Abstract
Bacteria were first detected in human tumors more than 100 years ago, but the characterization of the tumor microbiome has remained challenging because of its low biomass. We undertook a comprehensive analysis of the tumor microbiome, studying 1526 tumors and their adjacent normal tissues across seven cancer types, including breast, lung, ovary, pancreas, melanoma, bone, and brain tumors. We found that each tumor type has a distinct microbiome composition and that breast cancer has a particularly rich and diverse microbiome. The intratumor bacteria are mostly intracellular and are present in both cancer and immune cells. We also noted correlations between intratumor bacteria or their predicted functions with tumor types and subtypes, patients' smoking status, and the response to immunotherapy.
More than 16% of cancer incidence worldwide has been attributed to infectious agents (1). Intratumor bacteria have been reported in many tumor types (2-19), but these bacteria have not been characterized in a comprehensive manner (20). The gut microbiome has been shown to have multiple effects on tumor biology, such as the transformation process, tumor progression, and the response to anticancer therapies including immunotherapy (21-29). Thus, characterization of the tumor microbiome may be an essential step in unraveling the effects that tumor bacteria have on different cancer hallmarks.
Bacterial DNA, RNA, and lipopolysaccharide are present in many human solid tumors
Because the tumor microbiome has a relatively low biomass, contamination of the tumor samples with bacteria or bacterial DNA can be problematic (30, 31). Therefore, it is critical to include multiple measures to avoid, or at least detect, any possible contamination in the process of profiling the tumor microbiome (supplementary note) (32, 33). For next-generation sequencing applications, it is also important to use mechanical tissue shearing in the DNA isolation protocol to ensure the complete recovery of DNA from within the cell wall of Gram-positive bacteria—a step not included in most standard DNA isolation protocols (6, 7). To characterize and visualize intratumor bacteria, we applied an extensive combination of methods to a large cohort of solid human tumor samples to detect bacterial DNA, RNA, and bacterial outer membrane or cell wall components.
We focused on seven solid tumor types that represent either common cancer types or cancer types for which the tumor microbiome is largely unknown, such as melanoma, bone, and brain tumors (Fig. 1A). To address laboratory-borne contaminants, we introduced 643 negative controls that were processed with the samples, including 437 DNA extraction controls and 206 polymerase chain reaction (PCR) no-template controls (NTCs). To address contamination that might have occurred before the samples reached our laboratory, we also included 168 paraffin-only samples taken from the margins of the paraffin blocks (without tissue) that were used in the study (Fig. 1A).
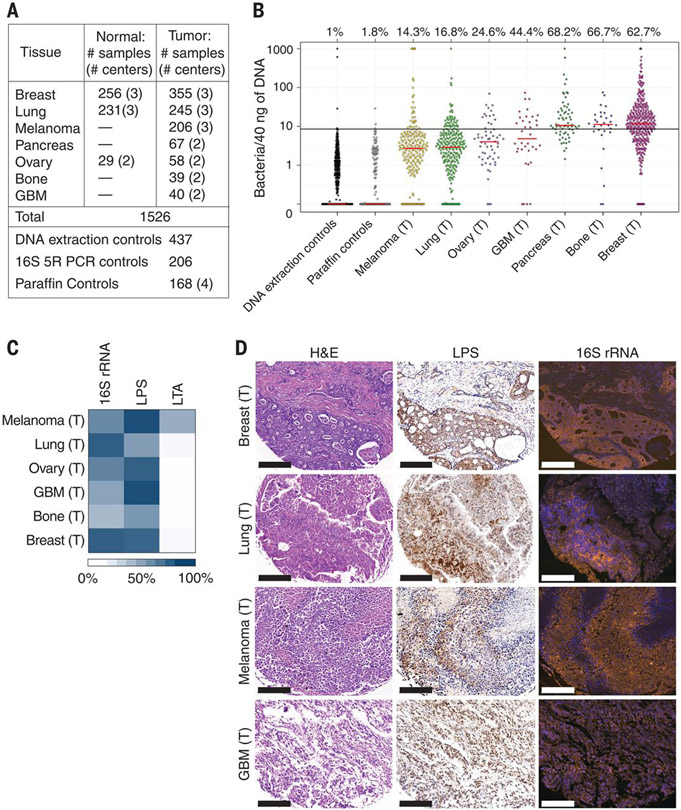
Fig. 1. Bacterial components are detected in human tumors.
(A) Number of human samples analyzed in the study. Normal samples include both normal tissue samples and normal adjacent tissue (NAT) to tumor samples, as detailed in table S1. Dashes indicate data not available. GBM, glioblastoma multiforme. (B) The presence of bacterial DNA in human tumors was assessed by bacterial 16S rDNA qPCR. A calibration curve, generated by spiking bacterial DNA into human DNA, was used to estimate bacterial load, which was then normalized against batch-specific qPCR NTCs. Values were floored to 0.1. Red bars represent the median. The proportion of samples of each cancer type that had more bacteria than the 99th percentile of the DNA extraction control samples (black bar) is depicted above each cancer type. (C) Heatmap representing the proportion of tumors that stained positively for 16S rRNA, LPS, or LTA. n = 40 to 101 tissue cores per tumor type. (D) Consecutive slices from four human tumor types were stained with H&E, anti-LPS antibody (LPS), or with FISH probes against bacterial 16S rRNA. Scale bars, 200 μm. The letter (T) indicates samples originating from tumors.
Overall, we profiled 1010 tumor samples and 516 normal samples, including normal adjacent tissues (NATs) from the same patients (Fig. 1A and table S1). In the case of ovarian cancer, our normal samples came from the ovaries or uteruses of the patients or from normal fallopian tube fimbria of unmatched healthy subjects (tables S1 and S2). To quantify bacterial DNA, we used a real-time quantitative PCR (qPCR) assay with universal primers 967F and 1064R specific for the bacterial ribosomal 16S gene [16S rDNA (ribosomal DNA)] (34). Levels of bacterial DNA in all tumor types were significantly higher than those found in both DNA extraction and paraffin controls (Fig. 1B; P value <10−10 for each tumor type, Wilcoxon rank sum test). We found that different cancer types vary in the proportion of tumors that are positive for bacterial DNA, ranging from only 14.3% in melanoma to >60% in breast, pancreatic, and bone tumors. Bacterial DNA was also detected in solid tumors that have no direct connection with the external environment, such as ovarian cancer, glioblastoma multiforme (GBM), and bone cancer.
To validate the presence of bacteria in human tumors, we stained >400 additional tumors (not related to the 1526 samples described above), representing six of our seven profiled tumor types, for the presence of bacteria. We conducted immunohistochemistry (IHC) using antibodies against bacterial lipopolysaccharide (LPS) and lipoteichoic acid (LTA) to detect Gram-negative and Gram-positive bacteria, respectively (35, 36). We also used RNA fluorescence in situ hybridization (FISH), with a universal probe against bacterial 16S ribosomal RNA (rRNA), to detect bacterial RNA in these tumors (37). To control for nonspecific staining, IHC-negative controls (no primary antibody) and FISH-negative controls (nonspecific complement probe) were also applied to the samples (figs. S1 and S2). Bacterial LPS and 16S rRNA were frequently detected in all tumor types (Fig. 1C) and demonstrated a similar spatial distribution (Fig. 1D and fig. S3). LTA was detected mainly in melanomas and was largely absent in other tumor types. Generally, more tumors were found to be positive for bacteria using visualization methods than by using qPCR. This disparity may be because of some limitation in the sensitivity of our qPCR assay, or it might be caused by our strict cutoff for confirming a sample as positive.
Intratumor bacteria are mostly intracellular and are present in both cancer and immune cells
Pathological examination of tumor cores indicated that LPS and bacterial 16S rRNA were localized mainly in cancer cells and immune cells (Fig. 2A and fig. S4). In cancer cells, bacterial 16S rRNA was detected mostly in the cytoplasm, whereas LPS staining was associated with both the cytoplasm and the nucleus (Fig. 2B and fig. S5). CD45-positive leukocytes generally exhibited a stronger cytoplasmic bacterial staining by 16S rRNA staining than that exhibited by cancer cells (Fig. 2C and fig. S6A). LTA-positive bacteria were almost exclusively found in macrophages, as detected by hematoxylin and eosin (H&E) staining and verified by immunofluorescence (IF) for CD68 (Fig. 2D and fig. S6B). LTA was rarely detected in cancer cells or in CD45+/CD68− immune cells (Fig. 2). Although the intensity of bacterial LPS and LTA staining was very strong in CD45+/CD68+ cells, bacterial 16S rRNA was only rarely found in macrophages by FISH (Fig. 2, A and D, and figs. S4 and S6). This discrepancy may reflect macrophage ingestion of bacterial components rather than live bacteria, or it may result from the accumulation of LPS and LTA in macrophages long after the bacteria have been phagocytized and processed by the macrophages. It has been previously demonstrated that the processing of bacterial LPS by macrophages is very slow; therefore, LPS can be found in these cells months after the bacteria were ingested and processed (38).
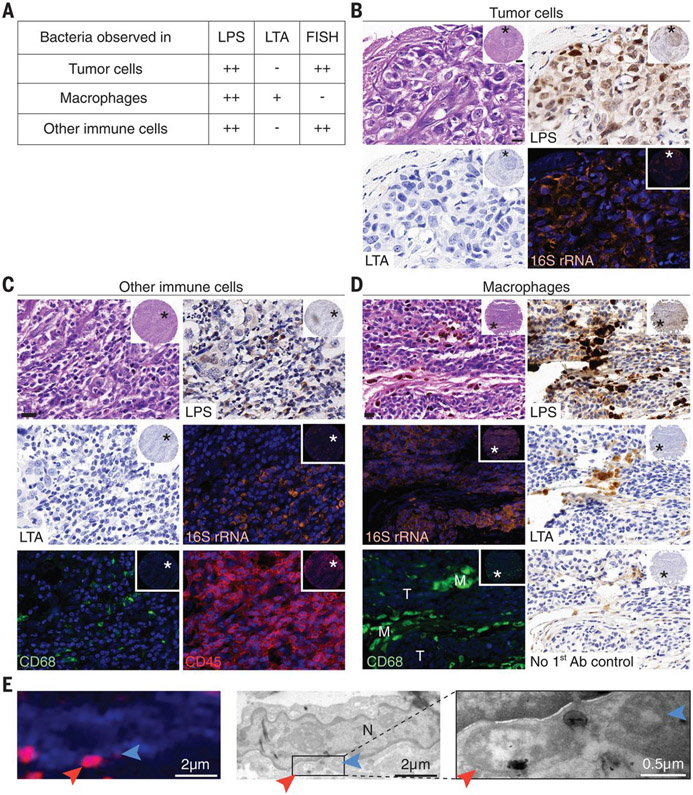
Fig. 2. Intratumor bacteria are found inside both cancer and immune cells.
(A) Summary of the staining patterns of LPS, LTA, and bacterial 16SrRNA in different cell types across 459, 427, and 354 tumor cores, respectively. CD45+/CD68+ cells are referred to as macrophages; CD45+/CD68− cells are referred to as other immune cells. (B to D) Representative cores are shown demonstrating the different staining patterns in human tumors. Asterisks mark the region that was selected for higher magnification. (B) Bacterial LPS and 16S rRNA are demonstrated in breast cancer cells. (C) Bacterial LPS and 16S rRNA are demonstrated in CD45+/CD68− cells of a highly inflamed breast tumor. (D) A melanoma tumor demonstrating typical staining of macrophage-associated bacteria (M), with positive LPS and LTA staining but no 16S rRNA staining. Nearby tumor cells (T) show the typical LPS and 16S rRNA staining, with negative LTA staining. Each inset demonstrates a low magnification of the entire core. Scale bars in high-magnification images, 20 μm. (E) CLEM demonstrates intracellular bacteria in human breast cancer. IF image shows DAPI in blue and LPS in red. Two bacteria are marked with arrows. TEM images of the same cell are shown in grayscale. High-magnification image of the boxed area is shown on the right. The letter N marks the cell nucleus.
To further verify the presence of bacteria inside cancer cells, we performed correlative light and electron microscopy (CLEM) (39, 40) on four human breast tumors that were positive for bacterial LPS and 16S rRNA (fig. S7). Combined LPS fluorescence staining and transmission electron microscopy (TEM) imaging of the same cells clearly demonstrated the intracellular localization of bacteria in all four tumors (Fig. 2E and fig. S7). In many cases, the bacteria were found in close proximity to the nuclear membrane. Because we did not detect intranuclear bacteria by TEM, we suspect that the appearance of LPS nuclear localization in some tumors represents the staining of cytoplasmic perinuclear bacteria.
Whereas bacterial 16S rRNA FISH staining appeared as a diffused signal inside cells, typical bacterial rods or cocci were only rarely detected (in 3 of 426 cores). This observation, combined with the fact that no cell wall polymer LTA was detected in cancer cells—despite the detection of many Gram-positive bacteria in human tumors by 16S rDNA sequencing—suggests that bacteria in tumor cells may have altered their envelope, perhaps leading to a cell wall–deficient state, akin to L-forms (41). Cell wall–deficient bacteria are known to be found exclusively inside cells, where their morphology transforms into less-defined structures of highly variable sizes and shapes (42, 43). Our TEM images also suggest that many of the intracellular bacteria lack a cell wall (Fig. 2Eand fig. S7).
The microbiome of breast tumors is richer and more diverse than that of other tumor types
To characterize the intratumor microbiome, we developed a multiplexed 16S rDNA sequencing protocol that amplifies five short regions along the 16S rRNA gene: the 5R 16S rDNA sequencing method (Fig. 3A). By amplifying 68% of the bacterial 16S rRNA gene using short amplicons, this method increases the coverage and resolution of the detection of bacterial species compared with the widely used V4 or V3-V4 amplification (fig. S8). Moreover, it can be applied to partially degraded DNA originating from formalin-fixed paraffin-embedded tumors. Reads from 1526 samples and 811 negative controls (DNA extraction controls, 16S 5R PCR controls, and paraffin controls) were computationally combined into long amplicons, using Short MUltiple Regions Framework (SMURF) (44) and the Greengenes database as a reference. To improve taxonomic assignment, we used the Ribosomal Database Project (RDP) classifier to augment the Greengenes database by assigning a species-level taxonomy to 380,000 bacterial 16S rRNA sequences that originally lacked such taxonomy (45) (table S3 and materials and methods). Thirty-nine samples and 10 controls that had fewer than 1000 normalized reads were discarded from further analysis (materials and methods).
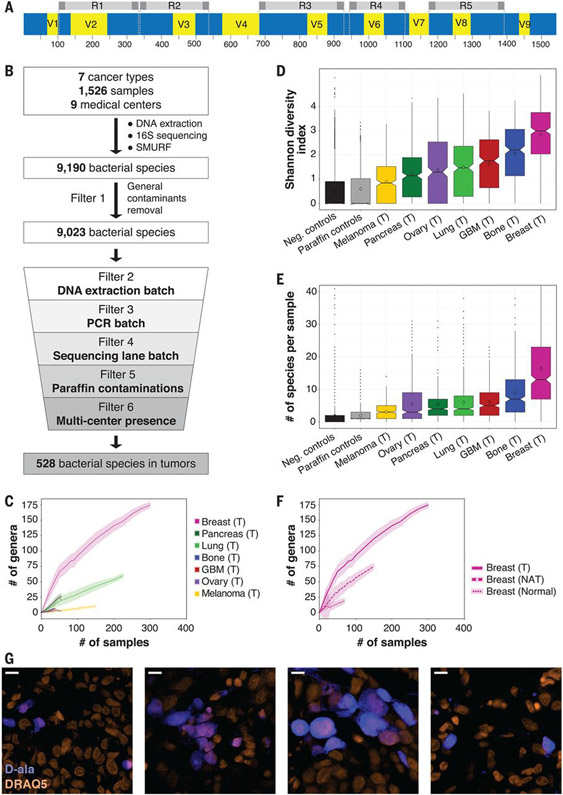
Fig. 3. The microbiome of breast tumors is richer and more diverse than that of other tumor types.
(A) Graphic representation of the bacterial 16S rRNA gene with its conserved (blue) and variable (yellow) regions. The sequence from Escherichia coli K-12 substrain MG1655 was used as a reference sequence. The five amplicons of the multiplexed 5R PCR method are depicted in gray. (B) Schematic representation of the analysis pipeline applied to 16S rDNA sequencing data. (C) Rarefaction plots showing the number of bacterial genera that passed all filters in the different tumor types per number of tumor samples that were selected for the analysis. Light color shading represents confidence intervals based on 100 random subsamplings for each number of tumor samples that was analyzed. (D) Box blot of Shannon diversity indexes of all samples, segregated by tumor type. Neg., negative. (E) Box blot of the numbers of bacterial species present in each tumor. For (D) and (E), values were calculated on rarefied data of 40 samples per tumor type, with 10 iterations. For each iteration, only bacteria that passed all filters in any of the tumor types were included in the analysis. (F) Rarefaction plots for the number of bacterial genera that passed all filters in breast tumor, breast NAT, and breast normal samples. Light color shading represents confidence intervals based on 100 random subsamplings for each number of samples that was analyzed. (G) Fluorescent images from four human breast tumors that were cultured ex vivo with fluorescently labeled D-alanine for 2 hours (blue). Nuclei were stained with DRAQ5 (orange). Scale bars, 10 μm.
Overall, we detected 9190 bacterial species across the different tumor or normal tissue types (Fig. 3B and table S2). Because some of these species may represent contamination of the samples, we applied a strict set of six filters to control for potential sources of contamination. To account for the most frequent general contaminants, filter 1 removed 167 bacterial species that were detected in >7.5% of our DNA extraction and NTC negative control samples or in the paraffin controls. This threshold demarcates the transition between most of the species that are absent or very rarely present in controls and the species that appear much more commonly in controls (fig. S9). We then applied three filters to control for batch effects that originate from DNA extraction, PCR amplification, or sequencing lane using hundreds of negative controls as a background for laboratory-borne contamination (filters 2 to 4). Filters 5 and 6 were added to control for contamination that might have been introduced to the samples before their processing in the laboratory. Filter 5 uses paraffin-only samples (without tissue) from the margins of the same paraffin blocks that were used in the study to control for contamination in the process of preparing and storing the paraffin blocks. Lastly, to account for other potential sources of medical center–specific contamination, filter 6 excluded bacteria that were not significantly enriched in a specific tumor type across multiple medical centers. Only bacteria that passed all six filters in a specific cancer type or its NAT were considered to be hits that are present in this cancer or NAT condition (Fig. 3B, table S4, materials and methods, and supplementary note).
We found that breast tumors had a richer and more diverse microbiome than all other tumor types tested (P value <10−15 for each tumor type, Wilcoxon rank sum test; Fig. 3, C and D, and figs. S10 and S11). An average of 16.4 bacterial species were detected in any single breast tumor sample, whereas the average was <9 in all other tumor types (P value <10−17 for each tumor type, Wilcoxon rank sum test; Fig. 3E and fig. S11). We also found that bacterial load and richness were higher in the breast tumor samples than those found in normal breast samples from healthy subjects. Tumor-adjacent normal breast tissue had an intermediate bacterial load and richness, between those of the breast tumor and normal samples (Fig. 3F and fig. S12). In contrast, we did not find a higher bacterial load in lung and ovarian tumors compared with their tumor-adjacent normal tissues (fig. S12).
To determine whether live bacteria are present in human tumors, we collected fresh breast tumor samples from five women undergoing breast surgery. All tissues were gently dissociated in sterile conditions, plated on 35 types of agar growth media, and incubated in both aerobic and anaerobic conditions, representing a broad span of growth conditions to accommodate a high diversity of bacteria (table S5) (46). In agreement with the positive staining of these tumors for LPS and 16SrRNA FISH (fig. S13), >1000 colonies were grown per tumor from four of the tumors, and 37 colonies were grown from one tumor. In contrast, applying the same steps of tissue dissociation and culturing protocol to five full sets of negative control plates (350 plates) using only phosphate-buffered saline (PBS) yielded only five colonies in total. Whole-genome sequencing of 474 representative colonies from all five tumors demonstrated that they represented 37 different bacterial species, 11 of which (29.7%) are bacteria that were previously detected as hits in our breast tumor cohort (table S5). Fifteen isolated species (40.5%) were detected in our breast tumor cohort but did not pass all filters. For 105 of the colonies, we could not identify the bacteria at the species level (table S5 and materials and methods). Overall, these results show that live bacteria from three main phyla—Proteobacteria, Firmicutes, and Actinobacteria—can be found in breast tumors.
To further validate the presence of live, metabolically active bacteria in human tumors, we cultured slices from four freshly resected human breast tumors ex vivo in the presence of fluorescently labeled D-alanine or dimethyl sulfoxide (DMSO) control. Although D-alanine is used by bacteria to generate peptidoglycan, an essential component of the bacterial cell wall, it is not used by mammalian cells (fig. S14) (47). We detected intracellular labeling in all four tumors, which supports the hypothesis that the tumors harbor live intracellular bacteria (Fig. 3G)
Different tumor types have distinct microbial compositions
Using a single sequencing methodology and platform for the characterization of the microbiome in multiple tumor types enabled us to directly compare the microbiomes of these tumors. Comparison of the beta-diversity between all pairs of samples within a given tumor type and across different tumor types revealed that the microbiomes of tumors of the same type tend to be more similar to each other than they are to the microbiomes of other tumor types (Fig. 4A and fig. S15). The distribution of order-level phylotypes revealed marked changes between the bacterial composition of the different tumor types (Fig. 4B and fig. S16). We added 22 colorectal tumors from one medical center to our cohort to help relate some of our findings to the known colorectal cancer microbiome (table S2) (11, 12). Consistent with previous reports, bacteria belonging to the Firmicutes and Bacteroidetes phyla were the most abundant species in colorectal tumors (Fig. 4B) (10). In contrast, Proteobacteria dominated the microbiome of pancreatic cancer, similarly to the normal duodenal microbiome makeup (16, 17, 48, 49). This may reflect a retrograde bacterial migration from the duodenum, to which the pancreatic duct opens, as we have previously reported (16). Although species belonging to the Proteobacteria and Firmicutes phyla accounted for most of the detected bacterial sequences in all cancer types, the Proteobacteria to Firmicutes (P/F) ratio appears to vary between tumor types (Fig. 4B). We also detected taxa of the Actinobacteria phylum, including the Corynebacteriaceae and Micrococcaceae families, mostly in nongastrointestinal tumors (Fig. 4B and fig. S16). These observations are in agreement with previous reports describing the microbiome of breast, lung, and ovarian cancer (2, 4, 6, 9, 14, 15, 18).
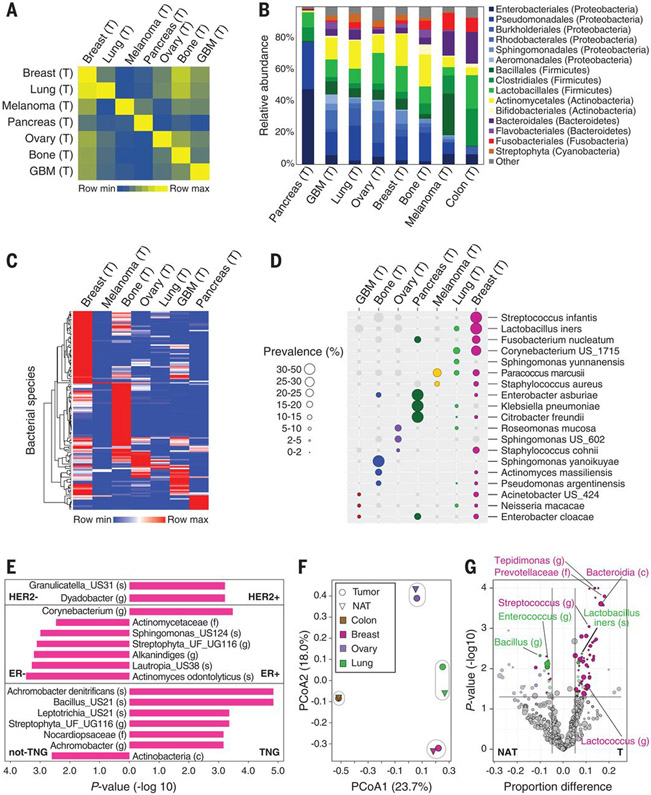
Fig. 4. Different tumor types have distinct microbial compositions.
(A) Jaccard similarity indexes were computed on the basis of profiles of bacterial species that passed all filters in tumors (n = 528) between all possible pairs of samples. The heatmap presents the average of all indexes between sample pairs from any two cancer types. (B) Distribution of order-level phylotypes across different tumor types. Relative abundances were calculated by summing up the reads of species that passed all filters in the different tumor types and belong to the same order. Orders are colored according to their associated phylum. (C) Unsupervised hierarchical clustering of the prevalence of 137 bacteria species that were hits in one of the tumor types and are present in 10% or more of the samples in at least one of the tumor types. (D) The prevalence of 19 bacteria from (C), displayed across the different tumor types. Only bacteria that are a hit in a given tumor type are represented with colored circles. Circle size indicates the prevalence level in samples. US, unknown species. (E) Bacterial taxa with a significant differential prevalence between different breast tumor subtypes are presented in a bar plot. P values were calculated using a two-sample proportion z test to compare between HER2+ (n = 61) and HER2− (n = 247), ER+ (n = 270) and ER− (n = 49), or triple negative (TNG) (n = 36) and non-TNG (n = 284) breast tumors. The direction of the bars indicates the enrichment direction. All bacteria presented had a false discovery rate (FDR)–corrected Q value <0.25. US, unknown species; UG, unknown genus; UF, unknown family; (s), species; (g), genus; (f), family; (c), class. (F) Principal coordinate analysis (PCoA) biplot on the Jaccard similarity indexes between bacterial species profiles of the different tissue types. Only bacteria that passed all filters for the specific tissue type were considered. Tumor types and their normal tissue are grouped. (G) Volcano plot demonstrating the differential prevalence of bacteria between tumors (T) and their NAT in breast, lung, and ovary samples. A two-sample proportion z test was used to calculate the P values. Sizes of dots reflect phylotype levels, gradually increasing from species to phylum. Bacteria are colored according to the tumor type (breast, pink; lung, green; and ovary, purple) if they passed significance thresholds (effect size >5%, P value <0.05, and FDR-corrected Q value <0.25).
A tumor-type distinctive microbiome composition was also apparent at the species level. Unsupervised clustering of the most prevalent intratumor bacterial species (n = 137 species) demonstrated that many of these species are enriched in certain tumor types (Fig. 4, C and D, and fig. S17). Fusobacterium nucleatum, previously reported to be enriched in colorectal tumors, was also a hit in our breast and pancreatic tumor cohorts (fig. S17). We also observed a distinct microbiome across subtypes of the same tumor type. For example, when comparing different subtypes of breast cancer according to their estrogen receptor (ER), progesterone receptor (PR), and HER2 status, we found multiple bacterial taxa whose prevalence was different between the subtypes (Fig. 4E and table S6). Lastly, although the overall microbial composition of the different tumor types was relatively similar to their NAT microbiome (Fig. 4F), we also detected bacteria with a different prevalence in tumors and in their NAT (Fig. 4G and table S7). Consistent with our observation that bacterial load and richness of breast tumors are higher than those in breast NAT (Fig. 3F and fig. S12), we found many bacteria that are significantly enriched in breast tumors compared with their NAT (Fig. 4G).
Metabolic functions encoded by intratumor bacteria are associated with clinical features of certain tumor subtypes
Our results demonstrate that intratumor bacteria span a wide spectrum of the bacterial kingdom. To investigate the functional activities of intratumor bacteria, we used the PICRUSt2 tool (50-52) to map the 16S sequences to the genes and pathways that these bacterial species may harbor (fig. S18 and tables S8 and S9).
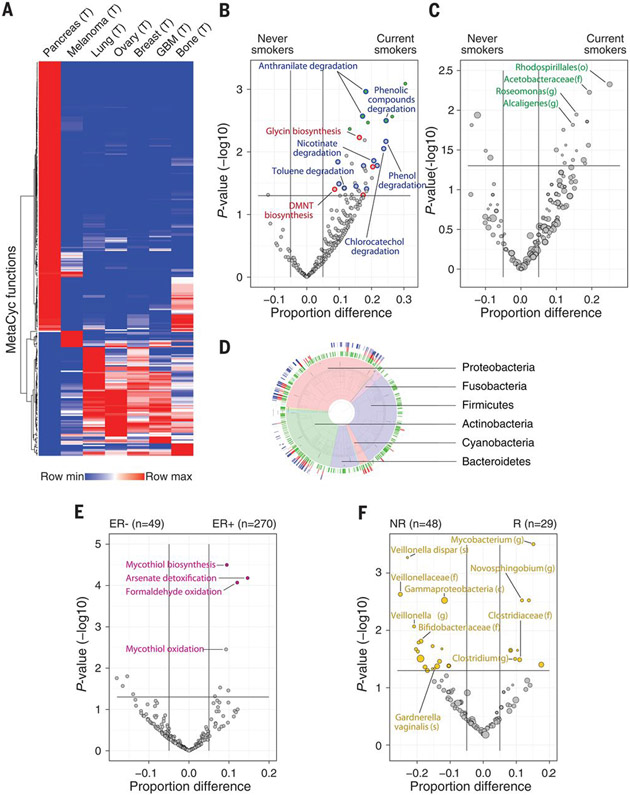
Unsupervised clustering analysis of 287 predicted metabolic MetaCyc pathways that showed the greatest variability between the tumor types revealed that certain microbiome metabolic pathways were relatively specific to certain tumor types (Fig. 5A). We found a few tumor type–specific enrichments of bacterial pathways that can degrade metabolites known to be enriched in these same tumor types (table S10). For example, degradation of hydroxyprolines by bacteria (MetaCyc PWY-5159) was enriched in bone tumors (effect size 14.6%, P value <0.01, proportion test). Bone collagen is a main source of hydroxyproline, and many bone pathologies, like bone tumors, have been shown to result in elevated hydroxyproline levels (53). In the case of lung cancer, MetaCyc pathways responsible for the degradation of chemicals in cigarette smoke, such as toluene, acrylonitrile, and aminobenzoates (TOLUENE-DEG-2-OH-PWY, P344-PWY, and PWY-6077), were significantly enriched in bacteria found in lung tumors compared with other tumor types (effect size 8.4, 8, and 7.2%, P value <0.001 for all, proportion test).
Fig. 5. Predicted bacterial metabolic functions are associated with clinical features.
(A) Heatmap demonstrating unsupervised hierarchical clustering of the frequencies of 287 MetaCyc pathways across the different tumor types. Only pathways that are abundant (frequency >10% in at least one tumor type) and variable (standard deviation divided by the average of frequencies >0.4) were included (table S10). (B and C) Volcano plots demonstrating bacterial MetaCyc pathways (B) and taxa (C) that are enriched in lung tumors from smokers (n = 100) versus never-smokers (n = 43). Effect size represents the difference in the proportion between the groups. A two-sample proportion z test was used to calculate the P values. Green filled circles indicate pathways with FDR-corrected Q values <0.25. Degrading pathways of smoke chemicals are indicated by blue circles in (B); plant-related metabolic pathways are indicated by red circles in (B). (o), order. (D) Taxonomy wheel plot of bacterial species that contributed to degradation of cigarette smoke metabolites (blue ring) and to the biosynthesis of plant metabolites functions (red ring) are indicated on the phylogenetic tree, along with all bacteria that are hits in lung tumors (green ring). (E) Volcano plot demonstrating enriched bacterial MetaCyc functions in ER+ versus ER− breast tumors. A two-sample proportion z test was used to calculate the P values. Colored circles indicate pathways with FDR-corrected Q values <0.25. (F) Volcano plot demonstrating the bacterial taxa enriched in melanoma patients who responded (R) to immune checkpoint inhibitors (ICI) versus nonresponders (NR). A binomial test was used to calculate the Pvalues for the enrichment or depletion of bacterial taxa in the responder cohort versus the nonresponder cohort. The size of dots reflects phylotype levels, gradually increasing from species to phylum. Colored circles indicate taxa with FDR-corrected Q values <0.25.
The enrichment for bacteria with the predicted capability to degrade cigarette smoke metabolites in lung tumors may suggest that high levels of these metabolites create a preferred niche for bacteria that can use these metabolites. To confirm this hypothesis, we compared the bacterial functions found in non–small cell lung cancers (NSCLCs) of 100 current smokers with those in NSCLCs of 43 people who had never smoked (never-smokers). We found that 17 of the 49 MetaCyc pathways that were significantly enriched in tumors of current smokers were pathways that degrade chemicals found in cigarette smoke, such as nicotine, anthranilate, toluene, and phenol (Fig. 5B, blue circles, and table S11). We also found eight MetaCyc pathways related to the biosynthesis of metabolites that can be used by plants—for example, for the biosynthesis of glycine, a key intermediate in plant photorespiration (Fig. 5B, red circles, and table S11). We speculate that some plant-associated bacteria, or their DNA, are present in cigarette tobacco and are thus enriched in the lung tumors of smokers.
To determine which bacteria contribute to the MetaCyc pathways that are enriched in the lung tumors of current smokers, we compared the proportion of all bacterial taxa found in lung tumors of current smokers (n = 100) with those in the tumors of never-smokers (n = 43). We found that most of the enriched taxa in the lung tumors of smokers belong to the Proteobacteria phylum. However, none of these bacteria reached significance after correction for multiple-hypothesis testing (Fig. 5C and table S12), which indicates that there was no homogeneous population of species conferring this functionality across samples. We reasoned that, although bacterial ecology differs between tumors, there is a shared functional signal related to the specific environment within the lungs of smokers. We were able to demonstrate that a very large number of heterogeneous bacteria contribute to the degradation functions of cigarette smoke metabolites and the biosynthesis of plant metabolites (Fig. 5D). Bacteria expressing these functions are found mainly in the Proteobacteria, Actinobacteria, and Cyanobacteria phyla, and they are depleted from the Firmicutes phylum (Fig. 5D).
We also found selective enrichment of bacterial functions in certain tumor subtypes. For example, multiple MetaCyc pathways were enriched in bacteria from 270 ER+ breast tumors compared with 49 ER− breast tumors (Fig. 5E and table S13). The most significantly enriched pathways in bacteria within ER+ breast tumors were arsenate detoxification and mycothiol biosynthesis. Arsenic is a Group 1 carcinogen that can increase the risk of breast cancer (54) and has been shown to induce expression of the estrogen receptor in human breast cancer (55). Mycothiol is used by bacteria to detoxify reactive oxygen species (56). Because ER+ breast tumors are known to have increased oxidative stress compared with ER− tumors (57), we hypothesize that bacteria with the ability to synthesize mycothiol can better survive in the ER+ tumor microenvironment. We also found enrichment of bacterial functions when comparing breast tumor with NAT samples (table S14). For example, enzymes related to anaerobic respiration were enriched in bacteria from breast cancer versus NAT. Overall, our analysis of MetaCyc pathways suggests a connection between the functions of bacteria present in the tumor and their tumor microenvironment.
Lastly, as our IF staining suggests (Fig. 2), bacteria can be found inside CD45+ immune cells, which indicates that they might influence or reflect the immune state of the tumor microenvironment. To determine whether a specific intratumor microbial signature is correlated with the response to immunotherapy, we compared metastatic melanomas from patients who responded to immune checkpoint inhibitors (ICI) (n = 29) with those from patients who did not respond (n = 48). Although we did not find significant changes in the load of bacteria between responders and nonresponders to ICI, we did find multiple taxa that were differentially more (n = 18) or less (n = 28) abundant in the melanomas of responders compared with nonresponders (Fig. 5F, fig. S19, and table S15). Taxa that were more abundant in tumors of responders included Clostridium, whereas Gardnerella vaginalis was more abundant in tumors of nonresponders. Notably, this is in line with differential abundances of taxa in the gut microbiome of melanoma patients responding to ICI (23-25).
Discussion
In the present study, we characterized the microbiome of 1526 samples from seven human tumor types. We took multiple measures to minimize and control for contamination (supplementary note) and used our 5R multiplexed bacterial 16S rDNA PCR sequencing technique to gain species-level resolution.
The exploration of multiple tumor types with a single platform allowed us to compare different tumor types and uncover cancer type–specific microbial signatures. This is consistent with a recent publication that demonstrated that reexamination of whole-genome and whole-transcriptome sequencing data from The Cancer Genome Atlas (TCGA) for microbial sequences identified associations between different cancer types and specific microbiota (19). Extending our analysis to the functional level demonstrated that, despite a very large variation in taxa levels, certain tumor environments are enriched for common, relevant bacterial functional traits. This observation is somewhat analogous to the relative stability of the human gut microbiome functions compared with its microbial taxa (58, 59). Using multiple visualization methods and culturomics, we were able to validate the presence of bacteria in the tumors and demonstrate their intracellular localization in both cancer and immune cells.
Our data do not establish whether intratumor bacteria play a causal role in the development of cancer or whether their presence simply reflects infections of established tumors (60, 61). As tumors develop, their disorganized, leaky vasculature may allow circulating bacteria to enter, and the immunosuppressed environment may provide a refuge for them (61, 62). Intratumor bacteria may also arise from the NAT, which can explain the high similarity we found between the tumor microbiome and its NAT microbiome. Whether or not bacteria play a causal role in tumorigenesis, it is of interest to further explore the effects that intratumor bacteria may have on different phenotypes of cancer cells and on the immune system and its interactions with tumor cells. Just as manipulation of the gut microbiome has been shown to affect the response of tumors to immune-checkpoint blockade therapy (23-25, 28), we speculate that manipulation of the tumor microbiome may also affect tumor immunity and the response to immune therapy. Thus, better understanding of these effects may pave the way for novel treatment options for cancer patients.
Supplementary Material
Acknowledgments:
We thank all members of the Straussman laboratory as well as E. Elinav, D. Douek, U. Gophna, and R. Kolter for fruitful discussions.
Funding: R.St. is funded by the Israel Science Foundation (grant no. 2044/17), the Binational Science Foundation (grant no. 2013332), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 818086), the Fabrikant-Morse Families Research Fund for Humanity, the Chantal d’Adesky Scheinberg Research Fund, the Moross Integrated Cancer Center, the Rising Tide Foundation, the International Collaboration Grant from the Jacki and Bruce Barron Cancer Research Scholars’ Program, and a partnership of the Israel Cancer Research Fund and City of Hope (COH), as supported by The Harvey L. Miller Family Foundation. R.St. is the incumbent of the Roel C. Buck Career Development Chair. N.S. is funded by the Ministry of Science, Technology and Space, Israel (grant no. 3-11174). Research reported in this publication included work performed in the COH Pathology Research Services Core supported by the National Cancer Institute of the National Institutes of Health under award no. P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A.P.C. is supported by the Cancer Prevention and Research Institute of Texas Research Training Program (RP170067).
Footnotes
Competing interests: R.St. received a grant from Merck EMD Serono and is a paid adviser to Biomica and BiomX. R.St., N.S., D.N., I.L., and G.F. are inventors on a U.S. provisional patent application (63/005,540) submitted by Yeda Research and Development, the Weizmann Institute of Science. J.A.W. and V.G. are inventors on a U.S. patent application (PCT/US17/53.717) submitted by the University of Texas MD Anderson Cancer Center that covers methods to enhance immune checkpoint blockade responses by modulating the microbiome. J.A.W. reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, Physician Education Resource, MedImmune, and Bristol-Myers Squibb. J.A.W. serves as a consultant and advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol-Myers Squibb, Merck, Biothera Pharmaceuticals, and Microbiome DX. J.A.W. also receives research support from GlaxoSmithKline, Roche/Genentech, Bristol-Myers Squibb, and Novartis.
Data and materials availability: Breast and colon samples from Sheba; melanoma samples from the Netherlands Cancer Institute–Antoni van Leeuwenhoekziekenhuis (NKI-AVL); and lung, breast, ovary, and GBM samples from the Israeli Biorepository Network for Research (MIDGAM) are available from R.St. under material transfer agreements with the Weizmann Institute. All processed data are available in the manuscript or the supplementary materials. The software package for applying SMURF using the 5R primers is available at Zenodo (63). The implementation code for the 16S sequencing analysis pipeline with filters can be found at Zenodo (64). Bacterial 16S sequencing data are available from the National Center for Biotechnology Information BioProject database under accession no. PRJNA624822.
References:
- 1.deMartel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M, Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 13, 607–615(2012). doi: 10.1016/S1470-2045(12)70137-7 pmid:22575588 [DOI] [PubMed] [Google Scholar]
- 2.Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, Lee DJ, Microbial dysbiosis is associated with human breast cancer. PLOS ONE 9, e83744 (2014).doi: 10.1371/journal.pone.0083744pmid:24421902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, Kandimalla KK, Johnson S, Yao JZ, Harrington SC, Suman VJ, Wang L, Weinshilboum RL, Boughey JC, Kocher J-P, Nelson H, Goetz MP, Kalari KR, A comprehensive analysis of breast cancer microbiota and host gene expression. PLOS ONE 12, e0188873 (2017). doi: 10.1371/journal.pone.0188873pmid:29190829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, DeMichele AM, Alwine JC, Robertson ES, Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front. Microbiol 9, 951 (2018). doi: 10.3389/fmicb.2018.00951pmid:29867857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costantini L, Magno S, Albanese D, Donati C, Molinari R, Filippone A, Masetti R, Merendino N, Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep 8, 16893 (2018). doi: 10.1038/s41598-018-35329-zpmid:30442969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, Polley EC, Bowman ED, Khan MA, Robles AI, Cooks T, Ryan BM, Padgett N, Dzutsev AH, Trinchieri G, Pineda MA, Bilke S, Meltzer PS, Hokenstad AN, Stickrod TM, Walther-Antonio MR, Earl JP, Mell JC, Krol JE, Balashov SV, Bhat AS, Ehrlich GD, Valm A, Deming C, Conlan S, Oh J, Segre JA, Harris CC,Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19, 123 (2018).doi: 10.1186/s13059-018-1501-6pmid:30143034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J, The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol. Biomark. Prev 28, 731–740 (2019). doi: 10.1158/1055-9965.EPI-18-0966pmid:30733306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apostolou P, Tsantsaridou A, Papasotiriou I, Toloudi M, Chatziioannou M, Giamouzis G, Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg 6, 137 (2011).doi: 10.1186/1749-8090-6-137pmid:21999143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, Coukos G, Robertson ES, The ovarian cancer oncobiome. Oncotarget 8, 36225–36245 (2017). doi: 10.18632/oncotarget.16717pmid:28410234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Z, Guo B, Gao R, Zhu Q, Qin H, Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol 6, 20 (2015). doi: 10.3389/fmicb.2015.00020pmid:25699023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA, Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012). doi: 10.1101/gr.126516.111pmid:22009989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M, Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012). doi: 10.1101/gr.126573.111pmid:22009990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sfanos KS, Sauvageot J, Fedor HL, Dick JD, De Marzo AM, Isaacs WB, A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 68, 306–320 (2008).doi: 10.1002/pros.20680pmid:18163428 [DOI] [PubMed] [Google Scholar]
- 14.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G, The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol 82, 5039–5048 (2016).doi: 10.1128/AEM.01235-16pmid:27342554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, Scott L, O’Hanlon DM, Burton JP, Francis KP, Tangney M, Reid G, Microbiota of human breast tissue. Appl. Environ. Microbiol 80, 3007–3014 (2014). doi: 10.1128/AEM.00242-14pmid:24610844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D,Gavert N, Zwang Y, Cooper ZA, Shee K, Thaiss CA, Reuben A, Livny J, Avraham R, Frederick DT, Ligorio M, Chatman K, Johnston SE, Mosher CM, Brandis A, Fuks G, Gurbatri C, Gopalakrishnan V, Kim M, Hurd MW, Katz M, Fleming J, Maitra A, Smith DA, Skalak M, Bu J, Michaud M, Trauger SA, Barshack I, Golan T, Sandbank J, Flaherty KT, Mandinova A, Garrett WS, Thayer SP, Ferrone CR, Huttenhower C, Bhatia SN, Gevers D, Wargo JA, Golub TR, Straussman R, Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017).doi: 10.1126/science.aah5043pmid:28912244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, Werba G, Zhang K, Guo Y, Li Q, Akkad N, Lall S, Wadowski B, Gutierrez J, Kochen Rossi JA,Herzog JW, Diskin B, Torres-Hernandez A, Leinwand J, Wang W, Taunk PS, Savadkar S, Janal M, Saxena A, Li X, Cohen D, Sartor RB, Saxena D, Miller G, The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 8, 403–416 (2018).doi: 10.1158/2159-8290.CD-17-1134pmid:29567829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, Caporaso NE, Goedert JJ, Ravel J, Landi MT, Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 17, 163 (2016). doi: 10.1186/s13059-016-1021-1pmid:27468850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, Kanbar J, Miller-Montgomery S, Heaton R, Mckay R, Patel SP, Swafford AD, Knight R, Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574 (2020). doi: 10.1038/s41586-020-2095-1pmid:32214244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman B, Gardner H, The microbiome and cancer. J. Pathol 244, 667–676 (2018).doi: 10.1002/path.5047pmid:29377130 [DOI] [PubMed] [Google Scholar]
- 21.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wu WKK, Zeng B, Chan FKL, Sung JJY, Wei H, Yu J, Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 153,1621–1633.e6 (2017). doi: 10.1053/j.gastro.2017.08.022pmid:28823860 [DOI] [PubMed] [Google Scholar]
- 22.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS, Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). doi: 10.1016/j.chom.2013.07.007pmid:23954159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA, Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). doi: 10.1126/science.aan4236pmid:29097493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, Gajewski TF, The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108(2018). doi: 10.1126/science.aao3290pmid:29302014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M,Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S,Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria J-C, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L, Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).doi: 10.1126/science.aan3706pmid:29097494 [DOI] [PubMed] [Google Scholar]
- 26.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther P-L, Eberl G, Bérard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Doré J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L, The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013).doi: 10.1126/science.1240537pmid:24264990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai R-M, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS, Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967 970 (2013). doi: 10.1126/science.1240527pmid:24264989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA, The microbiome, cancer, and cancer therapy. Nat. Med 25, 377–388 (2019). doi: 10.1038/s41591-019-0377-7pmid:30842679 [DOI] [PubMed] [Google Scholar]
- 29.Rutkowski MR, Stephen TL, Svoronos N, Allegrezza MJ, Tesone AJ, Perales-Puchalt A, Brencicova E, Escovar-Fadul X, Nguyen JM, Cadungog MG, Zhang R, Salatino M, Tchou J, Rabinovich GA, Conejo-Garcia JR, Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell 27, 27–40 (2015). doi: 10.1016/j.ccell.2014.11.009pmid:25533336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW, Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014). doi: 10.1186/s12915-014-0087-zpmid:25387460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS, Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol. 27, 105–117 (2019). doi: 10.1016/j.tim.2018.11.003pmid:30497919 [DOI] [PubMed] [Google Scholar]
- 32.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ, Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).doi: 10.1186/s40168-018-0605-2pmid:30558668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GCS, Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334 (2019). doi: 10.1038/s41586-019-1451-5pmid:31367035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ,Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A 103,12115–12120 (2006). doi: 10.1073/pnas.0605127103pmid:16880384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raetz CRH, Whitfield C, Lipopolysaccharide endotoxins. Annu. Rev. Biochem 71, 635–700 (2002). doi: 10.1146/annurev.biochem.71.110601.135414pmid:12045108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer W, in New Comprehensive Biochemistry, vol. 27, Ghuysen J-M, Hakenbeck R, Eds. (Elsevier, 1994), pp. 199–215. [Google Scholar]
- 37.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA, Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol 56, 1919–1925 (1990). doi: 10.1128/AEM.56.6.1919-1925.1990pmid:2200342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forestier C, Moreno E, Pizarro-Cerda J, Gorvel JP, Lysosomal accumulation and recycling of lipopolysaccharide to the cell surface of murine macrophages, an in vitro and in vivo study. J. Immunol 162, 6784–6791 (1999). pmid:10352299 [PubMed] [Google Scholar]
- 39.Tokuyasu KT, Application of cryoultramicrotomy to immunocytochemistry. J. Microsc 143, 139–149(1986). doi: 10.1111/j.1365-2818.1986.tb02772.xpmid:3531524 [DOI] [PubMed] [Google Scholar]
- 40.Abada A, Levin-Zaidman S, Porat Z, Dadosh T, Elazar Z, SNARE priming is essential for maturation of autophagosomes but not for their formation. Proc. Natl. Acad. Sci. U.S.A 114, 12749–12754 (2017).doi: 10.1073/pnas.1705572114pmid:29138318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klieneberger-Nobel E, Origin, development and significance of L-forms in bacterial cultures. J. Gen. Microbiol 3, 434–443 (1949). doi: 10.1099/00221287-3-3-434pmid:18147756 [DOI] [PubMed] [Google Scholar]
- 42.Errington J, L-form bacteria, cell walls and the origins of life. Open Biol. 3, 120143 (2013).doi: 10.1098/rsob.120143pmid:23303308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Errington J, Cell wall-deficient, L-form bacteria in the 21st century: A personal perspective. Biochem. Soc. Trans 45, 287–295 (2017). doi: 10.1042/BST20160435pmid:28408469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuks G, Elgart M, Amir A, Zeisel A, Turnbaugh PJ, Soen Y, Shental N, Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 6, 17 (2018).doi: 10.1186/s40168-017-0396-xpmid:29373999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB.Appl. Environ. Microbiol 72, 5069–5072 (2006). doi: 10.1128/AEM.03006-05pmid:16820507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, Bercik P, Surette MG, Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 8, 72 (2016).doi: 10.1186/s13073-016-0327-7pmid:27363992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR, (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol 8, 500–505 (2013).doi: 10.1021/cb3004995pmid:23240806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ou G, Hedberg M, Hörstedt P, Baranov V, Forsberg G, Drobni M, Sandström O, Nyunt Wai S, Johansson I, Hammarström M-L, Hernell O, Hammarström S, Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am. J. Gastroenterol 104,3058–3067 (2009). doi: 10.1038/ajg.2009.524pmid:19755974 [DOI] [PubMed] [Google Scholar]
- 49.Nistal E, Caminero A, Herrán AR, Arias L, Vivas S, de Morales JMR, Calleja S, de Miera LES, Arroyo P, Casqueiro J, Differences of small intestinal bacteria populations in adults and children with/without celiac disease: Effect of age, gluten diet, and disease. Inflamm. Bowel Dis 18, 649–656(2012). doi: 10.1002/ibd.21830pmid:21826768 [DOI] [PubMed] [Google Scholar]
- 50.Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI, PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 672295 [Preprint]. 15 June 2019. www.biorxiv.org/content/10.1101/672295v1. [Google Scholar]
- 51.Ye Y, Doak TG, A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLOS Comput. Biol 5, e1000465 (2009).doi: 10.1371/journal.pcbi.1000465pmid:19680427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louca S, Doebeli M, Efficient comparative phylogenetics on large trees. Bioinformatics 34, 1053–1055(2018). doi: 10.1093/bioinformatics/btx701pmid:29091997 [DOI] [PubMed] [Google Scholar]
- 53.Nakagawa M, Sugiura Y, Oshima T, Kajino G, Hirako H, Urinary hydroxyproline excretion in orthopedic disease, with special reference to systemic bone disease and bone tumor (secondary report). Nagoya J. Med. Sci 29, 345–367 (1967). pmid:6066099 [PubMed] [Google Scholar]
- 54.Khanjani N, Jafarnejad A-B, Tavakkoli L, Arsenic and breast cancer: A systematic review of epidemiologic studies. Rev. Environ. Health 32, 267–277 (2017). doi: 10.1515/reveh-2016-0068 pmid:28284039 [DOI] [PubMed] [Google Scholar]
- 55.Du J, Zhou N, Liu H, Jiang F, Wang Y, Hu C, Qi H, Zhong C, Wang X, Li Z, Arsenic induces functional re-expression of estrogen receptor α by demethylation of DNA in estrogen receptor-negative human breast cancer. PLOS ONE 7, e35957 (2012). doi: 10.1371/journal.pone.0035957pmid:22558281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reyes AM, Pedre B, De Armas MI, Tossounian M-A, Radi R, Messens J, Trujillo M, Chemistry and Redox Biology of Mycothiol. Antioxid. Redox Signal 28, 487–504 (2018).doi: 10.1089/ars.2017.7074pmid:28372502 [DOI] [PubMed] [Google Scholar]
- 57.Karihtala P, Kauppila S, Soini Y, Arja-Jukkola-Vuorinen, Oxidative stress and counteracting mechanisms in hormone receptor positive, triple-negative and basal-like breast carcinomas. BMC Cancer 11, 262 (2011).doi: 10.1186/1471-2407-11-262 pmid:21693047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). doi: 10.1038/nature11234 pmid:22699609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eng A, Borenstein E, Taxa-function robustness in microbial communities. Microbiome 6, 45 (2018).doi: 10.1186/s40168-018-0425-4pmid:29499759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pope JL,Tomkovich S, Yang Y, Jobin C , Microbiota as a mediator of cancer progression and therapy.Transl. Res 179, 139–154 (2017). doi: 10.1016/j.trsl.2016.07.021 pmid:27554797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cummins J, Tangney M, Bacteria and tumours: Causative agents or opportunistic inhabitants? Infect. Agent. Cancer 8, 11 (2013). doi: 10.1186/1750-9378-8-11 pmid:23537317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baban CK, Cronin M, O’Hanlon D, O’Sullivan GC, Tangney M, Bacteria as vectors for gene therapy of cancer. Bioeng. Bugs 1, 385–394 (2010). doi: 10.4161/bbug.1.6.13146 pmid:21468205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shental N, NoamShental/5R: First release, April 2020, version v1.0, Zenodo; (2020); 10.5281/zenodo.3740525. [DOI] [Google Scholar]
- 64.Livyatan I, ilivyatan/TMB: TMB for Science publication, version science_pub, Zenodo; (2020); 10.5281/zenodo.3740536. [DOI] [Google Scholar]
- 65.Barton HA, Taylor NM, Lubbers BR, Pemberton AC, DNA extraction from low-biomass carbonate rock: An improved method with reduced contamination and the low-biomass contaminant database. J. Microbiol. Methods 66, 21–31 (2006). doi: 10.1016/j.mimet.2005.10.005pmid:16305811 [DOI] [PubMed] [Google Scholar]
- 66.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD, Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4, 29 (2016). doi: 10.1186/s40168-016-0172-3pmid:27338728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weyrich LS, Farrer AG, Eisenhofer R, Arriola LA, Young J, Selway CA, Handsley-Davis M, Adler CJ, Breen J, Cooper A, Laboratory contamination over time during low-biomass sample analysis. Mol. Ecol. Resour 19, 982–996 (2019). doi: 10.1111/1755-0998.13011 pmid:30887686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ, Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 8, 24 (2016). doi: 10.1186/s13099-016-0103-7pmid:27239228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuperman AA, Zimmerman A, Hamadia S, Ziv O, Gurevich V, Fichtman B, Gavert N, Straussman R, Rechnitzer H, Barzilay M, Shvalb S, Bornstein J, Ben-Shachar I, Yagel S, Haviv I, Koren O, Deep microbial analysis of multiple placentas shows no evidence for a placental microbiome. BJOG 127, 159–169 (2020). doi: 10.1111/1471-0528.15896 pmid:31376240 [DOI] [PubMed] [Google Scholar]
- 70.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW, QuPath: Open source software for digital pathology image analysis. Sci. Rep 7, 16878 (2017). doi: 10.1038/s41598-017-17204-5pmid:29203879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). doi: 10.1038/nmeth.2019 pmid:22743772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruifrok AC, Johnston DA, Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol 23, 291–299 (2001). pmid:11531144 [PubMed] [Google Scholar]
- 73.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF, Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541 (2009). doi: 10.1128/AEM.01541-09 pmid:19801464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marco-Sola S, Sammeth M, Guigó R, Ribeca P, The GEM mapper: Fast, accurate and versatile alignment by filtration. Nat. Methods 9, 1185–1188 (2012). doi: 10.1038/nmeth.2221pmid:23103880 [DOI] [PubMed] [Google Scholar]
- 75.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W, MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). doi: 10.1093/bioinformatics/btv033 pmid:25609793 [DOI] [PubMed] [Google Scholar]
- 76.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N, Compact graphical representation of phylogenetic data and metadata with GraPhlAn.PeerJ 3, e1029 (2015). doi: 10.7717/peerj.1029 pmid:26157614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.