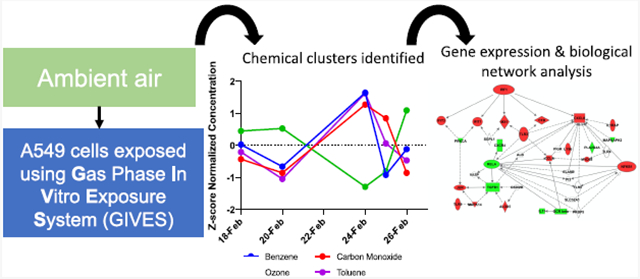
Abstract
Atmospheric pollution represents a complex mixture of air chemicals that continually interact and transform, making it difficult to accurately evaluate associated toxicity responses representative of real-world exposure. This study leveraged data from a previously published article and reevaluated lung cell transcriptional response induced by outdoor atmospheric pollution mixtures using field-based exposure conditions in the industrialized Houston Ship Channel. The tested hypothesis was that individual and co-occurring chemicals in the atmosphere relate to altered expression of critical genes involved in inflammation and cancer-related processes in lung cells. Human lung cells were exposed at an air–liquid interface to ambient air mixtures for 4 h, with experiments replicated across 5 days. Real-time monitoring of primary and secondary gas-phase pollutants, as well as other atmospheric conditions, was simultaneously conducted. Transcriptional analysis of exposed cells identified critical genes showing differential expression associated with both individual and chemical mixtures. The individual pollutant identified with the largest amount of associated transcriptional response was benzene. Tumor necrosis factor (TNF) and interferon regulatory factor 1 (IRFN1) were identified as key upstream transcription factor regulators of the cellular response to benzene. This study is among the first to measure lung cell transcriptional responses in relation to real-world, gas-phase air mixtures.
Graphical Abstract
1. INTRODUCTION
Ambient air pollution exposure is a significant global public health problem, associated with an estimated 4.2 million premature deaths per year.1 Air pollution is a complex mixture that can be broadly categorized into gas-phase pollutants and particulate matter (PM). For both PM and gas-phase pollutants, associations between exposure and a diverse array of adverse health outcomes are generally well established,2,3 including impaired respiratory function,4-6 multiple types of cancers,6-9 cardiovascular disease,10,11 and neurological impairment.12 However, much remains to be elucidated regarding exact toxicological mechanisms underlying these associations, particularly regarding which individual and co-occurring gas-phase chemicals in real-world atmospheric mixtures drive critical molecular-level changes.
A major challenge for studies evaluating toxicity of gas-phase air pollutants is that the atmosphere contains a large number of chemicals that are continuously transforming and interacting under the influence of co-occurring chemicals, PM and surrounding climatic conditions. Consequently, air pollution exposure conditions that are present in the real-world consist of a dynamic mixture spanning both freshly emitted species and secondary phase pollutants. For instance, alkanes can react with OH and other oxidants within the atmosphere, forming products that include diverse hazardous air pollutants (HAPs).13,14 As a specific example, n-butane, a common volatile organic compound produced from petrochemical industry emissions (namely, liquified petroleum gas), once oxidized, can result in butanone, acetaldehyde, butanal, 1-butyl nitrate, and 4-hydroxybutanal, and the subsequent oxidation of these products can result in the formation of formaldehyde and peroxyacetyl nitrate (PAN).15 To more fully understand the toxicity induced by gas-phase air pollutants, it is therefore advantageous to study cellular responses to complex gaseous mixtures that more accurately capture true atmospheric compositions than single-pollutant, lab-based approaches.
Previous in vitro toxicogenomic studies have shown exacerbated cellular response to aged air pollution mixtures, compared to specific freshly emitted species using exposure atmospheres generated in the laboratory-setting.16-20 As an example, our prior work evaluating laboratory-generated air pollutant mixtures present in urban atmospheres found that primary emitted air pollutants altered the expression levels of 19 genes, while photochemically aged mixtures, which include both primary and secondary species, altered the expression of 37-fold more genes (n = 709).16 In taking the next step to corroborate these laboratory findings, the present study presents an approach to measuring gene expression responses of in vitro human lung cells exposed to a real-world ambient mixture of gas-phase pollutants while employing statistical methods common in genomics studies to evaluate the high-dimensional data. The tested hypothesis was that individual and co-occurring chemicals in the atmosphere would relate to altered expression of critical genes involved in inflammation and cancer-related processes in lung cells.
This study draws upon the Benzene and other Toxics Exposure (BEE-TEX) field campaign, which focused on gas-phase pollution in the Houston Ship Channel area, Houston, Texas.21 Houston is located in Harris country, which contains the largest number of petrochemical facilities of all urban regions within the United States.22 In this study we aimed to: (1) expose human lung cells in vitro to ambient, gas-phase air pollution mixtures in the Houston Ship Channel using an in vitro exposure system designed to evaluate gaseous air pollutants, (2) conduct real-time monitoring of 26 freshly emitted and secondary gaseous species in the Ship Channel throughout the cell-exposure window, and (3) quantify transcript expression changes associated with individual and co-occurring air pollutant levels.
2. MATERIALS AND METHODS
2.1. In Vitro Exposure Conditions.
This study utilized data previously published in Vizuete et al. and added two new exposure days which were previously not available.21 Human epithelial lung cells (A549), representative of human alveolar type II cells, were exposed in vitro to ambient atmospheres on five different days: February 18, 20, 24, 25, and 26, 2015.23 Each exposure lasted 4 h, starting at 12:00 pm and ending at 4 pm Central Standard Time (CST). Exposures were conducted at the University of Houston (“UH”) field site, located in the Houston Ship Channel area, at the precise location of 9700 Manchester Street (29°43′00.18″ N, 95°15′21.83″ W).
The Gas Phase In Vitro Exposure System (GIVES), an airtight, 8 L, modular cell-exposure chamber was used to carry out all exposures (Billups- Rothenberg, MIC-101TM, Del Mar, CA). The GIVES has been previously developed and tested by our team.16,18-21,24 It represents an exposure system developed to expose cells to gases within varying atmospheric conditions at an air–liquid interface (ALI) while maintaining cell viability. In this experiment the GIVES was set up in an incubator that housed the 8 L exposure chamber holding two Transwell plates of cells (Costar, Cambridge, MA). Air samples were drawn through a humidification unit to reach and maintain a minimum humidity of 60% prior to entering the GIVES to maintain cell viability. To collect ambient air samples, a Teflon sample line was connected to ambient air outside of the trailer with an inlet that was 3 m above the ground and covered by an opened cap. Inside the instrument, a thermostat-controlled heat source maintained a uniform temperature of 37 °C and a gas cylinder provided 5% CO2. It has been previously shown that there is ~9% deposition of 50 nm and 1 μm diameter particles within the GIVES; however, the extent of particle deposition is so limited as to elicit no biomarker changes, making the GIVES a virtually gas-only sampler.25,26
As negative and positive controls, a separate set of lung cells were exposed to clean air or ozone, respectively, using similar conditions as the aforementioned ambient exposures. For the clean air exposure, the GIVES was connected at the field site to medical grade clean air and a total of three exposures were completed on three separate days, Feb 17, 22, and 27 2015. The ozone exposure was also completed at the field site using an ozone generator (Dynamic NO─NO2─O3 Calibrator Series 101, Thermo Electron Corporation) at ozone concentrations of 400 ppb on 2 days, 21 and 23 February 2015. After the control exposures, the cells were incubated for 16 h, after which supernatants were collected and used for cytotoxicity assessment, as further detailed below.
2.2. Ambient Measurements.
Ambient measurements were collected for the 5 days during which in vitro exposures occurred. Two locations were used to measure a total of 64 pollutants: the UH field site and the Texas Commission on Environmental Quality’s Continuous Ambient Monitoring Station (“TCEQ”) (Figure S1). To ensure adequate data quality, pollutants were included in the analysis if they were detected above detection limits on at least three of five experimental days and if their overall average concentration was above 0.1 ppbV. UH provided a mobile lab directly next to the field site where the GIVES was housed. The UH mobile lab was equipped with a proton-transfer-reaction mass spectrometer (PTRMS) and provided measurements for 17 air pollutants, 14 of which met inclusion criteria aforementioned. The instrument provided 5 min measurements of ozone and 3–6 s measurements for the other 13 species listed in Table 1. UH reported a measurement deviation of benzene, toluene, and C2-benzene of 5.2, 6.6, and 3.6%, respectively, and the measurement of the other species had a similar precision range. In addition to the UH data, we also used measurements provided by the surface network operated by the Texas Commission on Environmental Quality (TCEQ). Specifically, we downloaded data from the TCEQ website for the Continuous Ambient Monitoring Station (CAMS) 403 (29°44′1.00″ N, 95°15′27.00″ W) to obtain hourly averaged concentrations for an additional 47 pollutants, 12 of which met the aforementioned inclusion criteria.27 All air pollutant concentrations are reported in parts per billion by volume (ppbV) other than carbon monoxide, which is reported in parts per million by volume (ppmV).
Table 1.
Descriptive Statistics for the (A) Included Air Pollutants (in ppbV, Unless Noted) and (B) Atmospheric Conditions
| measure | mean | min | max | cluster | monitoring data location |
|---|---|---|---|---|---|
| (A) Air Pollutants | |||||
| methanol | 1.243 | 0.245 | 2.217 | cluster 1 | UH field site |
| acetonitrile | 0.101 | 0.062 | 0.145 | cluster 2 | UH field site |
| acetaldehyde | 0.669 | 0.325 | 0.913 | cluster 1 | UH field site |
| acetone | 1.505 | 0.781 | 2.150 | cluster 2 | UH field site |
| benzene | 0.225 | 0.157 | 0.346 | cluster 3 | UH field site |
| toluene | 0.305 | 0.187 | 0.490 | cluster 3 | UH field site |
| C2 benzenes | 0.288 | 0.207 | 0.350 | cluster 4 | UH field site |
| C3 benzenes | 0.241 | 0.094 | 0.413 | cluster 4 | UH field site |
| styrene | 0.137 | 0.065 | 0.383 | cluster 1 | UH field site |
| methyl ethyl ketone | 0.236 | 0.142 | 0.407 | cluster 2 | UH field site |
| carbon monoxide (ppmV) | 0.240 | 0.200 | 0.300 | cluster 3 | UH field site |
| nitric oxide | 5.004 | 0.040 | 10.100 | cluster 2 | UH field site |
| nitrogen dioxide | 13.212 | 5.140 | 21.200 | cluster 2 | UH field site |
| ozone | 26.000 | 16.400 | 34.200 | cluster 3 | UH field site |
| ethane | 8.836 | 6.856 | 10.786 | cluster 5 | TCEQ |
| ethylene | 0.825 | 0.470 | 1.480 | cluster 1 | TCEQ |
| propane | 4.846 | 3.748 | 6.258 | cluster 5 | TCEQ |
| propylene | 0.365 | 0.140 | 1.026 | cluster 1 | TCEQ |
| isobutane | 1.265 | 0.784 | 2.944 | cluster 1 | TCEQ |
| n-butane | 3.068 | 2.040 | 6.164 | cluster 1 | TCEQ |
| acetylene | 0.496 | 0.434 | 0.574 | cluster 4 | TCEQ |
| 1-butene | 0.169 | 0.074 | 0.492 | cluster 1 | TCEQ |
| isopentane | 1.030 | 0.636 | 2.076 | cluster 1 | TCEQ |
| n-pentane | 0.692 | 0.526 | 1.148 | cluster 1 | TCEQ |
| n-hexane | 0.232 | 0.152 | 0.442 | cluster 1 | TCEQ |
| m/p-xylene | 0.115 | 0.078 | 0.168 | cluster 1 | TCEQ |
| (B) Atmospheric Conditions | ||||
|---|---|---|---|---|
| wind speed (MPH) | 9.11 | 4.70 | 18.02 | TCEQ |
| outdoor temperature (°F) | 52.94 | 39.86 | 68.10 | TCEQ |
| dew point temperature (°F) | 37.63 | 28.40 | 58.22 | TCEQ |
| relative humidity (%) | 58.95 | 30.96 | 75.26 | TCEQ |
| solar radiation (W/m2) | 0.53 | 0.21 | 0.82 | TCEQ |
| ultraviolet radiation (mW/cm2) | 0.02 | 0.01 | 0.04 | TCEQ |
| barometric pressure (millibar) | 1018.680 | 1010.48 | 1026.14 | TCEQ |
| precipitation (millimeters) | 0 | 0 | 0 | TCEQ |
2.3. Statistical Identification of Co-occurring Chemicals.
Clusters of atmospheric chemicals that correlated in concentration across experimental days were identified using a weighted co-expression approach, a method that is classically implemented in the genomics field. Specifically, weighted gene co-expression analysis (WGCNA) was employed which represents a method for describing the correlation patterns among different types of data profiles, historically across gene expression profiles;28 though we have more recently broadened its application across integrated genomic and toxicity profiles.29 The statistical methods involved in WGCNA have been summarized elsewhere.28,30 In brief, this experiment used the WGCNA R package (v1.68) to first calculate Pearson correlation coefficients for all pairwise comparisons between chemical concentrations, averaged daily across each 4 h exposure. The resulting Pearson correlation matrix was transformed into an adjacency matrix resulting in a weighted network describing connection strengths between chemicals. An unsigned network was constructed such that chemicals with both negative and positive correlations were grouped together. Modules (referred to as “clusters”) were then identified as groups of chemicals with concentrations that were highly interconnected in the constructed network and thus correlated across experimental days. To confirm general trends in co-occurrence, the concentrations of individual chemicals were also tested for correlation using the Spearman Rank Correlation test. All statistical analyses were conducted in R (v3.6.1).
2.4. Cell Culture.
Human A549 type II lung epithelial cells, derived from a human lung adenocarcinoma, were used for experimental consistency to ensure viability and facilitate comparison to other relevant studies.16-20,23,24,31-33 Cells were grown and maintained at the Baylor College of Medicine (BCM) according to standard protocol (ATCC, Manassas, VA) using Ham’s F-12K growth media containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin, and plated onto 6-well Millicell plates consisting of 24-mm-diameter collagen-coated membranes with 0.4 μm pores (Trans-CLR; Costar, Cambridge, MA). Four hours prior to exposure, apical media were removed to create direct ALI culture conditions, and cells were supplied media through the basolateral compartment containing phenol red-free F-12K nutrient mixture without FBS, consistent with our previous methods.16,17,24 Immediately post-exposure, plates were taken to the BCM lab where cells were incubated for 16 h. This incubation period was selected to meet study logistical demands while still providing time for toxicological responses to occur, and is under the 24 h maximum allowed period that typically coincides with over confluency conditions, as has been described previously.21 The cells were then scraped and shipped on dry ice at −80 °C in TRIzol Reagent (Invitrogen Life Technologies, Carlsbad, CA), and the supernatants collected and shipped at −80 °C before being stored in −80 °C laboratory freezer at the University of North Carolina at Chapel Hill.
2.5. Cytotoxicity Assessment.
The cytotoxicity resulting from each of the ambient and control exposures was measured using the formazan dye-based Pierce lactate dehydrogenase (LDH) Cytotoxicity Assay Kit (Thermo Fisher Scientific). In this kit, the intensity of formazan dye is directly proportional to amount of LDH that is released from cells that are damaged. Samples were analyzed in triplicate wells according to manufacturer instructions, and the intensity of formazan dye was measured using the Thermo/LabSystems 352 Multiskan MS Microplate Reader. For each plate, one well was treated with Triton X-100 to kill all seeded cells as a positive control. This well was also evaluated in triplicate within the LDH assay and provided LDH quantification for 100% cell death as a reference. The relative cell death in the ambient, clean air, and ozone exposure sample groups were analyzed for statistical significance using a t-test, considered statistically significant at p < 0.05.
2.6. RNA Extraction and Measurement of Gene Expression.
Total RNA was extracted from the exposed cell samples using the RNeasy RNA extraction kit, according to the manufacturer’s protocol (Qiagen, Valencia, CA). RNA (50 ng) extracted from each sample was evaluated using the nCounter Inflammation Panel, comprising 249 genes, and the nCounter PanCancer Pathway Panel, comprising 730 genes (Nanostring Technologies). These panels were chosen as they represent major pathways and categories known to be activated by air pollutant mixtures from our previous studies.16 Exposures were completed with three to four replicates, and the sample with the highest quality RNA was used for the gene expression analysis.
2.7. Statistical Analysis of Gene Expression Profiles in Relation to Air Pollutants.
Nanostring nCounter count data were processed using R software, v3.6.1. The nCounter Inflammation panel and the nCounter PanCancer Pathway Panel were evaluated separately. For analysis of differential gene expression in response to ambient air pollution exposure, each of the five samples (from each of the 5 days) had 26 associated exposure measurements, representative of the 4 h mean concentration (in ppbV) between 12 and 4 pm of the 26 air pollutants included.
Count data were previously normalized within Partek Genomics Suite (Partek Inc., St. Louis, Missouri) in a two-step process as per the manufacturer’s specifications and as detailed previously.21,34 Specifically, positive control normalization was carried out which was followed by housekeeping gene normalization. The panels included 40 and 6 housekeeping genes, for the PanCancer and Inflammation panel, respectively.
Normalized count data were then filtered to exclude universally lowly expressed transcripts, requiring that, for a gene to be included in the statistical analysis, >25% of samples have that gene expressed at signals above the overall median signal intensity of all genes. This minimizes bias of low-counts and follows our previous analyses.35-37 Principal components analysis (PCA), specifically calculation and visualization of principal components via the prcomp function, was used to evaluate data distributions and identify outliers, and no outliers were identified.
The DESeq2 package (v3.9) was then used to scale the count data using median signal intensity and to statistically evaluate the count data for differential expression associated with air pollutant exposure conditions.38,39 To account for sources of sample heterogeneity, such as batch effect or atmospheric conditions, Remove Unwanted Variation (RUV) analysis was carried out using the RUVg function in the RUV R package, which estimates factors of unwanted variation to be used as surrogate variables in the model.40,41 For the PanCancer panel, 40 housekeeping genes were used as control genes in the RUV procedure. For the Inflammation panel, empirical control genes were calculated from nondifferentially expressed genes, which act effectively as endogenous negative controls, as recommended when there are few housekeeping genes.40,41
The DeSeq2 method for identifying differentially expressed genes utilizes a negative binomial generalized linear model. This method calculates shrunken logarithmic-fold changes in expression, which were then divided by their standard error values to produce z-statistics. Resulting z-statistics were compared against standard normal distribution curves to generate Wald test p-values. To account for multiple testing, these p-values were then adjusted using the Benjamini and Hochberg (BH) procedure.42 In this analysis, logarithmic-fold changes are representative of the average change in normalized gene expression for each one ppbV increase in air pollutant evaluated. Differentially expressed mRNAs were defined as those with a BH false discovery rate (FDR) ≤ 0.05.
2.8. Biological Network Analysis.
Network analysis was performed to identify enriched biological pathways among the mRNAs with differential expression if the air pollutant had five or more genes that were significantly differentially expressed. Additionally, network analysis was conducted on the aggregate of differentially expressed genes from cluster chemicals if the cluster contained more than five chemicals.
Utilizing the Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA), networks were constructed based on known protein–protein interactions and other molecular interactions to elucidate biological signaling involved in genomic response to individuals and mixtures of air pollutants. Networks were ranked based on right-tailed Fisher’s exact test p-values, indicating the likelihood of observing a network containing at least the same number of proteins encoded by the air pollutants-associated genes by chance within that network in comparison to random selections of other genes within the genome, as detailed previously.35,43
3. RESULTS
3.1. Exposure Characterization and Identification of Co-occurring Chemicals.
The mean, minimum and maximum concentrations for each pollutant and for atmospheric conditions across the exposure window of all five experimental days were calculated (Table 1).
Both freshly emitted and oxidized pollutants were present. Carbon monoxide had the highest total mean concentration of 0.24 ppmV and acetonitrile had the lowest total mean concentration of 0.101 ppbV of the included chemicals. There was no precipitation over the 5 days, the mean temperature was 53.0 °F, and the mean relative humidity was 59.0%. Daily and hourly measurements are summarized in Table S1. A summary of the original data has also been previously presented for three of the five experimental days.21
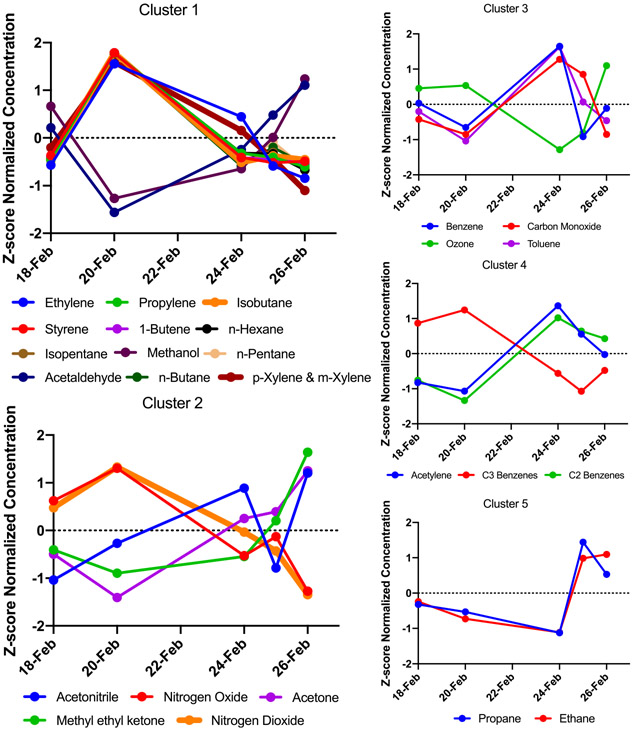
Five distinct clusters of co-occurring chemicals were identified using a weighted network analysis approach, which spanned from containing 2–12 chemicals (Table 1 and Figure 1). For example, cluster 1 contained ethylene, propylene, isobutane, styrene, 1-butene, hexane, isopentane, methanol, n-pentane, acetaldehyde, n-butane, p-xylene, and m-xylene. Within this cluster, there are significant positive correlations between ethylene, styrene, propylene, 1-butene, isopentane, n-hexane, p-xylene, and m-xylene. These positive correlations are plausible as these are all primary species associated with emissions generally known to originate from the oil and gas industry emissions. Their correlation suggests a common source, which is likely the many industrial sources located near the sampling site. Cluster 1 also contains a significantly negative correlation between the aforementioned primary species and acetaldehyde, which is probable given that acetaldehyde is an oxidation product of alkanes and alkenes, such as n-butane. Cluster 2 includes both oxides of nitrogen (NOx) as well as products of photochemical oxidation. NOx sources and photochemical oxidation products have clustered likely because they have many sources and reflect regional influences. The remaining clusters contain a mixture of both primarily emitted species and secondary products like ozone. In nearly all of the clusters the primary emitted species are negatively correlated with secondary products, as expected. These clusters of air mixtures with significant correlations in both directions underscore the complex, dynamic mixture to which the cells were exposed and are suggestive of significant photochemical aging of the ambient mixture.
Figure 1.
Plots of the average concentration of air pollutant in the 4 h exposure windows for each of the exposure days organized by clusters of co-occurring chemicals. Pollutant concentrations are standardized for visualization.
To validate the weighted network analysis, the 26 air pollutants were also evaluated for patterns of correlated concentrations across experimental days utilizing the Spearman rank correlation test (Table S2A,B and Figure S2). Chemicals within clusters from the weighted network analysis were also found in this separate Spearman rank correlation analysis to have significantly correlated concentrations.
3.4. Cytotoxicity Results.
All exposure conditions were evaluated for potential changes in cell death, based on the amount of LDH released in the supernatant in reference to the amount of LDH released upon treatment with cell death-inducing Triton X-100. Ambient air exposure conditions did not induce changes in cytotoxicity: no significant difference in cell death of the ambient air exposures compared to the incubator control was observed (p > 0.05). As a measure of quality assurance/quality control, ozone treatment was found to induce a significant increase in cell death compared to the incubator control (p = 0.005) (Figure S3), as expected based on previous experimentation.21 As an additional measure of quality assurance/quality control, the clean air treatment did not change the percentage of cell death significantly compared to the incubator control (p > 0.05) (Figure S3).
3.5. Gene Expression Modifications Associated with Air Pollutants.
Analysis of cancer- and inflammation-related gene (n = 979) expression in relation to 26 air pollutants identified differential expression of 88 unique genes in relation to 16 unique air pollutants (Table S3). Benzene exposure was associated with the most robust genomic response, with 78 differentially expressed genes. The four most significantly modified genes by any air pollutant were Complement C1r (C1R) (adjusted p value < 0.01), Interleukin 1 Receptor type 1 (IL1R1) (adjusted p value < 0.01), Interleukin 1 Receptor Accessory Protein (IL1RAP) (adjusted p value < 0.01), and Fibronectin 1 (FN1) (adjusted p value < 0.01), all in response to benzene. Additionally, FN1 displayed modification to the highest number of air pollutants (n = 12), followed by BCL2 Associated X, Apoptosis Regulator (BAX), and DNA Polymerase Delta 4, Accessory Subunit (POLD4) (n = 4). The 16 air chemicals that were associated with at least one significantly differentially expressed gene include acetaldehyde, butane, butene, hexane, isobutane, isopentane, pentane, propylene, styrene, xylene, acetone, methyl ethyl ketone, nitrogen dioxide, benzene, toluene, and ethane. Of these 16 air chemicals, 8 are in cluster 1. In sum, cluster 1 chemicals and benzene induce notable changes in gene expression of inflammatory- and cancer-related genes. Perhaps surprisingly, ozone was not found to initiate differential expression of any genes, although this may be due to the relatively low concentrations recorded (all measurements were below the NAAQS standard of 70 ppb).44
3.6. Pathway Enrichment of Genes with Modified Expression.
Based on the a priori defined cut-off of 5 or more genes with differential expression based on exposure, benzene was identified as a candidate for pathway analysis. Genes differentially expressed in response to exposure from chemicals in cluster 1 were also analyzed because cluster 1 contained over 5 air chemicals, the a priori defined cut-off.
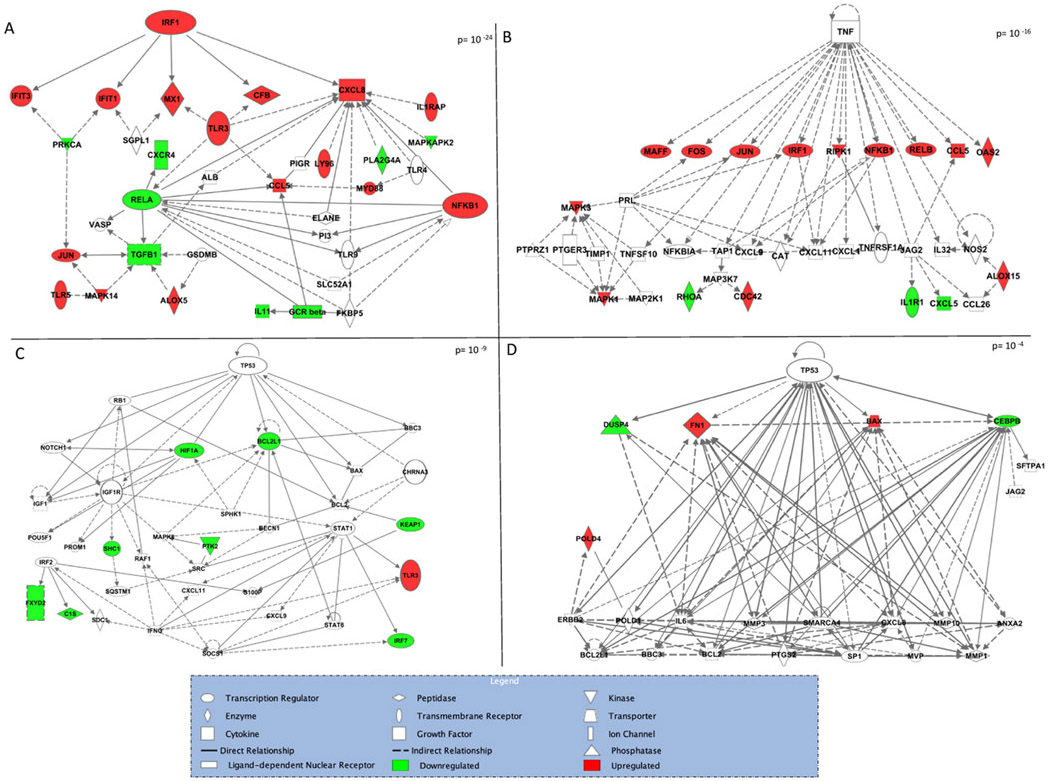
Among genes that were differentially expressed in response to benzene, tumor necrosis factor (TNF) and interferon regulatory factor 1 (IRF1) were identified as top upstream regulators with p-values of p = 1.09 × 10−6 and p = 5.7 × 10−9, respectively. Three significant molecular networks were identified among benzene-responsive genes (Figure 2A-C). The two most significant of these networks contain IRF1 and TNF as central nodes, highlighting the role of these regulators in the genomic response to benzene. The third of these molecular networks contains tumor protein p53 (TP53) as a central node. A number of other genes known for their roles in regulating cancer-processes and inflammation-process are included across these networks including Nuclear Factor Kappa B Subunit 1 (NFKB1) and Transforming Growth Factor β 1 (TGFB1). In genes differentially expressed in response to chemicals in cluster 1, a significant molecular network with TP53 as a central node was identified (Figure 2D).
Figure 2.
Network of genes significantly differentially expressed in response to benzene exposure (A, B, C) and cluster 1 chemicals (D).
4. DISCUSSION
In the present study, we set out to evaluate biological response of lung cells to ambient gaseous air pollutant mixtures using an in vitro exposure system coupled with real-time monitoring of outdoor exposures and mixtures modeling approaches. This study is unique because the composition of ambient air was simultaneously assessed alongside lung cell genomic responses resulting from these direct exposures. There were three major findings. First, we demonstrate that field-based, in vitro studies evaluating real-world atmospheric conditions are feasible and provide critical information toward understanding biological responses associated with air pollution mixtures, especially when combined with statistical methods adapted for high-dimensional data. Second, the most robust genomic response observed within lung cells was identified in association with benzene concentrations and found to be enriched for signaling patterns related to IRF1, TNF, and TP53. Third, similar pathway-level perturbations were identified in relation to a mixture of co-occurring chemicals (referred to as cluster 1 herein).
One of the greatest challenges for understanding the mechanisms driving air pollution toxicity is generating exposure scenarios that accurately reflect real-world atmospheres.21 In reality, humans are exposed simultaneously to complex mixtures of air pollutants. In fact, in the United States there are 187 (HAPs) that the EPA is required to control.45 Despite individuals likely being exposed simultaneously to these HAPs and other air chemicals, the current paradigm is to assess toxicity in laboratory settings one pollutant at a time. Doing so likely underestimates the impact of air pollution as there is growing recognition of the synergistic adverse health impacts of multipollutant exposures.46-48 Laboratory-based studies have shown that mixtures generate an amplified inflammatory response in lung cells.16,20,49 Yet, to date, translating these laboratory-based studies to field-based in vitro studies that more accurately capture real-world mixtures has been limited by logistical and statistical challenges, such as adjusting for background concentrations and the number of pollutants being quantified.18,20 To our knowledge, only a few other studies have exposed lung cell cultures at the ALI to ambient air, including prior work published using the data presented herein.21,50,51 Bisig et al. used a multicellular lung model and a mobile cell-exposure system to evaluate seasonal variation in toxicity of ambient air.50 They found winter air to have an exacerbated effect in terms of expression of three pro inflammatory genes assessed which were also validated through ELISA.50 Gualtieri et al. specifically evaluated the effects of PM using ambient exposure at the ALI interface of BEAS-2B cells.51 Here, we add to these studies and show that, with the deployment of GIVES, field-based in vitro exposure is feasible and can yield highly informative data. As the discipline shifts to a multipollutant-based approach, this study provides an effective design to achieve these goals.
A benefit of employing a multipollutant approach is that evaluation of simultaneous exposures can elucidate the primary drivers of the biological response. Among the 26 chemicals evaluated in the present study, benzene demonstrated the most robust genomic response, with 78 genes identified as responsive. Benzene, which initiated genomic responses enriched for IRF1, TNF, and TP53 signaling pathways, is a recognized HAP with carcinogenic properties and long-term exposure to benzene is associated with the development of cancers, including leukemia, and cardiovascular disease.52-55 Of the 78 benzene-responsive genes, the most significant responses were observed by C1R, IL1R1, IL1RAP, and FN1. Overall, the 78 genes demonstrated enrichment in pathway signaling involving IRF1, TNF, and TP53, which have each been separately associated with cancer or inflammation in the lung.56-62 For example, TNF is a well-recognized mediator of the systemic inflammatory response caused by persistent air pollutant exposure.60,61 Additionally, IRF1 is also known to play key functions in cancer progression, is involved in suppression of immunity to respiratory infections exacerbated by air pollution exposure and there is evidence that it regulates the autophagic response to lung injury.57,62-66 Lastly, TP53 is one of the most recognized oncogenes, playing a key role in tumor suppression, and TP53 mutations are frequently studied with regard to cancer risk.58,59,67 In fact, TP53 has been associated with PM-induced apoptosis in alveolar epithelial cells during the pathogenesis of lung cancer.68 In this study, we show that benzene most potently activates inflammatory- and cancer- related genes associated with adverse health outcomes, even among a photochemically aged mixture, and should continue to be a priority public health concern.
Field-based in vitro studies such as the present study not only better capture true human exposure, but they yield high-dimensional data, ideal for employing new analytic approaches. Through a combination of a field-based design and a bioinformatic analytic approach, co-occurring chemicals within the complex atmospheric mixture were identified. Clusters were classified as groups of chemicals that were highly interconnected in a weighted network analysis. Cluster 1, containing ethylene, propylene, isobutane, styrene, 1-butene, hexane, isopentane, methanol, n-pentane, acetaldehyde, n-butane, p-xylene, and m-xylene, was associated with the largest number of gene-level expression alterations. Similar to benzene exposure, genes responsive to cluster 1 chemicals demonstrated a genomic response that was enriched for FN1 and TP53. Other studies that have also investigated health effects of air pollutant mixtures have utilized varied analytical approaches; however, it is generally agreed that no common analytical approach yet exists for analyzing such high-dimensional air pollution data.46 A multipollutant approach involves estimating the total health effect associated with the exposure to multiple pollutants, which, while more representative of real-world human exposure, poses unique analytical challenges. Traditionally, interaction terms in regression models have been used to asses divergence from additive effects in the case of joint exposures; however, correlation among air pollutions reduces the capacity of regression models to capture the total effect.46 Other approaches involving data reduction techniques have been employed to generate a set of key predictors of outcomes of interest, utilizing principal components analysis, regression shrinkage or penalization methods.46 Approaches developed in the genomics field to handle high-dimensional data may be of particular use to characterizing high-dimension air pollution data.46 The weighted network approach employed is commonly applied in genomics and here we show that its use in air pollution studies can yield important information about mixtures of exposures and biological response to co-occurring chemicals.28 As the EPA and scientific community move toward a multipollutant approach, development of mixtures-based techniques for in vitro studies of air pollutant mixtures will be important to continue to develop alongside the field-laboratory apparatus.69,70
While this study demonstrates the exciting potential of in vitro field-based studies combined with mixtures modeling approaches, it is not without limitations. First, while the A549 cell line is a standard model for studying lung epithelium, it is a tumor cell line that responds differently than a primary cell line, which may affect the toxicogenomic findings presented herein. There is evidence that A549 cells are more vulnerable to inflammatory response to ambient air than a multicellular model.71 However, its common use allows for ease of comparison to other relevant studies.16-20,24,31-33 Second, while this study is more representative of real-world exposures than single-pollutant assessments, it does not capture the important effects that PM can have on air pollution induced cellular changes in real-world exposures and it studied only 4 h exposure windows, which is not necessarily representative of chronic exposure. Future studies could perhaps replicate the experimental setup while analyzing more air pollutants, integrating methods to evaluate PM at the air–liquid interface (as demonstrated by Gualtieri et al.) or exposing cells for a longer time period to more accurately reflect chronic exposure.51 Third, we were unable to conduct further validation of the results at the cytokine or protein level. Finally, given the inherent challenges of extrapolating in vitro findings to the entire human body system, future research could incorporate alternative cell models for a wider biological analysis of how air toxicants affect human health. Some relevant models that should be considered for incorporation include lung-on-a-chip or three-dimensional (3D) tissue models.72
In conclusion, in this study, we directly evaluated lung cell biological response using in vitro methods within an environment that exposed cells to ambient gas-phase air pollutants. This research method created a scenario that was highly reflective of real-world conditions. It was hypothesized that individual and co-occurring chemicals in the atmosphere related to altered expression of critical genes involved in cancer-related processes within lung cells. Clusters of chemicals were identified and correlated with gene expression levels of inflammation- and cancer-related genes. Among several clusters, benzene was a prominent pollutant that was enriched for relevant pathway signaling of three genes (FN1, TNF, and TP53) that have been shown to be associated with lung cancer. These findings contribute to understanding how individual and combinations of gaseous air pollutants alter biologic pathways on the exposure-disease continuum.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank everyone involved with the Benzene and other Toxics Exposure (BEE-TEX) study for their assistance and the communities of Manchester, Galena Park, and the Milby Park neighborhoods for their support. BEE-TEX was funded by the Fish and Wildlife Service of the U.S. Department of the Interior through Harris County, Texas. This research was funded in part by the Institute for Environmental Health Solutions, UNC-Chapel Hill.
Footnotes
The authors declare that they have no conflict of interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.0c02250.
Locations of the field site where the A549 cells were exposed and the CAMS 403 surface monitor (Figure S1); correlations of air pollutants used to further evaluate and confirm co-occurring chemicals in the atmosphere throughout the five exposure days (Figure S2); cytotoxicity assessment (Figure S3) (PDF)
Hourly and daily (12–4 pm LST) average measurements of (A) 26 included chemicals and (B) atmospheric conditions for the 4 days of exposure (Table S1); (A) correlation values of significantly correlated concentrations between ambient air chemicals; (B) p-values of correlation between concentrations of ambient air chemicals (Table S2); and significantly differentially expressed genes in response to ambient air chemicals; logarithmic-fold changes are representative of the average change in normalized gene expression for each one ppbV increase in air pollutant evaluated (Table S3)(XLSX)
The authors declare no competing financial interest.
Contributor Information
Lauren A. Eaves, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
Hang T. Nguyen, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States
Julia E. Rager, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, Curriculum in Toxicology, and The Institute for Environmental Health Solutions, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
Kenneth G. Sexton, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States
Thomas Howard, The Institute for Environmental Health Solutions, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
Lisa Smeester, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health and The Institute for Environmental Health Solutions, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
Anastasia N. Freedman, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States
Kjersti M. Aagaard, Department of Obstetrics & Gynecology, Division of Maternal Fetal Medicine, Baylor College of Medicine, Houston, Texas 77030, United States
Cynthia Shope, Department of Obstetrics & Gynecology, Division of Maternal Fetal Medicine, Baylor College of Medicine, Houston, Texas 77030, United States.
Barry Lefer, Department of Earth and Atmospheric Sciences, University of Houston, Houston, Texas 77004, United States; Tropospheric Composition Program, Earth Science Division, NASA, Washington, District of Columbia 20546, United States.
James H. Flynn, Department of Earth and Atmospheric Sciences, University of Houston, Houston, Texas 77004, United States
Mathew H. Erickson, Department of Earth and Atmospheric Sciences, University of Houston, Houston, Texas 77004, United States
Rebecca C. Fry, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, Curriculum in Toxicology, and The Institute for Environmental Health Solutions, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
William Vizuete, Department of Environmental Sciences and Engineering, Gillings School of Global Public Health and Curriculum in Toxicology, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
REFERENCES
- (1).WHO Ambient (outdoor) Air Pollution: Fact Sheet. https://www.who.int/en/news-room/fact-sheets/detail/ambient-(outdoor)-air-quality-and-health (accessed June 23, 2020).
- (2).Anderson JO; Thundiyil JG; Stolbach A Clearing the Air: A Review of the Effects of Particulate Matter Air Pollution on Human Health. J. Med. Toxicol 2012, 8, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Manisalidis I; Stavropoulou E; Stavropoulos A; Bezirtzoglou E Environmental and Health Impacts of Air Pollution: A Review. Front. Public Health 2020, 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gehring U; Gruzieva O; Agius RM; Beelen R; Custovic A; Cyrys J; Eeftens M; Flexeder C; Fuertes E; Heinrich J; Hoffmann B; Jongste JC; Kerkhof M; Klumper C; KOrek M; Molter A; Shultz ES; Simpson A; Sugiri D; Svarterngren M; von Berg A; Wijga AH; Pershagen G; Brunekreef B Air Pollution Exposure and Lung Function in Children: The ESCAPE Project. Environ. Health Perspect. 2013, 121, 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ierodiakonou D; Zanobetti A; Coull BA; Melly S; Postma DS; Boezen HM; Vonk JM; Williams PV; Shapiro GG; McKone EF; Hallstrand TS; Koenig JQ; Schildcrout JS; Lumley T; Fuhlbrigge AN; Koutrakis P; Schwartz J; Weiss ST; Gold DR Childhood Asthma Management Program Research, G., Ambient air pollution, lung function, and airway responsiveness in asthmatic children. J. Allergy Clin. Immunol 2016, 137, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nafstad P; Haheim LL; Oftedal B; Gram F; Holme I; Hjermann I; Leren P Lung cancer and air pollution: a 27 year follow up of 16 209 Norwegian men. Thorax 2003, 58, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hystad P; Demers PA; Johnson KC; Carpiano RM; Brauer M Long-term residential exposure to air pollution and lung cancer risk. Epidemiology 2013, 24, 762–772. [DOI] [PubMed] [Google Scholar]
- (8).Raaschou-Nielsen O; Ketzel M; Harbo Poulsen A; Sorensen M Traffic-related air pollution and risk for leukaemia of an adult population. Int. J. Cancer 2016, 138, 1111–1117. [DOI] [PubMed] [Google Scholar]
- (9).Parent ME; Goldberg MS; Crouse DL; Ross NA; Chen H; Valois MF; Liautaud A Traffic-related air pollution and prostate cancer risk: a case—control study in Montreal, Canada. Occup. Environ. Med 2013, 70, 511–518. [DOI] [PubMed] [Google Scholar]
- (10).Simkhovich BZ; Kleinman MT; Kloner RA Air Pollution and Cardiovascular Injury: Epidemiology, Toxicology, and Mechanisms. J. Am. Coll. Cardiol 2008, 52, 719–726. [DOI] [PubMed] [Google Scholar]
- (11).Brunekreef B; Holgate ST Air pollution and health. Lancet 2002, 360, 1233–1242. [DOI] [PubMed] [Google Scholar]
- (12).Xu X; Ha SU; Basnet R A Review of epidemiological Research on Adverse Neurological effects of exposure to Ambient Air Pollution. Front. Public Health 2016, 4, No. 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Calvert JG; Derwent RG; Orlando JJ; Tyndall GS; Wallington TJ Mechanisms of Atmospheric Oxidation of the Alkanes; Oxford Univeristy Press: USA, 2008. [Google Scholar]
- (14).Warneck P Chemistry of the Natural Atmosphere; Academic Press, 1999; Vol. 71. [Google Scholar]
- (15).Montero-Montoya R; Lopez-Vargas R; Arellano-Aguilar O Volatile Organic Compounds in Air: Sources, Distribution, Exposure and Associated Illnesses in Children. Ann. Glob Health 2018, 84, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rager JE; Lichtveld K; Ebersviller S; Smeester L; Jaspers I; Sexton KG; Fry RC A toxicogenomic comparison of primary and photochemically altered air pollutant mixtures. Environ. Health Perspect 2011, 119, 1583–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rager JE; Smeester L; Jaspers I; Sexton KG; Fry RC Epigenetic changes induced by air toxics: formaldehyde exposure alters miRNA expression profiles in human lung cells. Environ. Health Perspect 2011, 119, 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Doyle M; Sexton KG; Jeffries HE; Jaspers I Atmospheric photochemical transformations enhance 1,3-butadiene-induced inflammatory responses in human epithelial cells: The role of ozone and other photochemical degradation products. Chem.-Biol Interact 2007, 166, 163–169. [DOI] [PubMed] [Google Scholar]
- (19).Doyle M; Sexton KG; Jeffries HE; Bridge K; Jaspers I Effects of 1–3-butadiene, isoprene, and the photochemical degradation products on human lung cells. Environ. Health Perspect 2004, 112, 1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sexton KG; Jeffries HE; Jang M; Kamens RM; Doyle M; Voicu I; Jaspers I Photochemical products in urban mixtures enhance inflammatory responses in lung cells. Inhalation Toxicol. 2004, 16, 107–114. [DOI] [PubMed] [Google Scholar]
- (21).Vizuete W; Sexton KG; Nguyen H; Smeester L; Aagaard KM; Shope C; Lefer B; Flynn JH; Alvarez S; Erickson MH; Fry RC From the Field to the Laboratory: Air Pollutant-Induced Genomic Effects in Lung Cells. Environ. Health Insights 2015, 9, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Leuchner M; Rappenglu B VOC source–receptor relationships in Houston during TexAQS-II. Atmos. Environ 2010, 44, 4056–4067. [Google Scholar]
- (23).Lieber M; Smith B; Szakal A; Nelson-Rees W; Todaro G A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int. J. Cancer 1976, 17, 62–70. [DOI] [PubMed] [Google Scholar]
- (24).Jaspers I; Flescher E; Chen LC Ozone-induced IL-8 expression and transcription factor binding in respiratory epithelial cells. Am. J. Physiol.: Lung Cell. Mol. Physiol 1997, 272, L504–L511. [DOI] [PubMed] [Google Scholar]
- (25).Zavala J; Higuchi MA In Understanding Air-Liquid Interface Cell Exposure Systems: A Comprehensive Assessment of Various Systems Under Identical Conditions, Society of Toxicology Annual Meeting, San Antonio, Texas, 2018. [Google Scholar]
- (26).Ebersviller S; Lichtveld K; Sexton KG; Zavala J; Lin YH; Jaspers I; Jeffries HE Gaseous VOCs rapidly modify particulate matter and its biological effects—Part 1: Simple VOCs and model PM. Atmos. Chem. Phys 2012, 12, 5065–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).TCEQ CAMS 403 Monitor Site, 2020. https://www.tceq.texas.gov/airquality/airmod/data/site. [Google Scholar]
- (28).Langfelder P; Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rager JE; Auerbach SS; Chappell GA; Martin E; Thompson CM; Fry RC Benchmark Dose Modeling Estimates of the Concentrations of Inorganic Arsenic That Induce Changes to the Neonatal Transcriptome, Proteome, and Epigenome in a Pregnancy Cohort. Chem. Res. Toxicol 2017, 30, 1911–1920. [DOI] [PubMed] [Google Scholar]
- (30).Langfelder P; Horvath S WGCNA: An R Package for Weighted Correlation Network Analysis. https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/ (accessed Oct 20, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ebersviller S; Lichtveld K; Sexton K; Zavala j.; Lin Y; Jaspers I; Jeffries H Gaseous VOCs Rapidly Modify Particulate Matter and Its Biological Effects: Part 2, Complex Urban VOCs and Model PM. Atmos. Chem. Phys 2012, 12, 12293–12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Baldridge KC; Zavala J; Surratt J; Sexton KG; Contreras LM Cellular RNA is chemically modified by exposure to air pollution mixtures. Inhalation Toxicol. 2015, 27, 74–82. [DOI] [PubMed] [Google Scholar]
- (33).Lin YH; Sexton KG; Jaspers I; Li YR; Surratt JD; Vizuete W Application of chemical vapor generation systems to deliver constant gas concentrations for in vitro exposure to volatile organic compounds. Environ. Sci.: Processes Impacts 2014, 16, 2703–2710. [DOI] [PubMed] [Google Scholar]
- (34).Gene Expression Data Analysis Guidelines; Nanostring, 2017. [Google Scholar]
- (35).Rager JE; Bailey KA; Smeester L; Miller SK; Parker JS; Laine JE; Drobna Z; Currier J; Douillet C; Olshan AF; Rubio-Andrade M; Styblo M; Garcia-Vargas G; Fry RC Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen 2014, 55, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Rager JE; Moeller BC; Miller SK; Kracko D; Doyle-Eisele M; Swenberg JA; Fry RC Formaldehyde-associated changes in microRNAs: tissue and temporal specificity in the rat nose, white blood cells, and bone marrow. Toxicol. Sci 2014, 138, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rager JE; Ring CL; Fry RC; Suh M; Proctor DM; Haws LC; Harris MA; Thompson CM High-Throughput Screening Data Interpretation in the Context of In Vivo Transcriptomic Responses to Oral Cr(VI) Exposure. Toxicol. Sci 2017, 158, 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Anders S; Huber W Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Love MI; Huber W; Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Molania R; Gagnon-Bartsch JA; Dobrovic A; Speed TP A new normalization for Nanostring nCounter gene expression data. Nucleic Acids Res. 2019, 47, 6073–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Risso D RUVSeq: Remove Unwanted Variation from RNA-Seq Data. http://bioconductor.org/packages/release/bioc/vignettes/RUVSeq/inst/doc/RUVSeq.pdf (accessed October 31, 2015).
- (42).Benjamini Y; Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc 1995, 57, 289–300. [Google Scholar]
- (43).Fry RC; Navasumrit P; Valiathan C; Svensson JP; Hogan BJ; Luo M; Bhattacharya S; Kandjanapa K; Soontararuks S; Nookabkaew S; Mahidol C; Ruchirawat M; Samson LD Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007, 3, No. e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).EPA NAAQS Table. https://www.epa.gov/criteria-air-pollutants/naaqs-table#3 (accessed June 17, 2020).
- (45).EPA Pollutants and Sources. https://www3.epa.gov/airtoxics/pollsour.html (accessed June 23, 2020).
- (46).Dominici F; Peng RD; Barr CD; Bell ML Protecting human health from air pollution: shifting from a single-pollutant to a multipollutant approach. Epidemiology 2010, 21, 187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Billionnet C; Sherrill D; Annesi-Maesano I; study G Estimating the health effects of exposure to multi-pollutant mixture. Ann. Epidemiol 2012, 22, 126–141. [DOI] [PubMed] [Google Scholar]
- (48).Snowden JM; Reid CE; Tager IB Framing air pollution epidemiology in terms of population interventions, with applications to multipollutant modeling. Epidemiology 2015, 26, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Weitekamp CA; Stevens T; Stewart MJ; Bhave P; Gilmour MI Health effects from freshly emitted versus oxidatively or photochemically aged air pollutants. Sci. Total Environ 2020, 704, No. 135772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Bisig C; Petri-Fink A; Rothen-Rutishauser B A realistic in vitro exposure revealed seasonal differences in (pro-)inflammatory effects from ambient air in Fribourg, Switzerland. Inhalation Toxicol. 2018, 30, 40–48. [DOI] [PubMed] [Google Scholar]
- (51).Gualtieri M; Grollino MG; Consales C; Costabile F; Manigrasso M; Avino P; Aufderheide M; Cordelli E; Liberto LD; Petralia E; Raschella G; Stracquadanio M; Wiedensohler A; Pacchierotti F; Zanini G Is it the time to study air pollution effects under environmental conditions? A case study to support the shift of in vitro toxicology from the bench to the field. Chemosphere 2018, 207, 552–564. [DOI] [PubMed] [Google Scholar]
- (52).McMichael AJ Carcinogenicity of benzene, toluene and xylene: epidemiological and experimental evidence. IARC Sci. Publ 1988, 85, 3–18. [PubMed] [Google Scholar]
- (53).Strum M; Scheffe R National review of ambient air toxics observations. J. Air Waste Manage. Assoc 2016, 66, 120–133. [DOI] [PubMed] [Google Scholar]
- (54).Abplanalp W; DeJarnett N; Riggs DW; Conklin DJ; McCracken JP; Srivastava S; Xie Z; Rai S; Bhatnagar A; O’Toole TE Benzene exposure is associated with cardiovascular disease risk. PLoS One 2017, 12, No. e0183602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Baan R; Grosse Y; Straif K; Secretan B; El Ghissassi F; Bouvard V; Benbrahim-Tallaa L; Guha N; Freeman C; Galichet L; Cogliano V A review of human carcinogens—Part F: chemical agents and related occupations. Lancet Oncol. 2009, 10, 1143–1144. [DOI] [PubMed] [Google Scholar]
- (56).Wajant H The role of TNF in cancer. Results Probl Cell Differ. 2009, 49, 1–15. [DOI] [PubMed] [Google Scholar]
- (57).Shao L; Hou W; Scharping N; Delgoffe GM; Sarkar SN The intrinsic pro-tumorigenic role of IRF1. J. Immunol 2018, 200, No. 178.3. [Google Scholar]
- (58).Olivier M; Hollstein M; Hainaut P TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harbor Perspect. Biol 2010, 2, No. a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Mogi A; Kuwano H TP53 Mutations in Nonsmall Cell Lung Cancer. J. Biomed. Biotechnol 2011, 2011, No. 583929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).van Eeden SF; Tan WC; Suwa T; Mukae H; Terashima T; Fujii T; Qui D; Vincent R; Hogg JC Cytokines involved in the systemic inflammatory response induced by exposure to particulate matter air pollutants (PM(10). Am. J. Respir. Crit. Care Med 2001, 164, 826–830. [DOI] [PubMed] [Google Scholar]
- (61).Tsai D-H; Amyai N; Marques-Vidal P; Wang J-L; Riediker M; Mooser V; Paccaud F; Waeber G; Vollenweider P; Bochud M Effects of particulate matter on inflammatory markers in the general adult population. Part. Fibre Toxicol 2012, 9, No. 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Alsamman K; El-Masry OS Interferon regulatory factor 1 inactivation in human cancer. Biosci. Rep 2018, 38, No. 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Ibironke O; Carranza C; Sarkar S; Torres M; Choi HT; Nwoko J; Black K; Quintana-Belmares R; Osornio-Vargas Á; Ohman-Strickland P; Schwander S Urban Air Pollution Particulates Suppress Human T-Cell Responses to Mycobacterium Tuberculosis. Int. J. Environ. Res. Public Health 2019, 16, 4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Ren Z; Wang Y; Liebenson D; Liggett T; Goswami R; Stefoski D; Balabanov R IRF-1 signaling in central nervous system glial cells regulates inflammatory demyelination. J. Neuroimmunol 2011, 233, 147–159. [DOI] [PubMed] [Google Scholar]
- (65).Zhang L; Cardinal JS; Bahar R; Evankovich J; Huang H; Nace G; Billiar TR; Rosengart MR; Pan P; Tsung A Interferon regulatory factor-1 regulates the autophagic response in LPS-stimulated macrophages through nitric oxide. Mol. Med 2012, 18, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wu D; Pan P; Su X; Zhang L; Qin Q; Tan H; Huang L; Li Y Interferon Regulatory Factor-1 Mediates Alveolar Macrophage Pyroptosis During LPS-Induced Acute Lung Injury in Mice. Shock 2016, 46, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Li Y; Chang SC; Niu R; Liu L; Crabtree-Ide CR; Zhao B; Shi J; Han X; Li J; Su J; Cai L; Yu S; Zhang ZF; Mu L TP53 genetic polymorphisms, interactions with lifestyle factors and lung cancer risk: a case control study in a Chinese population. BMC Cancer 2013, 13, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Soberanes S; Panduri V; Mutlu GM; Ghio A; Bundinger GR; Kamp DW p53 mediates particulate matter-induced alveolar epithelial cell mitochondria-regulated apoptosis. Am. J. Respir. Crit. Care Med 2006, 174, 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).The Multi-Pollutant Report: Technical Concepts and Examples; EPA: Washington, DC, USA, 2008. [Google Scholar]
- (70).Air Quality Management in the United States; National Research Council: Washington, DC, USA, 2004. [Google Scholar]
- (71).Zavala J; O’Brien B; Lichtveld K; Sexton KG; Rusyn I; Jaspers I; Vizuete W Assessment of Biological Responses of EpiAirway 3-D Cell Constructs vs. A549 Cells for Determining Toxicity of Ambient Air Pollution. Inhalation Toxicol. 2016, 28, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Zavala J; Freedman AN; Szilagyi JT; Jaspers I; Wambaugh JF; Higuchi M; Rager JE New Approach Methods to Evaluate Health Risks of Air Pollutants: Critical Design Considerations for In Vitro Exposure Testing. Int. J. Environ. Res. Public Health 2020, 17, 2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.