SUMMARY
The colonic microbiota exhibits cross-sectional heterogeneity, but the mechanisms that govern its spatial organization remain incompletely understood. Here we used Citrobacter rodentium, a pathogen that colonizes the colonic surface, to identify microbial traits that license growth and survival in this spatial niche. Previous work showed that during colonic crypt hyperplasia, type III secretion system (T3SS)-mediated intimate epithelial attachment provides C. rodentium with oxygen for aerobic respiration. However, we find that prior to the development of colonic crypt hyperplasia, T3SS-mediated intimate attachment is not required for aerobic respiration, but for hydrogen peroxide (H2O2) respiration using cytochrome c peroxidase (Ccp). The epithelial NADPH oxidase NOX1 is the primary source of luminal H2O2 early after C. rodentium infection and is required for Ccp-dependent growth. Our results suggest that NOX1-derived H2O2 is a resource that governs bacterial growth and survival in close proximity to the mucosal surface during gut homeostasis.
eTOC blurb
Miller et al. use Citrobacter rodentium to investigate how type III secretion system-mediated epithelial attachment provides a selective growth benefit prior to the onset of intestinal inflammation. This intimate attachment provides C. rodentium with access to host-derived hydrogen peroxide, which fuels pathogen growth through cytochrome c peroxidase-mediated anaerobic respiration.
Graphical Abstract
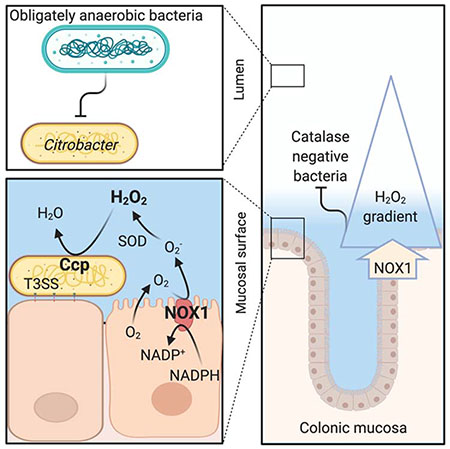
INTRODUCTION
By supplying the habitat, the host wields a prominent influence over microbial growth at body surfaces (Byndloss et al., 2018; Foster et al., 2017). Host-derived resources that limit the range of successful traits within the microbiota can be viewed as habitat filters (Costello et al., 2012; Tiffany and Baumler, 2019). These host control mechanisms are important for shaping the size, species composition, and spatial organization of microbial communities (Litvak and Baumler, 2019), but in many cases their identity remains to be determined.
One habitat filter that shapes the microbiota composition in the large intestine is epithelial hypoxia, which selects for microbial traits that permit survival and growth by maintaining anaerobiosis, thereby driving the species composition towards a dominance of obligate anaerobic bacteria (Litvak et al., 2018; Litvak et al., 2017; Rigottier-Gois, 2013; Rivera-Chavez et al., 2017). The importance of this host control mechanism for maintaining the microbiota composition was first identified during studies investigating how enteric pathogens use their virulence factors to invade the gut ecosystem (Lopez et al., 2016; Rivera-Chavez et al., 2016). Type III secretion systems (T3SSs) of the facultative anaerobic pathogens Salmonella enterica and Citrobacter rodentium (phylum Proteobacteria) shift epithelial metabolism from oxidative phosphorylation in the mitochondria to aerobic glycolysis (the conversion of glucose into lactate even in the presence of oxygen), thereby abrogating epithelial hypoxia in the colon (Carson et al., 2019; Gillis et al., 2018; Gillis et al., 2019; Lopez et al., 2016; Rivera-Chavez et al., 2016). The consequent increase in epithelial oxygenation drives a luminal pathogen expansion through aerobic respiration (Lopez et al., 2016; Rivera-Chavez et al., 2016), thereby causing a dysbiotic shift in the microbiota composition from obligate to facultative anaerobes (Lupp et al., 2007; Stecher et al., 2007). These findings suggest that pathogens are useful tools for microbiome research, because their virulence factors target host-derived habitat filters that are important for balancing the microbiota (Spiga et al., 2017).
In addition to functions that shape the species composition, the large intestine features host control mechanisms that structure the spatial organization of microbial communities. The lumen of the large intestine is not a homogenous environment, but the colonic microbiota exhibits cross-sectional heterogeneity in its spatial organization (Albenberg et al., 2014; Yasuda et al., 2015). The loose outer mucus layer is thought to be enriched in bacteria utilizing host-derived mucin, such as certain species belonging to the Verrucomicrobia and Bacteroidetes phyla (Berry et al., 2015; Li et al., 2015). Closer to the epithelium, host-derived oxygen radiates from the epithelial surface, which is thought to increase the proportion of “oxygen-tolerant” organisms of the Proteobacteria and Actinobacteria phyla (Albenberg et al., 2014), thereby helping to maintain an inner mucus layer that is largely devoid of bacteria (Johansson et al., 2008). However, our mechanistic understanding of the biogeography in the colon is still incomplete and it is likely that additional elements contributing to this process remain to be identified.
To elucidate spatial relationships between bacteria and their hosts, we used C. rodentium, a luminal pathogen that uses a T3SS to intimately attach to the epithelial surface in the cecum and colon of mice (Schauer and Falkow, 1993a, b). The T3SS is required for intestinal colonization by C. rodentium, suggesting that intimate attachment to the epithelial surface provides the pathogen with resources to compete with the gut microbiota (Kamada et al., 2012). The T3SS triggers a histopathological lesion, termed colonic crypt hyperplasia (Barthold et al., 1976), within a week after infection, thereby increasing epithelial oxygenation to enable the pathogen to respire oxygen using cytochrome bd oxidase (Lopez et al., 2016). However, prior to the development of intestinal lesions, the T3SS provides a growth advantage that is independent of cytochrome bd oxidase (Lopez et al., 2016). We reasoned that prior to the development of histopathological lesions, T3SS-mediated intimate attachment might provide C. rodentium access to unknown resources that license bacterial growth in close proximity to the epithelial surface. The goal of this study was to identify these resources.
RESULTS
The C. rodentium T3SS drives early growth using cytochrome c peroxidase
Salmonella enterica serovar (S.) Typhimurium uses its T3SSs to trigger intestinal inflammation (Tsolis et al., 1999), thereby orchestrating global changes in the environment inhabited by the microbiota (Rivera-Chavez et al., 2016; Winter et al., 2010). As a result, inflammation triggered by wild-type S. Typhimurium can rescue luminal growth of an avirulent S. Typhimurium mutant lacking both T3SSs (invA spiB mutant) when mice are inoculated with a mixture of both strains (Fig. 1A) (Stecher et al., 2007). In contrast, the T3SS system of C. rodentium provides the pathogen access to a spatial niche by mediating intimate attachment to the intestinal epithelium (Schauer and Falkow, 1993a, b). We reasoned that the T3SS might provide C. rodentium access to growth-limiting resources that are only available to bacteria that are intimately attached to the epithelial surface. However, these resources would no longer be accessible when intimate attachment is genetically ablated by inactivating escN, encoding a component of the T3SS. This hypothesis would predict that wild-type C. rodentium would not be able to rescue growth of an escN mutant when mice are inoculated with a mixture of both strains. To test this prediction, mice were inoculated with C. rodentium strain DSB100 (Barthold et al., 1976), an isogenic escN mutant (Lopez et al., 2016) or with a 1:1 mixture of both strains. Mice infected with the C. rodentium wild type shed the pathogen in significantly higher numbers than mice infected with the escN mutant (Fig. 1B), which was consistent with the idea that the T3SS is required for competing with the gut microbiota (Kamada et al., 2012). However, the C. rodentium wild type did not rescue the growth of an escN mutant when mice were infected with a 1:1 mixture of both strains (Fig. 1B and S1A), thus supporting the idea that the T3SS provided access to a spatial niche in close proximity to the epithelium.
Figure 1: Virulence factors provide C. rodentium access to a spatial niche in close proximity to the epithelium in which cytochrome c peroxidase provides an early fitness advantage.
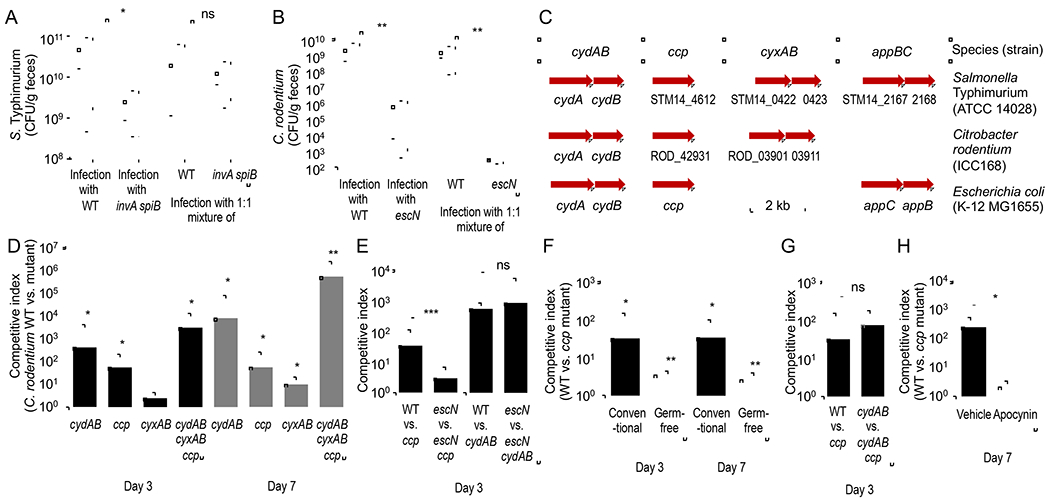
(A) Mice (C57BL/6J, N = 6) were infected intragastrically with S. Typhimurium wild type (WT) (Kingsley et al., 2003), an invA spiB mutant (Faber et al., 2016) or with a 1:1 mixture of both strains. The graph shows recovery from the feces at 5 days after infection. (B) Mice (C57BL/6J, N = 6) were infected intragastrically with C. rodentium wild type (WT), an escN mutant or with a 1:1 mixture of both strains. The graph shows recovery from the feces at 7 days after infection. (C) Schematic showing the presence of select respiratory enzymes in the genomes of C. rodentium, E. coli or S. enterica serovar Typhimurium. (D and E) Mice (C57BL/6J) were infected intragastrically with a 1:1 mixture of the C. rodentium wild type (WT) and one of the indicated mutant strains. (D) N=4. (E) N = 8. (F) Conventional mice (C57BL/6J, N = 4) or germ-free mice (Swiss Webster, N = 4) were infected intragastrically with a 1:1 mixture of the C. rodentium wild type (WT) and a ccp mutant. (G) Conventional mice (C57BL/6J, N = 6) were infected intragastrically with a 1:1 mixture of the indicated C. rodentium strain mixtures. (H) Conventional mice (C57BL/6J, N = 6) received water (Vehicle) or apocynin in their drinking water throughout the experiment and were infected intragastrically with a 1:1 mixture of the C. rodentium wild type (WT) and a ccp mutant. (D-H) The competitive index (ratio of WT to mutant) in the feces was determined at the indicated time points. (D and F) Statistical analysis indicates whether the competitive index was significantly different from 1. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ns, P > 0.05.
The marked differences in colony-forming units (CFU) recovered from colon contents of mice infected with the escN mutant compared to mice infected with C. rodentium wild type (Fig. 1B) prevent comparing the spatial localization of both strains. However, inoculation of germ-free mice results in similar recovery of wild-type and T3SS-deficient C. rodentium strains from colon contents (Kamada et al., 2012). To validate that inactivation of T3SS-mediated intimate attachment to epithelial cells by a mutation in escN would alter the spatial distribution of C. rodentium, germ-free mice were inoculated with either the C. rodentium wild type or an isogenic escN mutant. Whereas the C. rodentium wild type was detected in close proximity to the mucosal surface, the escN mutant did not colonize this niche (Fig. S1B), although both strains were recovered in similar numbers from colon contents (Fig. S1C). These data confirmed previous reports that the T3SS system provides C. rodentium access to a spatial niche in close proximity to the intestinal epithelium (Kamada et al., 2012).
Conventional wisdom holds that during gut homeostasis, an oxygen-gradient contributes to the cross-sectional heterogeneity in the spatial organization of the colonic microbiota (Donaldson et al., 2016; Tropini et al., 2017). Consistent with this idea, the presence of a functional T3SS enhances the growth advantage conferred by cytochrome bd oxidase at seven days after infection (i.e. after mice develop colonic crypt hyperplasia) (Lopez et al., 2016). However, prior to the development of colonic crypt hyperplasia (i.e. at three days after infection), cytochrome bd oxidase-mediated growth is T3SS-independent (Lopez et al., 2016), despite the fact that a functional T3SS provided a marked growth advantage at this time point (Fig. S1A). One possible explanation for this observation was that the spatial niche C. rodentium occupies provides access to oxygen, but the pathogen uses a different enzyme to respire oxygen prior to the development of colonic crypt hyperplasia. The C. rodentium genome does not contain the appBC genes, which encode a cytochrome bd-II oxidase present in Escherichia coli and Salmonella enterica (Dassa et al., 1991). In addition to the cydAB genes, which encode cytochrome bd oxidase, the C. rodentium genome contains the cyxAB genes, which encode a cytochrome bd-II oxidase that is predicted to enable the pathogen to respire oxygen under microaerophilic conditions (Fig. 1C). However, the cyxAB genes did not markedly enhance growth in the gut, as infection of mice with a 1:1 mixture of the C. rodentium wild type and a cyxAB mutant (Lopez et al., 2016) revealed only a small fitness advantage three days after infection (Fig. 1D). We thus entertained the idea that the spatial niche C. rodentium occupies provides access to a growth-limiting resource that is distinct from oxygen.
The conclusion that bacterial populations in close proximity to the epithelial surface are enriched in “oxygen tolerant” species is based on transcriptional profiling, which reveals an elevated expression of genes encoding catalase activity (Albenberg et al., 2014). However, catalase confers resistance to hydrogen peroxide (H2O2), which could also be responsible for the observed enrichment in “oxygen-tolerant” organisms. The C. rodentium genome encodes two catalases that confer “oxygen-tolerance” (Petty et al., 2010). In addition, the C. rodentium genome contains the ccp gene, which encodes a cytochrome c peroxidase (Fig. 1C). In E. coli, cytochrome c peroxidase transfers electrons from the quinone pool to H2O2 as an electron acceptor for anaerobic respiration (Khademian and Imlay, 2017). The ccp gene conferred a fitness advantage, because the C. rodentium wild type was recovered in approximately 20-fold higher numbers than a ccp mutant when mice were infected with a 1:1 mixture of both strains (Fig. 1D). Growth of the ccp mutant could be restored by introducing the cloned ccp gene on a plasmid (Fig. S2A). Notably, at three days after infection the growth advantage conferred by cytochrome c peroxidase required a functional T3SS, as it was no longer observed when mice were infected with a 1:1 mixture of an escN mutant and an escN ccp mutant (Fig. 1E and S2B). In stark contrast, a cydAB-mediated growth advantage was T3SS-independent at three days after infection, because it was still observed when mice were infected with a 1:1 mixture of an escN mutant and an escN cydAB mutant (Fig. 1E and S2B). These results suggested that prior to the development of colonic crypt hyperplasia, the T3SS systems provides a fitness advantage by providing access to H2O2, not oxygen.
NOX1-derived H2O2 promotes ccp-dependent growth prior to colonic inflammation
The ccp gene appeared to be more important for growth in conventional mice compared to growth in gnotobiotic mice, as indicated by decreased competitive index when gnotobiotic mice were inoculated with a 1:1 mixture of wild type and ccp mutant (Fig. 1F), indicative of ccp contributing to competition with the microbiota. A cydAB-dependent growth of C. rodentium in the intestinal lumen involves the tricarboxylic acid (TCA) cycle (Lopez et al., 2016), a metabolism that triggers endogenous production of H2O2 as a by-product in E. coli (Iuchi and Weiner, 1996). To test the possibility that ccp is required for growth in the mouse intestine because it detoxifies endogenous H2O2 generated as a byproduct of cydAB-mediated aerobic metabolism in the bacterial cytosol, mice were infected with a 1:1 mixture of a cydAB mutant and a cydAB ccp mutant. Notably, the growth advantage conferred by ccp was still observed when cytochrome bd oxidase-mediated aerobic respiration was genetically ablated (Fig. 1G), suggesting that the fitness advantage conferred by cytochrome c peroxidase was due to an exogenous source of H2O2. To determine whether host-derived H2O2 contributes to ccp-dependent growth, we inhibited host NADPH oxidase activity using apocynin (Kim et al., 2012). Apocynin treatment reduced the fitness advantage conferred by ccp (Fig. 1H), suggesting that the nutrient-niche C. rodentium occupies in the gut exposes the pathogen to H2O2 derived from host NADPH oxidases.
Next, we investigated possible cellular sources of H2O2. One source of H2O2 in the gut is the phagocyte NADPH oxidase (NOX2), as neutrophils migrating into the intestinal lumen clear C. rodentium between day 7 and day 14 after infection of mice (Kamada et al., 2015). Infection of mice with the C. rodentium wild type or a ccp mutant induced marked expression of inflammatory markers in the intestinal mucosa by day seven after infection, but not at three days after infection (Fig. 2A). Whereas no pathological changes were observed in sections of the cecal mucosa collected three days after infection with the C. rodentium wild type or a ccp mutant, colonic crypt hyperplasia and inflammatory infiltrates were observed by day seven after infection (Fig. 2B and 2C). Consistent with the idea that phagocytes alter the luminal environment by day seven after infection, the ccp-mediated growth advantage was reduced in mice lacking Cybb, a gene encoding a subunit of the phagocyte NADPH oxidase NOX2 (Fig. 2D). At 7 days after infection, concentrations of H2O2 were lower in colon contents of Cybb-deficient mice compared to congenic wild type mice (C57BL/6J) (Fig. 2E). However, at three days after infection, prior to the development of histopathological lesions (Fig. 2C), NOX2 did not contribute to cytochrome c peroxidase-mediated growth, as the fitness advantage conferred by ccp was not diminished in Cybb-deficient mice at this time point (Fig. 2D). Thus, in the absence of overt signs of inflammation (Fig. 2B and 2C), phagocyte NOX2 was not an important source of ccp-mediated C. rodentium growth.
Figure 2: Prior to C. rodentium-induced inflammation, phagocytes do not contribute to H2O2 levels in colon contents.
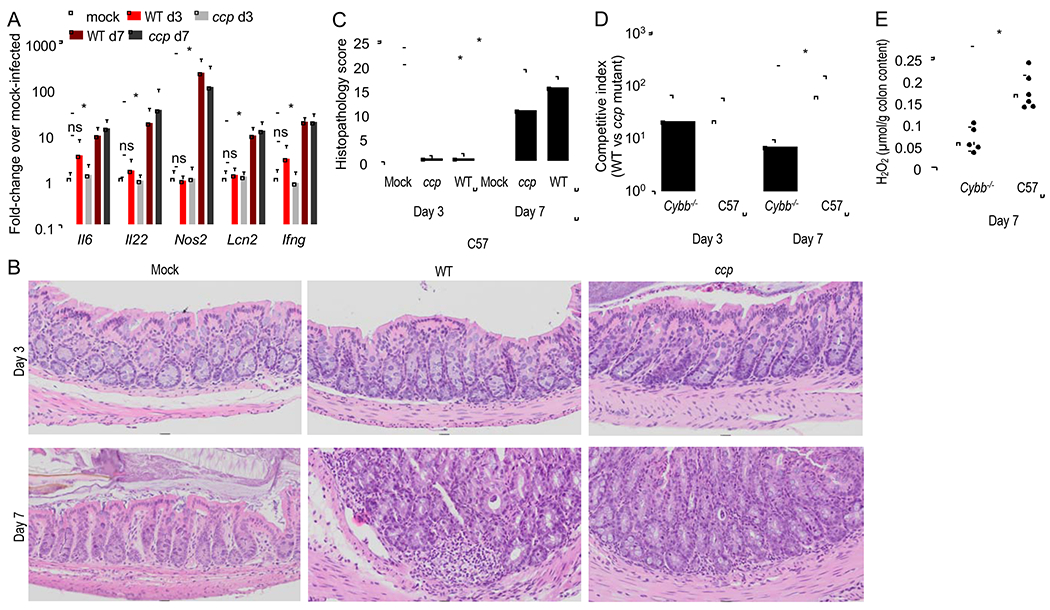
(A-C) Conventional mice (C57BL/6J, N = 6) were mock-infected or infected with the C. rodentium wild type (WT) or with a ccp mutant. (A) Expression of the indicated inflammatory genes was quantified by real-time PCR in RNA isolated from the colonic mucosa on days 3 (d3) and 7 (d7) after infection. (B-C) The severity of intestinal lesions was scored by a veterinary pathologist in blinded sections from the colon. (B) representative images are shown. (C) Combined histopathology score. (D-E) C57BL/6 mice (C57, N = 6) and congenic Cybb-deficient mice (Cybb−/−, N = 6) were infected with a 1:1 mixture of the C. rodentium wild type (WT) or with a ccp mutant. (D) The competitive index (ratio of WT to mutant) in the feces was determined at the indicated time points. (E) Hydrogen peroxide was measured in the supernatant of colon contents using Amplex Red and horseradish peroxidase on day 7 after infection. *, P < 0.05.
We then considered the epithelial surface as a possible source of H2O2 production at three days after C. rodentium infection. The Nox1 gene, encoding an epithelial NADPH oxidase, is expressed constitutively at high levels in the colonic epithelium and at lower levels in the ileal epithelium, but not in epithelial cells of the upper gastrointestinal tract (Matziouridou et al., 2018). We therefore investigated whether H2O2 produced by NOX1 was a factor influencing the growth of C. rodentium at three days after infection. Consistent with a constitutive expression of Nox1 in the colon (Matziouridou et al., 2018), Nox1-deficient mice had significantly lower concentrations of H2O2 in their colon contents compared to congenic wild type mice (C57BL/6J) (Fig. 3A). Infection with C. rodentium did not alter H2O2 levels in colon contents in wild type or Nox1-deficient mice (Fig. 3A). These data suggested that NOX1 was a significant source of colonic H2O2 under homeostasis. To determine whether NOX1 contributed to cytochrome c peroxidase-dependent fitness advantage early after infection, Nox1-deficient mice and congenic wild type mice (C57BL/6) were infected with a 1:1 mixture of the C. rodentium wild type and a ccp mutant. Notably, the fitness advantage conferred by cytochrome c peroxidase was no longer observed in mice lacking NOX1 (Fig. 3B).
Figure 3: The ccp gene provides a fitness advantage for C. rodentium that is dependent on the Nox1 mouse genotype, but independent of changes in the microbiota composition.
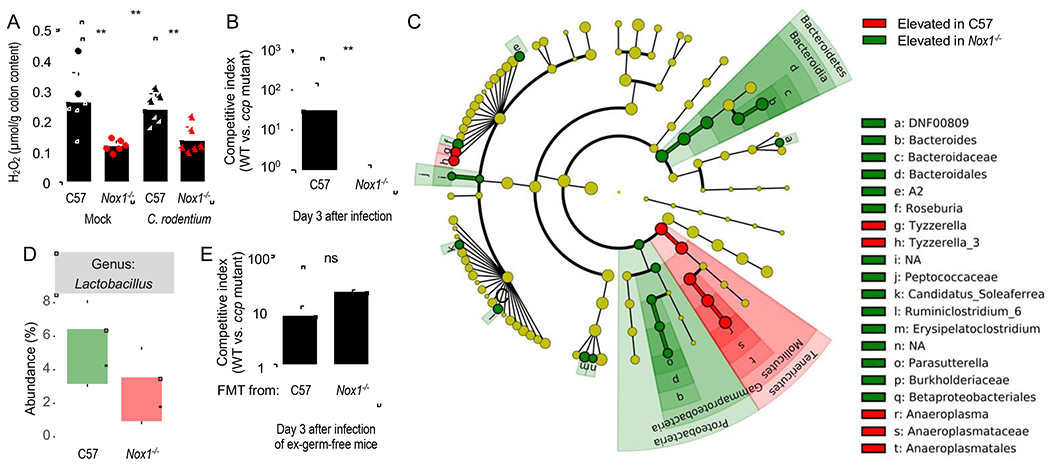
(A) C57BL/6 mice (C57, N = 6) and congenic Nox1-deficient mice (Nox1−/−, N = 6) were mock-infected or infected with C. rodentium wild type. Hydrogen peroxide was measured in the supernatant of colon contents using Amplex Red and horseradish peroxidase on day 3 after infection. (B) C57BL/6 mice (C57, N = 6) and congenic Nox1-deficient mice (Nox1−/−, N = 6) were infected with a 1:1 mixture of the C. rodentium wild type (WT) and a ccp mutant. The competitive index (ratio of WT to mutant) in the feces was determined at the indicated time point. (C and D) DNA was isolated from fecal contents of C57BL/6 mice (C57, N = 6) or congenic Nox1-deficient mice (Nox1−/−, N = 6) and the microbiota composition analyzed by 16S rRNA gene amplicon sequencing. (C) Results of a linear discriminant analysis of microbiota composition in C57BL/6 mice compared to Nox1-deficient mice are displayed as a cladogram, highlighting differences in the abundance of taxa (red and green highlighting) determined by 16S rRNA gene amplicon sequencing. Taxa are arranged on a circular phylogenetic tree. Green, taxa elevated in Nox1-deficient mice compared to C57BL/6 mice; Red, taxa reduced in Nox1-deficient mice compared to C57BL/6 mice. (D) Relative abundance of Lactobacillus species. (E) Germ-free mice (Swiss Webster, N = 4) received a fecal microbiota transplant from conventional mice (C57) or congenic Nox1-deficient mice (Nox1−/−) and seven days later mice were challenged with a 1:1 mixture of the C. rodentium wild type (WT) and a ccp mutant. The competitive index (ratio of WT to mutant) in the feces was determined at the indicated time point. **, P < 0.01; ns, P > 0.05.
Mice lacking the NOX dimerization partner CYBA exhibit lower C. rodentium colonization levels and an increased abundance of Lactobacillus species in their microbiota (Pircalabioru et al., 2016). Based on the observation that C. rodentium growth in the test tube can be inhibited by a cell-free culture supernatant of Lactobacillus reuteri it was postulated that increased H2O2 production by Lactobacillus species decreases growth of C. rodentium in Cyba-deficient mice (Pircalabioru et al., 2016). However, in a subsequent study the authors did not observe an expansion of Lactobacillus species in Cyba-deficient mice (Aviello et al., 2019). Similarly, microbiota analysis by 16S rRNA gene amplicon sequencing (microbiota profiling) did not reveal an elevated abundance of Lactobacillus species in the cecal microbiota of Nox1-deficient mice compared to wild type controls (Fig. 3C and 3D). The main differences in the microbiota composition were an increased abundance of the genera Bacteroides and Parasuterella in Nox1-deficient mice and an increased abundance of the genus Anaeroplasma in wild type (C57BL/6) mice (Fig. 3C). These data were consistent with the idea that the Nox1 genotype, rather than the abundance of Lactobacilli, governed H2O2 levels in the colon (Fig. 3A and 3D). To distinguish whether the ccp-mediated fitness advantage was caused by differences in the mouse genotype (Nox1-deficiency versus wild type) or by differences in the microbiota composition between mouse strains, germ-free mice received a fecal microbiota transplant from either Nox1-deficient mice or congenic wild type mice (Fig. S3A). Mice were then challenged with a 1:1 mixture of the C. rodentium wild type and a ccp mutant. Similar H2O2 levels were detected in colon contents of gnotobiotic mice receiving a fecal microbiota transplant from either Nox1-deficient mice or congenic wild type mice (Fig. S3B), suggesting that differences in the microbiota composition were not responsible for changes in H2O2 levels. Notably, engrafting germ-free mice with microbiota from Nox1-deficient mice did not abrogate the fitness advantage conferred by cytochrome c peroxidase (Fig. 3E), whereas the ccp-mediated fitness advantage was abrogated during infection of Nox1 deficient mice (Fig. 3B). Collectively, these data suggested that the ccp-mediated fitness advantage observed prior to development of colonic crypt hyperplasia was due to the constitutively synthesized epithelial NADPH oxidase NOX1.
Ccp promotes growth through anaerobic H2O2 respiration
A previous study suggests that reduced C. rodentium numbers recovered from Cyba-deficient mice were due to increased H2O2-mediated killing of the pathogen by Lactobacillus species (Pircalabioru et al., 2016). However, the hypothesis that H2O2-mediated killing was responsible for reducing C. rodentium numbers was not consistent with the finding that H2O2 levels were decreased, rather than increased, by Nox1-deficiency (Fig. 3A). Furthermore, in E. coli, cytochrome c peroxidase is ineffective as an enzyme defending against H2O2 toxicity, but it enables bacteria to respire using H2O2 as an anaerobic electron acceptor (Khademian and Imlay, 2017). To verify this conclusion, we investigated whether a mutation in ccp increases the susceptibility of C. rodentium to H2O2. C. rodentium was able to tolerate mM concentrations of H2O2 (Fig. 4A), presumably because its genome encodes catalase activity (Petty et al., 2010). However, we did not detect differences between the C. rodentium wild type and a ccp mutant in their ability to survive exposure to H2O2 under anaerobic or aerobic laboratory growth (Fig. 4A). The fact that H2O2 tolerance remained unchanged by genetic ablation of ccp might be due in part to a compensatory increase in expression of katG, encoding the periplasmic katalase (Fig. 4B). Nonetheless, these data were consistent with the idea that the ccp-mediated fitness advantage was not attributable to increased susceptibility to H2O2-mediated killing. Next. we wanted to investigate whether the fitness defect of a ccp mutant was due to reduced growth or enhanced killing in vivo when mice are infected with a single bacterial strain. We reasoned that Nox1-deficiency would increase numbers of a ccp mutant to levels of the C. rodentium wild type in case cytochrome c peroxidase functions in detoxifying H2O2. In contrast, Nox1-deficiency would be expected to lower the numbers of the C. rodentium wild type to those of the ccp mutant in case cytochrome c peroxidase enhances growth by using H2O2 for anaerobic respiration. Genetic ablation of cytochrome c peroxidase significantly reduced bacterial recovery from the feces of mice (C57BL/6) infected with a ccp mutant compared to mice infected with the C. rodentium wild type (Fig. 4C). Notably, Nox1-deficiency lowered the numbers of the C. rodentium wild type to those of the ccp mutant (Fig. 4C), thus providing additional support for the idea that cytochrome c peroxidase promotes growth through anaerobic H2O2 respiration rather than by preventing H2O2-mediated killing.
Figure 4: Cytochrome c peroxidase promotes growth by anaerobic H2O2 respiration but does not contribute to resistance against H2O2-mediated killing.
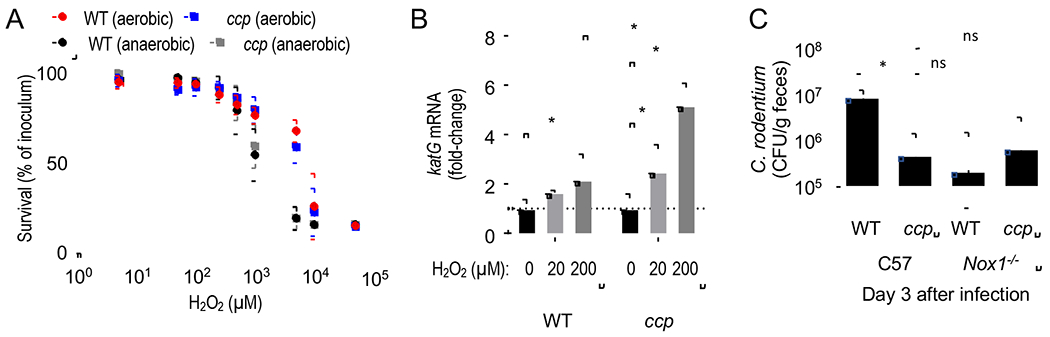
(A) Survival of the C. rodentium wild type (WT) and a ccp mutant was determined under aerobic or anaerobic conditions in the presence of the indicated H2O2 concentrations.(B) The C. rodentium wild type (WT) and an isogenic ccp mutant (ccp) were exposed to the indicated concentrations of H2O2 and katG expression determined by real-time PCR.(C) C57BL/6 mice (C57, N = 6) or congenic Nox1-deficient mice (Nox1, N = 6) were infected with the C. rodentium wild type (WT) or with a C. rodentium ccp mutant. Colony-forming units (CFU) of C. rodentium were determined in the feces at the indicated time point. *, P < 0.05; ns, P > 0.05.
DISCUSSION
An expansion of facultative anaerobic bacteria in the colon can be driven by elevated oxygen levels, which can arise because epithelial oxygenation is increased (Byndloss et al., 2017; Cevallos et al., 2019; Rivera-Chavez et al., 2016) or because mice are exposed to high atmospheric oxygen levels in a hyperbaric chamber (Albenberg et al., 2014). For example, respiration of oxygen by cytochrome bd oxidase contributes to T3SS-mediated growth of C. rodentium after mice develop colonic crypt hyperplasia, a lesion that leads to elevated epithelial oxygenation (Lopez et al., 2016)However, during conditions of homeostasis (i.e. in the absence of crypt hyperplasia or hyperbaric oxygen levels), the epithelial surface in the colon is hypoxic (<1% oxygen), which severely limits the amount of oxygen diffusing into the gut lumen (Zheng et al., 2015).
The idea that an oxygen gradient generates heterogeneity in the spatial organization of the colonic microbiota during homeostasis (Donaldson et al., 2016; Tropini et al., 2017) is supported by correlative evidence between the presence of an oxygen gradient and an increased proportion of “oxygen-tolerant” species in close proximity to the mucosal surface (Albenberg et al., 2014). This correlation assumes that genes encoding catalase activity make species “oxygen-tolerant” (Albenberg et al., 2014), rather than conferring resistance to H2O2 in the gut. However, constitutive synthesis of NOX1 in the epithelium of the large intestine (Matziouridou et al., 2018) supports an alternative explanation for the observed increased proportion of “oxygen-tolerant” species close to the epithelial surface (Albenberg et al., 2014). Rather than protecting bacteria from oxygen, catalases may protect bacteria from H2O2 produced by epithelial NOX1. The presence of catalase genes (Petty et al., 2010) likely accounts for the ability of C. rodentium to tolerate mM concentrations of H2O2. Thus, NOX1-derived H2O2 might exclude bacteria from the mucosal surface that lack required traits, such as catalase activity. Consistent with a spatial role of NOX1-derived H2O2 prior to the development of colonic crypt hyperplasia, T3SS-mediated intimate attachment provided C. rodentium with a ccp-dependent fitness advantage, by enabling the pathogen to access NOX1-derived H2O2 for growth by anaerobic H2O2 respiration. These findings provided experimental support for the idea that during homeostasis, H2O2 shapes bacterial growth in close proximity to the epithelial surface.
The picture emerging from these studies is that NOX1-derived H2O2 might serve as a habitat filter that shapes the spatial organization of the colonic microbiota. Since obligate anaerobic bacteria, which dominate the colonic microbiota, are generally sensitive to H2O2, this habitat filter might contribute to the inner mucus layer being largely devoid of bacteria (Johansson et al., 2008). Consistent with this idea, a variant of the NOX dimerization partner CYBA that generates reduced amounts of H2O2 exhibits profound mucus layer disruption with bacterial penetration into crypts (Aviello et al., 2019), despite the fact that NOX1-deficiency increases goblet cell differentiation and mucus production (Coant et al., 2010). An inability to keep the microbiota at a distance to the epithelial surface might thus contribute to early onset inflammatory bowel disease, which is observed in humans with NOX1-deficiency (Schwerd et al., 2018). However, additional work is needed to test whether and to which extend this habitat filter contributes to maintenance of a ‘demilitarized zone’ that protects the epithelial surface from the large community of obligate anaerobic bacteria in the colon.
STAR METHODS
Resource Availability
Lead Contact.
Andreas J. Bäumler (ajbaumler@ucdavis.edu).
Materials Availability.
The study did not create new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact.
Data and Code Availability:
The 16S rRNA gene amplicon sequencing data is available under BioProject accession numbers PRJNA660475 (gnotobiotic mice) and PRJNA607193 (conventional C57BL/6J and Nox1-deficient mice) through the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproiect/).
Experimental model and subject details
Animal experiments
The Institutional Animal Care and Use Committee at the University of California, Davis, approved all animal experiments. For experiments involving mice colonized with conventional specific pathogen free microbiota, female C57BL/6J, B6.129X1-Nox1tm1Kkr/J (Nox1−/−), or B6.129S-Cybbtm1Din/J (gp91phox-) strains (Jackson Laboratories) aged 5–7 weeks were inoculated with 1×109 colony forming units (CFU) per mouse of a single C. rodentium strain in LB broth intragastrically, or 0.1 ml sterile LB broth as mock infection. In competitive infections, a 1:1 ratio of two C. rodentium strains were given intragastrically at a combined final concentration of 1×109 CFU/mouse. Fecal pellets were collected on days 3 or 7, and mice were sacrificed between 3 and 7 days after infection. Colon contents were stored in phosphate-buffered saline (PBS) on ice. Colon tissue for histopathology was fixed in 10% buffered formalin phosphate overnight, while colon sections for murine RNA analysis were flash frozen and stored at –80°C. C. rodentium from feces and colon contents were enumerated by plating serial ten-fold dilutions of samples on MacConkey agar (BD Biosciences) containing the appropriate antibiotics based on the resistance markers listed for each strain in Table S1. Plates were incubated overnight at 37°C under atmospheric oxygen conditions. For competitive infections, the output ratio of recovered bacteria strains was divided by the input ratio of the inoculum to determine competitive index.
For infection of mice with S. Typhimurium, mice were pre-treated with an intragastric dose of 20 mg streptomycin 24 hours prior to infection. For single infections, mice were inoculated intragastrically with 1×109 CFU of the indicated S. Typhimurium strains. For competitive infection, mice were inoculated with 1×109 CFU of a 1:1 mixture of the indicated strains. Mice were euthanized 4 days after infection, and cecal contents were collected for enumeration of bacterial numbers. Bacterial numbers were determined by standard plate count on LB agar (BD Biosciences) containing the appropriate antibiotics. The ratio of recovered wild-type and mutant bacteria were normalized by the ratio of the inoculum to calculate the competitive index.
Germ-free Swiss Webster mice were bred and housed at the Teaching and Research Animal Care Services facilities at UC Davis. Male and female mice aged 5-7 weeks were used, and were infected and euthanized as described above.
To inhibit NADPH oxidase in vivo, mice were first mock infected or infected with a 1:1 mixture of the indicated C. rodentium strains as described above. Apocynin (Sigma) was initially dissolved in 100% ethanol and then further diluted in dPBS (Gibco). Mice given were 80 μg/ml of apocynin or ethanol vehicle control in sterile drinking water starting at day of infection. Fresh water containing apocynin and vehicle control were made daily continuing up to day 7 post-infection. Water intake was monitored, and no significant difference between consumption of apocynin and vehicle water was noted.
Bacterial strains and culture conditions
For the C. rodentium, E. coli and S. Typhimurium strains used in this study, see Table S1. Strains were grown routinely in Luria-Bertani (LB) broth (BD Biosciences #244620) or on LB plates unless otherwise indicated. Antibiotics were used at the following concentrations when required: carbenicillin (Carb), 0.1 mg/ml; chloramphenicol (Cm), 0.015 mg/ml; kanamycin (Kan), 0.1 mg/ml; tetracycline (Tet), 0.02 mg/ml; spectinomycin (Spec), 0.1 mg/ml.
Method details
Specific-pathogen free mouse husbandry
Prior to transport, mice at Jackson were fed LabDiet 5K52 formulation (6% fat). Upon arrival, mice from each cohort were randomly assigned into individually ventilated cages on one rack at a housing density of 3 to 4 animals per cage and allowed to acclimate in our vivarium for at least a week undisturbed. Feed was switched to irradiated TEKLAD GLOBAL 18% protein rodent diet 2918 (Envigo) and no breeding was performed. 70% ethanol was used to disinfect surfaces and gloves between groups. Clean (but not sterile) paper towels were utilized for fecal sample collections.
Hydrogen peroxide killing assay
Bacterial cultures were grown overnight at 37°C in LB broth under aerobic (atmospheric oxygen) conditions or in an anaerobic chamber (Coy Laboratories) with an environment of 5% CO2, 5% H2, and 90% N2. 105 bacterial cells were inoculated into PBS containing hydrogen peroxide of concentrations from 0 to 100 millimolar. The assay was allowed to run for 30 minutes at 37°C in aerobic or anaerobic conditions as stated above, after which each condition was plated on LB plates and enumerated by standard plate counting. Each condition was repeated in triplicate.
Construction of C. rodentium mutants
To construct an escN mutant, regions of approximately 500 bp in length flanking the escN gene were amplified via PCR using Q5 High Fidelity 2X Master Mix (NEB) and primers listed in Table S1, run on an agarose gel, and purified using the Zymoclean Gel DNA Recovery kit (Zymo Research). The kanamycin (KSAC) cassette from pUC4 (Lopez et al., 2016) was amplified, and this along with the escN flanking regions were ligated into pRDH10 (Kingsley et al., 1999) via the Gibson Assembly (NEB). The plasmid, named pBMM6, was propagated in E. coli DH5α λpir (Pal et al., 2005). pBMM6 was then transformed into E. coli S17-1 λpir (Simon et al., 1983), and this strain was conjugated with DBS100 (pSW172) (Spees et al., 2013). Double crossover events were selected via sucrose selection, and mutants were screened for kanamycin and carbenicillin resistance and loss of chloramphenicol resistance. PCR was used to confirm the deletion, and the temperature sensitive plasmid was cured after growth overnight at 40°C. The strain was then named BMM39.
To construct a ccp mutation, primers (Table S2) were designed that overlapped the 70 flanking base pairs on both ends of the C. rodentium ccp gene (ROD_42931) and 20 base pairs of the chloramphenicol cassette from pEP185.2 (Kinder et al., 1993). PCR was used to amplify the fragment with Q5 High Fidelity 2X Master Mix (NEB), run on an agarose gel, and gel purified via the Zymoclean Gel DNA Recovery kit (Zymo Research). The fragment was desalted via drop dialysis using nitrocellulose membrane filters (Millipore), and subsequently DpnI (NEB) treated. C. rodentium was electroporated with the temperature sensitive inducible lambda red recombinase plasmid pKD46 (Datsenko and Wanner, 2000) in which the ampicillin cassette has been replaced with a spectinomycin cassette. DBS100 (pKD46) was grown overnight at 30°C, and subcultured the next morning, and the plasmid recombinase was induced with 10 mM arabinose. DBS100 (pKD46) was then washed with distilled water, and the fragment was electroporated into DBS100 (pKD46). Mutants were selected via chloramphenicol resistance and confirmed for loss of pKD46 by loss of spectinomycin resistance. The mutant was verified via PCR screening, and then named BMM87.
To construct a cyxA mutant, primers (Table S2) were designed that overlapped the 70 flanking base pairs on both ends of the C. rodentium cyxA gene (ROD_03901) and 20 base pairs of the tetracycline cassette from pSPN23 (Raffatellu et al., 2009). Q5 High Fidelity 2X Master Mix (NEB) was used to amplify the fragment via PCR, the product was run on an agarose gel, and was gel purified using the Zymoclean Gel DNA Recovery kit (Zymo Research). The fragment was then prepared as described above. DBS100 (pKD46) was prepared as described above, followed by electroporation with the cyxA fragment. Mutants were selected via tetracycline resistance and confirmed for loss of pKD46 by loss of spectinomycin resistance. The mutant was verified via PCR screening, and was named BMM101.
To construct an escN ccp mutant, BMM39 was electroporated with the pKD46 plasmid and lambda red recombination was used as described above, using the ccp fragment. Mutants were selected via chloramphenicol and kanamycin resistance. Loss of pKD46 was confirmed via loss of spectinomycin resistance. The mutant was verified by PCR, and then named BMM109.
To construct a cydAB ccp mutant, lambda red recombination was used to introduce the ccp mutation into BMM86 as described above. Mutants were selected for kanamycin and chloramphenicol resistance, and were then screened via PCR. Loss of pKD46 was confirmed via loss of spectinomycin resistance. The mutant was named BMM88.
To construct a cydAB cyxAB ccp triple mutant, the pKD46 plasmid was introduced into BMM88. Lambda red recombination was used as described above to introduce the cyxA mutation. Mutants were selected for kanamycin, chloramphenicol, and tetracycline resistance. Loss of pKD46 was confirmed by loss of spectinomycin resistance. The mutant was screened via PCR and named BMM104.
To generate a plasmid for complementing the ccp mutation, primer pair ccpcomp Fwd (5′-ggtatcgataagcttgatTTTCCCGTTATCCCCTTTTTTAC-3′) and ccpcomp Rev (5′-gctgcaggaattcgatTTACTTGCCCTGCATATAAG-3′) was used to amplify the region containing coding sequence and 500 bp upstream of the C. rodentium ccp gene. The resulting PCR product was run on an agarose gel and gel purified using Zymoclean Gel DNA Recovery kit (Zymo Research). The fragment was then cloned into the EcoRV restriction site of the low copy number plasmid pWSK129 (Wang and Kushner, 1991) to generate the plasmid pBMM18 via Gibson Assembly Master Mix (NEB). The insert was confirmed via PCR. The plasmid was propagated in E. coli DH5α λpir and was then electroporated into BMM87.
Hydrogen peroxide measurements
Colonic contents and mucus scrapings were collected from infected mice for analysis. During necropsy, the middle 2-3cm of the colon was removed and cut longitudinally. Contents were removed carefully and stored in distilled H2O on ice. Forceps were then used to gently scrape the mucus layer from the underlying tissue, which was also stored in distilled H2O on ice. Samples were homogenized, centrifuged, and the supernatant was used for analysis. The Amplex Red/Horseradish Peroxidase assay kit was used (Thermofisher) according to manufacturer’s instructions. The absorbance was read on a microplate reader (BioRad) at 550nm.
Histopathology
Colon tissues were fixed by immersion in 10% neutral buffered formalin, embedded in paraffin, sectioned at 4 um, stained with hematoxylin & eosin (HE) using standard histologic techniques and examined by light microscopy. A board-certified veterinary pathologist scored histopathological changes of blinded samples. Inflammatory lesions were scored based on the degree of leukocyte infiltration, edema, crypt abscessation and accumulation of luminal exudate. Architectural changes to the colonic mucosa were scored based on the degree of mucosal epithelial hyperplasia, surface erosion, goblet cell depletion and extent of brush border bacterial colonization. Quantitative and semi-quantitative histopathological analyses of colonic lesions were completed using a defined set of scoring criteria listed in Table S2.
RNA isolation and quantitative real-time PCR
Murine colon tissue samples were homogenized in TRI Reagent (Molecular Research Center, Inc.) using a Mini-Beadbeater (BioSpec Products) and RNA was isolated following the manufacturer’s protocol. RNA was eluted in DNAse buffer and contaminating DNA was removed using the DNA-free kit (Applied Biosystems), and RNA was stored at −80°C.
RNA concentration and quality were measured spectrophotometrically using a Nanodrop (ND-1000, Nanodrop Technologies). Isolated RNA from murine samples was reverse transcribed using random hexamers and MuLV reverse transcriptase (Applied Biosystems). Quantitative real-time PCR (qPCR) was performed using SYBR green PCR mix (Applied Biosystems) and the primer sets listed in Table S1 to a concentration of 0.25 mM. Delta delta Ct was used to calculate fold change between groups.
Bacterial catalase expression
For in vitro measurements of catalase expression of C. rodentium, wild type and ccp mutant were grown overnight in 5mL LB under shaking conditions at 37°C. 109 bacteria were spun down in a microcentrifuge tube, washed with PBS twice, before being resuspended in PBS containing either: 0, 20, or 200 μM H2O2 in a 96 well plate. Bacteria were incubated at 37°C for 30 minutes, before being collected and washed twice in PBS as described above. The bacterial cell pellet was then resuspended in TRI Reagent. RNA was isolated and qPCR was conducted as described above.
Imaging of murine colon
For imaging of germ-free C57BL/6J mouse colons, sections of mid-colon were collected and fixed in 4% PFA overnight before being submerged in 40% sterile sucrose for 48 hours. Sections were then moved to Optimal Cutting Temperature (OCT) compound (Fisher HealthCare), and OCT was allowed to penetrate the sections for 3 hours, before sections were moved to molds and frozen in OCT on dry ice. Sections were cut in transverse orientation at a thickness of 7 μm. Slides were brought to room temperature, fixed with 4% PFA, and permeabilized with 0.2% Triton-X 100. C. rodentium were stained with rabbit anti-O152 primary antibody (Abcam ab78978), and a goat anti-rabbit Alexafluor 647 conjugate (Abcam ab 150079). Actin was stained with phalloidin-tetramethylrhodamine B isothiocyanate (Sigma Aldrich P1951), and nuclei were visualized with DAPI (Sigma Aldrich D9542). Sections were mounted with SlowFade Gold mounting media (Invitrogen S36938) and imaged.
16S rRNA gene amplicon sequencing sample preparation, library preparation, and sequencing
Nucleic acid extraction was done using the Powersoil kit from Qiagen following kit instructions. DNA concentration was recorded using Qubit a 2.0 flourometer. Samples were normalized to 20 ng/uL before library prep.
Primers 319F (TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG(spacer)GTACTCCTACGGGAGG CAGCAGT) and 806R (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG(spacer)CCGGACTACNVGGGT WTCTAAT) were used to amplify the V3-V4 domain of the 16S rRNA using a two-step PCR procedure. In step one of the amplification procedure, both forward and reverse primers contained an Illumina tag sequence (bold), a variable length spacer (no spacer, C, TC, or ATC for 319F; no spacer, G, TG, ATG for 806R) to increase diversity and improve the quality of the sequencing run, a linker sequence (italicized), and the 16S target sequence (underlined). Each 25 μl PCR reaction contained 1 Unit Kapa2G Robust Hot Start Polymerase (Kapa Biosystems), 1.5 mM MgCl2, 0.2 μM final concentration dNTP mix, 0.2 μM final concentration of each primer and 1ul of DNA for each sample. PCR conditions were: an initial incubation at 95°C for 3 min, followed by 25 cycles of 95°C for 45 s, 50°C for 30 s, 72°C for 30 s and a final extension of 72°C for 3 min. In step two, each sample was barcoded with a unique forward and reverse barcode combination using forward primers (AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTC) with an Illumina P5 adapter sequence (bold), a unique 8 nt barcode (N), a partial matching sequence of the forward adapter used in step one (underlined), and reverse primers (CAAGCAGAAGACGGCATACGAGATNNNNNNNNGTCTCGTGGGCTCGG)) with an Illumina P7 adapter sequence (bold), unique 8 nt barcode (N), and a partial matching sequence of the reverse adapter used in step one (underlined). The PCR reaction in step two contained 1 Unit Kapa2G Robust Hot Start Polymerase (Kapa Biosystems), 1.5 mM MgCl2, 0.2 mM final concentration dNTP mix, 0.2 μM final concentration of each uniquely barcoded primer and 1 ul of the product from the PCR reaction in step one diluted at a 10:1 ratio in water. PCR conditions were: an initial incubation at 95°C for 3 min, followed by 9 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 30 s and a final extension of 72°C for 3 min.
The final product was quantified on the Qubit instrument using the Qubit Broad Range DNA kit (Invitrogen) and individual amplicons were pooled in equal concentrations. The pooled library was cleaned utilizing Ampure XP beads (Beckman Coulter) then the band of interest was further subjected to isolation via gel electrophoresis on a 1.5% Blue Pippin HT gel (Sage Science). The library was quantified via qPCR followed by 300-bp paired-end sequencing using an Illumina MiSeq instrument in the Genome Center DNA Technologies Core, University of California, Davis.
16S rRNA gene amplicon sequencing analysis
Sequencing reads were demultiplexed using QIIME 1.8 (Caporaso et al., 2010), and non-biological nucleotides were trimmed using Trimmomatic (Bolger et al., 2014). 16S rRNA sequencing reads were subsequently processed and assembled into amplicon sequence variants (ASV) using dada2 (Callahan et al., 2016) in R. First, reads with more than 2 expected errors were removed. Dereplication and sample inference were then performed on forward and reverse reads, prior to merging. A sequence table was constructed from merged reads, and chimeric reads were subsequently removed. Taxonomy was assigned to reads to the genus level using the dada2 formatted rdp training dataset 14 found here https://zenodo.org/record/158955#.XJqlnxNKjUI. The R package phyloseq (McMurdie and Holmes, 2013) was then used in downstream analysis of the data, including the generation of a phyloseq object, relative abundance bar plots, and the principle coordinate analysis plot. For the weighted unifrac analysis, the R package msa (Bodenhofer et al., 2015) was used to generate a multiple sequence alignment from the assembled reads with the following parameters: method=“ClustalW”, type=“dna”, order=“input”. For linear discriminant analysis, data were reformatted in R, written to a tab separated text file, and then uploaded to the LEfSe galaxy server (Segata et al., 2011) where the default statistical parameters were used in the analysis to generate LDA scores and the LDA cladogram. Differential abundance analysis of taxa was performed using the R package deseq2 [25516281] with the parameters: test=“Wald”, fitType=“parametric”, cooksCutoff = FALSE.
The data have been deposited with links to BioProject accession numbers PRJNA660475 (gnotobiotic mice) and PRJNA607193 (conventional C57BL/6J and Nox1-deficient mice) in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
Statistical analysis
Data for all experiments displayed as bar graphs represent the geometric mean and the standard error of the mean. For most experiments, data points were first log transformed and differences between experimental groups were determined on the transformed data using a Student’s T-test (for comparing two groups) or ANOVA followed by Fisher’s LSD post hoc test (for comparison of more than two groups). A P-value of less than 0.05 was considered significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-E. coli O152 antibody | Abcam | ab78978 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor 647) | Abcam | ab150079 |
| Phalloidin-Tetramethylrhodamine B isothiocyanate | Sigma-Aldrich | P1951 |
| DAPI | Sigma-Aldrich | D9542-5MG |
| Bacterial and Virus Strains | ||
| For strains used in this study see Table S1 | see Table S1 | see Table S1 |
| Biological Samples | ||
| For plasmids used in this study see Table S1 | see Table S1 | see Table S1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hydrogen Peroxide | Sigma-Aldrich | H1009 |
| 4′-Hydroxy-3′-methoxyacetophenone (apocynin) | Sigma-Aldrich | A10809 |
| SlowFade Gold Antifade Mountant with DAPI | Thermo Fisher | S36938 |
| Critical Commercial Assays | ||
| TRI-reagent | Molecular Research Center | Cat#: RT111 |
| DNA-free DNA Removal Kit | Applied Biosystems | Cat#: AM1906 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#: 4309155 |
| Zymoclean Gel DNA Recovery Kit | Zymo Research | Cat#: D4001 |
| Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit | Invitrogen | Cat#: A22188 |
| QIAprep Spin Miniprep Kit | Qiagen | Cat#: 27106 |
| QIAGEN Plasmid Midi Kit | Qiagen | Cat#: 12143 |
| Gibson Assembly 2X Master Mix | New England Biolabs | Cat#: E5510S |
| Q5 Hot Start 2x Master Mix | New England Biolabs | Cat#: M0494S |
| DNAeasy Powersoil Kit | Qiagen | Cat#: 12888-100 |
| Experimental Models: Organisms/Strains | ||
| Mus musculus C57BL/6J | Jackson Labs | Cat#: 000664 |
| Mus musculus B6.129X1-Nox1tm1Kkr/J | Jackson Labs | Cat#: 018787 |
| Mus musculus B6.129S-Cybbtm1Din/J | Jackson Labs | Cat#: 002365 |
| Mus musculus Gnotobiotic Swiss Webster | Bred in house; originally acquired from Taconic | Cat#: SW-F and SW-M |
| Mus musculus Gnotobiotic C57BL/6J | Bred in house; originally acquired from UNC National Gnotobiotic Rodent Resource Center https://www.med.unc.edu/ngrrc/products-services/ | Gnotobiotic mice; C57BL/6J |
| Microbiota profiling data | ||
| 16S rRNA gene amplicon sequencing data | BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) | PRJNA660475 (gnotobiotic mice) and PRJNA607193 (conventional C57BL/6J and Nox1-deficient mice) |
| Oligonucleotides | ||
| For primers used in this study see Table S2 | This paper | see Table S2 |
| For qRT-PCR primers used in this study see Table S3 | This paper | see Table S3 |
| Software and Algorithms | ||
| Prism v8.0 | GraphPad | N/A |
| Microsoft Excel 2011 | Microsoft | N/A |
| MacVector v15.5.4 | MacVector | N/A |
| QIIME 1.8 | (Caporaso et al., 2010) | N/A |
| Trimmomatic | (Bolger et al., 2014) | N/A |
| dada2 | (Callahan et al., 2016) | N/A |
| phyloseq | (McMurdie and Holmes, 2013) | N/A |
| R package msa | (Bodenhofer et al., 2015) | N/A |
| LEfSe galaxy server | (Segata et al., 2011) | N/A |
| Other | ||
| Vinyl Anaerobic Chamber | Coy Laboratories | N/A |
Highlights.
Hydrogen peroxide drives Citrobacter growth in the non-inflamed gut
Citrobacter requires intimate epithelial attachment to respire hydrogen peroxide
Anaerobic hydrogen peroxide respiration by Citrobacter is NOX1-dependent
Epithelial-derived hydrogen peroxide shapes the epithelial surface environment
ACKNOWLEDGEMENTS
We would like to thank the UNC National Gnotobiotic Rodent Resource Center for kindly providing germ-free C57BL/6J mice.
Y.L. was supported by Vaadia-BARD Postdoctoral Fellowship FI-505-2014. Work in A.J.B.’s lab was supported by USDA/NIFA award 2015-67015-22930 (A.J.B), Crohn’s and Colitis Foundation of America Senior Investigator Award # 650976 and by Public Health Service Grants AI36309 (L.F.Z.), AI044170 (A.J.B.), AI096528 (A.J.B.), AI112445 (A.J.B.), AI146432 (A.J.B.) and AI112949 (A.J.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. (2014). Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063 e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviello G, Singh AK, O’Neill S, Conroy E, Gallagher W, D’Agostino G, Walker AW, Bourke B, Scholz D, and Knaus UG (2019). Colitis susceptibility in mice with reactive oxygen species deficiency is mediated by mucus barrier and immune defense defects. Mucosal Immunol 12, 1316–1326. [DOI] [PubMed] [Google Scholar]
- Barthold SW, Coleman GL, Bhatt PN, Osbaldiston GW, and Jonas AM (1976). The etiology of transmissible murine colonic hyperplasia. Lab Anim Sci 26, 889–894. [PubMed] [Google Scholar]
- Berry D, Mader E, Lee TK, Woebken D, Wang Y, Zhu D, Palatinszky M, Schintlmeister A, Schmid MC, Hanson BT, et al. (2015). Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci U S A 112, E194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhofer U, Bonatesta E, Horejs-Kainrath C, and Hochreiter S (2015). msa: an R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss MX, Olsan EE, Rivera-Chavez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, et al. (2017). Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss MX, Pernitzsch SR, and Baumler AJ (2018). Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol 11, 1299–1305. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson D, Barry R, Hopkins EGD, Roumeliotis TI, Garcia-Weber D, Mullineaux-Sanders C, Elinav E, Arrieumerlou C, Choudhary JS, and Frankel G (2019). Citrobacter rodentium induces rapid and unique metabolic and inflammatory responses in mice suffering from severe disease. Cell Microbiol, e13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevallos SA, Lee JY, Tiffany CR, Byndloss AJ, Johnston L, Byndloss MX, and Baumler AJ (2019). Increased Epithelial Oxygenation Links Colitis to an Expansion of Tumorigenic Bacteria. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, Ducroc R, Freund JN, Cazals-Hatem D, Bouhnik Y, Woerther PL, et al. (2010). NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol 30, 2636–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, and Relman DA (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa J, Fsihi H, Marck C, Dion M, Kieffer-Bontemps M, and Boquet PL (1991). A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol Gen Genet 229, 341–352. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Lee SM, and Mazmanian SK (2016). Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, et al. (2016). Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534, 697–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Schluter J, Coyte KZ, and Rakoff-Nahoum S (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, et al. (2018). Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe 23, 54–64 e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis CC, Winter MG, Chanin RB, Zhu W, Spiga L, and Winter SE (2019). Host-Derived Metabolites Modulate Transcription of Salmonella Genes Involved in 1-Lactate Utilization during Gut Colonization. Infect Immun 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, and Weiner L (1996). Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J Biochem 120, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, and Hansson GC (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105, 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, and Nunez G (2012). Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Sakamoto K, Seo SU, Zeng MY, Kim YG, Cascalho M, Vallance BA, Puente JL, and Nunez G (2015). Humoral Immunity in the Gut Selectively Targets Phenotypically Virulent Attaching-and-Effacing Bacteria for Intraluminal Elimination. Cell Host Microbe 17, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademian M, and Imlay JA (2017). Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc Natl Acad Sci U S A 114, E6922–E6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Moon KA, Jo HY, Jeong S, Seon SH, Jung E, Cho YS, Chun E, and Lee KY (2012). Anti-inflammatory effects of apocynin, an inhibitor of NADPH oxidase, in airway inflammation. Immunol Cell Biol 90, 441–448. [DOI] [PubMed] [Google Scholar]
- Kinder SA, Badger JL, Bryant GO, Pepe JC, and Miller VL (1993). Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R-M+ mutant. Gene 136, 271–275. [DOI] [PubMed] [Google Scholar]
- Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, and Baumler AJ (2003). Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun 71, 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, Everest P, Dougan G, Baumler AJ, Roberts M, and Williams PH (1999). Ferrioxamine-mediated Iron(III) utilization by Salmonella enterica. Appl Environ Microbiol 65, 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, et al. (2015). The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6, 8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak Y, and Baumler AJ (2019). Microbiota-Nourishing Immunity: A Guide to Understanding Our Microbial Self. Immunity 51, 214–224. [DOI] [PubMed] [Google Scholar]
- Litvak Y, Byndloss MX, and Baumler AJ (2018). Colonocyte metabolism shapes the gut microbiota. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak Y, Byndloss MX, Tsolis RM, and Baumler AJ (2017). Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol 39, 1–6. [DOI] [PubMed] [Google Scholar]
- Lopez CA, Miller BM, Rivera-Chavez F, Velazquez EM, Byndloss MX, Chavez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, and Baumler AJ (2016). Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, and Finlay BB (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe 2, 119–129. [DOI] [PubMed] [Google Scholar]
- Matziouridou C, Rocha SDC, Haabeth OA, Rudi K, Carlsen H, and Kielland A (2018). iNOS- and NOX1-dependent ROS production maintains bacterial homeostasis in the ileum of mice. Mucosal Immunol 11, 774–784. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Venkova-Canova T, Srivastava P, and Chattoraj DK (2005). Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. Journal of bacteriology 187, 7167–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, et al. (2010). The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. Journal of bacteriology 192, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircalabioru G, Aviello G, Kubica M, Zhdanov A, Paclet MH, Brennan L, Hertzberger R, Papkovsky D, Bourke B, and Knaus UG (2016). Defensive Mutualism Rescues NADPH Oxidase Inactivation in Gut Infection. Cell Host Microbe 19, 651–663. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. (2009). Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigottier-Gois L (2013). Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7, 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Lopez CA, and Baumler AJ (2017). Oxygen as a driver of gut dysbiosis. Free Radic Biol Med 105, 93–101. [DOI] [PubMed] [Google Scholar]
- Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, et al. (2016). Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 19, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer DB, and Falkow S (1993a). Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect Immun 61, 2486–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer DB, and Falkow S (1993b). The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun 61, 4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerd T, Bryant RV, Pandey S, Capitani M, Meran L, Cazier JB, Jung J, Mondal K, Parkes M, Mathew CG, et al. (2018). NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease. Mucosal Immunol 11, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, and Puhler A (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1, 784–791. [Google Scholar]
- Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, and Baumler AJ (2013). Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga L, Winter MG, Furtado de Carvalho T, Zhu W, Hughes ER, Gillis CC, Behrendt CL, Kim J, Chessa D, Andrews-Polymenis HL, et al. (2017). An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe 22, 291–301 e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. (2007). Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS biology 5, 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffany CR, and Baumler AJ (2019). Dysbiosis: from fiction to function. Am J Physiol Gastrointest Liver Physiol 317, G602–G608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Earle KA, Huang KC, and Sonnenburg JL (2017). The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe 21, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Adams LG, Ficht TA, and Baumler AJ (1999). Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67, 4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, and Kushner SR (1991). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199. [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, et al. (2015). Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Kelly CJ, and Colgan SP (2015). Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am J Physiol Cell Physiol 309, C350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene amplicon sequencing data is available under BioProject accession numbers PRJNA660475 (gnotobiotic mice) and PRJNA607193 (conventional C57BL/6J and Nox1-deficient mice) through the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproiect/).


