Abstract
Gibberellins (GAs) are a class of tetracyclic diterpenoid phytohormones that regulate many aspects of plant development, including seed germination, stem elongation, leaf expansion, pollen maturation, and the development of flowers, fruits and seeds. During the past decades, the primary objective of crop breeding programs has been to increase productivity or yields. ‘Green Revolution’ genes that can produce semidwarf, high-yielding crops were identified as GA synthesis or response genes, confirming the value of research on GAs in improving crop productivity. The manipulation of GA status either by genetic alteration or by exogenous application of GA or GA biosynthesis inhibitors is often used to optimize plant growth and yields. In this review, we summarize the roles of GAs in major aspects of crop growth and development and present the possible targets for the fine-tuning of GA metabolism and signaling as a promising strategy for crop improvement.
Keywords: Crop yield, Dormancy and germination, Gibberellins, Nitrogen use efficiency, Plant stature, Stress tolerance
Introduction
The improvement of yield is a challenge for modern agriculture due to a finite amount of arable land and an increasing world population. Consequently, improving traits related to yield and stress tolerance are urgently required to meet future demand (Sakamoto and Matsuoka 2004, Takeda and Matsuoka 2008). During the past few decades, tremendous progress has been made in understanding the molecular mechanisms underlying important agronomic traits. Gibberellins (GAs), a class of tetracyclic diterpenoid phytohormones, play an important role in modulating diverse processes throughout plant growth and development, including seed germination, stem elongation, leaf expansion, pollen maturation, and the development of flowers, fruits and seeds (Chhun et�al. 2007, Piskurewicz et�al. 2008, Aya et�al. 2009, Arnaud et�al. 2010, Fuentes et�al. 2012, Hauvermale et�al. 2012, Sakata et�al. 2014). Furthermore, the previous finding implies that GAs play a positive role in rice (Oryza sativa) heterosis at the seedling developmental stage (Ma et�al. 2011). In addition, GAs regulate plant adaptation to biotic and abiotic stresses (Dreher and Callis 2007, Colebrook et�al. 2014). As a consequence, manipulation of GA levels is often used in agricultural practice to optimize plant growth and yields.
GAs were initially identified in the pathogenic fungus Gibberella fujikuroi, the causal agent of the ‘foolish-seedling’ disease of rice, causing excessive stem elongation and falling over easily. Since its original discovery, at least 136 fully characterized GAs, named from GA1 to GA136, have been identified in plants, fungi and bacteria (MacMillan 2001, Tudzynski et�al. 2016). Among the GAs produced in plants, only a few of the GAs, such as GA1, GA3, GA4 and GA7, have biological activity as regulators of plant growth and development (Yamaguchi 2008, Hedden and Thomas 2012). Additional forms of GAs that exist in plants are precursors of the bioactive forms or deactivated metabolites. GA1 and GA4 are the two major active GAs in most plant species, although GA3 has also been identified in plants. The bioactivity of GA4 is much higher than that of GA1 in both Arabidopsis and rice, which is presumably attributed to their differences in binding affinity to the GA receptor (Cowling et�al. 1998, Ueguchi-Tanaka et�al. 2005, Magome et�al. 2013, Nomura et�al. 2013). Plants defective in GA biosynthesis or signaling show characteristic phenotypes, including dwarfism, small dark-green leaves, prolonged germination, root growth retardation, suppression of flowering, reduced seed production, and male sterility (Olszewski et�al. 2002, Lo et�al. 2008). Therefore, it is crucial for plants to tightly regulate the GA signaling pathway.
Different strategies have been employed for GA-mediated agronomic trait improvement: exogenous application of GAs or their inhibitors and genetic manipulation of their activities. These approaches have delivered promising results for improving agronomic performance in different crops. Due to the high cost of synthetic GA molecules and variability in the results, fine-tuning of the endogenous GA levels by genetic methods represents an efficient strategy for improving crop yield in a low-cost and predictable manner.
GA Metabolism and Signaling
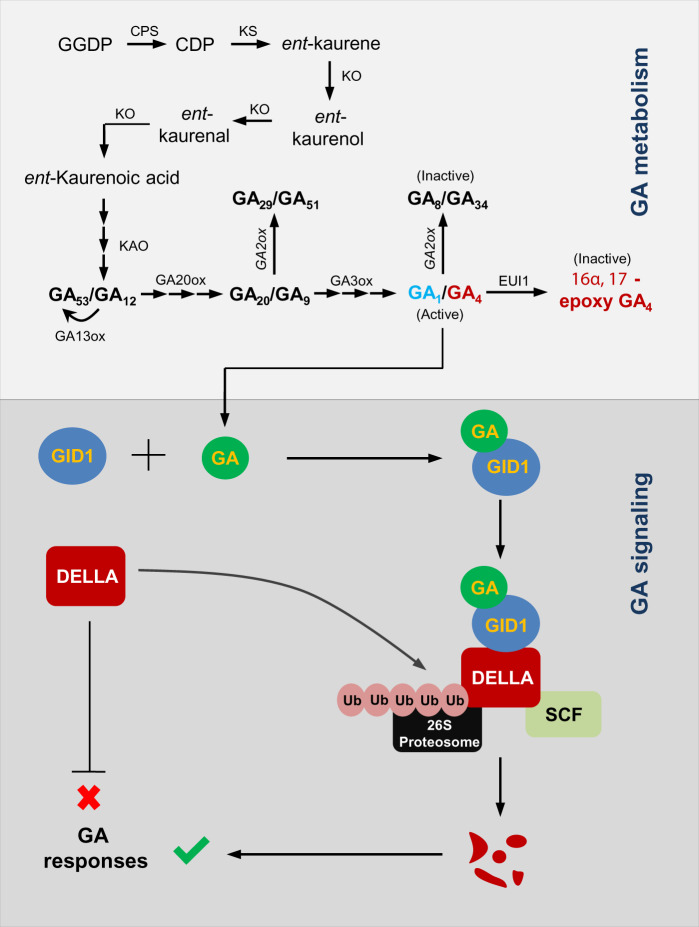
The power of molecular biology and genetics has dramatically advanced our understanding of GA synthesis and signaling (Sakamoto et�al. 2004, Sun and Gubler 2004, Ueguchi-Tanaka et�al. 2005, Chhun et�al. 2007, Xu et�al. 2014). Many genes encoding the components for GA biosynthesis and signaling pathways have been identified, and now more elaborate genetic screens are producing additional mutants that are providing new insights into this pathway (Fig.�1).
Fig. 1.
A simplified illustration of GA metabolism and signaling pathways in rice. GA biosynthesis starts from GGDP in the plastid and a portion of it is catabolized to inactive forms. Bioactive GAs are perceived by the soluble GA receptor GID1. DELLA protein acts as a negative regulator of the GA responses. In the GA signaling pathway, GA causes the destruction of DELLAs via the 26S proteasome machinery, resulting in GA responses. CDP, ent-copalyl diphosphate; CPS, ent-copalyl diphosphate synthase; GA2ox, GA2 oxidase; GA3ox, GA3 oxidase; GA20ox, GA20 oxidase; GGDP, geranylgeranyl diphosphate; KAO, ent-kaurenoic acid oxidase; KO, ent-kaurene oxidase; KS, ent-kaurene synthase; Ub, ubiquitin.
The signaling pathway from GA perception to transcriptional activation has been intensively studied over the past two decades, and its major components have been identified. The nuclear DELLA proteins, a subgroup of the GRAS transcription factors family, suppress GA signaling (Locascio et�al. 2013). Arabidopsis has five DELLA proteins, whereas rice, barley (Hordeum vulgare) and wheat (Triticum aestivum) each have only one, called SLENDER RICE1 (SLR1), SLENDER1 (SLN1) and Reduced height (Rht), respectively (Peng et�al. 1999, Ikeda et�al. 2001, Chandler et�al. 2002). The current model of GA action proposes that DELLA proteins restrain plant growth, whereas the GA promotes growth by overcoming DELLA-mediated growth restraint (Fu et�al. 2002, Ueguchi-Tanaka and Matsuoka 2010, Davi�re and Achard 2016). In rice, GA binds to the soluble receptor protein GIBBERELLIN INSENSITIVE DWARF1 (GID1), which promotes the interaction between GID1 and the transcriptional repressor DELLA protein, SLR1 (Chhun et�al. 2007, Ueguchi-Tanaka et�al. 2007, Shimada et�al. 2008, Yoshida et�al. 2018). SLR1 protein is the hub repressor of GA signaling (Hirano et�al. 2008, Hirano et�al. 2012). Loss-of-function mutants of the rice SLR1 gene have a slender phenotype with elongated leaf and stem (Ikeda et�al. 2001, Itoh et�al. 2002). F-box protein GIBBERELLIN INSENSITIVE DWARF2 (GID2) is involved in the formation of the Skp–Cullin–F-box (SCF) E3 ubiquitin ligase complex that polyubiquitinates DELLA protein for its subsequent degradation by the proteasome (Sasaki et�al. 2003, Gomi et�al. 2004, Hirano et�al. 2010).
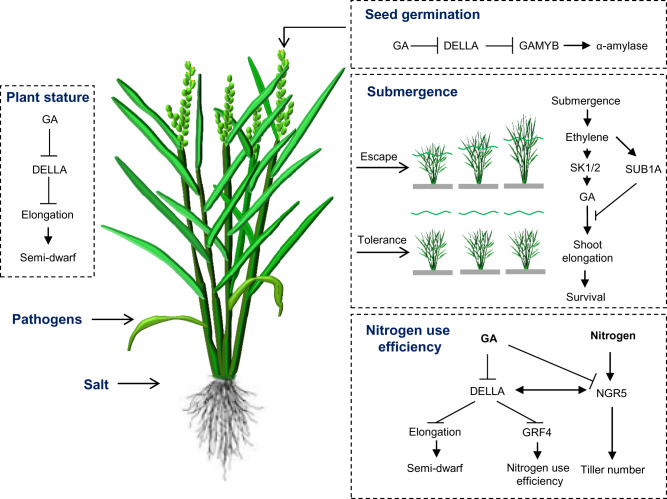
GA biosynthesis and signaling are critical to plant growth and development, and of vital importance in agriculture. In this review, we summarize the physiological effects of GAs on different rice agronomic traits, including plant stature, response to nutrient availability, seed dormancy and germination, and stress tolerance (Fig.�2).
Fig. 2.
GAs are crucial for agronomic traits and stress tolerance in crops. GAs act as growth-promoting phytohormones that play important roles throughout plant development, including plant stature, response to nutrient availability, seed germination, and stress tolerance.
The Roles of GAs in Agronomic Traits
Plant stature
The development of semidwarf varieties was the most significant achievement in the 20th century of agricultural practices and was one of the cornerstones of so-called ‘Green Revolution’, which led to a massive increase in both wheat and rice yields (Peng et�al. 1999, Silverstone and Sun 2000, Khush 2001, Ashikari et�al. 2002, Boss and Thomas 2002, Sasaki et�al. 2002, Spielmeyer et�al. 2002, Evenson and Gollin 2003, Ordonio et�al. 2014). The semidwarf habit is governed by GA biosynthetic gene semidwarf1 (sd1) in rice and GA signaling gene Rht in wheat. The sd1 gene was first identified in the Chinese variety Dee-geo-woo-gen (DGWG) and was crossed in the early 1960s with Peta (tall) to develop the semidwarf cultivar IR8, which produced record yields throughout Asia, and formed the basis for the development of new high-yielding, semidwarf plant types. Since the 1960s, sd1 is extensively used in modern rice cultivars (Spielmeyer et�al. 2002, Asano et�al. 2007, Tu and Wang 2019). Recessive semidwarfism sd1 gene in rice is the most important in a shortened culm with improved lodging and a greater harvest index, allowing for the increased use of nitrogen fertilizers (Jennings 1964). The sd1 gene was isolated and characterized to encode a GA20-oxidase isoenzyme (GA20ox2) that catalyzes the conversion of GA53 to GA20, leading to changes in GA content (Sasaki et�al. 2002). GA53 is over-accumulated, and GA20 is scarce in sd1 plants. Correspondingly, most mutants or knockdown lines of GA biosynthesis genes, including CPS, KS, KAO, KO, GA20oxs, and GA3oxs, also exhibit dwarfism phenotypes, which results in improved lodging resistance, a valuable trait for rice breeding under high inputs (Sakamoto et�al. 2004, Qin et�al. 2013). Meanwhile, overexpression of GA catabolism genes, GA2oxs, also leads to dwarfism (Lo et�al. 2008, Shan et�al. 2014). It was found that SPINDLY, participating in both BR and GA responses, regulates the elongation of lower internodes, which is correlated with levels of SPINDLY expression (Shimada et�al. 2006).
In wheat, the introduction of mutant dwarfing alleles at Rht-B1 and Rht-D1 loci led to large increases in grain yields worldwide in the 1960s, owing to the improvement in both harvest index and lodging resistance. Since then, Rht-1 dwarfism alleles are widely used in breeding modern wheat cultivars. The wheat Green Revolution genes Rht-B1 and Rht-D1 encode mutated DELLA proteins that have a reduced affinity for the GA receptor GID1 (Peng et�al. 1999). DELLA proteins act as repressors of GA signaling and are destabilized by GA. The mutant DELLA protein confers semidominant GA-insensitive dwarfism. Even though wheat has a hexaploid genome, these dominant alleles still cause obvious dwarf phenotypes. Similarly, mutations in the DELLA proteins were also identified as Sln1 in barley (Peng et�al. 1999, Chandler et�al. 2002).
In maize, genes defective in GA biosynthesis and signaling, including anther ear1, dwarf1 (d1), d3, Dwarf8 (D8) and D9, show various levels of plant height reduction (Bensen et�al. 1995, Winkler and Helentjaris 1995, Peng et�al. 1999, Cassani et�al. 2009, Teng et�al. 2013). The expression domains of GA2ox1 and KN1 (a maize KNOX gene) overlap mainly at the base of the shoot apical meristem (Bolduc and Hake 2009), and the KN1 directly induces GA2ox1 expression in reproductive meristems (Bolduc and Hake 2009). Meristem activity and maintenance processes are regulated via KNOX-mediated GA and cytokinin crosstalk (Wu et�al. 2016).
Genetic and functional analyses of the semidwarf genes not only greatly improved our understanding of the biosynthesis and signaling of GAs but also provided a powerful strategy for manipulating the plant height of crops. Although widespread cultivation of GA-deficient semidwarf varieties was successful in breeding, developing new varieties for further increasing the yield of crops is once again faced with the challenge (Hirano et�al. 2017). It was found that higher lodging resistance could be achieved by improving the mechanical strength of the culm by identifying genes related to culm quality and introducing these genes into high-yielding rice cultivars (Li et�al. 2003, Hirano et�al. 2017).
In addition to semidwarf varieties, another contributor to rice yield increase is the application of heterosis. Interestingly, GA plays an indispensable role in hybrid rice seed production, since rice male sterile lines have a common defect in the elongation of the uppermost internode, leading to incomplete panicle exsertion, which blocks pollination (Luo et�al. 2006, Zhu et�al. 2006). Consequently, exogenous GA has to be used to amend the panicle enclosure. The rice Elongated Uppermost Internode1 (EUI1) gene was found to encode a cytochrome P450 monooxygenase that inactivates 13-H GAs by epoxidation in anthers and might regulate the influx of GA4 into the stems from panicles (Luo et�al. 2006, Zhu et�al. 2006, Gao et�al. 2016, Gao and Chu 2018). Because of its prospective application to amend the panicle enclosure, the EUI1 locus has been incorporated into many male sterile lines, and this recessive trait, along with the male sterile line, the maintainer line and the restorer line, is referred to as four genetic elements for hybrid rice seed production. Therefore, fully understanding the molecular mechanism through which EUI1 activity is regulated may greatly help breeders in improving hybrid rice seed production (Gao et�al. 2016, Gao and Chu 2018, Xie et�al. 2018). Consistent with the involvement of GAs in the control of plant architecture, GA biosynthesis inhibitors paclobutrazol and uniconazole are also often used to modulate plant growth responses (Ikeda et�al. 2001, Gao et�al. 2017).
It should be pointed out that the GA dwarf mutants tend to exhibit negative pleiotropic effects, which limit their breeding application (Lo et�al. 2008). As changes in GA content cause a mix of positive and negative effects on traits of agronomic importance (e.g. reduced seed yield in rice), it is of utmost importance to determine how one GA effect could be uncoupled from another. When GA catabolism gene OsGA2ox1 was expressed under the control of the constitutively expressed Actin promoter, the resulting plants were dwarf with reduced grain yields, because GA is required for flower development (Sakamoto et�al. 2003). By expressing OsGA2ox1 under the promoter of the GA biosynthesis gene OsGA3ox2, which expressed specifically in the upper internode, dwarf, high-yielding rice could be produced (Sakamoto et�al. 2003). This result showed that tissue and cell-specific expression patterns of biosynthesis or deactivating enzymes can be used for the fine-tuning of plant growth. Thus, novel GA-related mutants with mild mutation effects and the breeding potential need to be further explored.
Response to nutrient availability
The Green Revolution represented by the breeding and wide application of semidwarf crops greatly promotes agricultural yield, but it also unfortunately led to the problem of reduced nitrogen use efficiency (NUE) of crops (Wu et�al. 2020). Nitrogen is the most imperative element for the proper growth and development of plants. Crop yields are heavily dependent on sufficient nitrogen nutrition that is provided primarily through the application of inorganic fertilizers. Because Green Revolution varieties are relatively insensitive to nitrogen, which is adverse in efficient nitrogen utilization of crops, thus, these varieties require more nitrogen fertilizer input. How to reduce the input of nitrogen fertilizer in agricultural production and continuously increase the grain yield has become a major challenge, which needs to be addressed in sustainable agriculture.
Rice GROWTH-REGULATING FACTOR 4 (GRF4) transcription factor was initially isolated as a major QTL in controlling grain size in three independent groups (Che et�al. 2015, Duan et�al. 2015, Hu et�al. 2015). It has been shown to participate in brassinosteroid signaling and can interact with DELLA protein (Che et�al. 2015). Later on, it has been further proved to play an important role in the GA signaling pathway (Li et�al. 2018). GRF4ngr2 allele is semidominant for increasing uptake. This allelic site was introduced into the current high-yield rice variety 9311 and the high-yielding wheat variety Kenong 199. The near-isogenic line 9311-OsGRF4ngr2 exhibited increased leaf and culm widths, enhanced NUE and grain yield, but plant height was not significantly changed. Similarly, GRF4ngr2 transgenic wheat plants also exhibited increased NUE and grain yield but no change in plant height (Li et�al. 2018). Therefore, the high accumulation of GRF4 protein enables high yielding with increased NUE.
APETALA2-domain transcription factor NITROGEN-MEDIATED TILLER GROWTH RESPONSE 5 (NGR5) is a key gene controlling tiller number changes under different nitrogen conditions (Wu et�al. 2020). NGR5 is a target of GA receptor GID1-promoted proteasomal destruction. DELLA protein can competitively bind the GA receptor, GID1 protein, inhibits the GA-mediated degradation of NGR5 protein and then increases the stability of NGR5 protein (Wu et�al. 2020). In addition to being negatively regulated by GA and its receptor GID1, NGR5 is responsive to nitrogen and overexpression of NGR5 in the semidwarf background significantly improves rice yields under low nitrogen conditions. NGR5 suppresses branching inhibitory genes, such as D14 and OsSPL14, through nitrogen-dependent recruitment of polycomb repressive complex 2 that promotes histone H3 lysine 27 tri-methylation in the regions harboring the branching suppressors (Wu et�al. 2020). The high-level accumulation of NGR5 does not change the semidwarf traits of the ‘Green Revolution’. Instead, it can increase the tiller number of rice and promotes the uptake and utilization of nitrogen fertilizer (Wu et�al. 2020). Thus, these studies uncover a new mechanism of GA signaling and enlighten novel breeding strategies for Green Revolution by breeding high-yield crops with enhanced NUE.
In addition, recent advances indicate that GAs substantially modulate the responses to other nutrient availabilities (Wild et�al. 2016, Eggert and von Wiren 2017). In Arabidopsis, e.g. the spatial distribution of GA-regulated DELLA growth repressors in roots adapts the root system architecture and the iron-uptake machinery to the plant’s iron demand (Wild et�al. 2016). Phosphate starvation root architecture is modulated by the GA-DELLA signaling pathway (Jiang et�al. 2007). In addition, DELLAs participate in potassium uptake under conditions of potassium scarcity (Oliferuk et�al. 2017).
Seed dormancy and germination
Seed dormancy and germination are two distinct developmental steps in the life cycle of plants and important for agricultural production. Seed dormancy is a mechanism by which seeds can inhibit their germination to wait for more favorable conditions. A high level of dormancy in cereal seeds can cause slow and uneven postharvest germination of seeds. Therefore, for cultivated crops, domestication of low dormancy cultivars over a long period ensures a higher and uniform emergence rate after sowing. However, lack of adequate seed dormancy leads to preharvest sprouting, which severely reduces both grain yield and quality (Fang et�al. 2008). Hence, the suitable dormancy extent allows uniformity of postharvest germination of seeds as it prevents the problems of sprouting and is generally regarded as a desirable trait (Fang et�al. 2008, Tuan et�al. 2018). Abscisic acid (ABA) and GAs act antagonistically in the control of seed dormancy and germination (Xie et�al. 2006, Fang et�al. 2008, Fang and Chu 2008, Penfield 2017, Du et�al. 2018, Xu et�al. 2019). ABA positively regulates the induction and maintenance of dormancy, while GAs are known as promoters of germination (Koornneef et�al. 2002, Shu et�al. 2016).
In barley, GA acts by promoting the activity of the GAMYB transcription factor, which induces the expression of genes, including α-amylase, β-glucanase, cathespin protease and β-glucosidase, promoting nutrient mobilization and cell wall loosening (Gubler et�al. 1995, Gubler et�al. 2002, Woodger et�al. 2003). Inhibition of SLN1 leads to derepression of α-amylase even in the absence of GA (Zentella et�al. 2002), and the sln1 mutant is characterized by nondormant seeds with high amylase activity in aleurone (Chandler 1988). The degradation of DELLA by GA activates GAMYB, which in turn induces the transcription of α-amylase in the aleurone of barley and rice seeds via binding to the GA-responsive elements present in its promoter (Gubler et�al. 1995, Gubler et�al. 1999, Sun and Gubler 2004). GAMYB can also interact synergistically with other transcription factors and regulate the expression of α-amylase in barley (Zou et�al. 2008). A recent study suggests that GAMYB might regulate GATA-type zinc finger transcription factor GATA12 that acts as a positive regulator of seed dormancy (Ravindran et�al. 2017). Moreover, reactive oxygen species (ROS) have been reported to have effects on ABA and GA metabolism, which eventually influence seed dormancy in barley (Bahin et�al. 2011).
Similarly, seed dormancy and germination in wheat are also reported to be associated with changes in seed sensitivity to GAs, which is influenced by the expression of genes encoding the GA signaling components (Penfield 2017, Guo et�al. 2018). Inhibition of the germination of nondormant seeds of wheat is associated with increased expression of Rht1, suggesting the role of GA signaling or seed GA sensitivity in the induction of dormancy (Izydorczyk et�al. 2018). Wheat allelic variations in the Rht1 loci are able to produce different levels of seed dormancy (Izydorczyk et�al. 2018). Dormancy decay in wheat seeds due to imbibition at lower than optimal temperature is related to enhanced expression of TaGA3ox2 and increased levels of bioactive GA in the embryo. While inhibition of germination at suboptimal temperature is associated with decreased expression of TaGA20oxs and TaGA3ox2 and reduced bioactive GA levels in both embryo and endosperm (Kashiwakura et�al. 2016, Izydorczyk et�al. 2018).
In rice, ABA can inhibit GA-inducible responses, such as the expression of α-amylase by regulating WRKY transcription factors (Xie et�al. 2006). AP2 domain-containing transcription factor OsAP2-39 in rice directly promotes transcription of the ABA biosynthesis gene OsNCED1 and expression of the GA deactivation gene EUI1, thus enhancing ABA biogenesis and impairing GA accumulation (Yaish et�al. 2010, Shu et�al. 2018). Overexpression of OsAP2-39 leads to increased seed dormancy, indicating that OsAP2-39 plays a pivotal role in regulating the ABA/GA ratio. Likewise, the expression of the GA biosynthetic genes (GA20ox and GA3ox2) is induced while that of GA-inactivating gene (GA2ox) is repressed during the imbibition of nondormant rice seeds, and this has been shown to be associated with an increased level of bioactive GA (Ye et�al. 2015, Magwa et�al. 2016). OsGA20ox2 and OsGA2ox3�were identified as candidate genes for controlling seed germination, and loss-of-function mutation in OsGA20ox2 leads to reduced seed GA level and enhanced dormancy (Ye et�al. 2015, Magwa et�al. 2016). However, further genetic analyses are required for a detailed understanding of the physiological roles of GA metabolic genes in the regulation of seed dormancy in cereals.
Abiotic stress tolerance
There is increasing evidence for the involvement of GA signaling in growth suppression or promotion, depending on the response to a particular abiotic stress (Colebrook et�al. 2014, Illouz-Eliaz et�al. 2019). By modifying GA levels and signaling, plants are able to regulate and coordinate both growth and stress tolerance to better survival under unfavorable conditions.
The involvement of GA in response to abiotic stress is supported by rice varieties adapted to escape or tolerate flooding. Unanticipated flooding challenges plant growth and fitness in natural and agricultural ecosystems. Rice varieties adapted to shallow, long-lived floods commonly employ an escape strategy, in which submergence triggers rapid internode elongation. Upon submergence, these varieties have evolved an ethylene-driven and GA-enhanced module that regulates the growth of submerged organs. Internode elongation is triggered by the upregulation of the ethylene response factors SNORKEL1 and SNORKEL2, which directly or indirectly leads to an increase in bioactive GA levels (Hattori et�al. 2009). GA promotes rapid growth in dark-grown plant tissues and in deepwater rice during flooding. This response allows the shoot to outgrow the floodwaters. On the other hand, the rice Sub1 locus controls the tolerate strategy of rice varieties that are adapted to short-lived, deep floods. Upon submergence, rice plants carrying the Sub1A gene will not activate an escape response. Instead, shoot elongation is restricted and carbohydrate resources are conserved for utilization in regrowth when the flood recedes. This restriction of elongation growth is associated with increased levels of the rice DELLA protein SLR1 and the negative regulator of GA signaling, SLR1-LIKE1 (Fukao et�al. 2006, Xu et�al. 2006). Breeding programs have introduced the submergence tolerance trait into high-yielding rice varieties, which are expected to produce enough grains to feed millions of people in flood-prone areas. More recently, a transcriptional gain-of-function allele of the SD1 gene can trigger rapid stem elongation in deepwater rice, enabling it to survive under adverse flooding conditions (Kuroha et�al. 2018). Interestingly, the SD1 gene has been co-opted several times to permit rice cultivation in highly contrasting production systems via decreasing the enzymatic activity in one case and enhancing the transactivation in another case. The capacity of SD1 to function in such diverse roles in cultivated rice highlights the intrinsic complexity and molecular plasticity of plant adaptation strategies (Kuroha et�al. 2018).
GAs also can respond to salt stress. Under high salinity, plants rapidly reduce the growth and developmental program to enhance the survival rate. Salt stabilizes DELLA proteins, and DELLA-deficient plants are less tolerant to salt stress (Achard et�al. 2006, Achard et�al. 2008). Analysis of growth parameters and salt tolerance in a range of DELLA mutants indicated a strong correlation between plant height, time to flowering transition, and susceptibility to severe salt stress, suggesting that DELLA proteins may restrain growth and enhance stress tolerance through a common mechanism. In rice, the ubiquitin-binding protein OsDSK2a interacts with EUI1 and promotes its degradation to maintain GA homeostasis under normal conditions, thereby contributing to vegetative growth. Under salt stress, OsDSK2a protein levels decrease, thereby releasing EUI1 protein and allowing it to decrease bioactive GA levels, leading to inhibited plant growth and increased survival rate (Wang et�al. 2020).
Thus, it is important at times for a plant to have enough GA to promote developmental changes, and important for a plant to limit GA production or response to survival under unfavorable environments. Further characterization of the molecular mechanisms regulating GA synthesis, signaling and action will facilitate the modification in GA biosynthetic pathways to breed crops with enhanced abiotic stress tolerance.
Biotic stress tolerance
Pathogen attack is one of the major limiting factors to crop productivity and ultimately food security. GAs also play important roles in triggering the plant immune signaling network (De Bruyne et�al. 2014). In rice, GAs enhance susceptibility against the pathogens Xanthomonas oryzae pv. oyrzae (Xoo, causing bacterial blight) and Magnaporthe oryzae (Mo, causing rice blast) (Yang et�al. 2008). Mutants deficient in GA biosynthesis and signaling show resistance phenotypes when plants are treated with Xoo or Mo (Yang et�al. 2008, Qin et�al. 2013, De Vleesschauwer et�al. 2016). Transgenic rice overexpressing EUI1 accumulates low levels of GA and SA and displays enhanced resistance to Mo and Xoo, whereas loss-of-function mutations in EUI1 are more vulnerable to these pathogens (Yang et�al. 2008). The OsGA20ox3 knockdown lines show enhanced resistance against rice pathogens Mo and Xoo and increased expression of defense-related genes. On the contrary, OsGA20ox3 overexpressing plants are more susceptible to these pathogens compared with the wild-type plants. The susceptibility of plants to Xoo is increased by exogenous application of GA3 (Qin et�al. 2013).
It has also been shown that foliar spray of plants with a low concentration of GA enhanced nematode infection (Yimer et�al. 2018). GAs antagonize jasmonate (JA)-induced defense against Meloidogyne graminicola in rice. GA signaling suppresses JA-mediated defense against M. graminicola, and likewise, the JA-induced defense against M. graminicola requires SLR1-mediated repression of the GA pathway (Yimer et�al. 2018). GAs are positive players in resistance against the necrotrophic root pathogen Pythium graminicola (De Vleesschauwer et�al. 2012). SLR1-overaccumulating mutants show increased susceptibility to the necrotroph P. graminicola but enhanced resistance to the (hemi)biotrophs Xoo and Mo (Tanaka et�al. 2006, De Vleesschauwer et�al. 2012). SLR1 serves as the main target of JA-mediated growth inhibition and immunity and is required for the expression of JA-inducible rice genes (Yang et�al. 2012).
Conclusions and Perspectives
Over the past decades, significant progress has been made in elucidating the GAs’ metabolism and signaling pathways in both model plants and crops. Accumulating results indicate that GAs are strongly involved in tuning a number of agronomically important traits, including plant stature, nutrient use efficiency, seed dormancy and germination, and tolerance to various abiotic and biotic stresses. These have continuously delivered the promising targets for seed yield and stress tolerance improvement of crops. Elucidation of the role of GA signaling in these responses would be an important step toward understanding and improving crop growth and stress responses under adverse environmental conditions. A recent report that natural variations in the maximal quantum yield of photosystem II also associate with the GA signaling pathway, indicating that GAs also participate in photosynthesis, adding to the list of potentially innovative applications in agriculture (Hamdani et�al. 2019). GA is a mobile signal in plants, and developmental and adaptive growth processes require the mobility of GA to take place properly (Regnault et�al. 2015, Regnault et�al. 2016, Binenbaum et�al. 2018). Hence, spatial control is an additional layer of regulation that plants exert over their GA signaling, which can be manipulated to improve plant growth and stress responses (Regnault et�al. 2015, Binenbaum et�al. 2018). The molecular basis of GA mobility and long-distance coordination are needed to be investigated further.
The relatively high cost of synthetic GA molecules and variability in the results have discouraged the use of exogenous GA in agriculture. By contrast, modulation of endogenous GA levels by genetic engineering represents an efficient strategy for improving crop yield in a uniform and predictable manner. New approaches, such as functional genomics and CRISPR/Cas (clustered regularly interspaced short palindromic repeat/CRISPR-associated protein) genome editing systems, promise even more rapid progress in unraveling the physiology and biochemistry of GA-regulated processes (Chen et�al. 2019, Hu et�al. 2019). Using CRISPR-based editing, de novo mutations could be generated, which facilitates the directed evolution of GA biosynthesis and signaling genes and accelerates the trait development of crops. In addition, the identified GA effectors can be engineered to boost crop productivity and food security, which is an important priority to cope with a constantly changing world. Therefore, combining with new technologies, such as genome-wide association study, genome editing and other genetic engineering techniques, for improved productivity in agronomically important crops will have great potential.
Funding
The Major Program of Guangdong Basic and Applied Research (2019B030302006) and the Chinese Academy of Sciences (QYZDJ-SSW-SMC014).
Acknowledgments
We thank Dr. Jiuyou Tang for assistance in the figure preparation. We apologize to colleagues whose work could not be discussed and cited owing to space limitations.
Disclosures
The authors have no conflicts of interest to declare.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94. [DOI] [PubMed] [Google Scholar]
- Achard P., Renou J.-P., Berthom� R., Harberd N.P., Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660. [DOI] [PubMed] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T.A., Lawrenson T., et al. (2010) Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24: 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Takashi T., Miura K., Qian Q., Kitano H., Matsuoka M., et al. (2007) Genetic and molecular analysis of utility of sd1 alleles in rice breeding. Breed. Sci. 57: 53–58. [Google Scholar]
- Ashikari M., Sasaki A., Ueguchi-Tanaka M., Itoh H., Nishimura A., Datta S., et al. (2002) Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘green revolution’. Breed. Sci. 52: 143–150. [Google Scholar]
- Aya K., Ueguchi-Tanaka M., Kondo M., Hamada K., Yano K., Nishimura M., et al. (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahin E., Bailly C., Sotta B., Kranner I., Corbineau F., Leymarie J. (2011) Crosstalk between reactive oxygen species and hormonal signalling pathways regulates grain dormancy in barley. Plant Cell Environ. 34: 980–993. [DOI] [PubMed] [Google Scholar]
- Bensen R.J., Johal G.S., Crane V.C., Tossberg J.T., Schnable P.S., Meeley R.B., et al. (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binenbaum J., Weinstain R., Shani E. (2018) Gibberellin localization and transport in plants. Trends Plant Sci. 23: 410–421. [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss P.K., Thomas M.R. (2002) Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416: 847–850. [DOI] [PubMed] [Google Scholar]
- Cassani E., Bertolini E., Badone F.C., Landoni M., Gavina D., Sirizzotti A., et al. (2009) Characterization of the first dominant dwarf maize mutant carrying a single amino acid insertion in the VHYNP domain of the dwarf8 gene. Mol. Breed. 24: 375–385. [Google Scholar]
- Chandler P.M. (1988) Hormonal regulation of gene expression in the ‘‘slender” mutant of barley (Hordeum vulgare L.). Planta 175: 115–120. [DOI] [PubMed] [Google Scholar]
- Chandler P.M., Marion-Poll A., Ellis M., Gubler F. (2002) Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Chen X., Tian X., Xue L., Zhang X., Yang S., Traw M.B., et al. (2019) CRISPR-based assessment of gene specialization in the gibberellin metabolic pathway in rice. Plant Physiol. 180: 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T., Aya K., Asano K., Yamamoto E., Morinaka Y., Watanabe M., et al. (2007) Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19: 3876–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. (2014) The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Bot. 217: 67–75. [DOI] [PubMed] [Google Scholar]
- Cowling R.J., Kamiya Y., Seto H., Harberd N.P. (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 117: 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davi�re J.-M., Achard P. (2016) A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9: 10–20. [DOI] [PubMed] [Google Scholar]
- De Bruyne L., H�fte M., De Vleesschauwer D. (2014) Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 7: 943–959. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Seifi H.S., Filipe O., Haeck A., Son Nguyen H., Demeestere K., et al. (2016) The DELLA Protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice. Plant Physiol. 170: 1831–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I.-R., et al. (2012) Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99: 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Xu F., Fang J., Gao S., Tang J., Fang S., et al. (2018) Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. Plant J. 95: 545–556. [DOI] [PubMed] [Google Scholar]
- Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., et al. (2015) Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2: 15203. [DOI] [PubMed] [Google Scholar]
- Eggert K., von Wiren N. (2017) Response of the plant hormone network to boron deficiency. New Phytol. 216: 868–881. [DOI] [PubMed] [Google Scholar]
- Evenson R.E., Gollin D. (2003) Assessing the impact of the green revolution, 1960 to 2000. Science 300: 758–762. [DOI] [PubMed] [Google Scholar]
- Fang J., Chai C., Qian Q., Li C., Tang J., Sun L., et al. (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 54: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Chu C. (2008) Abscisic acid and the pre-harvest sprouting in cereals. Plant Signal. Behav. 3: 1046–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Ait-Ali T., Hynes L.W., Ougham H., Peng J., et al. (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S., Ljung K., Sorefan K., Alvey E., Harberd N.P., �stergaard L. (2012) Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 24: 3982–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Xu K., Ronald P.C., Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Chu C. (2018) Fine-tuning of Eui1: breaking the bottleneck in hybrid rice seed production. Mol. Plant 11: 643–644. [DOI] [PubMed] [Google Scholar]
- Gao S., Fang J., Xu F., Wang W., Chu C. (2016) Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 28: 680–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Zhang Y., He Z., Fu X., Li J., Li C., et al. (2017) Gibberellins. In Hormone Metabolism & Signaling in Plants. Vol. 4, pp. 107–160. Elsevier. [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., et al. (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37: 626–634. [DOI] [PubMed] [Google Scholar]
- Gubler F., Chandler P.M., White R.G., Llewellyn D.J., Jacobsen J.V. (2002) Gibberellin signaling in barley aleurone cells. control of SLN1 and GAMYB expression. Plant Physiol. 129: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Kalla R., Roberts J.K., Jacobsen J.V. (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7: 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Raventos D., Keys M., Watts R., Mundy J., Jacobsen J.V. (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17: 1–9. [DOI] [PubMed] [Google Scholar]
- Guo G., Liu X., Sun F., Cao J., Huo N., Wuda B., et al. (2018) Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell 30: 796–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdani S., Wang H., Zheng G., Perveen S., Qu M., Khan N., et al. (2019) Genome-wide association study identifies variation of glucosidase being linked to natural variation of the maximal quantum yield of photosystem II. Physiol. Plant. 166: 105–119. [DOI] [PubMed] [Google Scholar]
- Hattori Y., Nagai K., Furukawa S., Song X.-J., Kawano R., Sakakibara H., et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030. [DOI] [PubMed] [Google Scholar]
- Hauvermale A.L., Ariizumi T., Steber C.M. (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol. 160: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012) Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Hirano K., Asano K., Tsuji H., Kawamura M., Mori H., Kitano H., et al. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Kouketu E., Katoh H., Aya K., Ueguchi-Tanaka M., Matsuoka M. (2012) The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 71: no–453. [DOI] [PubMed] [Google Scholar]
- Hirano K., Ordonio R.L., Matsuoka M. (2017) Engineering the lodging resistance mechanism of post-Green Revolution rice to meet future demands. Proc. Jpn. Acad. Ser. B: Physic. Biol. Sci. 93: 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Ueguchi-Tanaka M., Matsuoka M. (2008) GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13: 192–199. [DOI] [PubMed] [Google Scholar]
- Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., et al. (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8: 1455–1465. [DOI] [PubMed] [Google Scholar]
- Hu X., Cui Y., Dong G., Feng A., Wang D., Zhao C., et al. (2019) Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces. Sci. Rep. 9: 19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., et al. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illouz-Eliaz N., Ramon U., Shohat H., Blum S., Livne S., Mendelson D., et al. (2019) Multiple gibberellin receptors contribute to phenotypic stability under changing environments. Plant Cell 31: 1506–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izydorczyk C., Nguyen T.N., Jo S., Son S., Tuan P.A., Ayele B.T. (2018) Spatiotemporal modulation of abscisic acid and gibberellin metabolism and signalling mediates the effects of suboptimal and supraoptimal temperatures on seed germination in wheat (Triticum aestivum L.). Plant. Cell Environ. 41: 1022–1037. [DOI] [PubMed] [Google Scholar]
- Jennings P.R. (1964) Plant type as a rice breeding objective. Crop Sci. 4: 13–15. [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145: 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwakura Y-I., Kobayashi D., Jikumaru Y., Takebayashi Y., Nambara E., Seo M., et al. (2016) Highly sprouting-tolerant wheat grain exhibits extreme dormancy and cold imbibition-resistant accumulation of abscisic acid. Plant Cell Physiol. 57: 715–732. [DOI] [PubMed] [Google Scholar]
- Khush G.S. (2001) Green revolution: the way forward. Nat. Rev. Genet. 2: 815–822. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Bentsink L., Hilhorst H. (2002) Seed dormancy and germination. Curr. Opin. Plant Biol. 5: 33–36. [DOI] [PubMed] [Google Scholar]
- Kuroha T., Nagai K., Gamuyao R., Wang D.R., Furuta T., Nakamori M., et al. (2018) Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 361: 181–185. [DOI] [PubMed] [Google Scholar]
- Li S., Tian Y., Wu K., Ye Y., Yu J., Zhang J., et al. (2018) Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Qian O., Zhou Y.H., Yan M.X., Sun L., Zhang M., et al. (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.-F., Yang S.-Y., Chen K.-T., Hsing Y.-I., Zeevaart J.A., Chen L.-J., et al. (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A., Bl�zquez M.A., Alabad� D. (2013) Genomic analysis of DELLA protein activity. Plant Cell Physiol. 54: 1229–1237. [DOI] [PubMed] [Google Scholar]
- Luo A., Qian Q., Yin H., Liu X., Yin C., Lan Y., et al. (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47: 181–191. [DOI] [PubMed] [Google Scholar]
- Ma Q., Hedden P., Zhang Q. (2011) Heterosis in rice seedlings: its relationship to gibberellin content and expression of gibberellin metabolism and signaling genes. Plant Physiol. 156: 1905–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan J. (2001) Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20: 387–442. [DOI] [PubMed] [Google Scholar]
- Magome H., Nomura T., Hanada A., Takeda-Kamiya N., Ohnishi T., Shinma Y., et al. (2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA 110: 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwa R.A., Zhao H., Xing Y. (2016) Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.). BMC Genet. 17: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Magome H., Hanada A., Takeda-Kamiya N., Mander L.N., Kamiya Y., et al. (2013) Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes. Plant Cell Physiol. 54: 1837–1851. [DOI] [PubMed] [Google Scholar]
- Oliferuk S., Rodenas R., Perez A., Martinez V., Rubio F., Maria G.E.S. (2017) How DELLAs contribute to control potassium uptake under conditions of potassium scarcity? Hypotheses and uncertainties. Plant Signal. Behav. 12: e1366396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N., Sun T-P., Gubler F. (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonio R.L., Ito Y., Hatakeyama A., Ohmae-Shinohara K., Kasuga S., Tokunaga T., et al. (2014) Gibberellin deficiency pleiotropically induces culm bending in sorghum: an insight into sorghum semi-dwarf breeding. Sci. Rep. 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2017) Seed dormancy and germination. Curr. Biol. 27: R874–R878. [DOI] [PubMed] [Google Scholar]
- Peng J., Richards D.E., Hartley N.M., Murphy G.P., Devos K.M., Flintham J.E., et al. (1999) ‘ Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Liu J., Zhao W., Chen X., Guo Z., Peng Y. (2013) Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol. Plant Microbe Interat. 26: 227–239. [DOI] [PubMed] [Google Scholar]
- Ravindran P., Verma V., Stamm P., Kumar P.P. (2017) A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Regnault T., Davi�re J.-M., Achard P. (2016) Long-distance transport of endogenous gibberellins in Arabidopsis. Plant Signal. Behav. 11: e1110661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnault T., Davi�re J.-M., Wild M., Sakvarelidze-Achard L., Heintz D., Bergua E.C., et al. (2015) The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat. Plants 1: 15073. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Matsuoka M. (2004) Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 15: 144–147. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Miura K., Itoh H., Tatsumi T., Ueguchi-Tanaka M., Ishiyama K., et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134: 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Morinaka Y., Ishiyama K., Kobayashi M., Itoh H., Kayano T., et al. (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat. Biotechnol. 21: 909–913. [DOI] [PubMed] [Google Scholar]
- Sakata T., Oda S., Tsunaga Y., Shomura H., Kawagishi-Kobayashi M., Aya K., et al. (2014) Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164: 2011–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Shan C., Mei Z., Duan J., Chen H., Feng H., Cai W. (2014) OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One 9: e87110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., et al. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523. [DOI] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Sakamoto T., Fujioka S., Takatsuto S., Yoshida S., et al. (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48: 390–402. [DOI] [PubMed] [Google Scholar]
- Shu K., Liu X.-D., Xie Q., He Z.-H. (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9: 34–45. [DOI] [PubMed] [Google Scholar]
- Shu K., Zhou W., Yang W. (2018) APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 217: 977–983. [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Sun T. (2000) Gibberellins and the green revolution. Trends Plant Sci. 5: 1–2. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W., Ellis M.H., Chandler P.M. (2002) Semidwarf (sd-1), ‘‘green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.-P., Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223. [DOI] [PubMed] [Google Scholar]
- Takeda S., Matsuoka M. (2008) Genetic approaches to crop improvement: responding to environmental and population changes. Nat. Rev. Genet. 9: 444–457. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Matsuoka M., Kitano H., Asano T., Kaku H., Komatsu S. (2006) gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant. Cell Environ. 29: 619–631. [DOI] [PubMed] [Google Scholar]
- Teng F., Zhai L., Liu R., Bai W., Wang L., Huo D., et al. (2013) ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J. 73: 405–416. [DOI] [PubMed] [Google Scholar]
- Tu C., Wang F. (2019) Sixty years’ devotion to green revolution and a life time commitment to food security—review on the academic achievements of Huang Yaoxiang, father of semi-dwarf rice breeding. Guangdong Agric. Sci. 46: 1–7. [Google Scholar]
- Tuan P.A., Kumar R., Rehal P.K., Toora P.K., Ayele B.T. (2018) Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 9: 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B., Studt L., Rojas M.C. (2016) Gibberellins in fungi, bacteria and lower plants: biosynthesis, function and evolution. Annu. Plant Rev. 49: 121–151. [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Matsuoka M. (2010) The perception of gibberellins: clues from receptor structure. Curr. Opin. Plant Biol. 13: 503–508. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M. (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198. [DOI] [PubMed] [Google Scholar]
- Wang J., Qin H., Zhou S., Wei P., Zhang H., Zhou Y., et al. (2020) The ubiquitin-binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 32: 414–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M., Davi�re J.-M., Regnault T., Sakvarelidze-Achard L., Carrera E., Lopez Diaz I., et al. (2016) Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Dev. Cell 37: 190–200. [DOI] [PubMed] [Google Scholar]
- Winkler R.G., Helentjaris T. (1995) The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell 7: 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger F.J., Millar A., Murray F., Jacobsen J.V., Gubler F. (2003) The role of GAMYB transcription factors in GA-regulated gene expression. J. Plant Growth Regul. 22: 176–184. [Google Scholar]
- Wu K., Wang S., Song W., Zhang J., Wang Y., Liu Q., et al. (2020) Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367: eaaz2046. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang Y., Mi X.-F., Shan J.-X., Li X.-M., Xu J.-L., et al. (2016) The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 12: e1006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhang Y., Han J., Luo J., Li G., Huang J., et al. (2018) The intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of ELONGATED UPPERMOST INTERNODE1 in rice. Mol. Plant 11: 720–735. [DOI] [PubMed] [Google Scholar]
- Xie Z., Zhang Z.L., Zou X., Yang G., Komatsu S., Shen Q.J. (2006) Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46: 231–242. [DOI] [PubMed] [Google Scholar]
- Xu F., Tang J., Gao S., Cheng X., Du L., Chu C. (2019) Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 100: 1036–1051. [DOI] [PubMed] [Google Scholar]
- Xu H., Liu Q., Yao T., Fu X. (2014) Shedding light on integrative GA signaling. Curr. Opin. Plant Biol. 21: 89–95. [DOI] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., et al. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708. [DOI] [PubMed] [Google Scholar]
- Yaish M.W., El-Kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.-M., et al. (2010) The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yang D.-L., Li Q., Deng Y.-W., Lou Y.-G., Wang M.-Y., Zhou G.-X., et al. (2008) Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant 1: 528–537. [DOI] [PubMed] [Google Scholar]
- Yang D.-L., Yao J., Mei C.-S., Tong X.-H., Zeng L.-J., Li Q., et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Feng J., Zhang L., Zhang J., Mispan M.S., Cao Z., et al. (2015) Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169: 2152–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer H.Z., Nahar K., Kyndt T., Haeck A., Van Meulebroek L., Vanhaecke L., et al. (2018) Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in rice. New Phytol. 218: 646–660. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Tanimoto E., Hirai T., Miyanoiri Y., Mitani R., Kawamura M., et al. (2018) Evolution and diversification of the plant gibberellin receptor GID1. Proc. Natl. Acad. Sci. USA 115: E7844–E7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Yamauchi D., Ho T-hD. (2002) Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14: 2289–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nomura T., Xu Y., Zhang Y., Peng Y., Mao B., et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Neuman D., Shen Q.J. (2008) Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol. 148: 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]