkyne-Phenol Probe
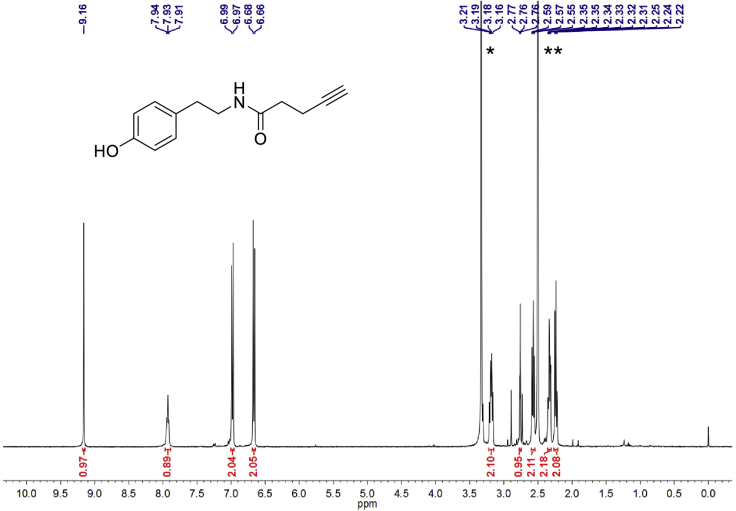
Alkyne-phenol probe was obtained by a two-step synthesis with overall yield probe of 79.4%. 1H-NMR (400 MHz, d6-DMSO): 9.16 (s, 1H), 7.93 (t, J = 5.7 Hz, 1H), 7.04–6.95 (m, 2H), 6.71–6.62 (m, 2H), 3.24–3.14 (m, 2H), 2.81–2.71 (m, 1H), 2.57 (t, J = 7.5 Hz, 2H), 2.33 (td, J = 6.8, 2.4 Hz, 2H), 2.24 (dd, J = 7.7, 6.1 Hz, 2H). NMR spectra were shown in Figure 2. Calculated m/z for C13H15NO2: [M+H]+, 218.27; found 218.51.
Figure 2.
1H-NMR Spectral of Alkyne-Phenol Probe
Solvent peaks are labeled with “∗” (HDO) and “∗∗” (DMSO). Figure from Li et al. (2020).
Western Blotting Analysis of APEX Labeling
The expected western blotting characterizations were shown in Figure 3.
Figure 3.

Streptavidin-HRP Blot Analysis of APEX2 Labeling in the Yeast Mitochondria with Alk-Ph Probe
Alkyne-modified proteins were ligated with azide-(PEG)3-biotin via click reaction. Molecular weight standards are shown in kDa. Arrows indicate endogenously biotinylated proteins. Bottom: α-Flag western blot showing the expression of Su9-APEX2 (∗) and Su9-eGFP (∗∗). Figure from Li et al., (2020).
In-Gel Fluorescence Analysis of APEX Labeling
The expected in-gel fluorescence characterization is shown in Figure 4.
Figure 4.

In-Gel Fluorescence Analysis of APEX2-Mediated Protein Labeling with Alk-Ph
Yeast cells expressing NLS-APEX2 were labeled with Alk-Ph at concentrations in the range of 0.5–5 mM. Alkyne-modified proteins were derivatized with azide-Cy5 fluorophore via click chemistry, and analyzed with in-gel fluorescence. Figure from Li et al. (2020).
Quantification and Statistical Analysis
Protein Identification and Quantification
Timing: 2–3 h
Use the software package pFind studio (ver. 3.0.11) (http://pfind.ict.ac.cn/) (Chi et al., 2015; Chi et al., 2018; Li et al., 2005; Wang et al., 2007) to analyze raw data files obtained from LC-MS/MS.
-
1.
Major parameters used for database searching and peptide quantification are listed in Table 2.
Table 2.
Parameters for the Identification and Quantification of Dimethyl Labeled Peptides
| Parameters | Settings |
|---|---|
| Database | Yeast (Saccharomyces cerevisiae), canonical, UniProt, 6729 entries |
| Enzyme | Trypsin KR_C (Full specific) |
| Maximum number of missed cleavages | 3 |
| Precursor tolerance | 10 p.p.m. |
| Fragment tolerance | 20 p.p.m. |
| Dynamic modification | Dimethyl (Light, +28.0313 Da) Dimethyl (Heavy, +34.0631 Da) |
| Quantitation type | Labeling - SILAC etc |
| Multiplicity | 2 |
| FDR | <1% at peptides level |
| Peptide mass | [600, 10,000] |
| Peptide length | [6, 100] |
| Open search | FALSE |
-
2.
For generating the final protein-level quantification ratio (heavy versus light), use an in-house algorithm building upon Python3 (ver. 3.7.3) to process the peptide-level quantification results obtained from the pQuant (Liu et al., 2014) module in pFind studio. The algorithm and the detailed user manual can be downloaded at https://github.com/morpheusliu/Proetein-level-post-processing-program-for-pfind.
Quantification of APEX RNA Labeling in Yeast
To characterize the relative abundance of representative genes in each sample, cytoplasmic marker gene GAPDH or ACT1 is set as the negative control. The relative level of enrichment of each gene is obtained from the value of 2ˆ(-ΔΔΔCt). ΔΔΔCt is calculated as follows: 1) For each gene in APEX2-labeled samples and samples omitting probes, ΔCt values are calculated as the difference between Ct values of enriched samples and the matched input samples; 2) ΔΔCt values are calculated as the difference between ΔCt values from APEX2-labeled sample versus those of control samples omitting the probe; 3) ΔΔΔCt values of MT-mRNA are calculated by the difference between ΔΔCt of each gene and that of reference gene, GAPDH or ACT1.
Limitations
APEX2 is transformed and overexpressed into yeast cells within pCTCON2 plasmid in this experiment. Low expression of APEX2, such as knocking in the genome, would lead to weak labeling signal.
To remove residual Alk-Ph in the cell lysate and access click reagents, two rounds of protein precipitation with methanol are typically required, which may lead to loss of material.
Troubleshooting
Problem 1
Protein degradation during cell lysis.
Potential Solution
Ensure the “shake and cool” cycle is performed at 4°C.
Add protease inhibitor to the lysis buffer.
Problem 2
Poor labeling efficiency shown on the streptavidin western blot or Cy5 in-gel fluorescence
Potential Solution
Check with western blot to see if APEX2 is expressed well.
Problem 3
Poor labeling efficiency but APEX2 is expressed well.
Potential Solution
Check the azide-(PEG)3-biotin reagent. Multiple thaws could result in the degradation of the stock.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peng Zou (zoupeng@pku.edu.cn).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
All data presented are available in the main text.
Acknowledgments
We thank Prof. Alice Ting (Stanford University) for providing the pCTCON2 vector and for helpful discussions. This work was supported by the Ministry of Science and Technology (2018YFA0507600, 2017YFA0503600), the National Natural Science Foundation of China (91753131, 21673009, and 21727806), Natural Science Foundation of Beijing Municipality (5182011), the Interdisciplinary Medicine Seed Fund of Peking University (BMU2017MC006), and State Key Laboratory of Proteomics (SKLP-K201804). P.Z. was supported by Li Ge-Zhao Ning Life Science Junior Research Fellowship and Bayer Investigator Award. We thank Profs. Tao Liu (Peking University) and Ping Wei (Peking University) for sharing W303 yeast strains.
Author Contributions
P.Z. conceived the project and supervised APEX labeling and protein-centric proteomic experiments. J.Y. supervised proteomic data analysis. Y.L. synthesized the probe and performed yeast proteomic experiments. Y.L., C.T., K.L., J.Y., and P.Z. analyzed the proteomic data. Y.Z. performed yeast RNA labeling experiments. Y.L., Y.Z., and P.Z. analyzed RNA labeling data. Y.L., J.Y., and P.Z. wrote the paper with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Contributor Information
Jing Yang, Email: yangjing54@hotmail.com.
Peng Zou, Email: zoupeng@pku.edu.cn.
References
- Chen C.-L., Hu Y., Udeshi N.D., Lau T.Y., Wirtz-Peitz F., He L., Ting A.Y., Carr S.A., Perrimon N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. U S A. 2015;112:12093. doi: 10.1073/pnas.1515623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H., He K., Yang B., Chen Z., Sun R.-X., Fan S.-B., Zhang K., Liu C., Yuan Z.-F., Wang Q.-H., Liu S.-Q., Dong M.-Q., He S.-M. pFind–Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Chi H., Liu C., Yang H., Zeng W.-F., Wu L., Zhou W.-J., Wang R.-M., Niu X.-N., Ding Y.-H., Zhang Y., Wang Z.-W., Chen Z.-L., Sun R.-X., Liu T., Tan G.-M., Dong M.-Q., Xu P., Zhang P.-H., He S.-M. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018;36:1059–1061. doi: 10.1038/nbt.4236. [DOI] [PubMed] [Google Scholar]
- Hung V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 2017;6:e24463. doi: 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Zou P., Rhee H.-W., Udeshi Namrata D., Cracan V., Svinkina T., Carr Steven A., Mootha Vamsi K., Ting Alice Y. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Espenshade Peter J. Proximity-dependent biotin labelling in yeast using the engineered ascorbate peroxidase APEX2. Biochem J. 2016;473:2463–2469. doi: 10.1042/BCJ20160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Fu Y., Sun R., Ling C.X., Wei Y., Zhou H., Zeng R., Yang Q., He S., Gao W. pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics. 2005;21:3049–3050. doi: 10.1093/bioinformatics/bti439. [DOI] [PubMed] [Google Scholar]
- Li Y., Tian C., Liu K., Zhou Y., Yang J., Zou P. A clickable APEX probe for proximity-dependent proteomic profiling in yeast. Cell. Chem. Biol. 2020;27:858–865.e8. doi: 10.1016/j.chembiol.2020.05.006. [DOI] [PubMed] [Google Scholar]
- Liu C., Song C.-Q., Yuan Z.-F., Fu Y., Chi H., Wang L.-H., Fan S.-B., Zhang K., Zeng W.-F., He S.-M., Dong M.-Q., Sun R.-X. pQuant improves quantitation by keeping out interfering signals and evaluating the accuracy of calculated ratios. Anal. Chem. 2014;86:5286–5294. doi: 10.1021/ac404246w. [DOI] [PubMed] [Google Scholar]
- Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.-C., Krach F., Yang D., Sen A., Fulzele A., Wozniak J.M., Gonzalez D.J., Kankel M.W., Gao F.-B., Bennett E.J., Lécuyer E., Yeo G.W. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A.W., Mak R., Troemel E.R., Bennett E.J. In vivo mapping of tissue- and subcellular-specific proteomes in Caenorhabditis elegans. Sci. Adv. 2017;3:e1602426. doi: 10.1126/sciadv.1602426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H.-W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Krüger B., Fröhlich T., Franz-Wachtel M., Nalpas N., Macek B., Jansen R.-P. APEX2-mediated proximity labeling resolves protein networks in Saccharomyces cerevisiae cells. FEBS J. 2020;287:325–344. doi: 10.1111/febs.15007. [DOI] [PubMed] [Google Scholar]
- Wang L.H., Li D.Q., Fu Y., Wang H.P., Zhang J.F., Yuan Z.F., Sun R.X., Zeng R., He S.M., Gao W. pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:2985–2991. doi: 10.1002/rcm.3173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data presented are available in the main text.