Summary
Cell-free extract derived from the eggs of the African clawed frog Xenopus laevis is a well-established model system that has been used historically in bulk aliquots. Here, we describe a microfluidic approach for isolating discrete, biologically relevant volumes of cell-free extract, with more expansive and precise control of extract shape compared with extract-oil emulsions. This approach is useful for investigating the mechanics of intracellular processes affected by cell geometry or cytoplasmic volume, including organelle scaling and positioning mechanisms.
For complete details on the use and execution of this protocol, please refer to Geisterfer et al. (2020).
Subject Areas: Biophysics, Cell Biology, Cell isolation, Microscopy, Model Organisms
Graphical Abstract

Highlights
-
•
Xenopus laevis cell-free extracts can be isolated as discreet volumes typical of cells
-
•
Approach provides enhanced control over the shape and position of encapsulated extract
-
•
Improved signal-to-noise during fluorescence imaging versus oil-emulsion extract droplets
Cell-free extract derived from the eggs of the African clawed frog Xenopus laevis is a well-established model system that has been used historically in bulk aliquots. Here, we describe a microfluidic approach for isolating discrete, biologically relevant volumes of cell-free extract, with more expansive and precise control of extract shape compared with extract-oil emulsions. This approach is useful for investigating the mechanics of intracellular processes affected by cell geometry or cytoplasmic volume, including organelle scaling and positioning mechanisms.
Before you begin
Previously established microfluidic approaches have proven successful in segmenting cell-free extract into droplets with volumes typical of cells (Hazel et al., 2013; Good et al., 2013; Oakey and Gatlin, 2018; Jimenez et al., 2011). Here, we have used many of the same principles and techniques while establishing a new method that employs hydrogel photolithography as a means of confining cell-free extracts.
This protocol was designed and implemented for the encapsulation of Xenopus laevis cell-free extracts. For more information on Xenopus laevis cell-free extracts, see (Good and Heald, 2018). However, this protocol can be extended to any cell-free extract, provided there is enough extract volume to fill the specific microfluidic devices used.
Preparing microfluidic devices
Timing: 6–7 h
Note: The steps listed below for preparing microfluidic devices were adapted from a previous protocol (Oakey and Gatlin, 2018).
-
1.
Thoroughly mix the two-part polydimethylsiloxane (PDMS) elastomer kit in a 1:10 (hardener:base) ratio in a clean plastic weighing boat.
-
2.
Place the microchannel master in a containment reservoir, such as a Petri dish, that allows the master to be completely covered by PDMS. Fasten the master securely to the bottom of the reservoir with vinyl laboratory tape so that it cannot shift or float away from the surface.
Note: For more information on the construction of microchannel masters or PDMS replicas, see (Whitesides and Stroock, 2001).
-
3.
Pour the mixed PDMS over the master until completely submerged, with 2–4 mm of excess PDMS covering the master (Figure 1A).
-
4.
Place the PDMS-covered master into a vacuum chamber, seal, and evacuate until bubbles emerge from the PDMS. Allow the PDMS to degas until bubbles no longer appear. This process may take 1–2 h.
-
5.
Place the degassed, but still fluid PDMS-covered master for 4 h into a 70°C oven. Care must be taken to level the oven and ensure that the reservoir sits flat in the oven, otherwise final devices will be of uneven thickness which may result in device-to-device variability in gas/fluid exchange through the PDMS replica (Figure 1A).
-
6.
Remove the now-cured PDMS from the oven. Using a sharp scalpel or X-Acto knife carefully cut through the PDMS, completely circumnavigating the channel network. With tweezers, remove the PDMS replica and set the master aside for subsequent replication. Excess PDMS need not be removed from the master before reuse, provided the replica was removed cleanly.
-
7.
Place the PDMS replica upon a clean, firm surface with network side facing up. Use the scalpel to cut and trim the device to fit upon a coverslip.
-
8.
With a sharpened 20 G blunt syringe tip and light pressure, punch holes through the PDMS at the microchannel inlet and outlet (Figure 1B). After cutting the replica out and punching holes for the inlet and outlet, the PDMS may be cleaned of dust and debris by dabbing the surface with Scotch tape.
-
9.
Place coverslip and clean PDMS device, network facing up, into the oxygen plasma cleaner.
CRITICAL: It is important to clean coverslips prior to device fabrication to ensure removal of any polishing particles or other types of debris. This helps ensure even and high efficiency of glass surface modifications and coatings (such as acrylation) important for device function. This can be achieved by several techniques such as acid washing, sonication, or plasma cleaning, all of which are sufficient to ensure removal of manufacturing oils or small debris.
-
10.
Expose the PDMS device and glass coverslip to an oxygen plasma under moderate power for about 1 min.
CRITICAL: The optimal oxygen plasma exposure time varies greatly based upon oxygen partial pressure, RF power, and ambient atmospheric conditions (temperature and humidity, for instance). Therefore, the exposure time must be determined empirically, by trial and error, for each plasma cleaner and laboratory setting. Successful oxygen plasma exposure of the PDMS replica and the glass coverslips results in a robust bond between the PDMS and the coverslip. In the event of an unsuccessful bond, the PDMS replica may lift off of the coverslip, or fluid introduced into the microfluidic chamber may extend beyond the confines of the channel.
-
11.
Remove the PDMS and coverslip from the plasma cleaner, taking care not to touch the bonding surfaces. Bring the PDMS replica into contact with the coverslip and gently form a seal by applying pressure with tweezers (Figure 1B).
-
12.
Place the hybrid device into a 70°C oven for 10 min to complete bonding.
Note: If PDMS fragments are found flowing through the network, their introduction is likely from the punched holes. To remedy this, the PDMS replicas may be cleaned prior to plasma bonding by sonicating the replica in ethanol for 15 min. PDMS swells considerably in ethanol, so devices treated in this way must be baked for an hour between 80°C and 100°C to volatilize all residual organic liquid before bonding to a glass coverslip (see “Troubleshooting”).
Pause Point: Microfluidic chambers can be stored for up to two months without observable changes in the elastic properties of the PDMS. It is best to store these chambers between 18°C and 22°C, and within a sealed container to prevent the accumulation of dust on the chamber.
Figure 1.
Preparation of microfluidic devices
Schematic showing (A) PDMS poured onto a silicon wafer containing multiple replicates of a microfluidic chamber positive and (B) device assembly after the PDMS has cured and a single chamber replicate has been cut from the cured slab.
Acrylation and PEGylation of the microfluidic chamber
Timing: 6–24 h
-
13.
Remove hybrid devices from the oven following bonding, and fill microfluidic chamber with 2% (v/v) 3-(Trimethoxysilyl)propyl methacrylate in 95% ethanol using a 22 G blunt needle, ensuring that any bubbles have been removed from within the chamber (Figure 1B). Let stand for 5 min.
Note: Treatment with 3-(Trimethoxysilyl)propyl methacrylate coats the coverslip glass with acryloyl chemical groups, which is required for later steps during surface passivation and the adherence of the hydrogel structures to the coverslip surface.
-
14.
Rinse chamber twice with 30 channel volumes of 95% ethanol using a 22 G blunt needle.
-
15.
Place the device for 15 min into a 70°C oven.
-
16.
Rinse chamber once with 30 channel volumes of 95% ethanol, followed by 10 channel volumes of ddH2O.
-
17.
Dry devices for 4 h in a 70°C oven or for 24 h between 20°C and 22°C.
CRITICAL: The process of drying or curing is also important for forming a covalent linkage between the 3-(Trimethoxysilyl)propyl methacrylate and the glass coverslip.
-
18.
After devices have been dried, place device on a reflective surface such as aluminum foil, fill chamber with PEGMA solution, and immediately expose to 10 mW/cm2 of UV light (352 nm) for 3 s using a UV lamp (Figure 1B).
CRITICAL: It is important that the coverslip side of the hybrid device be facing up when exposed to UV light. In addition, ensure that the entire channel is evenly illuminated. This will provide a biologically inert surface and prevent protein adsorption to the coverslip.
-
19.
Flush chamber with ten channel volumes of unexposed PEGMA solution and expose once more. Repeat this for three total exposures for each device.
-
20.
Flush chamber with 30 channel volumes of ddH2O and then submerge the entire device in ddH2O and keep at 4°C until use.
Pause Point: It is recommended that the devices be used within one week of preparation to ensure a consistent quality in the PEG coating.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| SYLGARD 184 silicone elastomer (PDMS elastomer kit) | Dow Corning | Material Number 99109937 |
| 3-(Trimethoxysilyl)propyl methacrylate | Sigma-Aldrich | Cat#440159 CAS: 2530-85-0 |
| Poly(ethylene glycol) methacrylate | Sigma-Aldrich | Cat#409537 CAS: 25736-86-1 |
| Poly(ethylene glycol) diacrylate | Sigma-Aldrich | Cat#455008 CAS: 26570-48-9 |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate | Sigma-Aldrich | Cat#900889 |
| Pico-Surf 1 (Novec 7500 with 2% surfactant) | Sphere Fluidics | SKU: C021 |
| Software and algorithms | ||
| Metamorph 7.7 software | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| Canvas 11 | Canvas | https://www.canvasgfx.com/en/support/canvas-11/ |
| Other | ||
| Inverted microscope | Olympus | IX71/IX81 |
| UV curing lamp | Excelitas Technologies | OmniCure S1500 |
| Digital micromirror device (DMD) | Mightex | Polygon400-G/Polygon1000-G |
| Plasma cleaner | Harrick Plasma | Cat# PDC-32G (115V) |
| Low flow or low-pressure syringe pump | Cetoni or Fluigent | neMESYS 290N Lineup FlowEZ |
| Microbore tubing (0.010-inch ID × 0.030-inch OD) | Saint Gobain Performance Plastics | 1204G81 |
| General protocol convection oven | Thomas Scientific | Cat# 1218W47 |
| 20 G blunt needle | CML Supply | SKU:901-20-050 |
| 22 G blunt needle | CML Supply | SKU:901-22-050 |
| 30 G blunt needle | CML Supply | SKU:901-30-050 |
Alternatives: RAN 008 (cat#: 008-FluoroSurfactant-2wtF-50G) from Ran Biotechnologies can be used in place of Pico-Surf.
Materials and equipment
Preparation of solutions
Note: For the total volumes of all solutions, plan on at least 30 channel volumes needed per a device (i.e., 270 μL for three devices featuring a 3 μL chamber)
Note: Due to the viscosity of the polyethylene glycol reagents, it is more accurate to make solutions on a weight basis.
Note: In Geisterfer et al. (2020), CSF-XB was used to dilute reagents to their final concentration, however this specific buffer may be exchanged for a buffer compatible with the cell-free extract being used or the cell cycle of interest.
Note: Solutions should be made the day they are required. For this, it is easiest to prepare stock solutions for each of the reagents. For example, 50% (w/w) solutions of poly(ethylene glycol) methacrylate (PEGMA) in ddH2O and 5% solutions (w/w) of Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) in ddH2O can be kept and stored for up to two weeks at 4°C away from light.
CRITICAL: When preparing each solution, add the photo-initiator (i.e., LAP) last, as the solution becomes light sensitive following its addition.
CSF-XB buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| XB salts (20×) | 1× | 25 mL |
| Sucrose (2 M) | 50 mM | 12.5 mL |
| K-HEPES (1 M, pH 7.7) | 10 mM | 5 mL |
| K-EGTA (0.5 M) | 5 mM | 5 mL |
| MgCl2 (1 M) | 1 mM | 0.5 mL |
| H2O | n/a | 452 mL |
| Total | n/a | 500 mL |
PEGMA solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Polyethylene glycol methacrylate | 10% w/w, or 0.14 M | 0.1 g |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate | 0.1% w/w, or 3.4 mM | 0.001 g |
| CSF-XB buffer | n/a | 0.899 g |
| Total | n/a | 1 g |
PEGDA solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Polyethylene glycol diacrylate | 20% w/w, or 0.29 M | 0.2 g |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate | 0.5% w/w, or 17 mM | 0.005 g |
| CSF-XB buffer | n/a | 0.795 g |
| Total | n/a | 1 g |
Alternatives: Other photo-initiators such as 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone may offer similar results, however LAP offers enhanced absorption at higher wavelengths and higher cytocompatability (Debroy et al., 2018; Fairbanks et al., 2009).
Step-by-step method details
Formation of PEGDA micro-enclosures
Timing: 1–2 h
A digital micromirror device (DMD) is used to photo-pattern the coverslip using a photo-labile hydrogel.
Note: The ability to place photomasks in an imaging plane that is conjugate to that of the coverslip is of paramount importance to this protocol. This can be achieved by the physical placement of photomasks into the light path of the microscope (LeValley et al., 2018). However, the use of a digital mask affords much more flexibility, is more robust, and is experimentally less cumbersome. We use a digital micromirror device (DMD; Polygon 400, Mightex) coupled with calibrated digital masks produced in Canvas 11 (Canvas).
Note: The size and feature resolution of digital masks is predominantly limited by the field of view and the numerical aperture (NA) of the objective lens, respectively. The diffusion of the photo-initiator will also negatively impact feature resolution. Anecdotally, this limited feature size to ∼ 5 um when we used a 20× objective with an NA of 0.75.
Note: Prior to beginning this step of the protocol, it is suggested that the inverted microscope, syringe pump, and DMD be turned on, with all driving software correctly set up and calibrated.
-
1.
Remove device from ddH2O and dab any droplets of water off the device. Clean the coverslip surface gently with a laboratory napkin and 100% ethanol.
-
2.
Using a syringe and a 22 G blunt needle, flush the channel with 30 channel volumes of the PEGDA solution and leave the device full. Dab away excess solution from the outlet. For this step, either inlet can be used.
-
3.
Immediately move the filled device onto the stage of an inverted microscope in a room with minimal ambient light.
-
4.
Select the desired exposure conditions based on the hydrogel-forming solution stoichiometry, as well as the digital mask for photo-patterning the coverslip surface. Begin patterning the coverslip (Figure 2A).
CRITICAL: Feature resolution and quality of the PEGDA microstructures is determined in part by the position of the focal plane and by the numerical aperture of the objective. For best results, a high NA objective is recommended, with the focal plane set just above (1–2 μm) the coverslip surface before exposure (Figure 3B). To help in structure formation, solutions can be supplemented with small inert beads that rest on the coverslip surface as a means of determining focal plane.
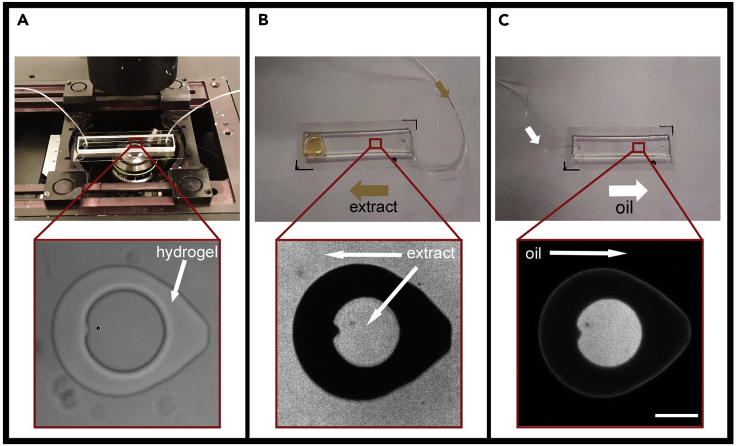
Figure 2.
Successive steps for the microfluidic encapsulation of cell-free extracts using hydrogels
(A) Photo-patterning of the coverslip surface in the microfluidic chamber using the DMD. A transmitted-light image of a hydrogel micro-enclosure is shown at the bottom in red boxes.
(B) Cell-free extract is pumped into the device using the inlet nearest the micro-enclosures. The flow through (large collection of extract on the left side of the device) is wiped away. Red ROI indicates the position of the filled micro-enclosure which is visualized below by a fluorescent image showing the cell-free extract (supplemented with a soluble fluorophore) contained within and surrounding the hydrogel micro-enclosure.
(C) After filling the micro-enclosures, an oil crossflow from the opposite inlet is used to isolate the micro-enclosure. After encapsulation, the fluorescent signal surrounding the hydrogel micro-enclosure is lost (bottom-right image). Scale bar, 50 μm. (Geisterfer et al., 2020).
Figure 3.
Hydrogel feature resolution
(A) Digital mask drawn using Canvas 11 and imported into the Mightex software to control the DMD.
(B) Transmitted-light image showing a hydrogel structure formed while the focal plane was just above the coverslip. The bottom schematic shows the position of the focal plane (in red) relative to the glass coverslip (dark blue).
(C) Hydrogel structure formed with the focal plane above the cover slip surface (microfluidic chamber bottom). This result is similar for focal planes used below the coverslip surface. Both images were collected at the same axial position for direct comparison. Scale bar, 50 μm.
Note: Exposure conditions can vary slightly per device, but in general, exposures of 0.8–1.5 mW/cm2 with 405 nm light for 5–10 ms at 17 mM LAP and 0.29 M PEGDA will result in sufficient polymerization and subsequent gelation of the hydrogel solution (Debroy et al., 2018).
Note: Devices are designed for one-time use only, however, PDMS chamber dimensions may allow for the formation of several hydrogel structures within one device.
Note: Flexible PDMS devices are subject to deformation under pressure-driven flow, so it is advised that top-filling micro-enclosures (such as those featured in Geisterfer et al., 2020) be formed nearest to the inlet that will be used to perfuse extract into the chamber. This will ensure that the tops of these PEGDA micro-enclosures are left open for filling with cell-free extract.
Note: To keep track of the position of micro-enclosures within device chambers, a fiduciary mark can be added to the PDMS side of the coverslip outside the chamber to indicate the location of the micro-enclosures.
-
5.
After patterning the coverslip with a desired micro-structure, use a syringe pump to flush out unpolymerized PEGDA solution. This is accomplished by carefully inserting microbore tubing into the outlet opposite the PEGDA microstructures. Once the tubing is securely positioned, a flow of ∼5–10 μL/min for a total of 50 channel volumes of CSF-XB buffer can be used to flush the channel of unpolymerized PEGDA solution.
CRITICAL: To prevent the introduction of bubbles into the device chamber, prime the microbore tubing (0.010-inch ID × 0.030-inch OD) with buffer prior to insertion into the PDMS inlet. This is most easily done by running buffer through the tubing at a high flow rate until a droplet forms at the end of the tubing. Inspect the length of tubing for any bubbles after priming.
Note: Flow rates used for flushing the device channels following hydrogel structure formation can vary depending on the dimensions of the channel. It is best to establish a mild flow rate that is amenable to the channel dimensions that does not deform the hydrogel structures or wash away any necessary experimental materials such as Aurora-A kinase coated beads. For more information regarding Aurora-A kinase coated beads see (Tsai and Zheng, 2005).
-
6.
Once the channel has been flushed with buffer, carefully remove the microbore tubing and store the device at 4°C submerged in CSF-XB buffer until use.
CRITICAL: Soaking the device in CSF-XB buffer after micro-enclosure formation saturates the PDMS and thus limits liquid permeation through channel walls upon filling the device with extract. In addition, the CSF-XB buffer limits the potential dilution of the extract from water stored in the hydrogel structure.
Pause Point: For best results, use devices within one week of PEG coating the coverslip (see “Acrylation and PEGylation of the microfluidic chamber”).
Encapsulation of cell-free extract
Timing: 20–30 min
The device chamber, and the micro-enclosures inside are filled with cell-free extract. An oil crossflow is then used to isolate the extract in the inner-reservoirs of the micro-enclosures.
Note: Prior to beginning the encapsulation process, it is important to have the cell-free extract prepared, supplemented with needed fluorophores, and induced into the correct phase of the cell cycle for the experimental question.
-
7.
In a 4°C cold room, remove the PDMS device from the CSF-XB buffer, and place near the syringe pump. Dab any excess buffer from the top of the PDMS device.
-
8.
Begin flushing device with cell-free extract. Be sure to insert the microbore tubing in the inlet nearest the hydrogel microstructures (Figure 2B). For those experiments in (Geisterfer et al., 2020), 50 channel volumes of extract were pumped through the device at 7 μL/min.
-
9.
After flushing the channel with extract, dab away fluid from the opposite port, and carefully remove the microbore tubing carrying extract using forceps. Insert the microbore tubing from the syringe containing the encapsulation oil and surfactant into the inlet opposite of your hydrogel structures (Figure 2C).
Note: For encapsulation, it is suggested a fluorocarbon oil and PEG-based surfactant be used to minimize interactions at the oil-extract interface. Geisterfer et al. used Pico-Surf 1 (Novec 7500 supplemented with 2% surfactant) to improve the encapsulation efficiency.
-
10.
Once the microbore tubing is securely inserted into the opposite port, begin flushing the channel with the encapsulation oil at a rate of ∼20–25 μL/min for short pulses (around 5 μL increments) separated by 5 s. Pulse the device with oil for a total of three times.
-
11.
Hydrogel structures should be isolated at this point (Figure 2C; bottom image). Using forceps, carefully remove the tubing and dab away any oil and extract from the top of the PDMS device. Inlets can be left open or sealed at this time using small sections of tubing with an overhand knot cinching one end. Salts or compounds dried on the bottom of the coverslip from the buffer soak can be removed with 100% ethanol.
-
12.
The device is now ready to be imaged and can be moved from the 4°C cold room to a temperature controlled room for imaging.
Expected outcomes
After encapsulation, hydrogel structures and the extract they contain should be isolated within the microfluidic device. This can be confirmed by supplementing the cell-free extract with a soluble fluorophore and visualizing structures using a fluorescent microscope; the viewer should only see fluorescent signal within the reservoir of the hydrogel micro-enclosure (Figure 2C; bottom image). It should be noted that there is often some signal on the peripheral margins of the hydrogel structure, this is most likely a thin layer of extract trapped between the oil and the hydrogel.
Limitations
Using hydrogel photolithography as a means of encapsulating cell-free extracts comes with inherent limitations. Notably, this technique is reliant on a calibrated digital micromirror device (DMD) or a physical photo-mask, which may limit the broad accessibility of this protocol.
The free-radical polymerization of the PEGDA used in this protocol is oxygen inhibited, and as a result the height of the hydrogel micro-enclosures is dependent on the microchannel depth, the rate of oxygen diffusion through the top of the PDMS microchannel replica, exposure conditions, and hydrogel-forming solution stoichiometry (Debroy et al., 2019; Dendukuri et al., 2008). Depending on the DMD used, resolution of the hydrogel structure and ultimately feature size may also vary. In addition, due to the 2D nature of the digital masks, researchers are limited to generating extruded micro-enclosures, i.e., those with dimensions that remain constant as a function of depth (i.e., no spherical enclosures).
Troubleshooting
Problem
Microfluidic channel contains large amounts of PDMS fragments reducing flow rate, or the device is not exchanging fluid at all.
Potential solution
If PDMS fragments are found flowing through the network, their introduction is likely from the punched holes.
-
1.
To limit the introduction of PDMS fragments throughout the channel, the PDMS replicas may be cleaned prior to plasma bonding by sonicating the replica in ethanol for 15 min. PDMS swells considerably in ethanol, so devices treated in this way must be baked for an hour between 80°C and 100°C to volatilize all residual organic liquid before bonding to a glass coverslip.
-
2.
A smaller blunt syringe tip (22G) can be passed through the punched hole to dislodge any large PDMS fragments left in PDMS inlet/outlet.
-
3.
If no fluid can be exchanged through the device, this is likely due to the PDMS plug created during hole punching being left in place while removing the 20G blunt syringe tip from the PDMS replica. To prevent this, ensure that the blunt syringe tip passes fully through the PDMS replica during the process of punching inlets/outlets, and with air pressure, or using a piece of scotch tape, remove the plug from the end of the syringe tip prior to pulling the syringe out of the PDMS replica.
Problem
During any of the channel flushing steps, after applying pressure-driven flow, fluid builds up around the inlet containing the microbore tubing.
Potential solution
Fluid droplets forming around the inlet containing the microbore tubing indicates an incomplete fit between the inlet and the microbore tubing. Typically, this can be addressed by the following considerations.
-
1.
While preparing the microfluidic chambers, if excess hardener (beyond the 1:10 ratio) is used when mixing the PDMS elastomer, the cured PDMS replica will be fairly brittle. This often results in cracks being introduced into the PDMS replica during the process of hole punching the inlets/outlets. These cracks can lead to inefficient coupling of the microbore tubing to the device inlet/outlet. To prevent this, ensure that the hardener is added at the correct ratio, or slightly reduce the amount of hardener used. It is worth noting that by reducing the amount of hardener, curing times are increased.
-
2.
If the microbore tubing is not inserted to a proper depth in the inlet, the pressure-driven flow may dislodge the microbore tubing, or fluid may preferentially travel up the inlet. To help ensure the proper seating of the microbore tubing into the device inlet, it often helps to cut the inserted end of the microbore tubing at a 45° angle to create a sharpened tip which is aids in insertion of the tubing.
Problem
Hydrogel micro-enclosures are deforming, or the micro-enclosures are moving within the microfluidic channel.
Potential solution
The deformation and movement of hydrogel micro-enclosures may indicate issues with flow rates or surface chemistry
-
1.
Large pressures created within the channel during fluid exchange by high flow, or by the physical depression of the PDMS during microbore tubing insertion, can lead to deformation of the hydrogel micro-enclosures. This can be avoided by reducing the flow rate during fluid exchange, or by ensuring the forceps do not press down on the microfluidic device during microbore insertion.
-
2.
If hydrogel structures are moving within the microfluidic channel during fluid exchange, this is an indication of insufficient surface treatment during the acrylation step of the “Acrylation and PEGylation of microfluidic chambers” section of this protocol. The presence of acryloyl groups on the coverslip surface ensures proper anchoring of the PEGDA micro-enclosures. Insufficient surface treatment can be remedied by ensuring the coverslip is properly cleaned prior to treatment, or that surface treatment is immediately followed by plasma bonding of the PDMS replica to the coverslip.
Problem
Incomplete oil encapsulation of hydrogel structure, or cell-free extract contained on the periphery of the hydrogel structure.
Potential solution
Inefficient oil encapsulation of the hydrogel structures could arise for several reasons.
-
1.
Insufficient oil crossflow could be one factor, with low rates of flow being unable to push the cell-free extract away from the outside of the hydrogel structure. This can be remedied by increasing the rate of oil crossflow (i.e., 23 μL/min to 30 μL/min).
-
2.
Hydrogel structures could be too close to one another, and as a result, create an area of high resistance to the oil crossflow that slows fluid velocities down and results in incomplete encapsulation. This can be avoided by leaving more than 0.2 mm of space between each hydrogel structure.
-
3.
The shape of the hydrogel structure will also determine to some extent the encapsulation efficiency, particularly at the leading and trailing edge during oil introduction. Tear drop shaped structures improve encapsulation efficiency if oriented with the point of the tear facing toward the inlet used to fill the device with cell-free extract.
Problem
Low feature resolution in the hydrogel micro-enclosures (Figure 3C).
Potential solution
A high NA objective, clean coverslips, and a properly calibrated DMD are required to achieve the best resolution during the hydrogel photolithography process. Beyond these considerations, a few experimental tips can be used to ensure consistent feature resolution.
-
1.
Ensure that the image focal plane is above the coverslip surface, or within the depth of field specified by the objective (Figure 3B). This will reduce any optical aberrations that might affect the feature resolution of the hydrogel structure.
-
2.
Longer exposure times will generate photo-initiator radicals that can diffuse beyond the illuminated portion of the coverslip. This results in broadened polymerization and gelation of the hydrogel solution beyond the confines of the mask which greatly affects feature resolution. To reduce this unwanted polymerization, use short exposure times (5–10 ms) at high intensity (1.7 mW/cm2).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jesse Gatlin (jgatlin@uwyo.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any unique datasets or code.
Acknowledgments
This work was made possible by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant no. 2P20GM103432. It was also supported by additional funding provided by the NIGMS under grant no. R01GM113028, the NSF Faculty CAREER Program under award no. BBBE 1254608, Whitman Center fellowships at the Marine Biological Laboratory, and the Biomedical Scholars program of the Pew Charitable Trusts. We thank Drs. Aaron Groen and Tim Mitchison for their intellectual contributions and involvement in some of the pioneering experiments that set the foundation for this approach.
Author contributions
Methodology, Z.G. and J.O.; Validation, Z.G., J.O., and J.C.G.; Resources, J.O. and J.C.G.; Writing – Original Draft, Z.G., J.O., and J.C.G.; Writing – Editing & Review, Z.G., J.O., and J.C.G.; Supervision, J.C.G.; Funding Acquisition, J.O. and J.C.G.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Zachary M. Geisterfer, Email: zgeister@uwyo.edu.
Jesse C. Gatlin, Email: jgatlin@uwyo.edu.
References
- Debroy D., Liu J., Li-Oakey K., Oakey J. Structured hydrogel particles with nanofabricated interfaces via controlled oxygen inhibition. IEEE Trans. Nanobiosci. 2019;18:253–256. doi: 10.1109/TNB.2019.2905489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debroy D., Oakey J., Li D. Interfacially-mediated oxygen inhibition for precise and continuous poly(ethylene glycol) diacrylate (PEGDA) particle fabrication. J Colloid. Interface Sci. 2018;510:334–344. doi: 10.1016/j.jcis.2017.09.081. [DOI] [PubMed] [Google Scholar]
- Dendukuri D., Panda P., Haghgooie R., Kim J.M., Hatton T.A., Doyle P.S. Modeling of oxygen-inhibited free radical photopolymerization in a PDMS microfluidic device. Macromolecules. 2008;41:8547–8556. [Google Scholar]
- Fairbanks B.D., Schwartz M.P., Bowman C.N., Anseth K.S. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisterfer Z.M., Zhu D.Y., Mitchison T.J., Oakey J., Gatlin J.C. Microtubule growth rates are sensitive to global and local changes in microtubule plus-end density. Curr. Biol. 2020;30:3016–3023. doi: 10.1016/j.cub.2020.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good M.C., Heald R. Preparation of cellular extracts from Xenopus eggs and embryos. Cold Spring Harb. Protoc. 2018 doi: 10.1101/pdb.prot097055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good M.C., Vahey M.D., Skandarajah A., Fletcher D.A., Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J., Krutkramelis K., Mooney P., Tomschik M., Gerow K., Oakey J., Gatlin J.C. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A.M., Roché M., Pinot M., Panizza P., Courbin L., Gueroui Z. Towards high throughput production of artificial egg oocytes using microfluidics. Lab. Chip. 2011;11:429–434. doi: 10.1039/c0lc00046a. [DOI] [PubMed] [Google Scholar]
- Levalley P.J., Noren B., Kharkar P.M., Kloxin A.M., Gatlin J.C., Oakey J.S. Fabrication of functional biomaterial microstructures by in situ photopolymerization and photodegradation. ACS Biomater. Sci. Eng. 2018;4:3078–3087. doi: 10.1021/acsbiomaterials.8b00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey J., Gatlin J.C. Microfluidic encapsulation of demembranated sperm nuclei in Xenopus egg extracts. Cold Spring Harb. Protoc. 2018;2018 doi: 10.1101/pdb.prot102913. [DOI] [PubMed] [Google Scholar]
- Tsai M.Y., Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr. Biol. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- Whitesides G.M., Stroock A.D. Flexible methods for microfluidics. Phys. Today. 2001;54:42–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.