Abstract
The past decade has witnessed a major increase in our understanding of the genetic underpinnings of childhood cancer. Genomic sequencing studies have highlighted key differences between pediatric and adult cancers. Whereas many adult cancers are characterized by a high number of somatic mutations, pediatric cancers typically have few somatic mutations but a higher prevalence of germline alterations in cancer predisposition genes. Also noteworthy is the remarkable heterogeneity in the types of genetic alterations that likely drive the growth of pediatric cancers, including copy number alterations, gene fusions, enhancer hijacking events, and chromoplexy. Because most studies have genetically profiled pediatric cancers only at diagnosis, the mechanisms underlying tumor progression, therapy resistance, and metastasis remain poorly understood. We discuss evidence that points to a need for more integrative approaches aimed at identifying driver events in pediatric cancers at both diagnosis and relapse. We also provide an overview of key aspects of germline predisposition for cancer in this age group.
Approximately 300,000 children from infancy to age 14 are diagnosed with cancer worldwide every year (1). Some of the cancer types affecting the pediatric population are also seen in adolescents and young adults (AYA), but it has become increasingly clear that cancers in the latter age group have unique biological characteristics that can affect prognosis and therapy (2). Pediatric and AYA cancer patients present with a heterogeneous set of diseases that can be broadly subclassified as leukemias, brain tumors, and non-central nervous system (CNS) solid tumors. These subgroups contain numerous distinct clinical entities, many of which are still poorly characterized from a molecular standpoint.
Recent large-scale genomic analyses have increased our understanding of the genetic drivers of pediatric cancer and have helped to identify new clinically relevant subtypes. These studies have also underscored the distinct nature of the genetic alterations in pediatric and AYA cancers versus adult cancers. Of particular note, the number of somatic mutations in most pediatric cancers is substantially lower than that in adult cancers (3, 4). Exceptions are tumors in children who carry germline mutations that compromise repair of DNA damage (5). For many pediatric cancers, driver events are conditioned on the developmental stage in which the tumor arises. For example, a mutation occurring in one developmental compartment (e.g., a muscle stem cell) may lead to cancer, whereas the same mutation in another compartment does not (6). Pediatric cancer genomes are also characterized by specific patterns of copy number alterations and structural alterations [chromoplexy (7), chromothripsis (8)] that are prognostic indicators in several cancer subtypes. Gene fusion events have long been recognized as oncogenic drivers in many pediatric cancers; however, advanced sequencing technologies have revealed that the number of fusion partners is greater than previously thought, and that previously undetected gene rearrangements may also function as drivers. Finally, germline mutations in a wide spectrum of genes that predispose to cancer appear to play a greater role in pediatric cancer than previously appreciated (9,10).
The lower mutational burden in pediatric cancer has been postulated to be due to a combination of (i) the embryonal origin of many of these cancers, (ii) dysregulation of developmental pathways, and (iii) a smaller contribution of environmental carcinogens. However, this low mutational burden should not be confused with simplicity in the underlying molecular mechanisms of pediatric cancer. Here, we discuss recent work that is beginning to elucidate the unique and biologically complex underpinning of pediatric cancers in both their somatic and germline genomes. We place particular emphasis on the emerging view that integrative analysis beyond DNA panels may have clinical relevance for both initial diagnosis and evaluation of relapse. The therapeutic implications of pediatric cancer genome analyses have been highlighted in other recent reviews (11–15).
Somatic alterations in pediatric cancers Genome landscape studies
Early large-scale sequencing studies of pediatric cancers identified novel driver genes while also underscoring the overall low mutational burden (16–19). Particularly surprising was the observation that even high-risk, highly aggressive cancers in many cases had no identifiable driver gene or pathway (16). Subsequent whole-genome sequencing (WGS) studies of Wilms tumor (20), T cell acute lymphoblastic leukemia (T-ALL) (21), and acute myeloid leukemia (AML) (22) identified subtype-specific driver events and emphasized the interplay between germline and somatic alterations in the development of pediatric cancer. These studies also revealed several mechanisms specific to pediatric cancer. For example, both adult and pediatric AML are characterized by an overall low mutational burden and a long “tail” of rare mutational events, whereas pediatric AML is uniquely characterized by frequent age-dependent gene fusion events and focal areas of gene deletion. In addition, it was found that the presence of specific co-occurring events—such as FLT3-ITD (internal tandem duplication) with the WT1 mutation, or the presence of a NUP98-NSD1 fusion—were characteristic of poor outcome specifically in pediatric AML (22). WGS of pediatric cancers also enabled the identification of enhancer hijacking events (alterations in the regulatory upstream regions of the gene that alter its expression without changing the coding sequence itself). For example, enhancer hijacking events in PRDM6 have been found in a subset of medulloblastomas (23), and similar events in TERT have been found in neuroblastoma (24).
More recent large-scale pan-cancer sequencing studies have begun to delineate additional mechanisms that appear to be uniquely important in the development of pediatric cancers (25, 26). In a study of 1699 patients drawn from six histotypes [T-ALL, B cell ALL (B-ALL), AML, osteosarcoma, Wilms tumor, and neuroblastoma], Ma et al. (26) identified 142 likely driver genes; only 45% matched those seen in adult cancers. The authors estimated that nearly 50% of these patients harbored at least one potentially targetable genetic event. The importance of tumor re-biopsy at relapse was highlighted by the fact that only one-third of tumors with a potentially targetable genetic event appeared to have retained the target upon serial analysis at later time points. This study also suggested the potential clinical utility of more expansive genomic analysis by WGS. Among neuroblastomas, at least one driver gene was identified in 72% of tumors analyzed by WGS compared to only 26% of samples analyzed by whole-exome sequencing (WES). WGS also demonstrated evidence for chromothripsis (massive rearrangements caused by a single catastrophic event) in 11% of samples. In another study, Gröbner et al. (25) analyzed 547 samples by WGS and 414 samples by WES from a much wider spectrum of pediatric cancers (24 different histotypes). They found an overall low incidence of single-nucleotide variants (SNVs). One exception was high-grade gliomas with biallelic germline mutations in the DNA repair genes MSH6 or PMS2; these rare tumors harbored more than 10 mutations per megabase. This study also underscored the value of germline analysis, as 7% of patients were found to carry a mutation in a cancer predisposition gene. As discussed below, this number is likely influenced by the specific subtypes of pediatric cancer in their cohort, because other reports have found a higher incidence of mutations in cancer predisposition genes (10, 27). The studies by Grobner et al. and Ma et al. also identified specific mutational signatures in pediatric cancer genomes. Our understanding of the clinical relevance of mutational signatures in cancer is still limited, although recent work suggests that certain signatures may predict therapeutic response to DNA-damaging agents (28).
Genomics of relapsed cancer
Relatively few studies have comprehensively evaluated the changes that occur in pediatric cancers in response to therapy and after relapse. Such studies are critical given what we have learned from adult cancers, which show a capacity to evolve rapidly and acquire new driver mutations. The most complete picture to date is for leukemias, where the ease of obtaining samples has accelerated our understanding of the mechanisms of relapse. The analysis of ALL samples obtained at diagnosis and relapse for the same patient has demonstrated the importance of clonal evolution, which can lead to acquisition of new genetic alterations and loss of other genetic alterations that were detected at diagnosis (29). Relapsed ALL is characterized by mutations in the NT5C3 nucleotidase gene, which renders leukemic cells resistant to 6-mercaptopurine, a backbone of leukemia treatment regimens (3, 30). The CREBBP gene, which encodes a transcriptional coactivator, is also frequently mutated in relapsed ALL, and these mutations confer resistance to glucocorticoids (31). Other studies have shown that many patients have multiple subclones at diagnosis but that a single subclone can acquire additional mutations that confer resistance to therapy (32).
For solid tumors, our understanding of the mechanisms leading to relapse is more limited. In many cases, the mechanistic basis for the increased prevalence of certain mutations at relapse is poorly understood. For example, the selective pressure for the emergence of RAS-MAPK pathway mutations in neuroblastoma is not clear, given that drugs targeting this pathway are not typically used to treat this disease (33). WGS analysis of diagnostic versus post-therapy medulloblastomas identified substantial divergence of the dominant clone after therapy, with fewer than 12% of diagnostic events retained at relapse. These studies demonstrate the need for further efforts to understand the genomic underpinning of pediatric cancers at relapse (34). An important question that is just beginning to be explored is the clonal heterogeneity between metastatic lesions, or between primary tumors and relapse tumors. A recent analysis of spatially and temporally distinct tumors identified several distinct patterns of evolutionary trajectory in childhood cancer. How these patterns ultimately influence progression and survival remains to be determined (35).
Gene fusions
Many pediatric cancers are characterized by gene fusion events that result in aberrant activity of the encoded proteins. Precision medicine approaches that were focused on gene fusion events were pioneered in adult and pediatric oncology in the context of Philadelphia chromosome-positive ALL. Leukemic cells in patients with this disease harbor the BCR-ABL1 fusion gene, which causes activation of the ABL1 tyrosine kinase. Treatment of pediatric patients in this molecular subgroup with the ABL1 kinase inhibitor imatinib has markedly improved their prognosis (36). Paired-end RNA sequencing has rapidly accelerated the discovery of new gene fusions in both adult and pediatric cancers (37). These studies have also helped to identify additional novel cancer subclasses, such as Ph-like ALL, which are characterized by distinct gene fusions and targetable kinase-activating genetic alterations (38).
The vast majority of gene fusions identified to date in pediatric cancer are rare and have not yet been functionally validated. In some instances, the fusion events involve genes that are known to be cancer drivers; this raises the intriguing possibility that some pediatric cancers are driven by “private” oncogenic fusions. Such a prospect has daunting implications for the development of precision medicines. Common gene fusion events, on the other hand, are important because they can be pathognomonic for a specific diagnosis (e.g., EWS-FLI1 in Ewing sarcoma, PAX-FOXO1 in rhabdomyosarcoma) and can help to determine optimal therapies. For example, CNS gliomas with the common BRAF V600E point mutation are more likely to respond to specific BRAF inhibitors, whereas gliomas with BRAF fusion genes are more likely to respond to drugs inhibiting the downstream MAPK pathway (39). Certain gene fusion events occur across a spectrum of cancer subtypes. The most striking examples are fusion genes involving NTRK (genes encoding neurotrophin receptors), which, although relatively rare, are seen in a wide variety of solid tumors in both children and adults. Their identification is highly relevant given the availability of an effective, FDA-approved drug (larotrectinib) that inhibits the tyrosine kinase activity of the fusion proteins (40, 41).
Many other novel gene fusion events identified by RNA sequencing (RNA-seq) have unique biologic and diagnostic relevance. Angiocentric gliomas are a rare subset of pediatric low-grade glioma (PLGG) that arise in the cerebral cortex and have histologic features in common with astrocytomas and ependymomas. The MYB-QK1 gene fusion is a common event in angiocentric gliomas (42). Interestingly, MYB-QK1 fusions are oncogenic through three distinct mechanisms: MYB activation by truncation, enhancer translocation driving increased expression of MYB, and hemizygous loss of the tumor suppressor QK1. Further complicating the fusion landscape, recent work indicates that some gene fusion events in childhood cancers arise as part of complex loop-like genomic rearrangements, a process called chromoplexy (43). Patients who have tumors with these complex rearrangements may have a worse prognosis, perhaps indicating a distinct evolutionary trajectory. Underscoring the value of fusion gene detection in pediatric cancer, a recent study indicated that fusion events accounted for 20% of the therapeutically actionable findings in pediatric cancer patients with relapsed or refractory disease (44).
Epigenetic alterations
Many pediatric cancers are characterized at diagnosis by mutations in genes encoding epigenetic regulators of gene expression. Chromatin remodeling is one key mechanism of epigenetic regulation. Several protein complexes are involved in chromatin remodeling; the best-characterized with respect to pediatric cancer is the SWI/SNF complex. The first clue that the SWI/SNF complex is important in pediatric cancers was provided by the identification of mutations in SMARCB1 (encoding a core component of the SWI/SNF complex) in rhabdoid tumors (45, 46). It is now appreciated that inactivation or disruption of this complex through deletions, mutations, or gene fusions involving almost all the components of the complex is important in a variety of pediatric and adult tumors (47), including synovial sarcoma, medulloblastoma, and renal cancers (48).
The first identification of recurrent mutations in a specific regulatory histone was in pediatric glioblastoma multiforme (GBM) (49). Although these tumors are histologically similar to their adult counterparts, they are distinct at the molecular level. Pediatric and young adult GBMs are characterized by frequent alterations in H3.3-ATRX-DAXX (the heterochromatin silencing complex containing histone variant H3.3 and the ATRX/DAXX chaperone). Similarly, diffuse intrapontine gliomas are characterized by frequent alterations in the histone gene H3F3A (50). Among the hematologic malignancies, almost 20% of relapsed ALL specimens contain mutations in CREB-binding protein, which is associated with alterations in histone acetylation (51). Alterations in SETD2, another epigenetic regulator, are also common in relapsed ALL (52). The high frequency of mutations in epigenetic regulators suggests a unique etiologic aspect of pediatric cancers that remains poorly understood.
“…integrating histology with genomic profiling and patient history…will refine risk assessment, optimize the selection of therapeutic strategies, and ultimately improve patient outcome.”
DNA methylation is another epigenetic mechanism that regulates gene expression. DNA methylation profiling studies of large cohorts of pediatric brain tumors including medulloblastoma, supratentorial embryonal CNS tumors, glioma, and atypical teratoid/rhabdoid tumor (AT/RT) have been performed. When used in combination with histology and other molecular assays, DNA methylation profiling provides a powerful method for subclassifying tumors with different genetic underpinnings and clinical outcomes. To date, this approach has primarily been used in the research setting, but it may ultimately provide an additional means of molecular stratification for treatment (53). Recently, it was shown that a DNA methylation-based classification of CNS tumors can indeed be used in a routine diagnostic setting (54).
Integrative studies of WGS and RNA-seq
Few studies to date have systematically integrated WGS and RNA-seq for the purpose of discovering genetic alterations in pediatric cancer. Comprehensive studies of ependymoma (a CNS tumor) illustrate how this information can help to improve diagnosis and prognosis (55). Recent work integrating WGS and RNA-seq has led to the identification of 11 subgroups of ependymoma, including one subgroup characterized by a c11orf95-RELA fusion gene (56). At present, this fusion appears to be relatively specific for supratentorial ependymomas, which suggests that anatomic location may correlate with distinct molecular features. Another recent study of ependymomas identified superenhancers linked to cancer-associated genes including PAX6, SKI, FGFRL1, and FGFR1. Notably, knockdown of 60% of the tested superenhancers in mice had a positive impact on survival (57), providing support for strategies that target non-protein-coding sequences as an avenue for therapeutic intervention. Combined DNA and RNA analysis has also contributed to a more refined genomic-based classification of medulloblastoma (58). This information in turn is being used to identify medulloblastoma patients who are at greatest risk of experiencing the adverse side effects of intensive chemotherapy and radiation (59). Overall, it remains to be defined how best to integrate WES, WGS, and RNA-seq studies, although recently published work suggests that WGS provides important additional clinically relevant information (60).
Data sharing and big data
The advancement of genomic analysis in pediatric cancer will require more data sharing (61). Most pediatric genomic studies to date have been relatively small, making it necessary to pool data to increase analytic power. Fortunately, several important resources for large-scale analysis of pediatric cancer data are now available. These include portals for probing discovery genomic data sets and data repositories that host clinical-grade “real world” sequencing data. A public portal was recently made available by St. Jude Research Hospital and currently provides access to discovery sequencing data from more than 4300 patients across 17 diagnoses (https://pecan.stjude.cloud/). With respect to clinical-grade sequencing, Foundation Medicine has made available a public portal that currently includes 3000 genetic alterations derived from 1215 samples for patients 0 to 18 years of age (https://pediatricdata.foundationmedicine.com/). Similarly, the GENIE effort is a public portal (62) that currently has more than 1400 sequenced tumors from patients <18 years of age and more than 1900 sequenced tumors from patients between ages 18 and 30. A potential advantage of this dataset is the ability to extract clinical metadata linking mutations to vital statistics and other measures of outcome. Another emerging effort is the Gabriella Miller Kids First Data Resource Portal, which includes sequence data from ~8000 DNA and RNA samples from patients with childhood cancer or structural birth defects and their families (https://kidsfirstdrc.org/). An Editorial in this issue provides a more in-depth perspective on data sharing (63).
Targeted therapy and response monitoring
To date, no prospective study has systematically evaluated the clinical utility of sequencing approaches to define new therapies for pediatric cancer. In the United States, the National Cancer Institute MATCH (Molecular Analysis for Therapy Choice) trial, initially designed for adult patients, is an ongoing effort to prospectively evaluate the clinical utility of matching cancer patients to drugs according to genomic analysis of the patients’ tumors. The Pediatric MATCH clinical trial currently has 10 arms evaluating targeted therapies for relapsed solid tumors in children. A key challenge in applying this adult cancer trial approach to pediatric cancers is that the number of patients with a targetable alteration is much smaller than in adults. The rate of match is projected to be 20%, illustrating the need to develop other approaches to pediatric cancer that better reflect the underlying mechanisms of this disease.
Emerging technologies
One of the most rapidly growing areas of research in precision medicine for pediatric cancer is the effort to detect minimal residual disease by next-generation sequencing (NGS)-based methods. For leukemia patients, this information can be derived from blood or bone marrow samples; for patients with solid tumors, it can be derived from analyses of circulating tumor cells or cell-free tumor DNA in cerebral spinal fluid, blood, or urine—so-called “liquid biopsies.” Several reports have demonstrated the feasibility of detecting tumor DNA in liquid biopsies to predict early relapse and/or poor outcome using NGS or droplet digital polymerase chain reaction, including for pediatric cancers (64, 65). These studies are still at an early stage, however. As discussed above, recent studies have shown that the genetic alterations present in the initial surgical specimens may not be present in the drug-resistant cancer cells responsible for disease relapse, which could potentially yield false negative results in targeted liquid biopsy assays. NGS-based liquid biopsy assays will have greater utility once they can be optimized as screening tools for detecting minimal residual disease and for evaluation of tumor progression. These assays will also need to be optimized for smaller volumes, as the blood volumes that can be safely obtained from young children are lower than the typical 10 to 20 ml of blood used for cell-free DNA assays in adults.
Genetic predisposition to childhood cancer
A critical component of the implementation of precision medicine in pediatric oncology is determining whether a patient has a genetic predisposition to cancer. As noted above, prior pan-cancer studies suggested that 7 to 8% of children with hematologic or solid tumors carry germline mutations in cancer predisposition genes (9, 10, 25). However, these analyses were not optimized to detect low-level mosaicism or structural variants (intragenic or whole-gene deletions or duplications) that might lead to loss of the encoded protein. In addition, epigenetic alterations—such as loss of imprinting in the chromosome 11p15 region, or promoter hypermethylation of tumor suppressor genes such as CDKN2A—were not evaluated. Recent genomic data mining studies of more than 10,000 cases of 33 cancer types in adults revealed germline sequence variant and copy number alterations in 8% of cases, including variants in genes that have not been fully evaluated in pediatric cohorts (66). It is therefore likely that the true incidence of cancer predisposition in children is higher than that suggested by previous reports.
Studies of medulloblastoma have illustrated the power of large-scale sequencing approaches in refining the frequency of germline alterations in specific tumor subtypes. In current clinical practice, medulloblastoma patients are classified into four subgroups [WNT, sonic hedgehog (SHH), group 3, and group 4] based on a consensus genomic and clinical stratification (58). Waszak et al. (27) analyzed a retrospective and prospective cohort of 673 and 349 patients, respectively, using WES or WGS to detect mutations and deletions in 110 cancer predisposition genes. Although the overall incidence of germline pathogenic variants in this study was only 5 to 6%, the frequency of germline alterations (notably, those that affect the SUFU, PTCH1, TP53, PALB2, and BRCA2 genes) in patients with the SHH subtype was as high as 20%. In contrast, germline APC mutations were seen almost exclusively in the WNT subgroup, specifcally in tumors without the common CTNNB1 exon 3 somatic mutation that characterizes this subgroup. Fewer than half of the patients with germline alterations had a personal or family history of cancer or evidence for a genetic syndrome that would have prompted germline testing. These studies strongly support the view that integrating histology with genomic profiling and patient history across the spectrum of pediatric cancers will refine risk assessment, optimize the selection of therapeutic strategies, and ultimately improve patient outcome.
Guidelines for germline testing in pediatric cancer
Identifying patients with germline alterations is required to optimize the initial therapy; to determine whether germline alterations are inherited or de novo, so that cascade testing for the family can be performed; and finally, to develop appropriate cancer surveillance protocols (Fig. 1). The recommendation to perform germline testing, and the use of clinically validated genetic tests, varies widely among medical providers. This is due to a combination of factors, including health insurance coverage, level of genetics experience among the clinical team, and the type of pre-test counseling that is provided to the family.
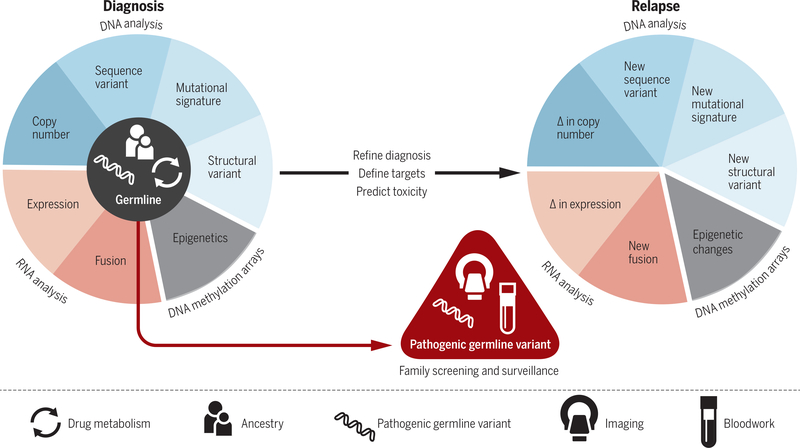
Fig. 1. Model for integrative genomic analysis of pediatric cancers in clinical practice.
Comprehensive cytogenomic and molecular assays are performed at diagnosis to determine genomic or epigenetic alterations in the tumor. Pathogenic germline variants, as well as DNA variants that reflect ancestry and influence drug metabolism (pharmacogenomics), are determined by analysis of normal tissues, typically blood or fibroblasts. Results from these assays are integrated to refine diagnosis and select therapy. Focused gene-specific assays are performed for a subset of patients to determine whether pathogenic germline variants are de novo or inherited from a parent. Carriers should be counseled regarding recurrence risk and enrolled in a surveillance program (imaging, blood work) for early detection of cancer. In the relapse setting, focused assays may be used to determine whether the same genetic events that were detected at diagnosis are present. In addition, comprehensive cytogenomic and molecular assays should be used to identify novel genetic alterations that would dictate the need for alternative therapy.
For the vast majority of pediatric patients, the need for germline testing is initially indicated by the identification of a pathogenic genetic alteration (mutation or deletion) in the tumor that affects a gene known to be associated with germline cancer predisposition. Targeted testing for that specific mutation or deletion of interest is cost-effective and is typically covered by insurance plans. Analysis of paired tumor and normal tissues at diagnosis streamlines the reporting process; in particular, it reduces the number of variants of unknown significance that exponentially increase with large panels and WES or WGS approaches.
If diagnostic molecular genetic studies of the tumor have not been performed, indications for germline testing are typically based on the age and phenotype of the patient (dysmorphic features, multiple café au lait macules, growth or intellectual differences); family history of cancer; the presence of two or more primary tumors in the patient; or the diagnosis of a rare tumor that is highly associated with a cancer predisposition gene. Notable examples of the latter include choroid plexus carcinoma (67) and adrenal cortical carcinoma (68), in which 50% of patients have germline mutations in TP53, and rhabdoid tumors, in which 25 to 35% of patients have mutations or copy number alterations in SMARCB1 (69, 70).
How to screen a child who is at risk for cancer
For the child who is at risk for cancer, options for testing include well-established cytogenetic and molecular assays as well as more recently available whole-genome and transcriptome approaches. A variety of genetic disorders lead to increased risk for hematologic malignancies and solid tumors, including chromosomal disorders [trisomy 21; constitutional 47,XXY karyotype (Klinefelter syndrome)], chromosome breakage disorders (Fanconi anemia and ataxia telangiectasia), overgrowth syndromes (Beckwith-Wiedemann syndrome), and the RASopathies (NF1 and the Noonan spectrum disorders) (71). Single-gene tests that include sequencing and exon-level copy number analysis are often sufficient to confirm a molecular diagnosis [e.g., for neurofibromatosis 1 (NF1) or tuberous sclerosis (TSC1/2)]. However, a child who has clinical signs of NF1 and has a negative test for NF1 alterations (including DNA and RNA sequencing and deletion/duplication analysis) should be tested to rule out a germline mutation in a larger panel of genes that includes GNAS, MLH1, MSH2, MSH6, NF2, PMS2, PTPN11, SOS1, and SPRED1 (72). Patients with suspected Fanconi anemia should be screened with a chromosomal breakage test and comprehensive DNA sequencing using targeted gene panels, as there are at least 20 genes that may increase the risk for hematologic malignancies and solid tumors in this disorder (73).
In contrast to single-gene or multigene panel approaches, comprehensive analysis using WES or WGS is appropriate when the patient presents with tumors affecting multiple tissues or with a rare tumor in which the chance for novel gene discovery is possible. However, these tests increase the number of sequence variants that need to be curated, and they identify numerous variants of unknown significance that are ultimately included in the clinical report. This may result in increased stress for the family, additional cascade testing, and potentially unnecessary long-term interventions. The frequency of reporting such variants will decrease over time with rapidly improving access to publicly available databases of curated genetic variants from normal and affected individuals with genetic disorders and cancer.
Screening for genes that modify risk of cancer development or treatment response
Single-nucleotide polymorphisms or other DNA variants that may modify cancer risk are not typically reported in clinical WES or WGS studies because they are not considered to be actionable with respect to treatment or surveillance. However, because both nuclear and mitochondrial DNA variants are known to affect metabolic state, immune competence, and response to drugs, there is a need for further research on pharmacogenomics predictors in pediatric cancer (74). As the genetic variants that are associated with drug response are, by nature and design, variants present in the normal population, they are typically not included in DNA sequencing panels and are filtered out in WES or WGS bioinformatics pipelines. Targeted pharmacogenomic testing, using single-gene assays or gene panels, has been introduced on a limited basis in the setting of pediatric cancer treatment to help reduce the adverse side effects of chemotherapy. For example, patients with ALL may be screened for variants in genes that encode drug-metabolizing enzymes, such as TPMT and NUD15, as specific genotypes are known to increase the risk of toxic side effects of thiopurine-based cancer treatment (75).
Demographic and laboratory-based research studies of childhood leukemia have provided proof of principle that integrating assays aimed at determining genetic ancestry can have a positive impact on the survival of children with cancer. For example, Yang et al. reported that Native American ancestry is significantly associated with higher risk of relapse in childhood ALL (76). This increased risk of relapse could be mitigated by the addition of an extra phase of chemotherapy. Genetic determination of ancestry by single-nucleotide polymorphism-based array profiling at the time of diagnosis could therefore reduce ethnic disparities in the outcome of the most common form of childhood acute leukemia.
Risk-adapted therapy in the setting of germline predisposition to cancer
Treatment strategies for children with germline alterations in cancer predisposition genes, particularly those involving chromosome instability or DNA repair loci, should, whenever possible, minimize the use of radiation or drugs that increase the risk for second malignancies. For example, the risk for second malignancies (osteosarcoma and pineoblastoma) in retinoblastoma patients with germline RB1 alterations approaches 40% in patients who receive radiation therapy compared with 20% in those who do not (77). Clinical protocols were adapted to eliminate radiation therapy in an attempt to decrease this risk; however, long-term studies have shown that patients who are treated with surgery and chemotherapy alone nonetheless have an increased risk for sarcomas in distal anatomic locations. Pediatric patients with predisposing germline NF1 mutations or deletions and gliomas have a better prognosis than children with sporadic gliomas, and chemotherapy can often be avoided if the tumors are stable by imaging (78). In contrast, for glioma patients with germline mutations in MLH1, MSH2, MSH6, and PMS2, whose tumors typically demonstrate a high tumor mutation burden, aggressive chemotherapy—potentially with the addition of immune checkpoint inhibitors—may be required (79).
Surveillance in children with genetic predisposition to cancer
The ways in which children with a genetic predisposition to cancer are monitored depends on multiple factors, including age at diagnosis, location of the tumor, and the genetic basis for disease. The pediatric working group of the American Association for Cancer Research has developed guidelines for the most common pediatric cancers (80). These strategies often involve serial ultrasounds and magnetic resonance imaging (MRI) but exclude computed tomography (CT) scans in an effort to decrease radiation exposure in patients already at risk for cancer (81). The guidelines for screening of children with an inherited predisposition to Li-Fraumeni syndrome are well established, and cohort studies have demonstrated that early detection improves survival (82). The intervals for screening for other tumor types are not as clear. For example, although the majority of rhabdoid tumors are diagnosed in children under 4 years of age, the median age of onset in children with a germline mutation or deletion in SMARCB1 is 6 months (69, 70). Foulkes et al. (83) have provided guidelines for screening during the first 5 years of life for children at risk for rhabdoid tumor. However, we and others have observed second primary rhabdoid tumors (or sarcomas) in patients more than 8 years after their initial presentation (84), which suggests that extended tumor surveillance is required.
Conclusions
Pediatric cancers have helped to broaden our understanding of tumorigenic mechanisms across all age groups. The introduction of technologies such as massively parallel DNA sequencing and RNA sequencing has vastly increased our ability to analyze individual pediatric cancers and has deepened our understanding of the complexity of these diseases. Precision medicine approaches that encompass risk assessment, surveillance, and diagnostic testing are ushering in a new era of care for children with cancer. As discussed throughout this Review, several challenges remain (see Box 1) before this vision of precision medicine is fully realized. While the field works to address these challenges, an integrative genomics approach that includes evaluation of both germline and somatic alterations at diagnosis and relapse will play a central role in advancing treatment of childhood cancers in the genomics era.
Box 1. Current challenges in precision medicine for pediatric cancer.
Most potential therapeutic targets have not been functionally validated.
The number of genomic targets is much higher than the number of targeted agents.
There is an urgent need for better screening methods for early detection and prevention.
There is limited understanding of the genomics of relapse and metastasis.
There is limited understanding of germline predictors of toxicity and poor response to therapy.
ACKNOWLEDGMENTS
Artwork was conceptualized and designed by S. Pyle.
Funding: E.A.S.-C. was funded by National Cancer Institute grant 5R01CA211657.
Footnotes
Competing interests: The authors declare no competing interests.
REFERENCES AND NOTES
- 1.Steliarova-Foucher E et al. , Lancet Oncol. 18, 719–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tricoli JV et al. , Cancer 122, 1017–1028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence MS et al. , Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B et al. , Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell BB et al. , Cell 171, 1042–1056.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Pappo A, Dyer MA, Oncogene 34, 5207–5215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baca SC et al. , Cell 153, 666–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens PJ et al. , Cell 144, 27–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons DW et al. , JAMA Oncol. 2, 616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J et al. , N. Engl. J. Med. 373, 2336–2346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdach SEG, Westhoff MA, Steinhauser MF, Debatin KM, Mol. Cell Pediatr. 5, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mack SC, Northcott PA, J. Clin. Oncol. 35, 2346–2354 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Mody RJ, Prensner JR, Everett J, Parsons DW, Chinnaiyan AM, Pediatr. Blood Cancer 64, e26288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran TH, Shah AT, Loh ML, Clin. Cancer Res. 23, 5329–5338 (2017). [DOI] [PubMed] [Google Scholar]
- 15.DuBois SG, Corson LB, Stegmaier K, Janeway KA, Science 363, 1175–1181 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Pugh TJ et al. , Nat. Genet. 45, 279–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downing JR et al. , Nat. Genet. 44, 619–622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome Project, Nat. Genet. 44, 251–253 (2012).22286216 [Google Scholar]
- 19.Zhang J et al. , Nature 481, 157–163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadd S et al. , Nat. Genet 49, 1487–1494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y et al. , Nat. Genet. 49, 1211–1218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolouri H et al. , Nat. Med. 24, 103–112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northcott PA et al. , Nature 547, 311–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valentijn LJ et al. , Nat. Genet 47, 1411–1414 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Gröbner SN et al. , Nature 555, 321–327 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Ma X et al. , Nature 555, 371–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waszak SM et al. , Lancet Oncol. 19, 785–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, Setton J, Lee NY, Riaz N, Powell SN, Nat. Commun. 9, 3292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullighan CG et al. , Science 322, 1377–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzoneva G et al. , Nature 553, 511–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullighan CG et al. , Nature 471, 235–239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X et al. , Nat. Commun. 6, 6604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eleveld TF et al. , Nat. Genet. 47, 864–871 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissy AS et al. , Nature 529, 351–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlsson J et al. , Nat. Genet. 50, 944–950 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Schultz KR et al. , J. Clin. Oncol. 27, 5175–5181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertens F, Johansson B, Fioretos T, Mitelman F, Nat. Rev. Cancer 15, 371–381 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Roberts KG et al. , N. Engl. J. Med. 371, 1005–1015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maraka S, Janku F, Discov. Med. 26, 51–60 (2018). [PubMed] [Google Scholar]
- 40.Albert CM, Davis JL, Federman N, Casanova M, Laetsch TW, J. Clin. Oncol. JCO1800573 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Amatu A, Sartore-Bianchi A, Siena S, ESMO Open 1, e000023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandopadhayay P et al. , Nat. Genet. 48, 273–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson ND et al. , Science 361, eaam8419 (2018).30166462 [Google Scholar]
- 44.Mody RJ et al. , JAMA 314, 913–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Versteege I et al. , Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Lee RS et al. , J. Clin. Invest. 122, 2983–2988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biegel JA, Busse TM, Weissman BE, Am. J. Med. Genet. C 166, 350–366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadoch C et al. , Nat. Genet. 45, 592–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartzentruber J et al. , Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Vivanco I et al. , Cancer Discov. 2, 458–471 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malinowska-Ozdowy K et al. , Leukemia 29, 1656–1667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mar BG et al. , Nat. Commun. 5, 3469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar R, Liu APY, Orr BA, Northcott PA, Robinson GW, Cancer 124, 4168–4180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capper D et al. , Nature 555, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavalli FMG et al. , Acta Neuropathol. 136, 227–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker M et al. , Nature 506, 451–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mack SC et al. , Nature 553, 101–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor MD et al. , Acta Neuropathol. 123, 465–472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson GW et al. , Lancet Oncol. 19, 768–784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rusch M et al. , Nat. Commun. 9, 3962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahimzadeh V et al. , JAMA Pediatr 172, 476–481 (2018). [DOI] [PubMed] [Google Scholar]
- 62.AACR Project GENIE Consortium, Cancer Discov. 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaske OM, Haussler D, Science 363, 1125 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Volckmar AL et al. , Genes Chromosomes Cancer 57, 123–139 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Shulman DS et al. , Br. J. Cancer 119, 615–621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang KL et al. , Cell 173, 355–370.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabori U et al. , J. Clin. Oncol. 28, 1995–2001 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Wasserman JD et al. , J. Clin. Oncol. 33, 602–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bourdeaut F et al. , Clin. Cancer Res. 17, 31–38 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Eaton KW, Tooke LS, Wainwright LM, Judkins AR, Biegel JA, Pediatr. Blood Cancer 56, 7–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niemeyer CM, Haematologica 99, 1653–1662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Evans DGR et al. , Clin. Cancer Res. 23, e46–e53 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Walsh MF et al. , Clin. Cancer Res. 23, e23–e31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Relling MV et al. , Clin. Pharmacol. Ther. 102, 897–902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Relling MV et al. , Clin. Pharmacol. Ther 10.1002/cpt.1304 (2018). [DOI] [Google Scholar]
- 76.Yang JJ et al. , Nat. Genet. 43, 237–241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamihara J et al. , Clin. Cancer Res. 23, e98–e106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helfferich J et al. , Crit. Rev. Oncol. Hematol. 104, 30–41 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Bouffet E et al. , J. Clin. Oncol. 34, 2206–2211 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Brodeur GM, Nichols KE, Plon SE, Schiffman JD, Malkin D, Clin. Cancer Res. 23, e1–e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Greer MC, Voss SD, States LJ, Clin. Cancer Res. 23, e6–e13 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Villani A et al. , Lancet Oncol. 17, 1295–1305 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Foulkes WD et al. , Clin. Cancer Res. 23, e62–e67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bhatt MD et al. , Pediatr Blood Cancer 66, e27546 (2019). [DOI] [PubMed] [Google Scholar]