Abstract
Purpose
To assess the use of deep learning for high-performance image classification of color-coded corneal maps obtained using a Scheimpflug camera.
Methods
We used a domain-specific convolutional neural network (CNN) to implement deep learning. CNN performance was assessed using standard metrics and detailed error analyses, including network activation maps.
Results
The CNN classified four map-selectable display images with average accuracies of 0.983 and 0.958 for the training and test sets, respectively. Network activation maps revealed that the model was heavily influenced by clinically relevant spatial regions.
Conclusions
Deep learning using color-coded Scheimpflug images achieved high diagnostic performance with regard to discriminating keratoconus, subclinical keratoconus, and normal corneal images at levels that may be useful in clinical practice when screening refractive surgery candidates.
Translational Relevance
Deep learning can assist human graders in keratoconus detection in Scheimpflug camera color-coded corneal tomography maps.
Keywords: deep learning, Scheimpflug camera, corneal tomography
Introduction
Keratoconus is an ectatic corneal disorder characterized by progressive corneal stroma thinning with corneal protrusion.1,2 Recent advances in corneal imaging have allowed the discrimination of subclinical keratoconus (forme fruste) characterized by a slight bowing of the posterior corneal surface, which, in the absence of clinical signs or diminished visual acuity, is only detectable by tomography.3–6 A keratoconus diagnosis is important in patients considering refractive surgery because keratoconus weakens the corneal stroma, thus increasing the risk of iatrogenic ectasia.1,7–12 The Oculus Pentacam (Pentacam HR, V.1.15r4 n7; Oculus Optikgeräte GmbH, Wetzlar, Germany) is a noninvasive anterior segment tomographer that uses a rotating Scheimpflug camera to provide direct anterior and posterior elevations and pachymetry measurements of the cornea. The Pentacam HR tomographer, launched in 2005, has five times the image resolution of the basic model.13
Several studies have used machine learning for keratoconus detection,14–24 but most used either topographic numeric indices measured with a Placido disc-based corneal topographer or tomographic numeric indices measured with a scanning slit tomographer and a rotating Scheimpflug camera. Convolutional neural networks (CNNs) perform particularly well in pattern recognition and image classification tasks, making these algorithms a reasonable choice for the automated analysis of color-coded Scheimpflug images. Such analyses could provide new intrinsic insights on keratoconus detection and allow detection of the relative importance of each image feature to aid in the classification of CNNs via a more understandable interface for human observers.25,26 Recent studies used anterior segment optical coherence tomography (AS-OCT) with CNNs for whole-image segmentation and classification to distinguish keratoconic from healthy corneas.27,28 This study aimed to assess the accuracy of a specially designed, domain-specific CNN trained with corneal color-coded maps of whole Scheimpflug images to discriminate among normal, forme fruste, and keratoconic eyes.
Methods
Study Population
This study followed the tenets outlined in the Declaration of Helsinki, in compliance with applicable national and local guidelines. Ethical approval was obtained from the Ethics Committee of Assiut University Hospital. Our institution approved this single-center, retrospective review of high-quality Pentacam four-map selectable display images of non-consecutive refractive surgery candidates, patients with unilateral or bilateral keratoconus, and patients with subclinical keratoconus. All study participants provided written informed consent.
Imaging was performed between July 2014 and March 2019. The Pentacam four-map selectable display provides composite, color-coded images of maps of corneal front elevation, back elevation, thickness, and front sagittal curvature, with numerical and spatial annotations. Two experienced corneal specialists (KA and MMM), each with 8 years of experience, independently classified the anonymized images as keratoconus, subclinical keratoconus, or normal and attached clinical examination summaries. The keratoconus class (K) included those with a clinical diagnosis of keratoconus (e.g., presence of a central protrusion of the cornea with Fleicher ring, Vogt striae, or both, as determined by slit-lamp examination) or an irregular cornea (as determined by distorted keratometry mires or distortion of retinoscopic red reflex, or both). The keratoconus class also included the following topographic findings, as summarized by Piñero and colleagues2: focal steepening located in a zone of protrusion surrounded by concentrically decreasing power zones, focal areas with diopteric (D) values > 47.0 D, inferior–superior (I–S) asymmetry measured as >1.4 D, or angling of the hemimeridians in an asymmetric or broken bowtie pattern with skewing of the steepest radial axis.
The subclinical keratoconus class (S) included subtle corneal topographic changes in the aforementioned keratoconus abnormalities in the absence of slit-lamp or visual acuity changes typical of keratoconus (forme fruste, or asymptomatic, keratoconus). The normal class (N) included refractive surgery candidates and subjects applying for a contact lens fitting with a refractive error of less than 8.0 D sphere, with less than 3.0 D of astigmatism, and without clinical, topographic, or tomographic signs of keratoconus or subclinical keratoconus. After classification, the labeled images were then reviewed by a third party (HA), who identified images with conflicting labels and adjudicated their classes by consensus, withholding the two raters’ first-round group labels during adjudication.
Image Dataset Preprocessing Pipeline
All images were exported in a JPG format of 1024 × 1064 pixels without compression, and each image was labeled according to its relevant class: K, N, or S. The image stack was cropped using ImageJ (National Institutes of Health, Bethesda, MD), keeping only the square composite image showing the four maps without the color scale bars. The images were then scaled to 600 × 600 pixels. The trace of the cross that divided the image into four maps was cleaned by selecting each arm and filling it with a background color in all stacks. A copy of these processed images was cropped separately to obtain each of the four component images (front elevation, back elevation, corneal pachymetry, and front sagittal curvature maps) and resized to obtain four separate image stacks of 256 × 256 pixels. All images were then denoised to remove the image annotation by consecutively using the “Remove Outliers” tool for bright and dark outliers, leaving only the color codes. We used fixed parameters in the “Remove Outliers” tool, with radius = 15 pixels and threshold = 1, applying it first to remove dark outliers and then again to remove bright outliers. Applying this denoising allowed the use of pure color codes without annotations and with the least possible loss of details. At this step, five image categories (a composite of the initial four images plus each of the four component images) were ready to be used by the CNN. Each five-category file contained three subdirectories named after an image group: K, N, or S. Twenty percent of each image category was randomly selected as the test set and withheld from training the CNN. Figure 1 depicts the image preprocessing steps.
Figure 1.
Image data preprocessing pipeline. (A) Exported anonymized image (A1); cropped four-map image resized to 600 × 600 pixels (A2); removal of image overlay (denoising) (A3); and removal of background artifacts between the images (A4). (B) Cropped front elevation map before (B1) and after (B2) processing. (C) Cropped back elevation map before (C1) and after (C2) processing. (D) Cropped corneal pachymetry map before (D1) and after (D2) processing. (E) Cropped front sagittal curvature map before (E1) and after (E2) processing.
CNN Architecture
We designed a relatively small CNN that can easily adapt and learn the features of color-coded input images without the need for using publicly available pretrained networks for transfer learning that often use millions of parameters, requiring a great amount of unique training data to reach their full potential.29–31 The CNN architecture (Fig. 2) consists of two convolutional layers (Conv1 and Conv2), each utilizing rectified linear unit (ReLu) activation.32 The first convolutional layer (Conv1) is the visible layer that is fed image input as patches (patch size = 15); it contains a stack of 16 filters with a kernel size of 3 × 3, followed by a max-pooling layer of window size 2 × 2. The second convolutional layer (Conv2) contains a stack of 32 filters with a kernel size of 3 × 3, followed by a max-pooling layer of window size 2 × 2. A flatten layer is then used to allow feeding of the fully connected layer. This is followed by four similar stacks of fully connected (Dense1, 2, 3, and 4) and dropout layers. Each fully connected layer contains 128 fully connected neurons and utilizes the ReLU activation function, followed by dropout regularization with 20% probability (0.2). The final (classifying) layer of the architecture is a fully connected layer (Dense5) with softmax activation that contains three output neurons, resulting in the probability of classifying each of the image groups. The softmax regression classifier predicts the class with the highest estimated probability.33
Figure 2.

Diagram of the CNN architecture followed in this study.
Training
We trained five identical proposed models on each of the five training/validation datasets for 14 epochs (an epoch is an iteration over the entire input data provided). During training, we used 0.3 validation split. The model sets apart this fraction of the training data, does not train on it, and evaluates the loss and any model metrics on the data at the end of each epoch. Categorical cross-entropy was used as the loss function.31 The optimization was performed using the Adam optimizer with the default parameters suggested by Kingma et al.34 (learning rate = 0.0001, β1 = 0.9, β2 = 0.999, ε = 1e-08).
Models Testing and Data Augmentation
The five models were saved with their weights after training/validation and then reloaded to classify the test set. To prevent model overfitting to the smaller test dataset, we further implemented data augmentation to increase artificially the number of original images used to test the CNN, thereby increasing the robustness of the testing process. Image data augmentation was done carefully to prevent unrealistic image deformation and included minimal rotation, width shift, height shift, and zoom. Neither horizontal nor vertical flip was applied. All generated batches of image data were randomly generated in real time at the start of each mini-batch.
Support Vector Machine
To obtain benchmark performance metrics that allow comparison with our custom network performance, we used a support vector machine (SVM) that was trained using a one-versus-all strategy to classify the eyes into the three groups based on selected corneal topographic parameters: keratometric power in the flattest meridian (Kflat), keratometric power in the steepest meridian (Ksteep), thinnest corneal thickness (TCT), and inferior–superior asymmetry (I–S value).18 The one-versus-all strategy consists of fitting one classifier per class. For each classifier, the class is fitted against all the other classes. Because each class is represented by only one classifier, it is possible to gain knowledge about the class by inspecting its corresponding classifier with better interpretability and computational efficiency.35
Class Activation Maps
A class activation map (CAM) for a particular category indicates the discriminative region used by the CNN to identify the category, thus enabling an accessible analysis of CNN model results.36 We used an approach similar to that in the study by Oquab et al.37; however, instead of a global average pooling layer, we used a global max-pooling before the softmax activation function was applied to the final fully connected layer to produce the desired output by projecting back the weights of the output layer.
Computer Hardware and Software
The deep learning computation was performed on a personal computer with an Intel Core i5-8250U processor (Intel Corp., Mountain View, CA) at 1.60 GHz (base clock speed) and 1.80 GHz (boost clock speed) with an NVIDIA GeForce MX130 graphics card (driver version: 436.15; (NVIDIA, Santa Clara, CA). The deep neural network was implemented using Keras 2.3.1 and TensorFlow 2.0.0 libraries32,38 in Jupyter notebook 6.0.1 installed in an Anaconda environment running on Python 3.6. Throughout our work, we did not implement a graphics processing unit (GPU)-enabled TensorFlow.
Statistical Analysis
Patient data were presented as the mean ± SD. Model performance was assessed by estimating the false-positive rate, false-negative rate, precision, recall, F1 score, accuracy, and area under the curve (AUC),34,39,40 all measured using scikit-learn, version 0.21.3.35 The inter-rater agreement was assessed using Cohen's κ, also computed by scikit-learn. The one-versus-all approach was applied to extend the receiver operating characteristic (ROC) curve used in this three-class problem, in which each class was defined as the positive class and the other two classes were defined jointly as the negative class.
Results
The four maps display images of 3218 eyes of the 1669 non-consecutive refractive surgery candidates, patients with unilateral or bilateral keratoconus, and patients with subclinical keratoconus who were used in this study. Table 1 summarizes the characteristics of the study population. There were statistically significant differences in TCT and I–S values among the three study groups (P < 0.05). There were no statistically significant differences in Kflat or Ksteep between eyes in the N group and eyes in the S group (P > 0.05).
Table 1.
Characteristics of the Study Population and a Comparison of Normal, Subclinical Keratoconus, and Keratoconus Eyes
| Group, Mean ± SD | P | |||||
|---|---|---|---|---|---|---|
| Parameter | Ka | Nb | Sc | N vs. K | N vs. S | K vs. S |
| Age (y) | 31.5 ± 8.20 | 36.50 ± 9.50 | 31.80 ± 8.30 | 0.159 | 0.331 | 0.566 |
| Kflat (D) | 45.64 ± 1.20 | 42.96 ± 8.20 | 43.41 ± 1.80 | 0.001* | 0.101 | 0.026 |
| Ksteep (D) | 49.33 ± 3.30 | 43.89 ± 2.70 | 44.83 ± 4.90 | 0.000* | 0.095 | 0.035 |
| Astigmatism (D) | 3.02 ± 2.13 | 0.92 ± 0.56 | 1.01 ± 0.75 | 0.000* | 0.821 | 0.001 |
| TCT (µm) | 459.88 ± 16.44 | 532.44 ± 27.14 | 513.19 ± 77.45 | 0.000* | 0.042* | 0.000 |
| I–S value (D) | 5.12 ± 1.76 | 0.04 ± 0.20 | 0.82 ± 0.88 | 0.000* | 0.005* | 0.002 |
Bolded numbers indicate statistically significant difference (P < 0.05).
Keratoconus group included 544 subjects and 1038 eyes.
Normal group included 579 subjects and 1108 eyes.
Subclinical keratoconus group included 546 subjects and 1072 eyes.
The original dataset was comprised of 3218 images in each of the five image categories, with each category divided into three classes: K = 1038 (32.3%), N = 1108 (34.4%), and S = 1072 (33.3%) images. The training/validation set was comprised of 2574 randomly selected images from the original dataset in each of its five image category files. Each file contained three subdirectories representing a balanced number of images representing each of the three classes: K = 830 images (32.2%), N = 887 images (34.5%), and S = 857 images (33.3%). The remaining 644 images (20%) were assigned to the test set in each of the five image category files representing images of the three classes in a ratio similar to that in the training/validation set: K = 208 images (32.3%), N = 221 images (34.3%), and S = 215 images (33.4%). This ensured that the dataset was balanced among the three classes and accurately revealed the true performance of the classifiers. The inter-rater agreement was 0.95 with a Cohen's κ score of 0.88.
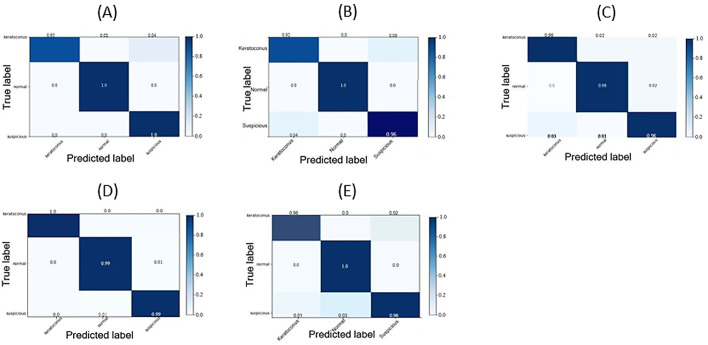
To record performance metrics, the training progress, confusion matrix, and ROC curves were plotted for each of the five map categories over the 10 cycles of training/validation. Each cycle started with different random seeds for the weight initialization (Tables 2, 3; Figs. 34–5).
Table 2.
Confusion Matrix Without Normalization
| Predicted Class | |||||
|---|---|---|---|---|---|
| Image Category | Actual Class | K | N | S | Total |
| Four-map selectable display | K | 789 | 8 | 33 | 830 |
| N | 0 | 887 | 0 | 887 | |
| S | 0 | 0 | 857 | 857 | |
| Front elevation map | K | 764 | 0 | 66 | 830 |
| N | 0 | 887 | 0 | 887 | |
| S | 34 | 0 | 823 | 857 | |
| Back elevation map | K | 796 | 18 | 16 | 830 |
| N | 0 | 869 | 18 | 887 | |
| S | 26 | 8 | 823 | 857 | |
| Pachymetry map | K | 830 | 0 | 0 | 830 |
| N | 0 | 878 | 9 | 887 | |
| S | 0 | 9 | 848 | 857 | |
| Front sagittal curvature map | K | 813 | 0 | 17 | 830 |
| N | 0 | 887 | 0 | 887 | |
| S | 9 | 26 | 822 | 857 | |
The numbers represent the average of 10 training/validation trials by the CNN using each image category and random seeds.
Table 3.
Sensitivity, Specificity, and Accuracy of the CNN Model After Normalization
| False- | False- | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Recall | |||||
| Image Category | Class | Rate | Rate | Precision | (Sensitivity) | F1 Score | Accuracy |
| Four-map selectable display | K | 0 | 0.05 | 1.00 | 0.95 | 0.98 | 0.98 |
| N | 0.01 | 0 | 0.99 | 1.00 | 0.99 | 0.99 | |
| S | 0.04 | 0 | 0.96 | 1.00 | 0.98 | 0.98 | |
| Front elevation map | K | 0.04 | 0.08 | 0.96 | 0.92 | 0.94 | 0.94 |
| N | 0 | 0 | 1.00 | 1.00 | 1.00 | 1.00 | |
| S | 0.08 | 0.04 | 0.92 | 0.96 | 0.94 | 0.94 | |
| Back elevation map | K | 0.03 | 0.04 | 0.97 | 0.96 | 0.96 | 0.97 |
| N | 0.03 | 0.02 | 0.97 | 0.98 | 0.98 | 0.98 | |
| S | 0.04 | 0.04 | 0.96 | 0.96 | 0.96 | 0.96 | |
| Pachymetry map | K | 0 | 0 | 1.00 | 1.00 | 1.00 | 1.00 |
| N | 0.01 | 0.01 | 0.99 | 0.99 | 0.99 | 0.99 | |
| S | 0.01 | 0.01 | 0.99 | 0.99 | 0.99 | 0.99 | |
| Front sagittal curvature map | K | 0.01 | 0.02 | 0.99 | 0.98 | 0.99 | 0.99 |
| N | 0.03 | 0.0 | 0.97 | 1.00 | 0.99 | 0.98 | |
| S | 0.02 | 0.04 | 0.98 | 0.96 | 0.97 | 0.97 |
The results show the average of 10 training/validation trials with random seeds.
Figure 3.
Epoch accuracy and training loss for each image category during training. During the training process, the accuracy increased whereas the loss, representing the error, decreased. Lighter colored curves show original data, and darker lines represent the corresponding smoothed curves.
Figure 4.
Confusion matrices after normalization by class support size (weights) according to the elements in each class. Every column of a confusion matrix represents a predicted class (keratoconus, normal, or subclinical keratoconus), and each row represents the instance of a class. This matrix shows the error rates of predictions by the classification model. Diagonal matrix elements represent the numbers of images for which the predicted label equals the true label, and the remaining elements represent images mislabeled by the classifier. The higher the diagonal values of a confusion matrix, the better the predictions were by the model. (A) Confusion matrix of the four-map display training. (B) Confusion matrix of the front elevation map training. (C) Confusion matrix of the back elevation map training. (D) Confusion matrix of the corneal pachymetry map training. (E) Confusion matrix of the front sagittal curvature map training.
Figure 5.

Comparisons of the ROC curves for the CNN using data from each of the five input image categories during training/validation of the CNN compared with the performance of the SVM classifier using some corneal topographic parameters. The one-versus-all approach was applied to extend the ROC curve used in this three-class problem, in which each class was defined as the positive class and the other two classes were jointly classified as the negative class. Classes: 0, keratoconus; 1, normal; 2, subclinical keratoconus. The plots labeled by lowercase letters are zoomed-in views of the upper left corner of the graph labeled by the corresponding uppercase letter. (A, a) ROC curves for four-map display images; (B, b) front elevation maps; (C, c) back elevation maps; (D, d) corneal pachymetry maps; (E, e) front sagittal curvature maps; (F) Kflat; (G) Ksteep; (H) TCT; and (I) I–S value. The micro-average ROC curve is defined as the precision (true positives [TPs]/TPs + false positives [FPs]) from individual TPs and FPs of each class (Precisionmicro = TP0 + TP1 + TP2 / TP0 + TP1 + TP2 + FP0 + FP1 + FP2). The macro-average ROC curve is defined as the average precision of the three classes (Precisionmacro = precision0 + precision1 + precision2/3).
Precision, recall, accuracy, and F1 score were excellent for all image categories. The N class was least prone to misclassification in all image categories, with only 2% and 1% of normal images misclassified as S class in back elevation and corneal pachymetry maps, respectively. The N class had the largest AUC in all image categories when the ROC curve for the N class was plotted versus the other two classes (one vs. all) showing that the CNN was more “confident” classifying features for the N class than the S and K classes. The CNN showed confusion mainly between K and S classes, especially in front elevation maps. Sensitivity, recall, and accuracy were highest when the pachymetry map image category was used. The five models were saved with their weights after training/validation and then reloaded to classify the test set. As deeper networks have a habit of overfitting during training when not enough training samples are provided, data augmentation was performed to prevent the model from overfitting to the relatively smaller test set data. At this step, the model should have the advantage of starting classification with weights it has learned from previous training while dealing with never before seen image data. By reviewing the plots of the model performance with each image category we applied “early stopping” callback. This callback monitor asks the model to stop training after a minimum of validation loss and validation squared errors and a maximum of validation accuracy have been reached for a certain number (three was used) of consecutive epochs (the “patience” argument).38 The average accuracy outcome after 10 repeated analyses was recorded for the CNN using each image set. The composite images showed the highest accuracy (0.989) after eight epochs, followed by the back elevation map (0.977) after 12 epochs, front sagittal curvature map (0.958) after 10 epochs, and the front elevation map (0.952) after 14 epochs. The corneal pachymetry map showed the lowest accuracy (0.914) after 22 epochs, indicating some degree of overfitting to the training data of this category during the training/validation phase. Among SVMs trained on corneal parameters, the one-versus-all ROC plots showed lower performance using the selected corneal parameters in discriminating eyes in the S group when compared with our custom model performance.
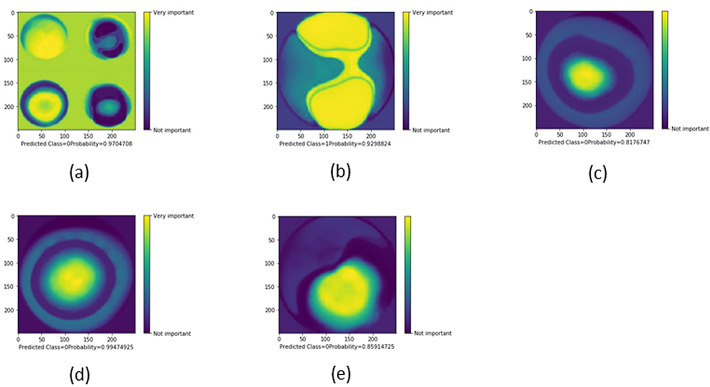
In Figure 6, we present five examples of CAMs obtained after training with each image category. The CNN appears to use the same clinically meaningful spatial regions to classify images. Figure 6A shows that the CNN was correctly influenced by the pixels covering the most important diagnostic areas in each of the four maps, ultimately succeeding in classifying the image as keratoconus with 0.97 probability. However, it skewed by paying more attention to the background pixels than the central pixels in the front and back elevation maps. This is probably because of the proximity of the highly important pixels at the front sagittal curvature map component to the background with consequent spillover so that the CNN regarded both as one unit. In Figure 6B, the CNN was correctly influenced by pixels overlying symmetric areas of steepening in the front elevation map, resulting in an incorrect classification of normal with 0.93 probability. Interestingly, the model showed a high degree of scrutiny when grading pixel importance so it assigned less weight to double arcs of pixels that symmetrically and concentrically encircle the upper and lower steep zones, representing a common color code step that should be spatially present in other image classes and thus gaining less classification importance. Figure 6C shows that the model used the pixels encoding a paracentral cone, classifying the image as keratoconus with 0.82 probability. Figure 6D shows that the model used the most central pixels encoding abnormally thin central cornea to yield a classification of keratoconus with 0.99 probability. This may lead to a false-positive diagnosis of keratoconus in cases with normal thin cornea if the position of the thinnest corneal location is not weighed by the model. In Figure 6E, the CNN was correctly influenced by pixels overlying inferior steepening in the front sagittal curvature map, resulting in an incorrect classification of keratoconus with 0.86 probability. All examples show that the CNN was correctly guided by meaningful spatial regions to classify images.
Figure 6.
Selected examples of CAMs showing the areas of an image that are most important for its CNN classification. Classes: 0, keratoconus; 1, normal; 2, subclinical keratoconus. CAMs of a four-map display image (A), a front elevation map (B), a back elevation map (C), a corneal pachymetry map (D), and a front sagittal curvature map (E).
Discussion
The present study showed that applying deep learning to color-coded corneal maps of Scheimpflug images can accurately and objectively classify eyes into the three studied categories: normal, keratoconus, and subclinical keratoconus. This classification is central when screening refractive surgery candidates to avoid the risk of postoperative ectasia.8–12
Several studies have confirmed the high accuracy of machine learning models for keratoconus screening using indices measured with Placido disk-based corneal topography or a Scheimpflug camera.14–24,26 These studies used topographic and numeric indices that describe the corneal shape for machine learning approaches. In accordance with Arbelaez et al.,18 the data in our study show high diagnostic accuracy of the SVM classifier with some limitation in performance in discriminating eyes with subclinical keratoconus. However, direct comparison with the work of Arbelaez et al.18 is not possible. Compared to these numeric values, the corneal color-coded maps obtained by Scheimpflug cameras add more spatial information to CNNs as the amount of image data is broken down into pixel values and summarized by the reading kernel. This allows picking characteristic image features in more detail, resulting in substantial improvement in classification performance compared to SVM. Kamiya et al.28 used transfer learning by applying the publicly available, pretrained network ResNet-18 to six color-coded AS-OCT maps (anterior elevation, anterior curvature, posterior elevation, posterior curvature, total refractive power, and pachymetry map) to discriminate between normal and keratoconic eyes with an accuracy of 0.99 and to further classify the keratoconus stage with an accuracy of 0.87. They used the arithmetic mean of the CNN output from each of the six AS-OCT component maps to classify the whole image; thus, their study cannot be directly compared with ours. In the present study, we used a simpler CNN with a less computationally intensive architecture of 13 layers which yielded the highest accuracy with initially different random seeds at the beginning of each training session. We also used different and larger datasets sourced from a more commonly used machine13 for early keratoconus detection. Also, we used different classifications, especially with the use of the whole four-map image for classification in addition to the component maps. We proved that when the whole four-map display was used, the CNN yielded accuracy values of 0.98, 0.99, and 0.98 for the K, N, and S classes, respectively, during training/validation; these values remained high (0.989) for the test set, indicating the added value of the four-map composite image compared with results yielded by each solitary map.
Our ROC analysis results suggest a lower value of anterior elevation maps for keratoconus detection. This result is consistent with that reported by Ishii et al.,41 who suggested a greater diagnostic value of the posterior elevation measurement, thus supporting the adoption of CNNs as a classification tool in clinical practice.33,37 This offers much-needed confidence in the procedure, permitting physicians to authenticate predictions made by the network and ensuring that predictions are not influenced by extraneous factors. Qualitative evaluation of model behavior via CAMs has provided insight into the most influential image pixels of the model to be used to guide classification decisions. These maps provided compelling evidence that the CNN classification is most easily influenced by clinically relevant spatial regions; however, they may generalize beyond the training area. These findings are consistent with those reported by Dunnmon et al.42 in their assessment of CNNs for automated classification of chest radiographs; they also noted that, although clinically meaningful spatial regions influence CNN classification, these models occasionally generalize beyond their triage task area meant for classification, resulting in false-positive or false-negative errors.
Our study has several limitations. The dataset used for both training/validation and testing was sourced from the same institution. Thus, generalizing our findings to other institutions should be considered with caution because of differences in image quality, data preprocessing, image labeling, sample weights, or other confounding factors that could lead to a higher error rate. Another limitation is ambiguity in defining subclinical keratoconus, forme fruste, and borderline cases, which should represent a corneal tomography spectrum including all patients who are at high postoperative risk for worsening ectasia.43 The balanced class distribution in this study is far from representing real-life prevalences and was selected to prevent model bias during training/validation and to facilitate portraying the class performance by all available metrics. We also noted a trend toward overfitting of the CNN to training data, but we did not assess the number of images needed to prevent this during training. Instead of using tens of thousands of images to prevent overfitting, we employed the well-known technique of image augmentation to assess the trained network on a small dataset with random perturbations. This provided satisfactory model performance while avoiding overfitting. However, our simple but powerful domain-specific CNN architecture ensures flexibility regarding the downsampling of high-resolution input images while optimizing the computational cost to be comparable to that of general-purpose central processing units. This would be challenging for standard transfer learning via pre-trained CNNs that usually require high-performance GPUs.
Conclusions
Our domain-specific CNN trained used prospectively labeled color-coded corneal images captured with a Scheimpflug camera to classify keratoconus, subclinical keratoconus, and normal corneal images at levels that may be useful in clinical practice. This was achieved not only with four-map selectable display images but also with the individual component images. Such a network does not require a great amount of computational resources and could be valuable in keratoconus detection and screening refractive surgery candidates. This approach could offer timely services in remote regions and reduce human error, and it is free from the bias and fatigue experienced by clinicians. We recommend making optional Scheimpflug images without numerical overlays available in the future to aid further research on this topic.
Acknowledgments
The authors thank Editage (www.editage.com) for the English language editing.
Disclosure: H. Abdelmotaal, None; M.M. Mostafa, None; A.N.R. Mostafa, None; A.A. Mohamed, None; K. Abdelazeem, None
References
- 1. Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998; 42(4): 297–319. [DOI] [PubMed] [Google Scholar]
- 2. Piñero DP, Nieto JC, Lopez-Miguel A. Characterization of corneal structure in keratoconus. J Cataract Refract Surg. 2012; 38(12): 2167–2183. [DOI] [PubMed] [Google Scholar]
- 3. Belin MW, Villavicencio OF, Ambrósio RR Jr. Tomographic parameters for the detection of keratoconus: suggestions for screening and treatment parameters. Eye Contact Lens. 2014; 40(6): 326–330. [DOI] [PubMed] [Google Scholar]
- 4. Rao SN, Raviv T, Majmudar PA, Epstein RJ. Role of Orbscan II in screening keratoconus suspects before refractive corneal surgery. Ophthalmology. 2002; 109(9): 1642–1646. [DOI] [PubMed] [Google Scholar]
- 5. Schlegel Z, Hoang-Xuan T, Gatinel D. Comparison of and correlation between anterior and posterior corneal elevation maps in normal eyes and keratoconus-suspect eyes. J Cataract Refract Surg. 2008; 34(5): 789–795. [DOI] [PubMed] [Google Scholar]
- 6. Mahon L, Kent D. Can true monocular keratoconus occur? Clin Exp Optom. 2004; 87(2): 126. [DOI] [PubMed] [Google Scholar]
- 7. Krachmer JH, Feder RS, Belin MW. Keratoconus and related noninflammatory corneal thinning disorders. Surv Ophthalmol. 1984; 28(4): 293–322. [DOI] [PubMed] [Google Scholar]
- 8. Ambrosio R Jr, Wilson SE. Complications of laser in situ keratomileusis: etiology, prevention, and treatment. J Refract Surg. 2001; 17(3): 350–379. [DOI] [PubMed] [Google Scholar]
- 9. Binder PS. Analysis of ectasia after laser in situ keratomileusis: risk factors. J Cataract Refract Surg. 2007; 33(9): 1530–1538. [DOI] [PubMed] [Google Scholar]
- 10. Binder PS. Risk factors for ectasia after LASIK. J Cataract Refract Surg. 2008; 34(12): 2010–2011. [DOI] [PubMed] [Google Scholar]
- 11. Randleman JB, Trattler WB, Stulting RD. Validation of the Ectasia Risk Score System for preoperative laser in situ keratomileusis screening. Am J Ophthalmol. 2008; 145(5): 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Randleman JB, Woodward M, Lynn MJ, Stulting RD. Risk assessment for ectasia after corneal refractive surgery. Ophthalmology. 2008; 115(1): 37–50. [DOI] [PubMed] [Google Scholar]
- 13. McAlinden C, Khadka J, Pesudovs K. A comprehensive evaluation of the precision (repeatability and reproducibility) of the Oculus Pentacam HR. Invest Ophthalmol Vis Sci. 2011; 52(10): 7731–7737. [DOI] [PubMed] [Google Scholar]
- 14. Maeda N, Klyce SD, Smolek MK. Neural network classification of corneal topography. Preliminary demonstration. Invest Ophthalmol Vis Sci. 1995; 36(7): 1327–1335. [PubMed] [Google Scholar]
- 15. Smolek MK, Klyce SD. Current keratoconus detection methods compared with a neural network approach. Invest Ophthalmol Vis Sci. 1997; 38(11): 2290–2299. [PubMed] [Google Scholar]
- 16. Accardo PA, Pensiero S. Neural network-based system for early keratoconus detection from corneal topography. J Biomed Inf. 2002; 35(3): 151–159. [DOI] [PubMed] [Google Scholar]
- 17. Souza MB, Medeiros FW, Souza DB, Garcia R, Alves MR. Evaluation of machine learning classifiers in keratoconus detection from Orbscan II examinations. Clinics (Sao Paulo). 2010; 65(12): 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arbelaez MC, Versaci F, Vestri G, Barboni P, Savini G. Use of a support vector machine for keratoconus and subclinical keratoconus detection by topographic and tomographic data. Ophthalmology. 2012; 119(11): 2231–2238. [DOI] [PubMed] [Google Scholar]
- 19. Smadja D, Touboul D, Cohen A, et al.. Detection of subclinical keratoconus using an automated decision tree classification. Am J Ophthalmol. 2013; 156(2): 237–246. [DOI] [PubMed] [Google Scholar]
- 20. Kovács I, Miháltz K, Kránitz K, et al.. Accuracy of machine learning classifiers using bilateral data from a Scheimpflug camera for identifying eyes with preclinical signs of keratoconus. J Cataract Refract Surg. 2016; 42(2): 275–283. [DOI] [PubMed] [Google Scholar]
- 21. Ruiz Hidalgo I, Rodriguez P, Rozema JJ, et al.. Evaluation of a machine-learning classifier for keratoconus detection based on Scheimpflug tomography. Cornea. 2016; 35(6): 827–832. [DOI] [PubMed] [Google Scholar]
- 22. Ruiz Hidalgo I, Rozema JJ, Saad A, et al.. Validation of an objective keratoconus detection system implemented in a Scheimpflug tomographer and comparison with other methods. Cornea. 2017; 36(6): 689–695. [DOI] [PubMed] [Google Scholar]
- 23. Yousefi S, Yousefi E, Takahashi H, et al.. Keratoconus severity identification using unsupervised machine learning. PLoS One. 2018; 13(11): e0205998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Issarti I, Consejo A, Jiménez-García M, Hershko S, Koppen C, Rozema JJ. Computer aided diagnosis for suspect keratoconus detection. Comput Biol Med. 2019; 109: 33–42. [DOI] [PubMed] [Google Scholar]
- 25. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; 521(7553): 436–444. [DOI] [PubMed] [Google Scholar]
- 26. Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2009: 248–255. [Google Scholar]
- 27. Dos Santos VA, Schmetterer L, Stegmann H, et al.. CorneaNet: fast segmentation of cornea OCT scans of healthy and keratoconic eyes using deep learning. Biomed Opt Express. 2019; 10(2): 622–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamiya K, Ayatsuka Y, Kato Y, et al.. Keratoconus detection using deep learning of colour-coded maps with anterior segment optical coherence tomography: a diagnostic accuracy study. BMJ Open. 2019; 9(9): e031313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rawat W, Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017; 29(9): 2352–2449. [DOI] [PubMed] [Google Scholar]
- 30. Shaha M, Pawar MM. Transfer learning for image classification. In: 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA). Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2018: 656–660. [Google Scholar]
- 31. Goodfellow I, Bengio Y, Courville A. Deep Learning. Cambridge, MA: MIT Press; 2016. [Google Scholar]
- 32. Abadi M, Barham P, Chen J, et al.. Tensorflow: a system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ’16). Berkeley, CA: USENIX; 2016: 265–283. [Google Scholar]
- 33. Zintgraf LM, Cohen TS, Welling M. A new method to visualize deep neural networks. Available at: https://arxiv.org/pdf/1603.02518.pdf. Accessed December 9, 2020.
- 34. Kingma D, Ba J. Adam: a method for stochastic optimization. Available at: https://arxiv.org/pdf/1412.6980.pdf. Accessed December 9, 2020.
- 35. Pedregosa F, Varoquaux G, Gramfort A, et al.. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12: 2825–2830. [Google Scholar]
- 36. Kim I, Rajaraman S, Antani S. Visual interpretation of convolutional neural network predictions in classifying medical image modalities. Diagnostics (Basel). 2019; 9(2): 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oquab M, Bottou L, Laptev I, Sivic J. Is object localization for free? – Weakly-supervised learning with convolutional neural networks. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2015: 685–694. [Google Scholar]
- 38. Chollet F. keras: GitHub. Available at: https://github.com/fchollet/keras. Accessed December 9, 2020.
- 39. Romanuke V. Appropriate number and allocation of ReLUs in convolutional neural networks. Res Bull NTUU. 2017; 1: 69–78. [Google Scholar]
- 40. de Boer P-T, Kroese DP, Mannor S, Rubinstein RY. A tutorial on the cross-entropy method. Ann Oper Res. 2005; 134: 19–67. [Google Scholar]
- 41. Ishii R, Kamiya K, Igarashi A, Shimizu K, Utsumi Y, Kumanomido T. Correlation of corneal elevation with severity of keratoconus by means of anterior and posterior topographic analysis. Cornea. 2012; 31(3): 253–258. [DOI] [PubMed] [Google Scholar]
- 42. Dunnmon JA, Yi D, Langlotz CP, Re C, Rubin DL, Lungren MP. Assessment of convolutional neural networks for automated classification of chest radiographs. Radiology. 2019; 290(2): 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Motlagh MN, Moshirfar M, Murri MS, et al.. Pentacam corneal tomography for screening of refractive surgery candidates: a review of the literature, part I. Med Hypothesis Discov Innov Ophthalmol. 2019; 8(3): 177–203. [PMC free article] [PubMed] [Google Scholar]