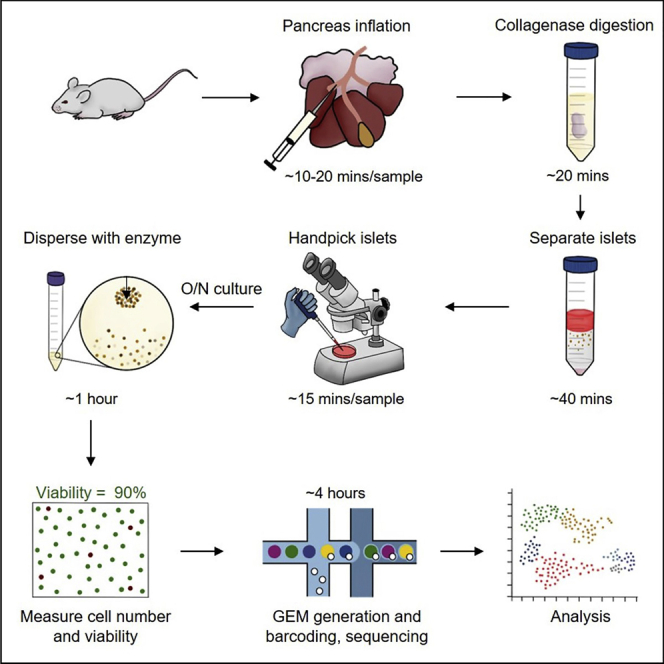
Summary
Pancreatic islets consist of several cell types, including alpha, beta, delta, epsilon, and PP cells. Due to cellular heterogeneity, it is challenging to interpret whole-islet transcriptome data. Single-cell transcriptomics offers a powerful method for investigating gene expression at the single-cell level and identifying cellular heterogeneity and subpopulations. Here, we describe a protocol for mouse pancreatic islet isolation, culturing, and dissociation into a single-cell suspension. This protocol yields highly viable cells for successful library preparation and single-cell RNA sequencing.
For complete details on the use and execution of this protocol, please refer to Lee et al. (2020).
Graphical Abstract
Highlights
-
•
A detailed protocol for the isolation and culture of pancreatic islets from mice
-
•
A procedure for dissociation of mouse islets into a single-cell suspension
-
•
Method consistently yields optimal cell viability (90%) for scRNA sequencing
Pancreatic islets consist of several cell types, including alpha, beta, delta, epsilon, and PP cells. Due to cellular heterogeneity, it is challenging to interpret whole-islet transcriptome data. Single-cell transcriptomics offers a powerful method for investigating gene expression at the single-cell level and identifying cellular heterogeneity and subpopulations. Here, we describe a protocol for mouse pancreatic islet isolation, culturing, and dissociation into a single-cell suspension. This protocol yields highly viable cells for successful library preparation and single-cell RNA sequencing.
Before You Begin
Preparation for Pancreas Inflation
Timing: 30 min
-
1.
Set water bath temperature to 37°C.
-
2.
Set centrifuge (capable of spinning 50 mL tubes) temperature to 4°C.
-
3.
Dissolve bovine serum albumin (BSA) (0.02% w/v) in 1× HBSS.
-
4.
Dissolve collagenase (0.5 mg/mL) in Hank's balanced salt solution (HBSS) and BSA solution.
-
5.
Prepare RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS).
-
6.
Prepare clean dissection tools.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 10× HBSS | Fisher Scientific | MT-200-23CV |
| Bovine serum albumin | Sigma-Aldrich | A8806 |
| Collagenase type XI | Sigma-Aldrich | C7657 |
| RPMI 1640 | Fisher Scientific | MT10040CV |
| Histopaque-1077 | Sigma-Aldrich | 10771 |
| Phosphate-buffered saline | Sigma-Aldrich | D8537 |
| Accutase | Innovative Cell Technologies | AT-104 |
| Antibiotic-antimycotic solution | Fisher Scientific | MT30004CI |
| Fetal bovine serum | Fisher Scientific | F0926 |
| Trypan blue | VWR | 97063-702 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3′ Library & Gel Bead Kit v2 | 10X Genomics | PN-120237 |
| Deposited Data | ||
| scRNA-seq data | GEO | GSE144471 |
| Experimental Models: Organisms/Strains | ||
| Mouse: NOD/ShiLtJ (5 weeks) | Jackson Laboratory | 001976 |
| Other | ||
| 5.5 inch surgical scissors | Roboz | RS-6782 |
| 3.5 inch small curved surgical scissors | Roboz | RS-5911 |
| 2 inch curved bulldog clamp | Roboz | RS-7439 |
| 4 inch tissue forceps | Roboz | RS-5153 |
| 30G needle | VWR | 76290-488 |
| 5 mL syringe | BD | 309647 |
| 50 mL conical tubes | Fisher Scientific | 12-565-270 |
| 15 mL conical tubes | Fisher Scientific | 12-565-268 |
| 40 μm cell strainer | Fisher Scientific | 087711 |
| Kimwipes | Kimberly-Clark | 34155 |
| 6 W LED Dual Goose-neck Illuminator | Amscope | LED-6W |
| 20× to 40× stereo microscope with bottom lighting | Amscope | SE306R-P-LED |
| 3.5× to 90× Zoom Trinocular Stereo Microscope with Table Pillar Stand | Amscope | SM-1TSZ-V203 |
| 1,000 μm Spectra Mesh Woven Filters, cut into 2 inch squares | Fisher Scientific | 08-670-183 |
| Water bath | Fisher Scientific | n/a |
| Refrigerated centrifuge | Fisher Scientific | n/a |
| Countess II automated cell counter | Fisher Scientific | AMQAX1000 |
Materials and Equipment
1× HBSS with 0.02% BSA (for approximately four mice)
-
•
50 mL 10× HBSS
-
•
0.1 g BSA
-
•
450 mL ddH2O
0.5 mg/mL collagenase solution (per mice)
-
•
Dissolve 15 mg collagenase in 30 mL 1× HBSS with 0.02% BSA
10% serum-containing RPMI 1640
-
•
50 mL FBS
-
•
450 mL RPMI 1640
Prepare all solutions fresh and keep on ice until ready to use.
Step-By-Step Method Details
Isolation of Pancreatic Islets
Timing: 3–6 h
Pancreatic islets approximately constitute only 1%–2% of the pancreas. This step digests the pancreatic exocrine tissue through a collagenase reaction. The pancreas is inflated with the collagenase solution in order to increase the efficiency of breakdown. The islets are then isolated through a density gradient before being handpicked to include only healthy-looking, intact islets.
-
1.Inflating mouse pancreas
-
a.Sacrifice mouse with CO2 asphyxiation or other approved methods. Confirmation of successful euthanasia can be done through assessment of heartbeat or toe pinch.
-
b.Place the mouse ventral side up and douse with 70% ethanol. Pinch the skin around the urethral opening and lift upwards slightly. Create an incision with the surgical scissors through to the abdominal cavity. Starting from the incision point, cut along the lateral side of the mouse until around the forelimbs. Repeat this on the opposite side.
-
c.Open the abdominal cavity to expose the internal organs.
-
d.Gently shift the intestinal organs to the left side of the mouse.
-
e.Under the dissecting microscope, locate the bile duct and follow it posteriorly until it reaches the small intestine. Clamp the bile duct close to the sphincter of Oddi with the bulldog clamp to prevent the collagenase solution from entering the intestine during pancreas inflation.
-
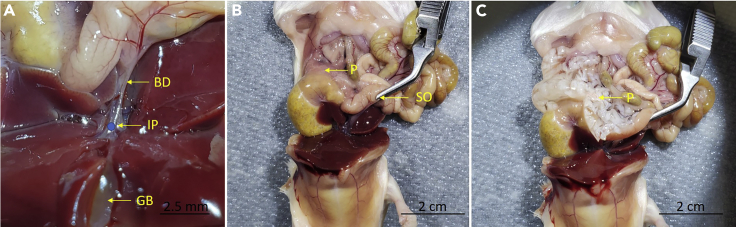
f.Gently lift the liver to locate the gallbladder. Under the dissecting microscope, follow the bile duct until the bifurcation and use the small curved surgical scissors to make a small incision (Figure 1A). The size of the incision should be just enough to accommodate a 30G needle.
-
a.
Figure 1.
Inflation of Pancreas
(A) Close-up image of the bile duct. The incision point for needle insertion is denoted by a blue circle.
(B) The abdominal cavity showing the pancreas before inflation.
(C) The inflated pancreas after collagenase injection through the bile duct. BD, bile duct; GB, gallbladder; IP, incision point; P, pancreas; SO, sphincter of Oddi.
-
g.Fill a 5 mL syringe with collagenase solution and use a 30G needle to inject the collagenase solution into the bile duct under the dissecting microscope. If the inflation is successful, the pancreas will start to expand immediately and turn from opaque (Figure 1B) to translucent (Figure 1C). If the needle was not inserted successfully, another attempt can be made by creating a new incision point further posterior on the intact portion of the duct. Continue to inflate the pancreas until it stops expanding.
-
h.Remove the bulldog clamp. Using the flat end of the curved scissors, carefully separate the attachments between the small intestine and the inflated pancreas by gliding the scissors between the two organs.
-
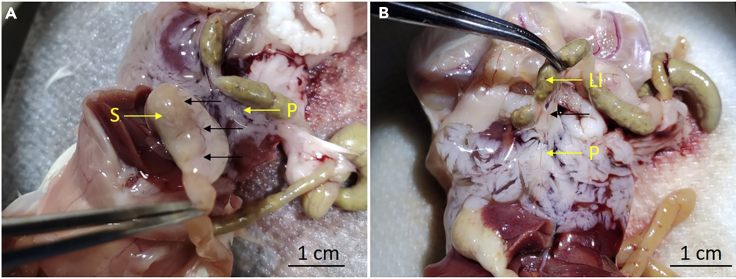
i.Cut the attachments between the inflated pancreas and the stomach (Figure 2A).
-
g.
Figure 2.
Detaching Pancreas from Surrounding Organs
Black arrows indicate the points to detach between the pancreas and (A) stomach or (B) large intestine. LI, large intestine; P, pancreas; S, stomach.
-
j.Cut the attachments between the inflated pancreas and the large intestine (Figure 2B).
-
k.Gently pick up the inflated pancreas and remove the final attachments with the spleen. Move the pancreas to a 50 mL tube filled with 5 mL of the collagenase solution for rinsing.
-
l.Proceed with pancreas inflation with the rest of the samples and place them in 5 mL collagenase before proceeding to the next step.
-
j.
CRITICAL: Take extra care to ensure that the whole circumference of the duct is not cut when making the incision point, as it will become extremely difficult to insert the needle.
CRITICAL: Dissections must be completed within 1 h to prevent excessive digestion in the collagenase solution.
-
2.
Move all of the pancreata into separate 50 mL tubes filled with 20 mL collagenase solution.
-
3.
Transfer the tubes to a 37°C water bath and set a timer for 20 min. When 10 min are remaining, shake the tubes vigorously every 2 min until the timer rings.
CRITICAL: Optimization of incubation time may be necessary as collagenase activity can vary by lot. Examine if islets are under- or over-digested during handpicking.
-
4.
Add 15 mL of RPMI 1640 medium with 10% FBS into each tube.
-
5.
Spin at 200 × g for 2 min at 4°C.
-
6.
Aspirate supernatant with a vacuum until 5 mL of the solution is left, taking care not to disturb the pellet.
-
7.
Add 25 mL of the 0.02% BSA solution in HBSS to wash the pellet and centrifuge again at 200 × g for 2 min. Repeat this wash for a total of three times.
-
8.
After the third wash, resuspend the pellet in 10 mL 0.02% BSA solution in HBSS and gently vortex to resuspend.
-
9.
Filter the suspension through a 1,000 μm Spectra Mesh woven filter. Rinse the tube with another 10 mL of 0.02% BSA solution in HBSS and pour through the 1,000 μm filter.
-
10.
Spin solution at 200 × g for 2 min at 4°C. Remove the supernatant by decanting and use a Kimwipe to wipe off the remaining HBSS on the inside of the tube.
-
11.
Resuspend the pellet in 5 mL Histopaque-1077 and add another 5 mL to the side of the tubes to prevent any islets from attaching to the tube wall.
Alternatives: Although Histopaque-1077 is an optimal component for the density gradient (McCall et al., 2011), lymphocyte separation medium 1.077 g/mL may also be used if Histopaque-1077 is unavailable.
-
12.
Carefully tilt the tubes at a 30° angle and very slowly add 10 mL serum-free RPMI 1640 medium to the side of the tubes to create a Histopaque-1077 and RPMI 1640 medium interface.
-
13.
Change the centrifuge settings to minimal acceleration with no braking and spin the tubes at 1,800 × g for 20 min. The islets will migrate to the interface, while islets with infiltration may be further down the tube.
-
14.
Use a disposable plastic dropper pipet to transfer the islets into a 50 mL tube filled with 20 mL RPMI 1640 medium with 10% FBS, while taking care not to disturb the pellet.
-
15.
Spin at 200 × g for 3 min at 4°C.
-
16.
Aspirate the supernatant and add 20 mL of serum-containing media. Repeat this wash for a total of three times.
-
17.
Resuspend the islets in 5 mL of serum-containing media and transfer the resuspended islets to a 60 mm petri dish. Rinse the tube with an additional 5 mL of serum-containing media and transfer to the petri dish.
-
18.
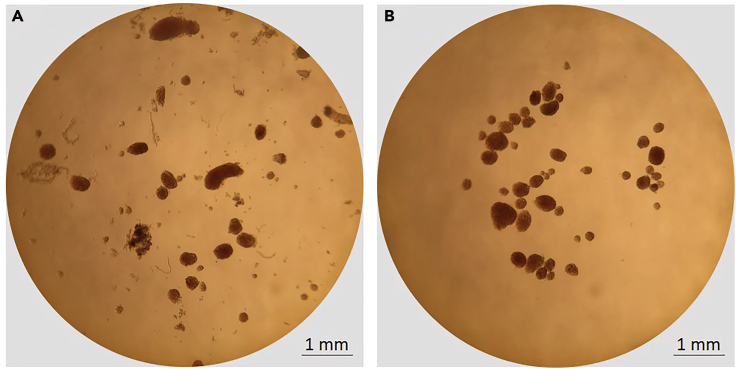
Carefully handpick islets with a 20 uL pipette using a 20× to 40× stereo microscope with bottom lighting (Figure 3) and transfer them to a 15 mL tube with 2 mL of serum-containing media.
Figure 3.
Microscopic Evaluation of Isolated Islets
Representative image of isolated islets of varying size from a 4-week-old female NOD mouse (A) before and (B) after handpicking. Images were taken under a stereo microscope with bottom lighting at 20× magnification.
-
19.
Once all of the islets are collected, add 10 mL of sterile serum-containing media with antibiotics/antimycotics to the tube.
-
20.
Spin at 200 × g for 3 min at 4°C. Repeat this wash for a total of three times.
-
21.
Transfer islets to a sterile 60 mm petri dish and incubate overnight (14–16 h) at 37°C and 5% CO2.
CRITICAL: Ensure that the dish is not tissue culture-treated as the islets have to stay in suspension.
Dissociation of Pancreatic Islets
Timing: 1–2 h
Acquiring single-cell suspensions with high viability from isolated pancreatic islets.
-
22.
Transfer islets and media to 15 mL tubes and centrifuge at 200 × g for 3 min at 22°C–23°C.
-
23.
Aspirate supernatant and wash islets with 10 mL of prewarmed phosphate-buffered saline.
-
24.
Spin at 200 × g for 3 min at 22°C–23°C. Carefully aspirate as much of the supernatant as possible.
-
25.
Add 2 mL of Accutase solution and put the tubes in a 37°C water bath. Set a timer for 30 min. When 15 min have passed, pipet up and down ten times every 5 min until time is up.
-
26.
Stop the reaction by adding 10 mL of prewarmed serum-containing media.
-
27.
Filter the cell solution through a 40 μm cell strainer. Rinse the tube and strainer with additional media and combine with the cell solution.
-
28.
Spin at 200 × g for 3 min at 22°C–23°C. Repeat this wash for a total of three times.
-
29.
Aspirate supernatant until around 100–300 μL of solution is left. Gently shake the tube to resuspend cells.
CRITICAL: Proper optimization of this step is crucial for obtaining a single-cell suspension with high viability. We strongly recommend optimizing this step before continuing forward with library preparation. As the enzymatic activity of Accutase may vary by lot, it may be necessary to optimize incubation time if the cells have low viability or are not in a single-cell suspension. Furthermore, ensure that pipetting force remains consistent throughout.
Library Preparation for Single-Cell Transcriptomics
Determine cell number and viability prior to preparing single-cell RNA-seq libraries.
-
30.
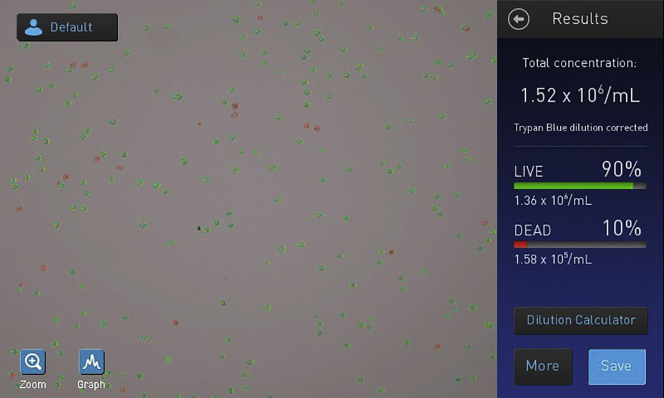
Take 10 μL of the cell solution and mix thoroughly with 10 μL of trypan blue. Load the mixture into a Countess II or other available automated cell counters (Figure 4).
Figure 4.
Representative Image of Cell Viability Assessed by an Automated Cell Counter
Measurement of cell viability following dissociation of pancreatic islets into a single-cell suspension was performed by mixing the cell suspension with trypan blue at a 1:1 ratio before being loaded onto the Countess II automated cell counter. Cells outlined in green are live, while cells outlined in red are dead.
-
31.
Prepare libraries for single-cell RNA-seq according to manufacturer's instructions for a single-cell RNA library preparation kit.
CRITICAL: Using an automated cell counter is preferred over manual counting to minimize the time the samples spend idling on ice.
Note: Following the single-cell RNA-seq, the data can be analyzed by Monocle (Qiu et al., 2017), Seurat (Stuart et al., 2019), or other available platforms.
Expected Outcomes
We routinely obtain approximately 100, 000 - 300, 000 islet cells per mouse using the above protocol with nondiabetic NOD mice. However, this can vary depending on the number and size of the isolated islets, as well as the strain and age of mice. High viability is necessary for successful library preparation in single-cell RNA-seq. In our experiments, we typically obtain approximately 90% cell viability (Figure 4).
Limitations
The isolation process for pancreatic islets is stressful for the cells. Not only is it a long process, but there are also multiple sources of stress throughout the procedure, such as chemical stress through Histopaque-1077 centrifugation and mechanical stress through shaking. At the end of the protocol, cells that were more vulnerable or stressed may not be intact in the single-cell suspension and will not be represented in the single-cell RNA-seq analysis.
Troubleshooting
Problem 1
Islets appear broken or too much exocrine tissue is attached to islets during handpicking (step 18).
Potential Solution
The enzymatic activity of collagenase might vary from lot to lot. It may be necessary to first optimize the protocol by changing the length of the collagenase digestion to see which time will lead to healthy and intact islets without any attached exocrine tissue. If islets appear broken, decrease the incubation time or be wary of shaking too vigorously. If there are exocrine tissues attached to the islets, increase the incubation time.
Problem 2
Low cell viability (step 30).
Potential Solution
It is critical to ensure that all solutions are cold during islet isolation, and that samples are on ice during processing. After the overnight culture, inspect the islets to ensure that they are healthy and are not hypoxic, which is indicated by a dark red spot within the islet. A larger petri dish may be necessary if there are too many islets for the 60 mm dish to avoid hypoxia. It may also be necessary to optimize the Accutase incubation time as its activity varies by lot.
Problem 3
Cells are not in a single-cell suspension (i.e., doublets) (step 30).
Potential Solution
Increase Accutase incubation time or pipet more vigorously. However, be extremely wary of decreasing cell viability during optimization of these steps.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Feyza Engin (fengin@wisc.edu).
Materials Availability
This study did not generate new unique reagents or mouse lines.
Data and Code Availability
The accession number for the single-cell RNA-seq data reported in this paper is GEO: GSE144471.
Acknowledgments
We thank Josie Mitchell Graphics and Visuals for creation of the graphical abstract. H.L. is supported by NIH National Research Service Award T32 GM007215. F.E. is supported by grants from the JDRF-5-CDA-2014-184-A-N and NIH 5K01DK102488-03.
Author Contributions
H.L. optimized the protocol, performed the experiments, and wrote the manuscript. F.E. conceived, supervised and supported the project, and revised the manuscript.
Declaration of Interests
The authors declare no competing interests.
References
- Lee H., Lee Y.S., Harenda Q., Pietrzak S., Oktay H.Z., Schreiber S., Liao Y., Sonthalia S., Ciecko A.E., Chen Y.G. Beta cell dedifferentiation induced by IRE1alpha deletion prevents type 1 diabetes. Cell Metab. 2020;31:822–836.e5. doi: 10.1016/j.cmet.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall M.D., Maciver A.H., Pawlick R., Edgar R., Shapiro A.M. Histopaque provides optimal mouse islet purification kinetics: comparison study with Ficoll, iodixanol and dextran. Islets. 2011;3:144–149. doi: 10.4161/isl.3.4.15729. [DOI] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession number for the single-cell RNA-seq data reported in this paper is GEO: GSE144471.