Summary
Protein lysine methylation mediates a variety of biological processes, and their dysregulation has been established to play pivotal roles in human disease. A number of these sites constitute attractive drug targets. However, systematic identification of methylation sites is challenging and resource intensive. Here, we present a protocol combining MethylSight, a machine learning model trained to identify promising lysine methylation sites, and mass spectrometry for subsequent validation. Our approach can reduce the time and investment required to identify novel methylation sites.
For complete information on the use and execution of this protocol, please refer to Biggar et al. (2020).
Graphical Abstract

Highlights
-
•
Identify high-confidence lysine methylation sites for experimental validation
-
•
Webserver interface allows for user-defined prediction threshold
-
•
Protocol has been used for the validation of 45 new histone methylation sites
-
•
Validation of candidate methylation sites by targeted mass spectrometry
Protein lysine methylation mediates a variety of biological processes, and their dysregulation has been established to play pivotal roles in human disease. A number of these sites constitute attractive drug targets. However, systematic identification of methylation sites is challenging and resource intensive. Here, we present a protocol combining MethylSight, a machine learning model trained to identify promising lysine methylation sites, and mass spectrometry for subsequent validation. Our approach can reduce the time and investment required to identify novel methylation sites.
Before You Begin
Preparing the Data for Submission to the MethylSight Web Server
Timing: 5 min
-
1.If your query proteins are of human origin and are indexed in the SwissProt database, make sure to have their Uniprot Accession ID on hand, as this will constitute the input to the MethylSight web server.
-
a.Uniprot Accession ID can be obtained at https://www.uniprot.org, where you may enter the name of the protein in the search bar, or any other identifier (e.g., Entrez, Ensembl, etc.).
-
b.If you have more than one protein of interest, prepare a .txt file with one Uniprot Accession ID per line.
-
a.
-
2.
If your query proteins are not indexed (e.g., truncated protein, mutants, predicted protein), or if the proteins are from any other organism, then you will need to provide a valid file in FASTA format containing the amino acid sequence (standard amino acids and unknown amino acid “X” only) of the proteins. To comply with the FASTA format, the sequences must be preceded by a tag on a separate line prefixed with a mandatory “>” character. A valid FASTA file (with a .fasta extension) may look as follows:
investigated_proteins.fasta.
>H2B
MPEPAKSAPAPKKGSKKAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAM
GIMNSFVNDIFERIAGEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVT
KYTSSK
>H3.1
MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTE
LLIRKLPFQRLVREIAQDFKTDLRFQSSAVMALQEACEAYLVGLFEDTNLCAIHAKRVTI
MPKDIQLARRIRGERA
Note: MethylSight was trained using sites extracted from human proteins. The quality of its predictions for other organisms is unknown.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tris-HCl | Sigma-Aldrich | Cat# 10812846001 |
| Acetonitrile with 0.1% Formic Acid (v/v), LC/MS Grade | Fisher Scientific | Cat# LS1201 CAS# 75-05-8 |
| Water with 0.1% Formic Acid (v/v), LC/MS Grade | Fisher Scientific | Cat# LS1181 CAS# 7732-18-5 |
| Formic Acid, 99.0+%, LC/MS Grade | Fisher Scientific | Cat# A11750 CAS# 64-18-6 |
| Sequencing-grade modified Trypsin | Promega | Cat# V5111 |
| Guanidine-HCl | Sigma-Aldrich | Cat# G3272 |
| Iodoacetamide | Sigma-Aldrich | Cat# I6125 |
| Ammonium Bicarbonate | Sigma-Aldrich | Cat# A6141 |
| Software and Algorithms | ||
| MethylSight | Biggar et al., 2020 | www.methylsight.com |
| Skyline | MacLean et al., 2010 | https://www.skyline.ms/ |
| ExPASy PeptideCutter | Gasteiger et al., 2003 | https://web.expasy.org/peptide_cutter/ |
Materials and Equipment
Recipe for Denaturation Buffer
| Reagent | Final Concentration |
|---|---|
| Guanidine-HCl | 6 M |
| Tris-HCl (pH 8) | 50 mM |
| DTT | 2 mM |
Recipe for 100 mL Tip Equilibration Solution
| Reagent | Amount |
|---|---|
| Acetonitrile + 0.1% FA | 100 mL |
Recipe for 100 mL Tip Wash Solution
| Reagent | Amount |
|---|---|
| H2O + 0.1% FA | 100 mL |
Recipe for 100 mL Peptide Elution Solution
| Reagent | Amount |
|---|---|
| Acetonitrile + 0.1% FA | 80 mL |
| H2O + 0.1% FA | 20 mL |
Step-By-Step Method Details
Predicting Lys-Methylation Sites with MethylSight
Timing: negligible for indexed human sequences (with a valid Uniprot ID), and approximately 1 s per lysine for non-indexed sequences
This step allows one to identify the most promising Lys-methylation sites for subsequent validation via targeted mass spectrometry or immunoblotting with methyl-specific antibodies. The researcher can then make an informed decision as to which proteins/sites are most likely to generate hits.
-
1.
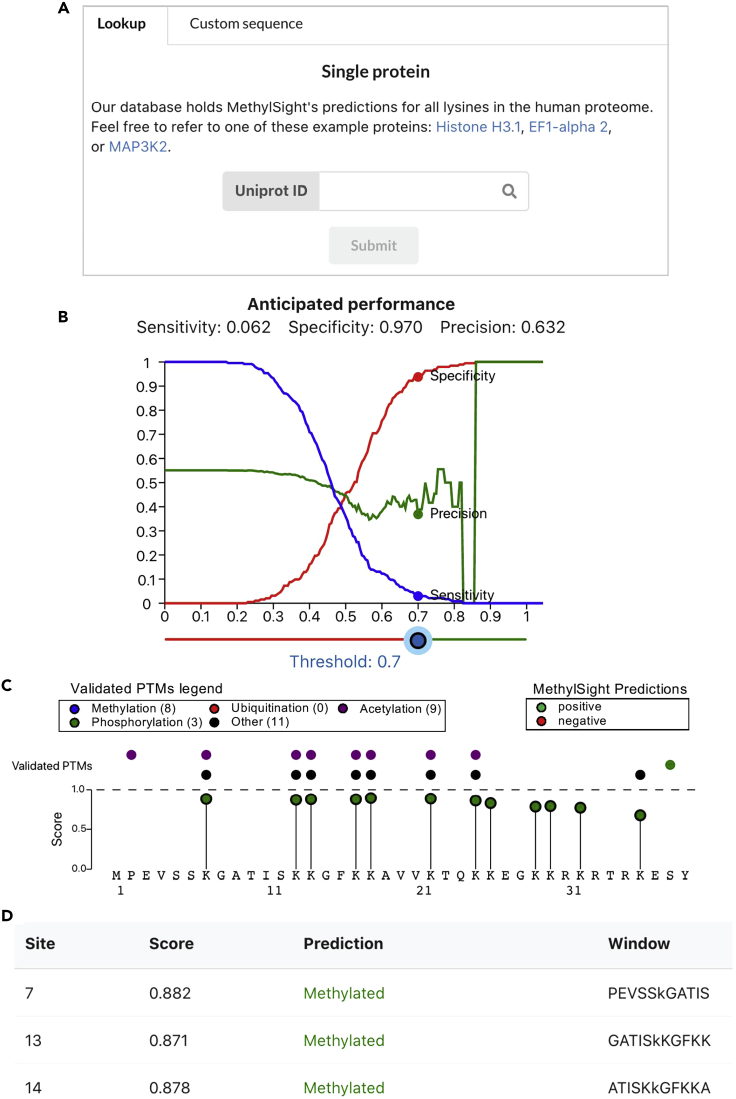
Navigate to https://www.methylsight.com, and proceed to the “predictor” page (Figure 1).
Figure 1.
Predictions Output by MethylSight
(A) Once the user’s query has been completed, the MethylSight web server displays a results page outlining the predictions made by the predictor.
(B) User-adjustable operating threshold for the predictor (i.e., to adjust the score cut-off above which predictions are considered to be positive and displays the anticipated accuracy of the predictions).
(C) Graphical display of prediction results along the length of the protein, showing predictions, along with other validated post-translational modifications.
(D) The user is offered the option to consult the predictions in a table format and to download a tab-separated value (tsv) file.
-
2.Submit your proteins of interest in the submission form (Figure 1A).
-
a.If the proteins are indexed in the SwissProt database, stay in the “Lookup” tab of the submission form select a .txt file containing the Uniprot ID.
-
b.If the proteins are not indexed, select the “Custom sequence” tab, and select the FASTA file containing the sequences.
-
a.
-
3.
Click on “Submit” (individual ID) or “Upload” (ID list or FASTA file) and wait for the prediction results to be generated.
Note: Uploading non-indexed sequences may lead to longer wait time – particularly if the FASTA file contains numerous sequences, as the MethylSight server needs to pre-process the sequences and to run the prediction algorithm.
CRITICAL: Do not close the browser tab while MethylSight is generating predictions.
-
4.
The “results” page will automatically load once predictions are complete. In this page, set an operating threshold based on your preference for sensitivity and precision of the predictions (Figure 1B). “Permissive” and “conservative” parameter presets are provided.
Note: Sensitivity and precision should be considered by the user. These parameters and the trade-off between them are described in greater detail in Quantification and Statistical Analysis – Interpreting the Predictions Made by Methylsight.
-
5.
Select candidates among the sites labeled as positives or with the highest prediction scores for experimental validation (Figures 1C and 1D).
CRITICAL: The decision threshold is the value that discriminates between positive and negative predictions. Sites with prediction scores below the threshold are labeled as negative, while those with scores greater or equal to the threshold value are considered positive. Setting an appropriate threshold drastically affects the predictions and how they should be interpreted – a topic discussed in Quantification and Statistical Analysis – Interpreting the Predictions Made by Methylsight. It is advised to set the threshold parameter (Figure 1B) before consulting the predictions.
Targeted Validation of Lysine Methylation Sites
Timing: 2–3 days
In this step, we discuss one approach to validate MethylSight predicted lysine methylation sites. To validate the status of predicted methylation sites, we suggest the use of a targeted mass spectrometry approach, such as parallel reaction monitoring (PRM-MS). Further, it is best to proceed with a quality source of semi-purified protein of interest, such as an immunoprecipitation preparation from a relevant cell line or tissue. PRM-MS can differentiate and relatively quantify peptides with isobaric post-translational modifications (PTMs). In yeast, in which the histone H3 K4 mono-methyltransferase SET1 was lacking, PRM-MS was used to quantify several newly identified methyl modifications (Lee et al., 2020).
-
6.Building a PRM-MS isolation list.
-
a.Download and install a local copy of Skyline software (MacLean et al., 2010).
-
b.Navigate to “Settings” and select “Peptide settings.”
-
c.Under the “Digestion” tab, select your protease of interest (e.g., Trypsin) and max missed cleavages (e.g., 1 missed cleavage).
-
d.Under the “Modifications” tab, select Carbamidomethyl (C) if iodoacetamide was used. Close the “Peptide Settings” window.
-
e.Next, enter the FASTA protein sequence of your target to the “Targets” window (copy & paste directly in the “Targets” panel, or File>Import>FASTA>select the desired FASTA file). The Skyline software will automatically perform an in silico digest of your protein and select the most likely proteotypic peptides and optimal transitions.
-
a.
Note: Peptides and transitions can also be manually selected by right clicking the peptide ion and selecting “Pick Children.”
Note: Skyline software currently has a large and active user group. Please refer to the Skyline Support Board for up to date information and assistance in developing your isolation list.
-
f.
Navigate to peptide(s) containing the candidate methylation site in the “Targets” panel. Right click on the peptide sequence and select “Modify” from the drop-down list to add a variable modification to lysine. A new “Modify” window will appear, on the modified lysine select either mono-, di-, and tri-methylation modifications representing the addition of 14.01565, 28.03130 and 42.04695 Da, respectively.
Note: Peptides can be copied and pasted within the Skyline program to maintain any manually selected transitions. This allows users to include several variants in the isolation list that represent several PTM m/z and maintain comparable transition lists.
Note: We apply validation criteria that are specific for methylated peptides when confirming modification sites identified by Methylsight. This criteria includes the inclusion of fragment ions that are diagnostic of a methylation event and that constrain the methyl group to a specific amino acid. Importantly, methylation differs from other modifications as any amino acid could be considered a potential site of methylation if exchanged for another amino acid by the addition of a methyl group (e.g., glycine, serine, etc.).
-
g.
Export your isolation list for PRM-MS analysis. Navigate to “File” and select “Export.”
Troubleshooting - Difficulty in selecting peptides that are diagnostic of the methyl-Lys site of interest
-
7.Sample preparation for mass spectrometry. Digest your isolated protein sample using mass-spec grade Trypsin (i.e., serine protease that specifically cleaves at the carboxylic side of lysine and arginine except when followed by a proline).
-
a.Dissolve target protein (e.g., immunopurified) in 50 μL of denaturation buffer. Scale volume, as necessary, up to a protein concentration of 1 mg/mL.
-
b.Heat at 95°C for 20 min and allow to cool.
-
c.Add iodoacetamide to a final concentration of 55 mM in darkness and incubate at room temperature for another 30 min.
-
d.Add 50 mM NH4HCO3 (pH 7.8) (such that the final concentration of guanidine-HCl is below 1 M).
-
e.Add trypsin protease to a final protease:protein ratio of 1:50 (w/w) and incubate at 37°C overnight.
-
a.
Pause Point: At this stage, samples can be stored short-term (1–2 days) at 4°C and long-term (months) at −80°C. Dried protein digests can be stored at −80°C for years without compromising sample quality.
Note: Other proteases can be used other than Trypsin to create ionizable proteotypic peptides that contain your methylation site of interest. To determine an appropriate protease for use, perform an in silico digestion with Trypsin or other proteases using the ExPASy PeptideCutter webserver (https://web.expasy.org/peptide_cutter/) (Gasteiger et al., 2003). Follow recommended manufacturer guidelines for digestion reactions.
-
8.Sample preparation and desalting.
-
a.Pre-treat sample: Adjust sample from step 6e such that they contain approximately 0.1% formic acid (FA). Samples should be below 4.0 (check with pH paper). If needed, adjust the pH using formic acid.
-
b.Condition tip: Securely attach a Bond Elut OMIX C18 tip (Agilent; Cat# A57003100) and set pipettor to 100 μL. Aspirate 100 μL of tip equilibration solution and discard. Repeat 4 times. Keep plunger depressed and move to the next step.
-
c.Equilibrate: Aspirate 100 μL of tip wash solution and discard. Repeat 4 times. Keep pipette plunger depressed and move to the next step.
-
d.Bind: Aspirate 100 μL of pretreated digested sample into tip and cycle the sample across the C18 column bed 3 to 5 times to increase peptide binding efficiency. Depress pipette plunger and move to the next step.
-
e.Wash: Aspirate 100 μL of tip wash solution and discard solution to waste. Cycle 2–4 times. Keep plunger depressed and move to the next step.
-
f.Elute: Carefully draw 100 μL of peptide elution solution into the top.
-
a.
CRITICAL: Do not repeat this step or expel pipette plunger as your peptides will elute into your peptide elution solution.
-
g.
Elute the sample directly into a clean microcentrifuge tube and cycle the elution solution up to ten cycles across the C18 column bed to improve elution efficiency.
-
h.
Repeat steps f and g, eluting your sample into the same microcentrifuge tube.
-
i.
Dry samples using a Thermo Scientific Savant SpeedVac (or similar) to evaporate the sample solution.
-
j.
Resuspend in 20–30 μL of tip wash solution for mass spectrometry analysis (step 9).
Pause Point: Dried peptides may be stored at 4°C for 1 week or at −20°C for 1 month.
Note: Your MS facility will have guidelines on the optimal peptide amount to be injected into their particular LC-MS/MS system. Generally, 1 μg/μL is optimal but affinity purified samples are likely to be much lower.
-
9.
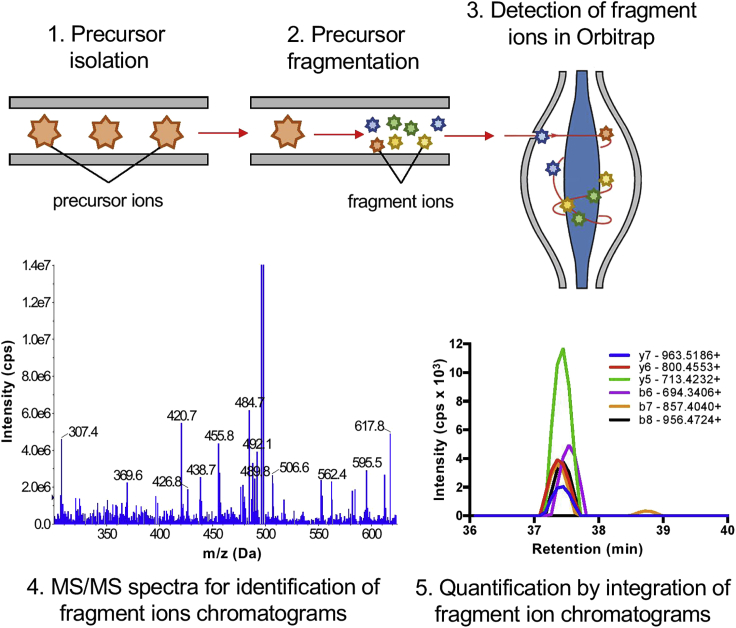
Mass spectrometry analysis. Analyze each sample collected in step 8j. Analysis of proteins is routine for most mass spectrometry facilities, but the sample preparation and procedure for LC-MS/MS analysis depends on the configuration of the mass spectrometer and liquid chromatography system. We analyze each fraction by using a Thermo EASY nano liquid chromatography (EASYnLC) system on a Thermo Q Exactive mass spectrometer (Figure 2).
Figure 2.
Validation of MethylSight Predicted Methyl-Lys Sites by Targeted Mass Spectrometry
Briefly, a target precursor ion containing the methylation site of interest is (1) isolated in the quadrupole analyzer and (2) fragmented in the HCD cell. The fragment ions are then (3) detected in the Orbitrap mass analyzer. A (4) MS/MS spectrum (representative) is used for the confident identification of the peptide, and (5) peak areas of fragment ions (representative) can be integrated across the elution profile for quantification.
-
a.
Peptides are first concentrated on a trapping-column (EASY-Column, 100 μm 2 cm, Thermo Scientific) using 0.1% formic acid at a flowrate of 20 μL/min and subsequently separated on a reverse phase main-column (EASY-Column, 75 μm 10 cm, Thermo Scientific) using a binary gradient consisted of A: 0.1% formic acid and B: 84% acetonitrile, 0.1% formic acid at a flowrate of 250 nL/ min. The gradient increased linearly from 3% A to 29.7% B over 75 min.
-
b.
Three washing steps at 95% of B at the end of the gradient are then applied to prevent carry-over into the next run.
-
c.
Each MS1 survey scan in the Orbitrap is followed by data-dependent MS/MS scans that is guided by the isolation list (designed in step 6) in the ion trap using collision-induced dissociation (CID) fragmentation.
-
d.
Obtain results in RAW file format.
Expected Outcomes
MethylSight produces a list of predictions for all the potential methylation sites in the proteins provided. These predictions are presented in a graphical format and in a tabular format (Figure 1).
The graphical output outlines all the predictions along the sequence of the proteins in the form of a lollipop plot (Figure 1C), wherein positive predictions are shown in green and negative predictions are shown in red, given an operating decision threshold. If the user provided a Uniprot ID, the graphic also displays the location of other known post-translational modifications and annotated protein regions and domains. This feature may help assess the relevance of a particular site, which may be located within a critical region of the protein such as a catalytic site or a binding domain. The user can move along the length of the protein by scrolling, and the exact score for a site is displayed when the user’s cursor hovers on a prediction.
The predictions table (Figure 1D), which can be downloaded as a tab-separated value file for further analysis, provides a prediction score along with a label ("methylated” or “not methylated”) based on the threshold set by the user. In addition, the amino acid window surrounding the site (+/- 5 amino acids) is provided.
Quantification and Statistical Analysis
Interpreting the Predictions Made by MethylSight
In order to adequately interpret the predictions made by MethylSight, one should be familiar with the metrics used to assess the performance of a binary predictor. In the context of novel methyl-Lys sites prediction, the most relevant metrics are sensitivity, specificity, and precision.
Sensitivity corresponds to the expected fraction of all methylation sites that are “captured” by the predictor. For example, at a decision threshold value of 0.5, MethylSight achieves a 0.54 sensitivity level. This implies that the positive predictions made by MethylSight at that threshold will capture 54% of all true methylation sites. The sensitivity of the predictions decreases as the threshold value increases, because the predictor will only label very high-scoring predicted sites as being positive sites. The opposite is true as the threshold value is decreased, leading to a prediction that makes positive predictions more liberally. Figure 3 summarizes how the threshold impacts the number of predicted methyl-Lys sites.
Figure 3.
Distribution of the Prediction Scores across the Human Proteome and Impact of the Threshold on the Number of Positive Predictions
(A) The prediction scores across the human proteome are approximately normally distributed.
(B) The relationship between the threshold and number of predicted methyl-Lys sites is inversely proportional. Permissive and conservative threshold values of 0.5 and 0.7, respectively, are labeled on the curve.
Specificity corresponds to the fraction of all lysine sites expected to not be methylated that are correctly predicted not to be. In other words, specificity measures the capacity of the predictor to retrieve negative sites.
Precision is arguably the most important performance metric. In this context, it is defined as the fraction of positive predictions expected to be true methylation sites. Precision typically varies proportionally with the threshold, as the fraction of true positives increases with the prediction score. MethylSight achieves a precision of 0.63 at a more conservative threshold of 0.7. In theory, this implies that if one were to experimentally validate by mass spectrometry 100 sites predicted to be methylated with a score of 0.7 or higher, approximately 63 sites would be shown to be truly methylated. Consistent with the no free lunch idea, this increase in performance is offset by a decrease in sensitivity. At this threshold, the sensitivity is approximately 0.06, meaning that the classifier will miss 0.94% of all actually methylated sites.
Unfortunately, a trade-off exists between precision and sensitivity. For almost any predictor, higher precisions come at the cost of a lower sensitivity and vice-versa. To make matters worse, there is no globally applicable “best” threshold. In general, the value of the threshold is set so as to achieve an acceptable balance between the number of positive predictions, and the quality of those predictions. For example, lower thresholds and lower precision can be acceptable in the context of high-throughput screening of sites. On the other hand, if resources are scarce, setting a higher threshold may be more appropriate, as the positive predictions are more likely to result in confirmed hits. In Biggar et al. (2020), we performed validation experiments and annotated the methyl-Lys proteome using a threshold value of 0.7, to obtain a specificity of 97%.
Analysis of PRM-MS Data by Skyline Software
Quantification by a label-free targeted method yields confident results particularly in cases where substantially similar samples are analyzed. To avoid potential stoichiometry issues with identifying new Lys-methylation events, Biggar et al. (2020) utilized a label-free targeted proteomic strategy to validate methylation sites that have been guided by MethylSight predictions. Below are steps involved in validating and relatively quantifying methylation events by label-free targeted proteomics.
-
1.
Open your PRM isolation method in Skyline software and import your RAW data file from step 9.
-
2.
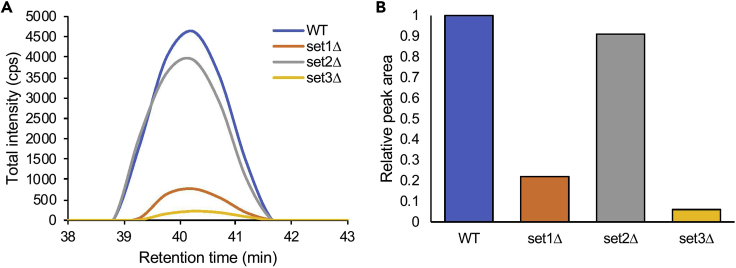
The data will be automatically processed and should be manually verified and product ions with interferences should be removed. Only the fragment ions showing symmetrical chromatographic shapes should be used for quantification (Figure 4). Skyline will automatically integrated boundaries for the peaks. These can be adjusted if the software does not reliably determine the peak boundaries. The boundaries must be the same for the target peptide and all corresponding samples.
Figure 4.
Relative Quantification of Novel Methyl-Lys Events by Targeted Mass Spectrometry
(A) Chromatogram of total fragment ions extracted from the mono-methylated peptide IFSPYGHIMQ[+1.0]IN[+1.0]IK[+14.0] (m/z 559.628731, 3+) from S. cerevisiae showing relative detection from Nab3 protein purified from WT versus set1Δ, set2Δ, or set3Δ strains.
(B) Relative total peak area of Nab3-K363me1 chromatographs in WT versus set1Δ, set2Δ, or set3Δ strains. Figure modified from Lee et al. (2020).
-
3.
Once chromatographic peaks (containing at least three diagnostic fragment ions) have been defined, peak area can be measured directly in the Skyline software by navigating to “View” and selecting “Peak Areas,” then “Replicate Comparison.”
-
4.If comparing methylation between experimental conditions or replicates, users must normalize for differences in protein abundance between samples (i.e., any experimental condition may alter the expression, and not only methylation, of your target protein).
-
a.To compare relative methyl-Lys detection between experimental conditions, divide median peak area of each experimental sample by control median peak area. This will provide you with an un-normalized relative detection of methylated peptides.
-
b.To normalize your samples to the abundance of your target protein, obtain the median peak area for 2–3 non-modified proteotypic peptides from within the target protein.
-
c.Average these median peak areas from the non-modified peptides (step 4b) and calculate the relative detection of non-modified peptides as done in step 4a.
-
d.To normalize your relative detection of methylated peptides, divide the relative detection of your methylated peptide (from step 4a) with the average median peak area of your non-modified peptides (from step 4c).
-
a.
Limitations
Limitation 1
One important limitation to consider is that the MethylSight algorithm was developed under the assumption that any lysine residue that is not currently known to be methylated, is indeed not truly methylated. Of course, it is expected that a number of these sites may, in fact, be methylated and simply not yet discovered by modern proteomics. As such, there will be a number of sites predicted to not be methylated by MethylSight will eventually be shown to be methylated.
Limitation 2
MethylSight was trained using validated methylation sites in human proteins. As such, applying MethylSight to other organisms may produce inaccurate predictions. Work aiming to evaluate the accuracy of MethylSight and similar models on yeast protein is under way.
Limitation 3
Many modification events are temporal and cell specific. Therefore, it may be necessary to optimize conditions that facilitate cellular methylation events for validation.
Troubleshooting
Problem
Difficulty in selecting peptides that are diagnostic of the methyl-Lys site of interest.
Potential Solution
Regardless whether SRM or PRM is used to acquire the data, selecting suitable peptides that represent the target methylation site is crucial. Only the correct set of peptides can yield reliable quantification of the selected proteins. Peptides that are always observed for a specific protein, regardless of whether they are suitable for quantification, are known as proteotypic peptides. The key feature of a proteotypic peptide is that its abundance must correlate with the abundance of the parent protein. Presence of a modified form of a peptide will decrease the level of its unmodified form. Hence, you should target both the unmodified and Lys-methylated peptides. Criteria for selecting proteotypic target peptides are listed below, however the Skyline software (MacLean et al., 2010) can also be used for predicting suitable proteotypic peptides that contain your methylation site of interest:
-
a.
Peptide length: m/z value of the peptides should be within the mass range of the instrument. Peptides of 8–25 amino acids are usually preferred.
-
b.
Uniqueness: Selected peptide should be unique to the proteins of interest. A search with the Basic Local Alignment Search Tool (BLAST) on the peptides can confirm if the peptide sequence is unique to the candidate protein.
-
c.
Missed cleavage: When using trypsin, selected peptides should be fully tryptic and ideally should not contain missed cleavage sites. However, the presence of a methylation modification may result in a resistance to trypsin cleavage. If possible, avoid peptides with ragged ends—series of arginine and lysine amino acids, e.g., KK, KR, RK, or RR.
-
d.
Precursor charge: The peptide charge state that fragments better and generates the most sensitive measurements should be selected. Doubly or triply charged precursor ions are favorable due to their measurable m/z ranges.
-
e.
Chromatographic peak: The shape of chromatographic peak of a peptide should be symmetrical with narrow width.
-
f.
Signal intensity: The peptide should ionize efficiently and provide a stable and intense signal.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kyle K. Biggar (kyle_biggar@carleton.ca).
Materials Availability
This study did not generate any new materials.
Data and Code Availability
The code for the MethylSight predictor and web server is available on GitHub: https://github.com/fcharih/MethylSight.
Acknowledgments
This work was supported by a National Science and Engineering Research Council (NSERC) Canada postgraduate scholarship to F.C. and NSERC Canada Discovery grants to K.K.B. and J.R.G.
Author Contributions
K.K.B. and F.C. wrote the manuscript with input from J.R.G. who also proofread and reviewed the manuscript.
Declaration of Interests
The authors declare no competing interests.
Contributor Information
James R. Green, Email: jrgreen@sce.carleton.ca.
Kyle K. Biggar, Email: kyle_biggar@carleton.ca.
References
- Biggar K.K., Charih F., Liu H., Ruiz-Blanco Y.B., Stalker L., Chopra A., Connolly J., Adhikary H., Frensemier K., Hoekstra M. Proteome-wide prediction of lysine methylation leads to identification of H2BK43 methylation and outlines the potential methyllysine proteome. Cell Rep. 2020;32:107896. doi: 10.1016/j.celrep.2020.107896. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hooglanbd C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Chopra A., Burke G.L., Chen Z., Greenblatt J.F., Biggar K.K., Meneghini M.D. A crucial RNA-binding lysine residue residue in the Nab3 RRM domain undergoes SET1 and SET3-responsive methylation. Nucleic Acids Res. 2020;48:2897–2911. doi: 10.1093/nar/gkaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code for the MethylSight predictor and web server is available on GitHub: https://github.com/fcharih/MethylSight.