Abstract
The coronavirus disease 2019 (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), warranting urgent study of the molecular mechanisms of SARS-CoV-2 infection and host immune response. Type I interferon (IFN-I) is a key component of host innate immune system responsible for eliminating the virus at the early stage of infection. In contrast, SARS-CoV-2 has evolved multiple strategies to evade innate immune response to facilitate viral replication, transmission, and pathogenesis. This review summarizes the recent progresses on SARS-CoV-2 proteins that antagonize host IFN-I production and/or signaling. These progresses have provided knowledge for new vaccine and antiviral development to prevent and control COVID-19.
Keywords: SARS-CoV-2, COVID-19, interferon, immune evasion
Introduction
The pandemic of coronavirus disease 2019 (COVID-19) first emerged in Wuhan, China, and has rapidly spread across the globe, leading to millions of infections (WHO 2020). The causative pathogen of COVID-19 is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus that belongs to β-coronavirus of the Coronaviridae family (Zhu and others 2020). SARS-CoV-2 infection, like previous severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), induces mild-to-severe respiratory illness, including fever, dry cough, breathing difficulties, and acute respiratory distress syndrome, which may lead to long-term reduction in lung function and death (Assiri and others 2013; Huang and others 2020; Wu and McGoogan 2020).
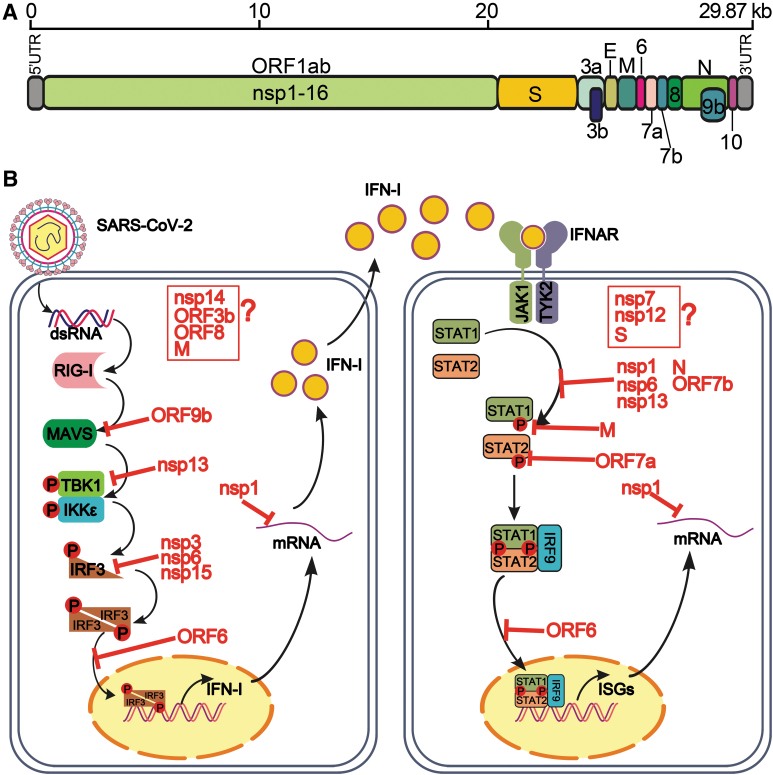
SARS-CoV-2 is an enveloped virus, containing a positive-sense single-strand ∼30 kb genome RNA, which encodes 16 nonstructural proteins (nsp1–16), 4 structural proteins [spike (S), envelop (E), membrane (M), and nucleocapsid (N)], and 7 accessory proteins and 8 accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, and ORF10) (Fig. 1A). Among them, the nsps are responsible for viral replication, the structural proteins for virion formation, and the accessory proteins for modulation of host response. The accessory proteins facilitate viral infection, but are not essential for viral replication.
FIG. 1.
SARS-CoV-2 proteins antagonize host IFN-I response. (A) Genome structure of SARS-CoV-2. The opening reading frames are shown, including ORF1ab (nonstructural proteins), structural proteins, and accessory proteins. (B) Evasion of IFN-I by SARS-CoV-2 proteins. The inhibitory targets of IFN-I production (left) and signaling (right) are indicated. Viral proteins with IFN-I inhibitory activities but with unknown targets are indicated in red boxes and question marks. See text for details. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; IFN-I, type I interferon.
Although intensive efforts are being made toward countermeasure development, only repurposed drugs (e.g., remdesivir as direct antiviral and dexamethasone as a steroid immune modulator for patients on ventilator) and convalescent plasma have been approved for emergency use authorization from the Food and Drug Administration. Promising therapeutic monoclonal antibodies are currently evaluated in clinical trials. Various vaccine candidates with different technological platforms are also being developed, many of which are in clinical trials (Mulligan and others 2020).
The innate interferon (IFN) response constitutes one of the first lines of defense against viral infections. Type I IFN (IFN-I) is a vital component of the early innate immune response that is initiated through multiple host pattern recognition receptors recognizing viral pathogen-associated molecular patterns, such as retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), Toll-like receptors (TLRs), cytoplasmic DNA receptors, and nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) (Meylan and others 2006; Acharya and others 2020; Li and others 2020a and 2020b; Park and Iwasaki 2020).
Upon SARS-CoV-2 infection, the stem loop in genomic RNA and/or double-strand RNA generated in replication are recognized by RIG-I or melanoma differentiation-associated gene 5 (MDA5), inducing a conformation change to expose the caspase activation recruitment domains (CARD) of RIG-I or MDA5. The exposure of CARD could interact with the CARD of the adapter mitochondrial antiviral signaling protein (MAVS), which subsequently recruits multiple downstream signaling components to the mitochondria, including inhibitor of κ-B kinase ɛ (IKKɛ) and TANK binding kinase 1 (TBK1). Both kinases phosphorylate and activate themselves, such activation leading to phosphorylate the IFN regulatory factor 3 (IRF3). The phosphorylated IRF3 forms homodimer and translocates into nucleus to stimulate IFN-I genes (IFN-α and IFN-β) expression (Fitzgerald and others 2003; Liu and others 2015).
Secreted IFN-I binds to the IFN-α and IFN-β receptors on cell surface, leading to activating Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which, in turn, phosphorylate the downstream components, signal transducer, and activator of transcription proteins (STAT1 and STAT2) (Levy and Darnell 2002). Phosphorylated STAT1 and STAT2 heterodimerize and interact with IRF9 to form the IFN-stimulated gene factor 3 (ISGF3). The ISGF3 complex undergoes nuclear translocation and binds to IFN-I–stimulated response elements (ISREs), inducing the expression of ISGs with antiviral functions, such as protein kinase R (PKR), 2′,5′-oligoadenylate synthetase (OAS), and RNase L (Balachandran and others 2000; Samuel 2001; Malathi and others 2007; Schneider and others 2014; Schoggins and others 2015).
In response to host immune system, coronaviruses have evolved diverse strategies to suppress the induction of IFN-I and antiviral functions of ISGs.
The outcome of any viral infection is determined by pathogen amplification and immune response. SARS-CoV-2 has been reported to be highly sensitive to IFN-I inhibition (Lokugamage and others 2020; Mantlo and others 2020). Extremely low levels of IFN-I expression or ISGs was detected in SARS-CoV-2–infected cells, animals, and patients (Blanco-Melo and others 2020; Hadjadj and others 2020; Israelow and others 2020). Early IFN-I administration is vital to protect host from lethal infection through limiting viral replication and spread (Channappanavar and others 2019; Goncalves and others 2020). Collectively, these studies indicate that SARS-CoV-2 could efficiently antagonize innate immune response through IFN-I induction and downstream IFN-1 signaling. In this review, we summarize the host IFN-I–mediated immune response in SARS-CoV-2 infection and the strategies the virus has employed to evade the immune response.
SARS-CoV-2 Proteins Antagonizing Host Innate IFN-I Response
SARS-CoV-2 utilizes various approaches to evade host IFN-I response, including suppression of IFN-I production and IFN-I signaling (Fig. 1B).
Nonstructural Proteins
SARS-CoV-2 encodes a larger ORF1ab polyprotein, which is processed into 16 nsps (nsp1–16). The first nsp1 has shown potent efficacy to block IFN-I production and signaling pathways (Mantlo and others 2020; Xia and others 2020). SARS-CoV nsp1 has been well documented to inhibit IFN-I through suppression of host gene expression (mRNA degradation and translation inhibition) and through reduction of STAT1 phosphorylation (Kamitani and others 2006, 2009; Narayanan and others 2008; Huang and others 2011). SARS-CoV-2 nsp1 shows 84% amino acid sequence identity with SARS-CoV nsp1, suggesting a similar mechanism to block IFN-I response.
The C-terminal region of SARS-CoV-2 nsp1 was shown to interact with the 40S subunit of ribosome, blocking the mRNA channel, leading a shutdown of host protein translation, including the expression of IFN-I and ISGs (Schubert and others 2020; Thoms and others 2020). After the SARS-CoV-2 outbreak, different groups have reported SARS-Cov-2 nsp1 is a potent IFN-I antagonist that significantly decreases >95% expression of IFN-I and ISGs (Lei and others 2020; Xia and others 2020; Yuen and others 2020). Besides shutting off host translational machinery, SARS-CoV-2 nsp1 has been found to inhibit IFN-I signaling by blocking STAT1 and STAT2 phosphorylation. In addition, SARS-CoV-2 nsp1 is more efficient than those of SARS-CoV and MERS-CoV to suppress phosphorylation of STAT1 and STAT2 (Xia and others 2020).
Nsp3, known as papain-like protease (PLpro), functions as a protease to cleave nsp1–4 and a potential deubiquitinase (DUB) based on motif sequence analysis. Owing to the large size (∼5.8 kb), it is challenging to express the full length of nsp3. Yuen and colleagues expressed the PLpro domain of nsp3 and reported that SARS-CoV-2 nsp3-PLpro marginally inhibited IFN-I production and signaling (Yuen and others 2020). However, compared with SARS-CoV-2 nsp3-PLpro, SARS-CoV nsp3-PLpro not only inhibited RIG-I–induced IFN-I production and IFN-I downstream signaling, but also strongly deubiquitinated host polyubiquitinated proteins, implying that the DUB activity of SARS-CoV-2 nsp3-PLpro plays a pivotal role in IFN-I antagonism and viral pathogenesis (Clementz and others 2010; Matthews and others 2014; Yuen and others 2020).
By contrast, Lei and others (2020) found that SARS-CoV-2 inhibited IFN-β production by 50% using Sendai virus (SeV) or RIG-I as a stimulator, but not MDA5. Moreover, Dikic and coworkers showed that SARS-CoV-2 nsp3-PLpro limited IFN-I production by cleaving the ubiquitin-like protein ISG15 and decreasing the phosphorylated IRF3 (Shin and others 2020). Furthermore, Moustaqil and others (2020) employed a cell-free cleavage assay to show that SARS-CoV-2 nsp3 was able to directly cleave IRF3, resulting in decreased IRF3 activation and IFN-I production.
Coronavirus nsp6 protein is a transmembrane protein with multiple functions. It participates in the formation of double-membrane vesicles for replication–transcription, affects host autophagy, and restricts IFN-I response (Cottam and others 2011; Angelini and others 2013; Xia and others 2020). In vitro IFN-β promoter-driven luciferase report assay shows that SARS-CoV-2 nsp6 interacts with TBK1 to inhibit the activation of IRF3, although such interaction does not affect TBK1 phosphorylation. In addition, nsp6 reduces the phosphorylation of STAT1 and STAT2 during IFN-I signaling. Notably, SARS-CoV-2 nsp6 exhibits more efficient suppression of RIG-I–induced IFN-I production and IFN-I–stimulated ISGs production than those nsp6 from SARS-CoV and MERS-CoV do, which confers higher viral replication in an IFN-I–stimulated transient replicon system (Xia and others 2020).
Nsp13 is a highly conserved superfamily 1 helicase, which has strong inhibitory effects on IFN-I production and signaling. Overexpression of SARS-CoV-2 nsp13 significantly suppresses IFN-β–driven luciferase activity when cells are continually stimulated by SeV, RIG-I, MAVS, IKKɛ, or TBK1, except IRF3–5D (a phosphor-mimic of the activated IRF3) (Lei and others 2020; Xia and others 2020; Yuen and others 2020). Coimmunoprecipitation and proteomic results indicated that nsp13 binds to TBK1, leading to decreased phosphorylation of TBK1 and inactivation of IRF3 (Gordon and others 2020; Hoffmann and others 2020; Xia and others 2020). Further studies are required to determine the molecular details of how TBK1 is inactivated by nsp13. In addition, nsp13 is identified as a potent antagonist of IFN-I signaling through inhibiting STAT1 and STAT2 activation, resulting in the retention of STAT1 in the cytoplasm and compromised stimulation of ISRE promoter (Lei and others 2020; Xia and others 2020; Yuen and others 2020).
Coronaviruses nsp14 and nsp15 have been identified as potent IFN-I antagonists (Chen and others 2009; Frieman and others 2009; Deng and others 2017). Nsp14 is a guanine-N7-methyltransferase, which is required for coronavirus replication and transcription by forming an RNA cap that mimics the host mRNA cap (Chen and others 2009). Overexpression of SARS-CoV-2 nsp14 significantly reduced the luciferase activities driven by IFN-β promoter or ISRE promoter (Yuen and others 2020). Our group and Lei and others (2020) also identified the inhibitory effect of nsp14 on IFN-I signaling, but suppression of IFN-I production induced by RIG-I was not observed. Further studies focusing on the mechanisms would address this discrepancy and reveal the underlying molecular details.
Nsp15 functions as an endoribonuclease, Yuen and others (2020) demonstrated nsp15 could inhibit IFN-I production and signaling effectively. Affinity-purification mass spectrometry results show nsp15 is involved in IFN-I induction by interaction with RNF41, an E3 ligase associated with activation of IRF3, but the exact mechanism remains to be determined (Gordon and others 2020).
In addition, nsp7 and nsp12 have been shown to weakly antagonize IFN-I response, in which the IFN-β and ISRE promoter retain >60% activity (Yuen and others 2020). Owing to the dual role of the nsps in viral replication and IFN-I antagonism, future investigations are needed to differentiate the 2 roles in the context of complete SARS-CoV-2. The availability of the infectious cDNA clone of SARS-CoV-2 makes it possible to perform such studies (Xie and others 2020a).
Structural Proteins
Coronavirus structural proteins have also been shown to inhibit IFN-I production and signaling through multiple mechanisms. SARS-CoV N protein antagonizes IFN-I production by suppressing TRIM25-mediated RIG-I activation, and M protein can directly interact with RIG-I, TBK1, IKKɛ, and TRAF3 to prevent the formation of TBK1-IKKɛ complex (Siu and others 2009; Hu and others 2017). However, little information is available about the function of SARS-CoV-2 structural proteins in antagonizing IFN-I response.
A recent report by Yuen and others (2020) shows that S and M inhibit IFN-β production to ∼10%, and that individual structural proteins marginally inhibit IFN-I signaling. Lei and others found that M protein significantly suppressed IFN-I induction when cells were stimulated by SeV, MDA5, or RIG-I. They also found that S protein suppressed IFN-I signaling, whereas N protein inhibited both IFN-I production (when cells were infected with SeV) and IFN-I signaling through decreasing the phosphorylation of STAT1 and STAT2 (Lei and others 2020; Mu and others 2020).
In our study, SARS-CoV-2 M protein was found to significantly block phosphorylation of STAT1, leading to a reduction of expression of ISGs (Xia and others 2020). These discrepancies could be due to various experimental systems, especially when the luciferase report assay was performed under different expression levels of viral proteins. Further mechanic studies are needed to define the molecular details of viral and host proteins that contributed to the observed antagonism of innate immune response.
Accessory Proteins
Although accessory proteins are dispensable for coronavirus infection and replication, they are evolved to modulate the host immune response and to facilitate viral infection and pathogenesis. SARS-CoV-2 accessory proteins were found to antagonize IFN-I response through different tactics, among which ORF6 was consistently identified as the most potent inhibitor of IFN-I response (Lei and others 2020; Xia and others 2020; Yuen and others 2020). We found that SARS-CoV-2 ORF6 could hijack the nuclear importin Karyopherin α 2 (KPNA2) to block IRF3 and ISGF3 nuclear translocation, leading to the silence of downstream IFN-I and ISGs gene expression levels. Lei and others (2020) made a series of truncations to show that the C-terminus of ORF6 was critical for its inhibitory effects.
Recently, ORF3b was reported to be a shorter variant, when compared with SARS-CoV ORF3b, due to early termination. SARS-CoV ORF3b potently inhibited IFN-I production through its C-terminus (Konno and others 2020). Because a low amount of ORF3b-expressing plasmid DNA (20 ng) was used in our transfection experiment, we did detect any inhibition of IFN-β promoter activity by ORF3b.
Both ORF7a and ORF7b strongly blocked IFN-I signaling in our screening system; interestingly, ORF7a only suppressed STAT2 phosphorylation, whereas ORF7b inhibited both STAT1 and STAT2 phosphorylation (Xia and others 2020). ORF8 was also found to block IFN-β production and signaling in a dose-dependent manner, but the underlying mechanism remains to be defined (Li and others 2020a and 2020b). In addition, ORF9b, an alternative ORF in the N protein, was reported to be recruited to MAVS through interacting with TOM70, leading to the inactivation of TOM70-mediated IFN-I production (Jiang and others 2020).
Conclusion
It is critical to understand pathogen–host interactions for any infectious disease. Such knowledge provides the rationale for therapeutics and vaccine development. Since IFN-I represents the most important first line of defense, it has been used to treat different viral infections in patients (Smith and others 1999; Hung and others 2020). SARS-CoV-2 was shown to be sensitive to IFN-I inhibition, suggesting its potential use for COVID-19 treatment, especially at the early stage of infection (Blanco-Melo and others 2020; Hadjadj and others 2020).
An increasing number of reports have been published on SARS-CoV-2 proteins that antagonize IFN-I response. Some viral proteins were consistently found to inhibit IFN-I response in different studies, whereas other viral proteins were not. As already discussed, the discrepancies were mainly caused by different experimental conditions in various studies (Jiang and others 2020; Konno and others 2020; Mu and others 2020; Schubert and others 2020; Xia and others 2020; Yuen and others 2020). To consolidate these results, it is important to validate the screen findings in multiple experimental systems, particularly in the context of authentic viral replication (Kim and others 2020).
To achieve such validation, reverse genetic systems of SARS-CoV-2 could be used to generate mutant viruses, chimeric virus (with viral proteins from SARS-CoV or MERS-CoV), and specific gene deletion viruses (Muruato and others 2020; Xia and others 2020; Xie and others 2020b). Such recombinant viruses could be examined for their IFN-I antagonism in cell culture and animal models. Viruses defective in antagonizing IFN-I response, in combination with replication-defective mutations, could potentially be developed as live-attenuated vaccine candidates. These studies may reveal a universal vaccine approach that is suitable for any future emerging coronaviruses.
Acknowledgments
We thank our colleagues for helpful discussions during the course of this study.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
P.-Y.S. was supported by NIH grants AI142759, AI134907, AI145617, and UL1TR001439, and awards from the Sealy & Smith Foundation, Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gillson Longenbaugh Foundation, and the Summerfield Robert Foundation.
References
- Acharya D, Liu G, Gack MU. 2020. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol 20(7):397–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. 2013. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 4(4):e00524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, et al. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13(9):752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13(1):129–141 [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5):1036–1045.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB Jr., Meyerholz DK, Perlman S.. 2019. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 129(9):3625–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106(9):3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, et al. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84(9):4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, Britton P, Ktistakis NT, Wileman T. 2011. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7(11):1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114(21):E4251–E4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4(5):491–496 [DOI] [PubMed] [Google Scholar]
- Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83(13):6689–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves A, Bertrand J, Ke R, Comets E, de Lamballerie X, Malvy D, Pizzorno A, Terrier O, Rosa Calatrava M, Mentre F, et al. 2020. Timing of antiviral treatment initiation is critical to reduce SARS-CoV-2 viral load. CPT Pharmacometrics Syst Pharmacol 9(9):509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583(7816):459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Pere H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369(6504):718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, Sanchez-Rivera FJ, Luna JM, Ashbrook AW, Soto-Feliciano YM, Leal AA, Le Pen J, Ricardo-Lax I, Michailidis E, et al. 2020. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. bioRxiv. [Epub ahead of print]; DOI: 10.1101/2020.09.11.291716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. 2017. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 91(8):e02143-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog 7(12):e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, Ng YY, Lo J, Chan J, Tam AR, et al. 2020. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 395(10238):1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, et al. 2020. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 217(12):e20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HW, Zhang HN, Meng QF, Xie J, Li Y, Chen H, Zheng YX, Wang XN, Qi H, Zhang J, et al. 2020. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol 17(9):998–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16(11):1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 103(34):12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181(4):914–921.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Sauter D, Gifford RJ, Consortium U-C, Nakagawa S, et al. 2020. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 32(12):108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, et al. 2020. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11(1):3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Darnell JE Jr.. 2002. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3(9):651–662 [DOI] [PubMed] [Google Scholar]
- Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X, et al. 2020a. Coronavirus infections and immune responses. J Med Virol 92(4):424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, Ge XY. 2020b. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 286:198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347(6227):aaa2630. [DOI] [PubMed] [Google Scholar]
- Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, Rajsbaum R, Menachery VD. 2020. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J Virol 94(23):e01410–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M Jr., Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448(7155):816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. 2020. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res 179:104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Schafer A, Pham A, Frieman M. 2014. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol J 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. 2006. Intracellular pattern recognition receptors in the host response. Nature 442(7098):39–44 [DOI] [PubMed] [Google Scholar]
- Moustaqil M, Ollivier E, Chiu H-P, Van Tol S, Rudolffi-Soto P, Stevens C, Bhumkar A, Hunter DJB, Freiberg A, Jacques D, et al. 2020. SARS-CoV-2 proteases cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species and the search for reservoir hosts. bioRxiv. [Epub ahead of print]; DOI: 10.1101/2020.06.05.135699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Fang Y, Yang Q, Shu T, Wang A, Huang M, Jin L, Deng F, Qiu Y, Zhou X. 2020. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, et al. 2020. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586(7830):589–593 [DOI] [PubMed] [Google Scholar]
- Muruato AE, Fontes-Garfias CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, Shi PY. 2020. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun 11(1):4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82(9):4471–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A, Iwasaki A. 2020. Type I and type III interferons—induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 27(6):870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14(4):778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2015. Corrigendum: a diverse range of gene products are effectors of the type I interferon antiviral response. Nature 525(7567):144. [DOI] [PubMed] [Google Scholar]
- Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muhlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27(10):959–966 [DOI] [PubMed] [Google Scholar]
- Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, et al. 2020. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587(7835):657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, Jin DY. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 284(24):16202–16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JK, Siddiqui AA, Krishnaswamy GA, Dykes R, Berk SL, Magee M, Joyner W, Cummins J. 1999. Oral use of interferon-alpha stimulates ISG-15 transcription and production by human buccal epithelial cells. J Interferon Cytokine Res 19(8):923–928 [DOI] [PubMed] [Google Scholar]
- Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, et al. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369(6508):1249–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Coronavirus disease (COVID-19) pandemic. 2020. Available at https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed October10, 2020)
- Wu Z, McGoogan JM. 2020. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 323(13):1239–1242 [DOI] [PubMed] [Google Scholar]
- Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33(1):108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, et al. (2020a). An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27(5):841–848.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Muruato AE, Zhang X, Lokugamage KG, Fontes-Garfias CR, Zou J, Liu J, Ren P, Balakrishnan M, Cihlar T, et al. (2020b). A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun 11(1):5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, Cai JP, Jin DY, To KK, Chan JF, et al. 2020. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect 9(1):1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382(8):727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]