Abstract
DELLA proteins are the negative regulators of the gibberellin (GA) signaling pathway. GAs have a pervasive effect on plant physiology, influencing processes that span the entire life cycle of the plant. All the information encoded by GAs, either environmental or developmental in origin, is canalized through DELLAs, which modulate the activity of many transcription factors and transcriptional regulators. GAs unlock the signaling pathway by triggering DELLA polyubiquitination and degradation by the 26S proteasome. Recent reports indicate, however, that there are other pathways that trigger DELLA polyubiquitination and degradation independently of GAs. Moreover, results gathered during recent years indicate that other post-translational modifications (PTMs), namely phosphorylation, SUMOylation and glycosylation, modulate DELLA function. The convergence of several PTMs in DELLA therefore highlights the strict regulation to which these proteins are subject. In this review, we summarize these discoveries and discuss DELLA PTMs from an evolutionary perspective and examine the possibilities these and other post-translational regulations offer to improve DELLA-dependent agronomic traits.
Keywords: DELLA, O-GlcNAcylation, O-Fucosylation, Phosphorylation, SUMOylation, Ubiquitination
Introduction
DELLAs are land plant-specific proteins that belong to the GRAS family of transcriptional regulators (Hern�ndez-Garc�a et�al. 2020). The interest in DELLA proteins arose due to their participation as master negative elements in the gibberellin (GA) signaling pathway. This role was confirmed after the isolation of loss-of-function mutants in DELLA genes in Arabidopsis and in rice, which rendered constitutive GA responses like enhanced growth (Dill and Sun 2001, Ikeda et�al. 2001, King et�al. 2001, Feng et�al. 2008). Currently, we have a good understanding of the mechanism through which the GA signal is transduced in angiosperms and the role of DELLAs therein. They are polyubiquitinated and degraded by the 26S proteasome after active GAs are perceived by the soluble receptor GA-INSENSITIVE DWARF 1 (GID1), as described below (Ueguchi-Tanaka et�al. 2005, Murase et�al. 2008, Shimada et�al. 2008). Consequently, DELLAs are only allowed to accumulate in the nucleus when GA levels are low. Nonetheless, it seems that DELLAs have not always been subject to GA regulation during evolution. While DELLAs are present in early-divergent plants, suggesting that they were present in the common ancestor of land plants (Hern�ndez-Garc�a et�al. 2019), GID1 proteins, which likely evolved from carboxylesterases (Yoshida et�al. 2018), appeared later coinciding with the origin of lycophytes (Yasumura et�al. 2007).
DELLAs modulate the activity of >300 transcription factors (TFs) and transcriptional regulators through protein–protein interactions (Mar�n-de la Rosa et�al. 2014, Van De Velde et�al. 2017, Lantzouni et�al. 2020). There are different modes of DELLA action (Thomas et�al. 2016). They can inactivate TFs and transcriptional regulators by a sequestration mechanism, preventing binding either to DNA or to the TFs they regulate, respectively. The interaction with other TFs, on the contrary, occurs in the vicinity of their target promoters, where DELLAs usually act as transcriptional activators. Although the role of DELLAs in early-divergent plants has not yet been determined, it has been proposed that they could act as transcriptional activators in these species as well (Hern�ndez-Garc�a et�al. 2019).
Thanks to the ability of DELLAs to interact with other proteins and to the high sensitivity of the GA metabolism to environmental changes (Sun 2011), it is thought that DELLAs act as signaling hubs, connecting the transcriptional pathways regulating growth and defense with the environment (Claeys et al. 2014). Indeed, in silico analyses of DELLA-associated gene coexpression networks suggest that DELLAs coordinate transcriptional programs (Briones-Moreno et�al. 2017). The signaling hubs are usually tightly regulated to prevent unwanted regulation of downstream pathways. For example, the stability and activity of p53 and c-Myc, two well-known hub proteins in animals, are controlled by multiple post-translational modifications (PTMs) (Hann 2006, Kruse and Gu 2009). The post-translational control to which DELLA levels are subject by polyubiquitination in response to GAs would be in line with the close regulation expected for a signaling hub. Therefore, the identification of different PTMs of DELLAs, including GA-independent DELLA polyubiquitination and destabilization pathways, adds novel layers of regulation to the control of DELLA activity (Figs�1, 2). In this review, we aim to critically summarize the studies leading to these discoveries and to discuss the evolutionary and biotechnological implications of these post-translational regulatory mechanisms of DELLAs.
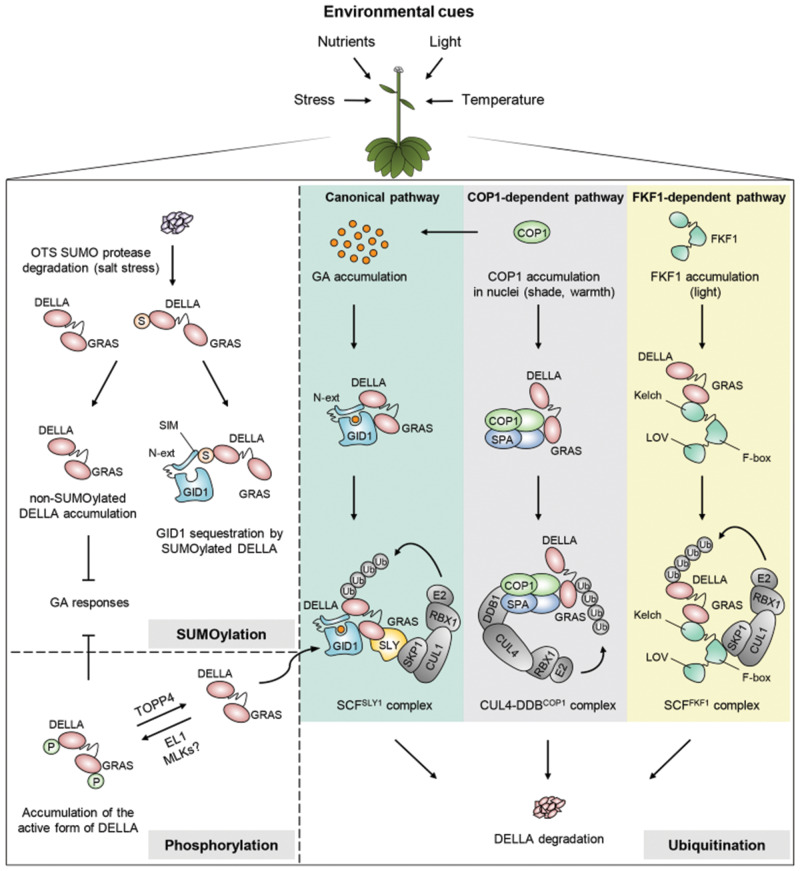
Fig. 1.
Regulatory mechanisms of DELLA stability by PTMs. Ubiquitination: (1) canonical pathway: high GA levels allow for the formation of the GA–GID1–DELLA ternary complex and the subsequent recruitment of the SCFSLY1/GID2 E3 ubiquitin complex for DELLA ubiquitination (Ub) and degradation. (2) COP1 pathway: under shade or warm temperature, COP1 is quickly re-accumulated in the nucleus to target DELLAs for 26S proteasome-mediated degradation by the CUL4–DDBCOP1 complex. Likewise, COP1 enhances GA levels, thereby promoting DELLA destabilization through the canonical pathway. (3) FKF1 pathway: under long-day photoperiods, FKF1 interacts directly with DELLAs to promote their polyubiquitination by the SCFFKF1 complex and proteolytic degradation. SUMOylation: salt stress induces OTS SUMO protease degradation and consequently leads to the accumulation of SUMOylated (S) DELLAs. SUMO-conjugated DELLAs are able to bind to the SIM site of GID1 independently of GAs. Thus, GID1 is sequestered, which allows for the accumulation of non-SUMOylated DELLAs to trigger defense responses. Phosphorylation: phosphorylation (P) mediated by EL1 (and probably by MLKs and/or other kinases) stabilizes DELLAs, whereas TOPP4-mediated dephosphorylation leads to GA/GID1-dependent DELLA degradation.
Ubiquitination: New Pathways Emerge to Regulate DELLA Stability
The covalent binding of the small protein ubiquitin to other proteins represents a major PTM in eukaryotes, one that determines protein activity or fate, including stability (Sadanandom et�al. 2012). Ubiquitin is attached to one or more lysine (K) residue of the target protein. Inter-ubiquitin chains can then be formed by the linkage of additional moieties to specific K residues at the first ubiquitin. In particular, polyubiquitin chains of at least four moieties formed by linkage at K48 lead the substrate protein to degradation by the 26S proteasome. The attachment of ubiquitin is achieved by the consecutive action of three enzymes, E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase, which is the one that recognizes the protein substrate. Among the seven types of E3 ubiquitin ligases, the RING and Skp1-Cullin-F-box (SCF) types are relevant for DELLA ubiquitination.
The first indications that DELLAs were the targets of ubiquitin-mediated degradation by the 26S proteasome came early in DELLA research. Key was the observation that their presence in the nucleus is reduced by GAs (Silverstone et�al. 2001, Itoh et�al. 2002) and that the reduction in DELLA levels is prevented by treatment with 26S proteasome inhibitors (Fu et�al. 2002). A pivotal player to understand the mechanism of DELLA destabilization was identified by various forward genetics strategies conducted in Arabidopsis and in rice (Wilson and Somerville 1995, Steber et�al. 1998, Sasaki et�al. 2003). Arabidopsis SLEEPY 1 (SLY1) and rice GID2 encode orthologous, plant-specific F-box containing proteins (McGinnis et�al. 2003, Sasaki et�al. 2003). F-box proteins are part of the SCF-type E3 ubiquitin ligases where they function as substrate adaptors (Lechner et�al. 2006). The GA insensitivity and DELLA overaccumulation found in gid2 and sly1 loss-of-function mutants (McGinnis et�al. 2003, Sasaki et�al. 2003), together with the presence of the F-box in SLY1/GID2 proteins, strongly suggested that they target DELLAs for polyubiquitination prior to degradation by the 26S proteasome. In fact, polyubiquitination of the only DELLA protein in rice, SLENDER RICE 1 (SLR1), in response to GAs was impaired in gid2 mutant plants (Sasaki et�al. 2003), while the GA insensitivity of gid2 and sly1 loss-of-function mutants was due to overaccumulation of DELLAs, as their dwarf phenotype was alleviated when combined with della loss-of-function alleles (Sasaki et�al. 2003, Dill et�al. 2004, Fu et�al. 2004). As bona fide F-box proteins, SLY1 and GID2 assemble in a SCFSLY1/GID2 complex in vivo, anchored through the F-box to the Skp1 subunit (Fu et�al. 2004, Gomi et�al. 2004), and interact with the substrate DELLA proteins (Dill et�al. 2004, Fu et�al. 2004, Gomi et�al. 2004). SLY1/GID2 is, thereby, instrumental for DELLA polyubiquitination in response to GAs.
The mechanism used by GAs to promote the polyubiquitination and degradation of DELLAs, however, was still unknown after identifying SLY1/GID2. The last piece of the puzzle was the identification of the GA receptor, GID1 (Ueguchi-Tanaka et�al. 2005). GID1 is a soluble protein with homology to hormone-sensitive lipases that binds active GAs (Ueguchi-Tanaka et�al. 2005, Nakajima et�al. 2006) and that interacts with DELLA proteins in a GA-dependent manner (Ueguchi-Tanaka et�al. 2005, Griffiths et�al. 2006, Nakajima et�al. 2006, Ueguchi-Tanaka et�al. 2007, Willige et�al. 2007). The formation of the GID1–GA–DELLA complex involves the DELLA/TVHYNP motifs of DELLAs and residues around the GA-binding pocket and lid of GID1 (Murase et�al. 2008). More importantly, it enhances the interaction between the DELLA protein and SLY1/GID2, thus favoring its recruitment to the SCFSLY1/GID2 complex for polyubiquitination and degradation (Fig.�1) (Griffiths et�al. 2006, Hirano et�al. 2010). This model is consistent with the GA resistance of DELLA mutants lacking the DELLA and/or TVHYNP motifs. In line with these results, in vitro assays showed that DELLAs promote GA binding to GID1 (Nakajima et�al. 2006) and they stabilize it as well (Ueguchi-Tanaka et�al. 2007). All together, these findings indicate that DELLAs contribute to their degradation in the presence of GAs through a feed-forward biochemical mechanism that facilitates their assembly into a protein complex suitable for their polyubiquitination and degradation by the 26S proteasome.
Recent studies have demonstrated, however, that GA-independent pathways exist for DELLA polyubiquitination and degradation. A crucial observation was that protein levels for the GA-resistant version of the Arabidopsis DELLA REPRESSOR OF ga1-3 (RGA), rga-Δ17, were reduced by the 26S proteasome when seedlings were placed in shaded or warm environments (Blanco-Touri��n et�al. 2020). The RING-type E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (Lau and Deng 2012) was considered as a prime candidate to define this new pathway for two reasons: RGA accumulates in plants with impaired COP1 function (Cagnola et�al. 2018) and, like GAs, COP1 promotes growth in shade or in warm conditions (Djakovic-Petrovic et�al. 2007, Stavang et�al. 2009, Pac�n et�al. 2013, Delker et�al. 2014, Park et�al. 2017). The reduction in rga-Δ17 levels in hypocotyls in response to shade or warm temperatures was abolished in cop1-4 mutant seedlings, indicating that it is COP1 dependent and independent of GAs (Blanco-Touri��n et�al. 2020). COP1, aided by its functional partner SUPPRESSOR of phyA-105 1 (SPA1) (Hoecker 2017), interacts with DELLAs RGA and GAI in nuclear bodies, leading to their polyubiquitination and degradation by the 26S proteasome, as demonstrated through in vitro and in vivo assays (Fig.�1) (Blanco-Touri��n et�al. 2020). What is the physiological relevance of the COP1 pathway compared to the canonical one? Do these two pathways act independently? Blanco-Touri��n et�al. (2020) proposed that, although both are required to support hypocotyl growth under these environmental conditions, the role of each pathway is different. COP1 leads to DELLA degradation, triggering it soon after the environmental change is perceived and, at the same time, it enhances GA biosynthesis (Fig.�1). On the contrary, the GA pathway sets the range of DELLA levels in the hypocotyl sensitive to COP1. The mechanism to connect this pathway with environmental changes might involve the red and far-red light photoreceptor phytochrome B (phyB), a known negative regulator of COP1 (Lu et�al. 2015, Sheerin et�al. 2015). The rapid inactivation of the phyB in response to temperature increases (Jung et�al. 2016, Legris et�al. 2016) or to shade (Casal 2013) would activate the pool of COP1 that is already in the nucleus previous to the environmental change (Park et�al. 2017), being thus available to derepress the growth restraints imposed by DELLAs. This initial growth derepression might represent an adaptive advantage for seedlings, which need to respond quickly to environmental challenges.
One recent study focused on a different physiological context, the regulation of flowering time by photoperiod, and led to the identification of another pathway triggering DELLA polyubiquitination and degradation by the 26S proteasome (Yan et�al. 2020). The Arabidopsis FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1) forms an SCFFKF1 E3 ubiquitin ligase (Song et�al. 2014), similar to SCFSLY1/GID2, which promotes flowering when the photoperiod lengthens (Imaizumi et�al. 2003). Notably, the work by Yan et�al. (2020) shows that the fkf1 mutant is less sensitive to GAs than the wild type regarding the promotion of flowering or the induction of target genes, while the opposite is observed in FKF1-overexpressing plants, this indicating a positive role for FKF1 in GA signaling (Yan et�al. 2020). In agreement with this finding, RGA protein levels overaccumulate in fkf1 mutants, while they are reduced in the overexpressing lines (Yan et�al. 2020). However, changes in RGA levels in plants with impaired FKF1 activity are independent of GAs, as demonstrated, for instance, by the additive effects of mutations ga1-3, which causes GA deficiency, and fkf1. FKF1, through its Kelch domain, is able to interact with the GRAS domain of DELLAs, leading to their polyubiquitination and degradation (Fig.�1) (Yan et�al. 2020). These results set FKF1 as a new GA-independent pathway triggering DELLA degradation by the 26S proteasome. Interestingly, despite the fact that FKF1 may act as a blue light photoreceptor (Imaizumi et�al. 2003), its role in DELLA degradation appears to be independent of the light quality (Yan et�al. 2020). Notably, FKF1 expression is positively correlated with DELLA activity, suggesting that it forms part of the feedback regulatory circuit controlling homeostasis of the GA pathway (Yan et�al. 2020).
Phosphorylation: Protecting DELLAs from Degradation
Phosphorylation of serine (S), threonine (T) and tyrosine (Y) residues is a common PTM of proteins that has a pervasive effect in cell physiology (Ubersax and Ferrell 2007). Phosphorylation of DELLAs has been studied for several years, although there is controversy over the effect of this PTM on their function. Early studies suggested that phosphorylation was a prerequisite for DELLA polyubiquitination and degradation, as demonstrated for other proteins. This hypothesis was supported by several observations: (i) treatment with Y kinase inhibitors prevented the GA-stimulated degradation of barley and Arabidopsis DELLAs (Fu et�al. 2002, Hussain et�al. 2007); (ii) GA treatments promoted the accumulation of phosphorylated SLR1 in gid2 mutants but not in wild-type plants (Sasaki et�al. 2003); and (iii) GID2 and SLY1 preferably interacted with phosphorylated forms of SLR1 and GAI in vitro (Gomi et�al. 2004) and in semi-in vivo assays (Fu et�al. 2004), respectively.
Results from other studies, however, provided contradicting information. Phosphorylation of SLR1 occurred in the wild type and also in GA-deficient rice callus (Itoh et�al. 2005). Versions of Arabidopsis RGL2 and RGA in which conserved S and T residues were changed either to the negatively charged aspartic/glutamic acid (phosphomimic) or to the conservative cysteine/alanine (de-phosphomimic) resulted in reduced or enhanced degradation in response to GA, respectively (Hussain et�al. 2005, Wang et�al. 2014). Accordingly, S/T phosphatase inhibitors blocked GA-induced DELLA degradation, whereas S/T kinase inhibitors did not have any obvious effect (Hussain et�al. 2005, Wang et�al. 2009). DELLA phosphorylation also seemed to affect DELLA activity. The overexpression of the phosphomimic RGA resulted in a GA-deficient phenotype, in agreement with the enhanced accumulation of this protein version, while it lacked the ability to activate the expression of GA biosynthesis genes (Wang et�al. 2014). Taken together, these observations suggest that phosphorylation promotes DELLA stability and also affects its activity. Nonetheless, these results should be interpreted with caution. We cannot rule out that phosphatase and/or kinase protein inhibitors affect the phosphorylation status of other regulatory elements involved in GA-induced degradation of DELLAs rather than phosphorylation of the DELLAs themselves. And, more importantly, it remains to be proven whether mutated sites in RGL2 and RGA are actually phosphorylated in vivo. To date, only RGA S18 and S21 have been shown to be phosphorylated (Fig.�3A) (Wang et�al. 2013) and they were not studied by Wang et�al. (2014). The identification of phosphorylated residues seems to be, therefore, a crucial step toward understanding the relevance of this modification for DELLA stability and activity.
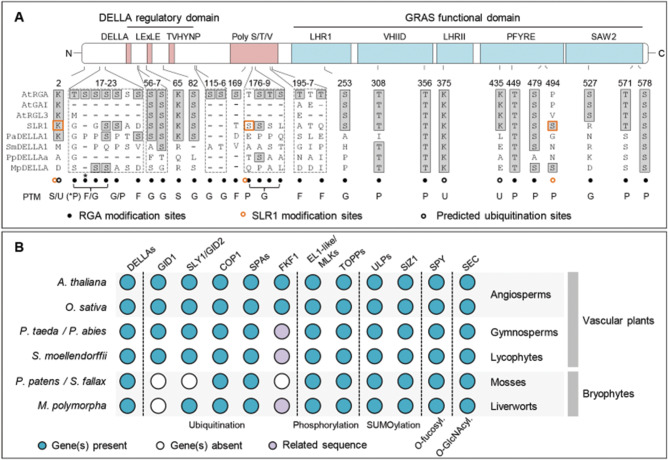
Fig. 3.
Evolutionary analysis of DELLA regulation by PTMs. (A) Position of DELLA amino acid residues that are post-translationally modified or susceptible to be. DELLA protein domains and motifs are shown above. The RGA sequence was used as a reference. Residue conservation was determined by multiple protein sequence alignment and highlighted with gray squares. All residues were experimentally analyzed in RGA (black dots) with the exception of K2, S176 and S494 (orange circles), which were identified in SLR1 (K2, S196 and S510 in the SLR1 sequence). The asterisk indicates that RGA S18 can be alternatively phosphorylated. RGA potential ubiquitination sites (black circles) were determined in silico using the MusiteDeep webtool (https://www.musite.net). (B) Presence of DELLA post-translational regulators, or their homologs, in land plants. Arabidopsis thaliana, Oryza sativa, Pinus taeda or Picea abies, Selaginella moellendorfii, P. patens or S. fallax and M. polymorpha were included in the analysis as representatives of the different land plant lineages. The presence/absence of DELLA, GID1, SLY1/GID2, COP1, SPAs, FKF1, EL1-like/MLKs and OTSs/FUG1 orthologs in most of these species was studied in previous works (Kubota et�al. 2014, Castro et�al. 2018, Liu et�al. 2018, Han et�al. 2019, Hern�ndez-Garc�a et�al. 2019, Kang and Wang 2020). Other homologs were identified by a BLASTP search and confirmed by phylogenetic analysis. Sequences were retrieved from the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) database and from both PLAZA (https://bioinformatics.psb.ugent.be/plaza/versions/gymno-plaza) and ConGenIE databases (http://congenie.org) for gymnosperms. Blue circles denote the presence of gene, purple circles denote the presence of a related sequence and white circles denote the absence of gene.
Genetic approaches in rice and Arabidopsis provided key advances in the field and led to the identification of protein kinases and phosphatases that contribute to define the phosphorylation status of DELLAs (Fig.�1). The characterization of the rice mutant early flowering 1 (el1) revealed that EL1/Hd16 (EL1 onwards), a rice ortholog of the S/T protein kinase Casein Kinase I (CKI), acts as a negative regulator of GA signaling (Dai and Xue 2010). The el1 loss-of-function mutant displayed enhanced GA responses and partially suppressed the dwarf phenotype caused by SLR1 overexpression. Dai and Xue (2010) showed that both the N- and the C-terminal domains of SLR1 were phosphorylated by EL1 in vitro, coinciding with the presence of CKI-predicted phosphorylation sites in each region (S196 and S510; Fig.�3A), and that the GA-dependent degradation of SLR1 was enhanced in the el1 mutant. Studies with phosphomimic and de-phosphomimic versions of SLR1 led these researchers to propose that EL1-mediated phosphorylation on the C-terminal domain sustains SLR1 activity, whereas the N-terminal phosphorylated form blocks the GA-mediated degradation (Fig.�1) (Dai and Xue 2010). The effect of EL1 on SLR1 phosphorylation levels in vivo remains to be determined. Four EL1-like proteins, also known as MUT9p-LIKE KINASEs (MLKs), have been identified in Arabidopsis, two of them direct interactors of RGA, although it is currently unknown whether these proteins phosphorylate DELLAs (Zheng et�al. 2018).
On the other hand, isolating the dwarf and GA-insensitive mutant topp4-1 in Arabidopsis allowed for the identification of TYPE-ONE PROTEIN PHOSPHATASE 4 (TOPP4), an ortholog of the animal protein phosphatase 1, as a phosphatase that dephosphorylates DELLAs (Qin et�al. 2014). The dwarf architecture of topp4-1 mutant plants was partially due to overaccumulated RGA and GAI, as demonstrated by the milder phenotype of topp4 rga gai mutants. Qin et�al. (2014) showed that the TOPP4 protein accumulates in response to GAs and that it dephosphorylates GAI and RGA upon interaction, this being required to destabilize both DELLAs. The TOPP4-mediated destabilization is likely dependent on the formation of the ternary GID1–GA–DELLA complex (Fig.�1) since TOPP4 overexpression could not suppress the dwarf phenotype of the GA-insensitive gai-1 mutant. TOPP4 belongs to a family of nine, highly similar members. The original topp4-1 mutation exerted a dominant negative effect that impaired GAI and RGA dephosphorylation, although its role as a positive element in GA signaling was confirmed by a knockdown line (Qin et�al. 2014). TOPP4 seems to represent a highly specific mechanism that operates only on certain DELLAs, GAI and RGA, and at certain developmental stages (Qin et�al. 2014, Yue et�al. 2016). Therefore, the set of protein phosphatases that controls the stability of the different DELLAs in specific developmental or physiological contexts must still be explored.
SUMOylation: Control of Both DELLA Stability and Activity
Protein modification by the SUMO (Small Ubiquitin-like Modifier) attachment is involved in many aspects of plant development and defense by altering the activity, localization or stability of proteins (Augustine and Vierstra 2018). SUMOylation provides the substrate with a new interaction interface that facilitates binding to proteins that contain a SUMO-interacting motif (SIM). SUMO is conjugated to a single or multiple K residues of the substrate, either as monomer or as poly-SUMO chain, following an enzymatic cascade similar to ubiquitination. In Arabidopsis, around 60% of SUMOylated proteins are modified at the consensus motif ψKxD/E (where ψ is a hydrophobic amino acid and x can be any) (Augustine and Vierstra 2018). SUMOylation is a reversible modification that may be cleaved by SUMO proteases (Morrell and Sadanandom 2019). The SUMOylation machinery is controlled by developmental and environmental signals, making SUMOylation a highly dynamic PTM that transmits information from these cues to hundreds of protein substrates. Notably, SUMO conjugates increase in response to different types of abiotic stress, such as cold, high salinity or drought (Lois and Benlloch 2018).
Several recent studies have identified a role for SUMO in the regulation of DELLA stability or activity (Conti et�al. 2014, Campanaro et�al. 2016, Gon�alves et�al. 2020). For example, salt stress induces the degradation of Arabidopsis OVERLY TOLERANT TO SALT 1 (OTS1) and OTS2 SUMO proteases (Conti et�al. 2008). The reduced root growth observed when the ots1 ots2 mutant is grown in high salt conditions led researchers to investigate if this response was caused by altered levels of DELLAs (Conti et�al. 2014). In effect, RGA accumulated to higher levels in the ots1 ots2 mutant than in the wild type, without significant changes in RGA gene expression or in GA levels, highlighting post-translational regulation by SUMOylation. Biochemical analyses proved that RGA and GAI are indeed SUMOylated at a K in the L/QKLE motif (K65 for RGA and K49 for GAI; Fig.�3A) and deSUMOylated by OTS1 (Fig.�1) and that a K49R amino acid substitution destabilizes GAI (Conti et�al. 2014). But how does SUMOylation affect DELLA stability? The GID1 receptor contains a conserved SIM site at the N terminus overlapping with the DELLA-interacting surface. This motif mediates interaction with SUMOylated RGA independently of GA, while the binding of non-SUMOylated RGA was strictly GA-dependent (Conti et�al. 2014, Nelis et�al. 2015). These observations led to a model in which salt stress promotes DELLA SUMOylation by destroying OTS1/2 proteases. SUMOylated DELLAs titrate GID1 receptors, allowing for the accumulation of non-SUMOylated DELLAs that inhibit growth and trigger defense responses (Fig.�1). A similar mechanism may operate to restrict plant organ growth under nonstress conditions. Thus, the ots1 ots2 mutant shows decreased fertility with shorter anther filaments, a phenotype that is fully restored to the wild type in the ots1 ots2 rga triple mutant (Campanaro et�al. 2016). Conversely, SUMOylation can also promote growth through DELLA destruction. For example, SUMO conjugation stabilizes SLY1 to reduce DELLA levels (Kim et�al. 2015) and it also increases COP1 activity (Lin et�al. 2016), although the relationship between COP1 SUMOylation and DELLA degradation has yet to be characterized.
SUMOylation of DELLAs also seems to modulate their interaction with other proteins. A recent study revealed that the rice SLR1 was SUMOylated at K2 but not at K60 (the position equivalent to K65 in RGA), suggesting that DELLA SUMOylation operates differently in monocots and eudicots. In turn, SLR1 can be deSUMOyated by the FUG1 SUMO protease (Gon�alves et�al. 2020). The fusion of SUMO to the K2 of SLR1 (SUMO–SLR1) disrupted SLR1 interaction with certain TFs but not with others (Fig.�2A). Rice plants overexpressing SUMO–SLR1 displayed improved tolerance to salt stress and altered expression of genes involved in the defense response and in GA metabolism (Gon�alves et�al. 2020).
Fig. 2.
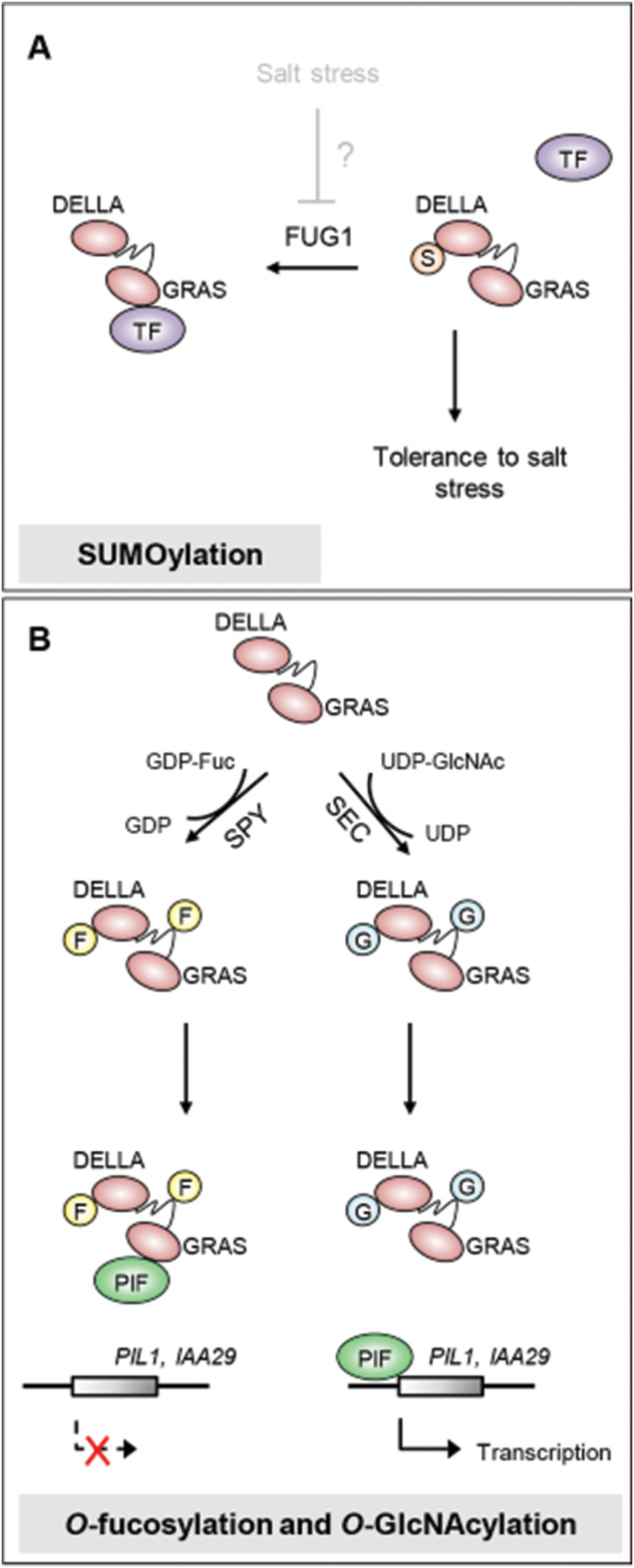
Proposed mode of action of PTMs that regulate DELLA interaction with transcription factors. (A) SUMOylation: environmental stress may promote the degradation of FUG1 SUMO protease and hence the accumulation of SUMOylated (S) SLR1 in rice. SUMO–SLR1 loses the capacity to interact with certain TFs, which results in improved tolerance to salt stress. (B) Glycosylation: SPY and SEC compete with each other to attach O-fucose (F) and O-GlcNAc (G) sugars, respectively, at the DELLA and poly S/T/V regions of RGA. O-Fucosylation promotes RGA binding to PIF and hence blocks PIF transcriptional activity. Conversely, O-GlcNAcylation impedes RGA binding to PIF, which is then allowed to activate the transcription of target genes.
Glycosylation: Sugars Adjust DELLA Activity
O-GlcNAcylation consists in the attachment of a single N-acetylglucosamine monosaccharide to the hydroxyl group of S or T residues by the O-GlcNAc transferase (OGT), which can be reversed by an O-GlcNAcase (OGA) (Yang and Qian 2017). O-GlcNAcylation serves as a nutrient and stress sensor in animals to regulate a wide variety of cellular processes, many of them through crosstalking with other PTMs (Yang and Qian 2017). A recent study has identified 262 O-GlcNAcylated proteins in Arabidopsis, indicating that this PTM is widespread in plants as well (Xu et�al. 2017).
O-GlcNAcylation regulates GA signaling by modifying DELLA proteins in Arabidopsis (Zentella et�al. 2016). Two potential OGTs were initially described in this species, SPINDLY (SPY) and SECRET AGENT (SEC) (Jacobsen et�al. 1996, Hartweck et�al. 2002), while no orthologs for OGA have been identified to date. In contrast to SEC, the role of SPY in GA signaling has been known for some time. spy loss-of-function mutants exhibit a GA-overdose phenotype and are able to germinate in the presence of paclobutrazol, an inhibitor of GA biosynthesis, pointing to a negative role in GA signaling (Jacobsen and Olszewski 1993). Accordingly, strong spy alleles are largely epistatic to GA-insensitive or deficient mutants (Jacobsen and Olszewski 1993, Wilson and Somerville 1995, Silverstone et�al. 1997, Peng et�al. 1999b, Silverstone et�al. 2007). Similar to Arabidopsis, knockdown of rice OsSPY increased internode elongation and partially rescued the dwarfism of GA-insensitive or GA-deficient mutants (Shimada et�al. 2006). Most of the current knowledge about how SPY and SEC modulate DELLA activity comes from two recent reports in Arabidopsis (Zentella et�al. 2016, Zentella et�al. 2017). For the first time in plants, Zentella et�al (2016) showed the O-GlcNAcylation of a DELLA protein, RGA (Fig.�3A highlights modified peptides detected by mass spectrometry in the aforementioned study). Although both SEC and SPY interacted with RGA in co-immunoprecipitation assays, biochemical analyses unambiguously showed that only SEC O-GlcNAcylates RGA. sec mutants displayed a semidwarf architecture and slightly reduced fertility, phenotypes that are consistent with diminished GA signaling. RGA gene expression, protein accumulation and localization were not affected in genetic backgrounds with altered SEC activity. Pull-down assays showed that O-GlcNAcylation prevents RGA interaction with the TFs BRASSINAZOLE RESISTANT 1 (BZR1), PHYTOCHROME INTERACTING FACTOR 3 (PIF3) and PIF4 that otherwise would be inactivated, i.e. not able to bind to target genes (Zentella et�al. 2016). Accordingly, genes normally upregulated by BZR1 or by PIF3/4 showed reduced expression in sec mutants. Therefore, SEC promotes GA signaling by inactivating DELLAs by O-GlcNAcylation (Fig.�2B) (Zentella et�al. 2016).
But what about SPY? Studies from the same group of researchers shed light on this, expanding the repertoire of DELLA regulation by glycosylation (Zentella et�al. 2017). Mass spectrometry analyses revealed a novel PTM of RGA, mono-O-fucosylation, consisting in the attachment of O-fucose to specific S or T residues. RGA O-fucosylation levels were decreased in spy but unaffected in sec mutants, and highly O-fucosylated RGA was recovered after the coexpression of SPY and RGA in tobacco leaves. These observations plus in vitro enzymatic assays demonstrated that SPY is an O-fucosyltransferase, which modifies RGA. O-fucosylated RGA displayed enhanced binding to BZR1, PIF3 and PIF4 and, accordingly, BZR1 or PIFs downstream targets were upregulated in spy mutants (Zentella et�al. 2017). Moreover, O-GlcNAcylation and O-fucosylation sites largely overlap in the DELLA and poly S/T/V regions of RGA (Zentella et�al. 2016, Zentella et�al. 2017). Transient expression assays in tobacco elegantly showed that SEC and SPY compete against each other to modify RGA (Zentella et�al. 2017). Overall, these observations support a model where O-GlcNAcylation by SEC and O-fucosylation by SPY antagonistically modulate DELLA function. Thus, O-GlcNAcylation decreases DELLA-binding affinity to TFs, which are now free to regulate gene expression (Fig.�2B). By contrast, O-fucosylation promotes DELLA sequestration of TFs and hence blocks downstream gene regulation (Zentella et�al. 2017). Likewise, it would be interesting to determine how glycosylation affects DELLA capacity to bind to TFs, e.g. members of the INDETERMINATE DOMAIN (IDD) family, which allow transcriptional coactivation of target genes.
Evolutionary Insights into DELLA Post-translational Regulation
Both the origin of the regulatory proteins and the conservation of the target residues are aspects to be considered in the evolution of PTMs in DELLAs. The proteins that add or remove DELLA PTMs are present in most land plant lineages (Fig.�3B). A relevant exception is the acquisition of the GA/GID1 system for DELLA ubiquitination, reported to coincide with the origin of lycophytes (Fig.�3B) (Yasumura et�al. 2007). By contrast, orthologs of COP1 and its functional partner SPA are already present in the genomes of the mosses Physcomitrella patens (Rensing et�al. 2008) and Sphagnum fallax (Leebens-Mack et�al. 2019) and of the liverwort Marchantia polymorpha (Bowman et�al. 2017). Indeed, the phytochrome signaling pathway, which would feed COP1 with environmental information, has also been demonstrated in Marchantia (Inoue et�al. 2016). The COP1 pathway might represent an ancient regulatory mechanism to establish DELLA levels before the acquisition of the GA/GID1 system. On the other hand, bona fide orthologous genes of FKF1 have not been found in gymnosperms or in early-divergent land plants (Fig.�3B) (Liu et�al. 2018), suggesting that DELLA regulation by FKF1 could operate specifically in angiosperms. Nonetheless, only the functional characterization of both the COP1 and FKF1 pathways in these plants will determine if they represent ancient mechanisms for DELLA degradation.
The regulation of DELLA activity by SUMOylation is another interesting mechanism from an evolutionary perspective. Notably, the SUMO attachment sites (K2 of rice SLR1 and K65 of Arabidopsis RGA) are conserved across angiosperms and gymnosperms, but not in early-divergent land plant lineages (Fig.�3A). By contrast, the SUMO proteases and E3 ligases are present in all land plants (Fig.�3B) and, specifically, the SIM site of GID1 is already present in lycophytes (Yoshida et�al. 2018). This suggests that the capacity to SUMOylate DELLAs was acquired in the ancestor of seed plants and then maintained during evolution as a tool to refine DELLA interactions with other proteins, including the GID1 receptor.
DELLA phosphorylation and glycosylation machineries are also conserved throughout the land plants’ lineage, making it likely that these modifications occur in early-divergent land plants as well (Fig.�3B). If this hypothesis was true, both sorts of modifications would modulate the activity of DELLAs, probably by affecting the interaction with TFs, as demonstrated for glycosylation in Arabidopsis (Zentella et�al. 2016, Zentella et�al. 2017). Regulation of DELLA stability by phosphorylation might have been recruited, eventually, coinciding with the acquisition of GID1 in lycophytes. On the other hand, the modified residues, either glycosylated or phosphorylated, are unevenly conserved (Fig.�3A). Indeed, although similar consensus O-GlcAcylation sites have been described for plant and animal proteins, the sequence surrounding the modified S/T is relatively ambiguous (Kao et�al. 2015). Therefore, glycosylation seems to be a highly specific protein modification that could depend on 3D structural cues as well.
In summary, the post-translational regulation of DELLA proteins appears to have gained in complexity throughout the evolution of land plants. However, the lack of large-scale studies into protein PTMs in different plants hinders comparative analysis from an evolutionary perspective. This information would pave the way to address fundamental questions on the evolution of DELLA functions.
Open Questions and Perspectives
As seen in the previous sections, our current knowledge about how DELLAs are regulated by PTMs is still quite fragmented. For instance, identifying which residues are ubiquitinated in response to the GID1/GA, COP1 or FKF1 pathways would provide crucial information to better dissect the relative relevance of each one for DELLA regulation. In addition, knowing the physiological context in which a post-translational regulation takes place is vital to clarify the role of DELLAs in particular processes. In this sense, we have a good understanding of the contributions of DELLA polyubiquitination or SUMOylation in response to salt stress or flowering. However, many other contexts under DELLA regulation have not been examined (e.g. response to drought, response to biotic stress, cell division, fertility, etc.). How O-GlcNAcylation and O-fucosylation are regulated by environmental or developmental cues are also issues that need to be addressed. Does SEC function as a sensor for the cellular nutrient status as OGT does in animals? And eventually, does it feed DELLAs with this information? Likewise, SPY expression is induced by drought (Qin et�al. 2011), a not surprising observation that should be further explored.
Mounting evidence indicates that crosstalk between different PTMs is crucial to fine tune protein activities (Zhang and Zeng 2020). The titration of GID1 receptors by SUMOylated DELLAs is an example of crosstalk between SUMOylation and ubiquitination (Conti et�al. 2014). Similarly, crosstalk between both PTMs at the target K might also be envisioned. The interaction of O-GlcNAcylation, and by extension O-fucosylation, with other PTMs, such as phosphorylation and ubiquitination that has been extensively studied in animals, should also be addressed in plants (Yang and Qian 2017). Indeed, among the peptides identified to be O-GlcNAcylated in Arabidopsis, 35% of them were alternatively phosphorylated (Xu et�al. 2017). Interestingly, the downregulation of OsSPY was associated with enhanced SLR1 phosphorylation in rice (Shimada et�al. 2006). In this regard, it is notable that no OGA orthologs have been identified in plants, raising questions about the reversibility of O-GlcNAcylation and the dynamics of interaction with phosphorylation and with other PTMs. The accumulation of residues susceptible to be post-translationally modified at the structurally disordered N-terminal region of DELLA proteins (Fig.�3A) also suggests the existence of heretofore unidentified crosstalk mechanisms.
An in-depth understanding of DELLA regulation by post-translational regulatory mechanisms will offer the possibility to rationally design novel DELLA alleles as tools to enhance crop yield. The inadvertent introduction of the ‘green revolution’ dwarfing DELLA alleles that boosted wheat and maize production in the 1960s (Peng et�al. 1999a) is only one example of the power of uncoupling DELLA function from a particular PTM, polyubiquitination in this case. The relevance of other PTMs in an agronomical context is just starting to emerge. Recently, it has been discovered that an OsSPY allele with reduced activity was inadvertently selected in the process of rice breeding (Yano et�al. 2019). However, the manipulation of post-translational modifiers of DELLA, or the over-stabilization of DELLAs caused by the dwarfing alleles, may impact other signaling pathways and cause undesired side effects. Therefore, the direct manipulation of DELLA residues targeted by PTMs is likely to be an appropriate strategy not only to improve the ‘green revolution’ alleles but also to design new ones with minimum side effects. Examples of this type of strategy are ‘overgrowth’ mutants that contain second-site mutations in the dwarfing Sln1d and Rht-B1 alleles of barley and wheat, respectively (Chandler and Harding 2013). One of these mutants, Sln1d.5 of barley, specifically separates DELLA effects on inflorescence size and stem elongation (Serrano-Mislata et�al. 2017). Another ‘overgrowth’ allele of wheat, Rht-B1c.17, includes an S258F substitution that results in a 30% increase in plant height, probably due to reduced DELLA activity (Chandler and Harding 2013). Of note, this substitution corresponds to S479 in RGA, a conserved residue that might be phosphorylated (Fig.�3A) (Wang et�al. 2014). These observations emphasize the interest to decode the regulation of DELLA proteins by PTMs to create ad hoc alleles to improve agriculture.
Funding
The Spanish Ministry of Science and Innovation (PID2019-109925GB-I00 to D.A.) and the European Union (H2020-MSCA-IF-2016-746396 to A.S.-M.). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Acknowledgment
The authors thank Debra Westall for proofreading the manuscript.
References
- Augustine R.C., Vierstra R.D. (2018) SUMOylation: re-wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 45: 143–154. [DOI] [PubMed] [Google Scholar]
- Blanco-Touri��n N., Legris M., Minguet E.G., Costigliolo-Rojas C., Nohales M.A., Iniesto E., et al. (2020) COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 117: 13792–13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Kohchi T., Yamato K.T., Jenkins J., Shu S., Ishizaki K., et al. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha. Genome Cell 171: 287–304. [DOI] [PubMed] [Google Scholar]
- Briones-Moreno A., Hern�ndez-Garc�a J., Vargas-Ch�vez C., Romero-Campero F.J., Romero J.M., Valverde F., et al. (2017) Evolutionary analysis of DELLA-associated transcriptional networks. Front. Plant Sci. 8: 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnola J.I., Cerd�n P.D., Pac�n M., Andrade A., Rodr�guez M.V., Zurbriggen M.D., et al. (2018) Long-day photoperiod enhances jasmonic acid-related plant defense. Plant Physiol. 178: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanaro A., Battaglia R., Galbiati M., Sadanandom A., Tonelli C., Conti L. (2016) SUMO proteases OTS1 and 2 control filament elongation through a DELLA-dependent mechanism. Plant Reprod. 29: 287–290. [DOI] [PubMed] [Google Scholar]
- Casal J.J. (2013) Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64: 403–427. [DOI] [PubMed] [Google Scholar]
- Castro P.H., Santos M.�., Freitas S., Cana-Quijada P., Louren�o T., Rodrigues M.A.A., et al. (2018) Arabidopsis thaliana SPF1 and SPF2 are nuclear-located ULP2-like SUMO proteases that act downstream of SIZ1 in plant development. J. Exp. Bot. 69: 4633–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P.M., Harding C.A. (2013) ‘ Overgrowth’ mutants in barley and wheat: new alleles and phenotypes of the ‘Green Revolution’ DELLA gene. J. Exp. Bot. 64: 1603–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., De Bodt S., Inze D. (2014) Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 19: 231–239. [DOI] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., et al. (2014) Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- Conti L., Price G., O'Donnell E., Schwessinger B., Dominy P., Sadanandom A. (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Xue H.W. (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29: 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., Sonntag L., James G.V., Janitza P., Iba�ez C., Ziermann H., et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9: 1983–1989. [DOI] [PubMed] [Google Scholar]
- Dill A., Sun T. (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T., de Wit M., Voesenek L.A., Pierik R. (2007) DELLA protein function in growth responses to canopy signals. Plant J. 51: 117–126. [DOI] [PubMed] [Google Scholar]
- Feng S., Mart�nez C., Gusmaroli G., Wang Y., Zhou J., Wang F., et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Ait-Ali T., Hynes L.W., Ougham H., Peng J., et al. (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Fleck B., Xie D., Burton N., Harberd N.P. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., et al. (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37: 626–634. [DOI] [PubMed] [Google Scholar]
- Gon�alves N.M., Fernandes T., Nunes C., Rosa M.T.G., Matiolli C.C., Rodrigues M.A.A., et al. (2020) SUMOylation of rice DELLA SLR1 modulates transcriptional responses and improves yield under salt stress. bioRxiv. 2020.03.10.986224.
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.-L., Powers S.J., et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Chang X., Zhang Z., Chen H., He H., Zhong B., et al. (2019) Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol. Plant 12: 847–862. [DOI] [PubMed] [Google Scholar]
- Hann S.R. (2006) Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin. Cancer Biol. 16: 288–302. [DOI] [PubMed] [Google Scholar]
- Hartweck L.M., Scott C.L., Olszewski N.E. (2002) Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hern�ndez-Garc�a J., Briones-Moreno A., Bl�zquez M.A. (2020) Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. doi:0.1016/j.semcdb.2020.04.009. [DOI] [PubMed] [Google Scholar]
- Hern�ndez-Garc�a J., Briones-Moreno A., Dumas R., Bl�zquez M.A. (2019) Origin of gibberellin-dependent transcriptional regulation by molecular exploitation of a transactivation domain in DELLA proteins. Mol. Biol. Evol. 36: 908–918. [DOI] [PubMed] [Google Scholar]
- Hirano K., Asano K., Tsuji H., Kawamura M., Mori H., Kitano H., et al. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. (2017) The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 37: 63–69. [DOI] [PubMed] [Google Scholar]
- Hussain A., Cao D., Cheng H., Wen Z., Peng J. (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 44: 88–99. [DOI] [PubMed] [Google Scholar]
- Hussain A., Cao D., Peng J. (2007) Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein. Planta 226: 475–483. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., et al. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306. [DOI] [PubMed] [Google Scholar]
- Inoue K., Nishihama R., Kataoka H., Hosaka M., Manabe R., Nomoto M., et al. (2016) Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell 28: 1406–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Sasaki A., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Hasegawa Y., et al. (2005) Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46: 1392–1399. [DOI] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Binkowski K.A., Olszewski N.E. (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Olszewski N.E. (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889. [DOI] [PubMed] [Google Scholar]
- Kang J., Wang Z. (2020) Mut9p-LIKE KINASE family members: new roles of the plant-specific Casein Kinase I in plant growth and development. Int. J. Mol. Sci. 21: 1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.-J., Huang C.-H., Breta�a N.A., Lu C.-T., Huang K.-Y., Weng S.-L., et al. (2015) A two-layered machine learning method to identify protein O-GlcNAcylation sites with O-GlcNAc transferase substrate motifs. BMC Bioinformatics 16: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Park B.S., Kim D.Y., Yeu S.Y., Song S.I., Song J.T., et al. (2015) E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signalling and plant development. Biochem. J. 469: 299–314. [DOI] [PubMed] [Google Scholar]
- King K.E., Moritz T., Harberd N.P. (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J.P., Gu W. (2009) Modes of p53 regulation. Cell 137: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A., Kita S., Ishizaki K., Nishihama R., Yamato K.T., Kohchi T. (2014) Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 5: 3668. [DOI] [PubMed] [Google Scholar]
- Lantzouni O., Alkofer A., Falter-Braun P., Schwechheimer C. (2020) GROWTH-REGULATING FACTORS interact with DELLAs and regulate growth in cold stress. Plant Cell 32: 1018–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. (2006) F-box proteins everywhere. Curr. Opin. Plant Biol. 9: 631–638. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J.H., Barker M.S., Carpenter E.J., Deyholos M.K., Gitzendanner M.A., Graham S.W., et al. (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M., Klose C., Burgie E.S., Rojas C.C., Neme M., Hiltbrunner A., et al. (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900. [DOI] [PubMed] [Google Scholar]
- Lin X.L., Niu D., Hu Z.L., Kim D.H., Jin Y.H., Cai B., et al. (2016) An Arabidopsis SUMO E3 Ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet. 12: e1006016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wu Y., Liao Z., Xiong J., Wu F., Xu J., et al. (2018) Evolutionary conservation and functional divergence of the LFK gene family play important roles in the photoperiodic flowering pathway of land plants. Heredity 120: 310–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois L.M., Benlloch R. (2018) Sumoylation in plants: mechanistic insights and its role in drought stress. J. Exp. Bot. 69: 4539–4554. [DOI] [PubMed] [Google Scholar]
- Lu X.D., Zhou C.M., Xu P.B., Luo Q., Lian H.L., Yang H.Q. (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- Mar�n-de la Rosa N., Sotillo B., Miskolczi P., Gibbs D.J., Vicente J., Carbonero P., et al. (2014) Large-scale identification of gibberellin-related transcription factors defines Group VII ERFs as functional DELLA partners. Plant Physiol. 166: 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., et al. (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell R., Sadanandom A. (2019) Dealing with stress: a review of plant SUMO proteases. Front. Plant Sci. 10: 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463. [DOI] [PubMed] [Google Scholar]
- Nakajima M., Shimada A., Takashi Y., Kim Y.C., Park S.H., Ueguchi-Tanaka M., et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46: 880–889. [DOI] [PubMed] [Google Scholar]
- Nelis S., Conti L., Zhang C., Sadanandom A. (2015) A functional Small Ubiquitin-like Modifier (SUMO) interacting motif (SIM) in the gibberellin hormone receptor GID1 is conserved in cereal crops and disrupting this motif does not abolish hormone dependency of the DELLA-GID1 interaction. Plant Signal. Behav. 10: e987528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pac�n M., Legris M., Casal J.J. (2013) COP1 re-accumulates in the nucleus under shade. Plant J. 75: 631–641. [DOI] [PubMed] [Google Scholar]
- Park Y.J., Lee H.J., Ha J.H., Kim J.Y., Park C.M. (2017) COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 215: 269–280. [DOI] [PubMed] [Google Scholar]
- Peng J., Richards D.E., Hartley N.M., Murphy G.P., Devos K.M., Flintham J.E., et al. (1999. a) ‘ Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Peng J., Richards D.E., Moritz T., Ca�o-Delgado A., Harberd N.P. (1999. b) Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol. 119: 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Kodaira K.S., Maruyama K., Mizoi J., Tran L.S., Fujita Y., et al. (2011) SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157: 1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Wang W., Guo X., Yue J., Huang Y., Xu X., et al. (2014) Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 10: e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H., et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Sadanandom A., Bailey M., Ewan R., Lee J., Nelis S. (2012) The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 196: 13–28. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Serrano-Mislata A., Bencivenga S., Bush M., Schiessl K., Boden S., Sablowski R. (2017) DELLA genes restrict inflorescence meristem function independently of plant height. Nat. Plants 3: 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., Zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., et al. (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., et al. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523. [DOI] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Sakamoto T., Fujioka S., Takatsuto S., Yoshida S., et al. (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48: 390–402. [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Mak P.Y., Martinez E.C., Sun T.P. (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Tseng T.S., Swain S.M., Dill A., Jeong S.Y., Olszewski N.E., et al. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Estrada D.A., Johnson R.S., Kim S.K., Lee S.Y., MacCoss M.J., et al. (2014) Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 111: 17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang J.A., Gallego-Bartolom� J., G�mez M.D., Yoshida S., Asami T., Olsen J.E., et al. (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60: 589–601. [DOI] [PubMed] [Google Scholar]
- Steber C.M., Cooney S.E., McCourt P. (1998) Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–45. [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Bl�zquez M.A., Alabad� D. (2016) DELLA proteins: master regulators of gibberellin-responsive growth and development In Annual Plant Reviews, Edited by Peter, H. and Stephen, G.T. Vol. 49 pp. 189–228. John Wiley & Sons, Ltd, Chichester, UK. [Google Scholar]
- Ubersax J.A., Ferrell J.E. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8: 530–541. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., et al. (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Velde K., Ruelens P., Geuten K., Rohde A., Van Der Straeten D. (2017) Exploiting DELLA signaling in cereals. Trends Plant Sci. 22: 880–893. [DOI] [PubMed] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., et al. (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang J., Qin Q., Yue J., Huang B., Xu X., et al. (2014) The six conserved serine/threonine sites of REPRESSOR OF ga1-3 protein are important for its functionality and stability in gibberellin signaling in Arabidopsis. Planta 240: 763–779. [DOI] [PubMed] [Google Scholar]
- Wang X., Bian Y., Cheng K., Gu L.-F., Ye M., Zou H., et al. (2013) A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteomics 78: 486–498. [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., et al. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.N., Somerville C.R. (1995) Phenotypic suppression of the gibberellin-insensitive (gai) mutant of Arabidopsis. Plant Physiol. 108: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.L., Chalkley R.J., Maynard J.C., Wang W., Ni W., Jiang X., et al. (2017) Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E1536–E1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Xinmei L., Zeng B., Zhong M., Yang J., Yang P., et al. (2020) FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis. J. Integr. Plant Biol. doi:10.1111/jipb.12971. [DOI] [PubMed] [Google Scholar]
- Yang X., Qian K. (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18: 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Morinaka Y., Wang F., Huang P., Takehara S., Hirai T., et al. (2019) GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc. Natl. Acad. Sci. USA 116: 21262–21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y., Crumpton-Taylor M., Fuentes S., Harberd N.P. (2007) Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr. Biol. 17: 1225–1230. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Tanimoto E., Hirai T., Miyanoiri Y., Mitani R., Kawamura M., et al. (2018) Evolution and diversification of the plant gibberellin receptor GID1. Proc. Natl. Acad. Sci. USA 115: E7844–E7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J., Qin Q., Meng S., Jing H., Gou X., Li J., et al. (2016) TOPP4 regulates the stability of PHYTOCHROME INTERACTING FACTOR5 during photomorphogenesis in Arabidopsis. Plant Physiol. 170: 1381–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Hu J., Hsieh W.P., Matsumoto P.A., Dawdy A., Barnhill B., et al. (2016) O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 30: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Sui N., Barnhill B., Hsieh W.P., Hu J., Shabanowitz J., et al. (2017) The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 13: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng L. (2020) Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 1: 100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Zhang F., Wang S., Su Y., Ji X., Jiang P., et al. (2018) MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 30: 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]