Abstract
Background
With the rapid development of new advanced molecular detection methods, identification of new genetic mutations conferring pathogen resistance to an ever-growing variety of antimicrobial substances will generate massive genomic datasets for public health and clinical laboratories. Keeping up with specialized standard coding for these immense datasets will be extremely challenging. This challenge prompted our effort to create a common molecular resistance Logical Observation Identifiers Names and Codes (LOINC) panel that can be used to report any identified antimicrobial resistance pattern.
Objective
To develop and utilize a common molecular resistance LOINC panel for molecular drug susceptibility testing (DST) data exchange in the U.S. National Tuberculosis Surveillance System using California Department of Public Health (CDPH) and New York State Department of Health as pilot sites.
Methods
We developed an interface and mapped incoming molecular DST data to the common molecular resistance LOINC panel using Health Level Seven (HL7) v2.5.1 Electronic Laboratory Reporting (ELR) message specifications through the Orion Health™ Rhapsody Integration Engine v6.3.1.
Results
Both pilot sites were able to process and upload/import the standardized HL7 v2.5.1 ELR messages into their respective systems; albeit CDPH identified areas for system improvements and has focused efforts to streamline the message importation process. Specifically, CDPH is enhancing their system to better capture parent-child elements and ensure that the data collected can be accessed seamlessly by the U.S. Centers for Disease Control and Prevention.
Discussion
The common molecular resistance LOINC panel is designed to be generalizable across other resistance genes and ideally also applicable to other disease domains.
Conclusion
The study demonstrates that it is possible to exchange molecular DST data across the continuum of disparate healthcare information systems in integrated public health environments using the common molecular resistance LOINC panel.
Keywords: Data Exchange Formats, Electronic Laboratory Reporting, Health Information Exchange, LOINC, Health Level Seven, Public Health Surveillance
Introduction
Background
With the rapid development of new advanced molecular detection methods, identification of new genetic mutations conferring pathogen resistance to an ever-growing variety of antimicrobial substances will generate massive genomic datasets for public health and clinical laboratories. Keeping up with specialized standard coding for these immense datasets will be extremely challenging. Even though Health Level Seven (HL7) International has developed and published implementation guides for electronic laboratory reporting (ELR) involving the use of the HL7 v2.5.1 messaging standard [1], many implementers of the standard have difficulty representing molecular drug susceptibility testing (DST) results in the HL7 v2.5.1 ELR message segments to achieve meaningful health information exchange. Molecular DST results refer to a single genetic testing result or a large panel of multiple results for different genes/loci and their implied susceptibility interpretations.
HL7 has developed an implementation guide known as “HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 1 (US Realm)” (represented as HL7 v2.5.1 ELR R1) [2] that is named in the Meaningful Use (MU) standards for electronic health record (EHR) technology among healthcare providers [3,4]. The HL7 v2.5.1 ELR R1 messaging standard is a national-level profile that is further constrained to accommodate agency-specific differences for the purposes of submitting laboratory test results to public health agencies. Test tools are available to validate conformance of implementations to the standard [5]. The optionality property of the HL7 v2 messaging standard (i.e., an inherent property that indicates whether a field within a message segment is required, optional, or conditional [1]) supports implementers’ systems that are using different levels of precoordinated or postcoordinated expressions, subsequently contributing to different ways of reporting the same battery of laboratory test results.
Likewise, ELR implementations existed in the United States, before the HL7 v2.5.1 ELR R1 messaging standard became adopted in the MU as the standard for nationally notifiable laboratory test results, and not all state and local systems have been upgraded. This adds to the variations in HL7 ELR message formats received at the public health agencies. These variations of HL7 ELR messages, coupled with fragmented, voluminous, and heterogeneous data sources from the public healthcare environment, make it difficult to transmit and translate notifiable disease cases to the U.S. Centers for Disease Control and Prevention (CDC) and other public health agencies.
The objective of this paper is to describe the development and use of a common molecular resistance Logical Observation Identifiers Names and Codes (LOINC) panel, developed in collaboration with CDC, Association of Public of Health Laboratories (APHL), California Department of Public Health (CDPH), New York State Department of Health (NYSDOH), and Regenstrief Institute, to pilot molecular DST data exchange in the U.S. National Tuberculosis Surveillance System (NTSS) using CDPH and NYSDOH as external partners. The NTSS contains reported tuberculosis (TB) cases provided by all 50 states, the District of Columbia (DC), Puerto Rico, and other U.S.-affiliated island jurisdictions [6,7].
Related Work
The health informatics literature contains limited research pertaining to data exchange formats for reporting molecular DST results. In a study conducted to formally define a comprehensive and minimal sets of data elements for molecular DST, Tornheim et al. [8] developed a framework for standardizing clinical laboratory reporting of Next Generation Sequencing (NGS) data for resistance-associated mutations in Mycobacterium tuberculosis complex (MTBC). The standardized reports were developed in consultation with participants from workshops and dozens of stakeholders worldwide [8]. The focus of the standardization efforts by Tornheim et al. [8], however, was not ELR-based but was primarily meant to be used in a paper-based format for reporting of NGS-derived molecular DST results for MTBC.
In 2019, we reported our first attempt to standardize molecular DST data elements in an ELR-based format [9]. We utilized a combination of standardization protocols to develop data exchange formats to capture, store, and monitor molecular DST results for MTBC. Those formats, however, were limited in their ability to support health information exchange for molecular DST results in other disease domains [9]. Moreover, most of the standardized vocabulary and value sets used in the study were restricted to the specifications published in the implementation guides by the HL7 Clinical Genomics workgroup [10-12], which are limited to human genetic testing. Hence, the need to develop a unified template for reporting molecular DST results for all microorganisms in different disease domains with generic vocabulary and value sets is of great importance.
Methods
Use of Implementation Guides and Reference Terminology Standards
The study utilized standardization protocols that involved many published implementation guides and reference terminology standards. Molecular DST data elements were standardized in alignment with the HL7 v2.5.1 ELR message specifications [13]. While the HL7 v2.5.1 ELR message specifications were used as the preferred data format for the molecular DST data elements, the common molecular resistance LOINC panel was used as the preferred terminology for identifying the laboratory tests or laboratory test results. Other preferred terminologies used for the study include: Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT) for microbiology related test results [13]; and clinical genomics coding systems [10-12] (i.e., 2.16.840.1.113883.6.340 (geneCodeNCBI) for the Mycobacterium tuberculosis resistance genes’ identifiers [14]; 2.16.840.1.113883.6.328 (HGVS.c) for the nucleic acid changes; and 2.16.840.1.113883.6.329 (HGVS.p) for the amino acid changes [15]).
Panel Design and Considerations
The current study used the common molecular resistance LOINC panel (i.e., 92254-2: Microorganism identification and resistance pattern determination panel by Molecular genetics method) developed as an extension of our previously described study [9]. LOINC is a freely available universal standard for identifying clinical and laboratory observations [16,17], with its panel being defined by a collector term that contains an enumerated set of discrete child elements [17]. Briefly, the goal of the panel design is to build on our previous work [9] and make it more generalizable. This will allow for the establishment of a common template to report sequence, resistance gene and gene mutation detection, and implied susceptibility interpretations across microorganisms, similar to the clinical genomics panel developed by the HL7 Clinical Genomics workgroup [10-12].
The common molecular resistance LOINC panel supports advanced molecular microorganism resistance testing and includes “terms to identify: 1) the microorganism identified; 2) the resistance genes, target sequences and/or mutations tested for and identified; and 3) the predicted susceptibility or resistance to one or more antimicrobials based on the genotyping results” [18]. More specifically, the panel includes a “representative set of LOINC terms for various antimicrobials, but any LOINC term for antimicrobial susceptibility by genotyping may be used as appropriate depending on genes and/or mutations tested for” [18]. The panel was first released in TRIAL status in the June 2019 LOINC version 2.66, pending feedback from public health and clinical laboratories regarding this approach for reporting the results of molecular resistance testing.
Several design considerations were made during the development of the panel. Notable among them was consideration of the preferred data format availability in Laboratory Information Management Systems (LIMS) and state/local epidemiologic data systems as well as how the data flow from LIMS to state/local epidemiologic data systems and then to CDC. It was also important that the incoming data fit CDC data requirements. More importantly, the panel is designed to be generalizable across other resistance genes and ideally also applicable to diseases other than TB.
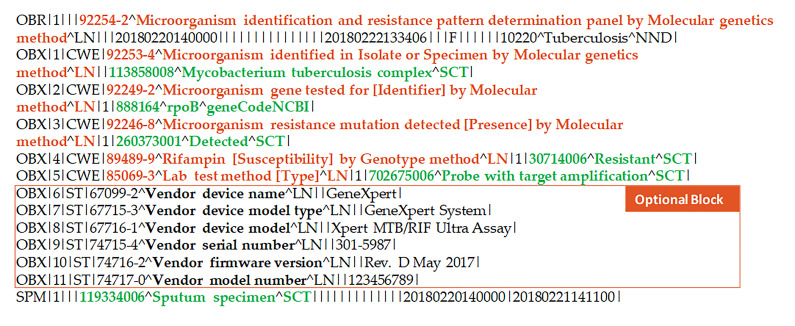
The common molecular resistance LOINC panel is intended to be used by public health and clinical laboratories to report the results of molecular DST. The design approach used in the panel is also “intended to be more sustainable over time compared to creating new LOINC codes for every microorganism + gene + antibiotic combination” [18]. The panel also includes vendor device information that is entirely optional. The panel is focused on microorganisms, not on human clinical genomics. For the purposes of this study, the panel was used to represent two types of molecular DST results: probe-based and sequence-based methods [19]. Whereas the Cepheid Xpert® MTB/RIF assay was used to illustrate results from a probe-based method, results provided back to states using CDC’s Molecular Detection of Drug Resistance (MDDR) service were used to illustrate specific genetic mutations detected using sequence-based methods [9].
Use of the Panel to Pilot Molecular DST Data Exchange
The common molecular resistance LOINC panel was used to pilot molecular DST data exchange within the NTSS. The piloting process consisted of (a) developing an interface to receive HL7 v2.5.1 ELR messages from the NTSS Case Reporting (NTSSCR) application via Orion Health™ Rhapsody Integration Engine v6.3.1 (Rhapsody) [20]; (b) creating data mapping routines using the common molecular resistance LOINC panel in Rhapsody to aid in transforming and constructing structured query language to load molecular DST data elements into the NTSS; and (c) executing test (plans) to verify that the interface satisfies the design requirements.
The pilot was conducted using sample molecular DST data from CDPH and NYSDOH. Whereas the CDPH data exchange involved the use of sample Cepheid Xpert® MTB/RIF test results from California Reportable Disease Information Exchange (CalREDIE), the NYSDOH data exchange involved the use of sample whole genome sequencing test results received from the NYSDOH laboratory. Essentially, all the sample molecular DST results from the two pilot sites were manually entered into the CDC NTSSCR application and then mapped to the common molecular resistance LOINC panel using the HL7 v2.5.1 ELR message specifications in Rhapsody.
Results
Data Representation of Rifampin Resistant Results from Cepheid Xpert® MTB/RIF
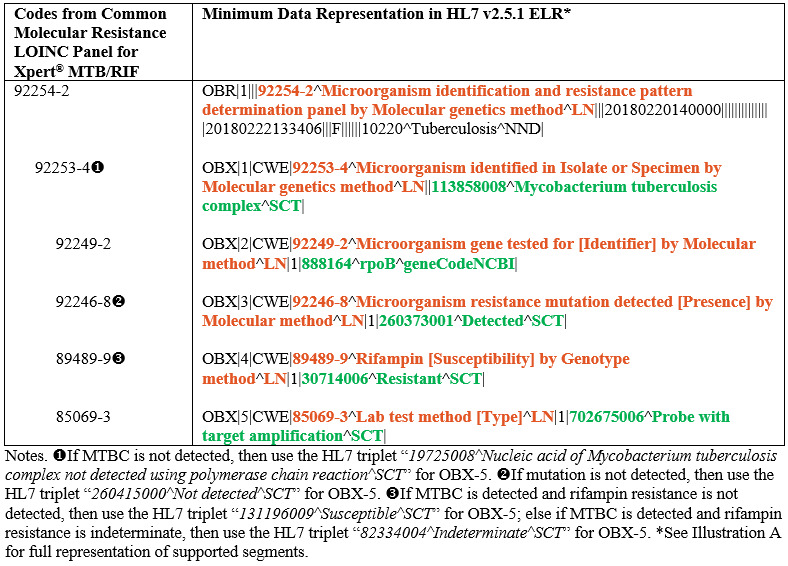
The Cepheid Xpert® MTB/RIF is a U.S. Food and Drug Administration (FDA) market-authorized method that uses a cartridge-based, fully automated nucleic acid amplification test (NAAT) for simultaneous detection of MTBC and mutations associated with rifampin resistance [9,21,22]. Keeping in alignment with our previous study [9], the key molecular DST data elements of interest for Xpert® MTB/RIF are MTBC identification; and if MTBC is identified, whether a mutation in the rifampin resistance determining region of the rpoB gene has been detected indicating resistance. Table 1 depicts the key codes from the common molecular resistance LOINC panel for Xpert® MTB/RIF and their associated minimum data representations in HL7 v2.5.1 ELR.
Table 1. Key codes from the common molecular resistance LOINC panel for Xpert® MTB/RIF and their associated minimum data representations in HL7 v2.5.1 ELR.
Data Representation of Specific Genetic Mutations Detected from DNA Sequencing Results
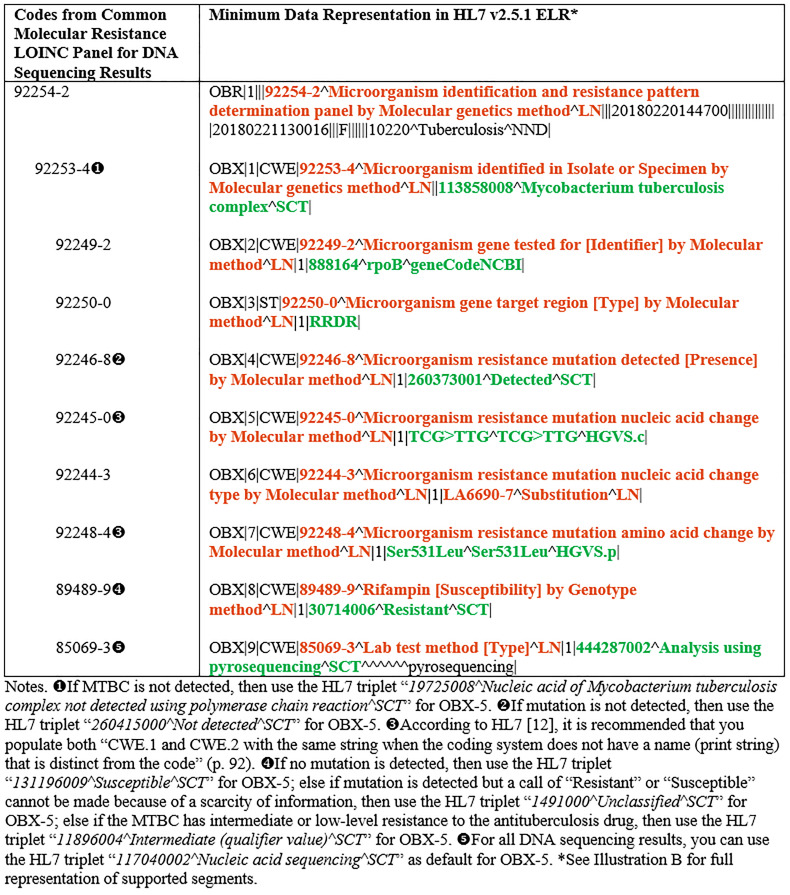
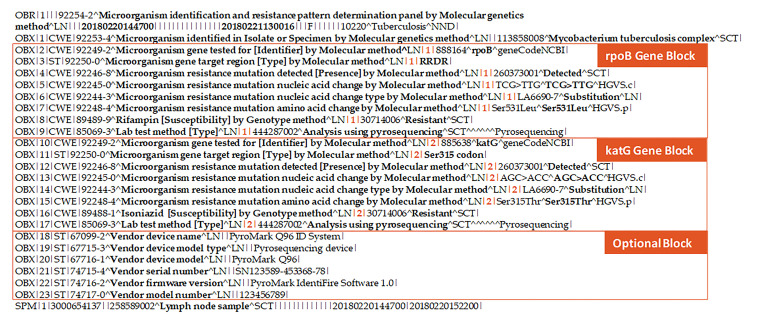
Public health and clinical laboratories performing sequencing-based assays report the identity of specific mutations in the gene [19,21]. For the purposes of this study, results from the CDC’s MDDR service were used as a guide to generalize the common molecular resistance LOINC panel for results from sequence-based assays. The CDC’s national MDDR service examines DNA from MTBC isolates or NAAT-positive sediments for MTBC at specific targets to determine the presence of mutations known to be associated with drug resistance [9]. The key molecular DST data elements of interest for the CDC’s MDDR service, therefore, focus on capturing and storing data elements relating to antituberculosis drug names, MTBC gene names, nucleic acid changes, amino acid changes, indels (nucleic acid change types), and results indicating if genetic mutation is detected or not at each relevant target gene [9]. Table 2 shows the key codes from the common molecular resistance LOINC panel for DNA sequencing results and their associated minimum data representations in HL7 v2.5.1 ELR.
Table 2. Key codes from the common molecular resistance LOINC panel for DNA sequencing results and their associated minimum data representations in HL7 v2.5.1 ELR.
Pilot Results
The common molecular resistance LOINC panel was piloted to assess its usability and impact in supporting molecular genetic data exchange within the NTSS. The piloting process resulted in the generation of four sample HL7 v2.5.1 ELR messages from NTSSCR that are compatible with the common molecular resistance LOINC panel. While two HL7 v2.5.1 ELR messages representing Cepheid Xpert® MTB/RIF test results were sent to CDPH for subsequent importing into CalREDIE, two HL7 v2.5.1 ELR messages representing whole genome sequencing test results were sent to NYSDOH for subsequent importing into that agency’s Electronic Clinical Laboratory Reporting System (ECLRS).
Both pilot sites were able to process and upload/import the standardized HL7 v2.5.1 ELR messages into their respective systems; albeit CDPH identified areas for system improvements and has focused efforts to streamline the message importation process. Specifically, CDPH is enhancing the current Drug Susceptibility Result module within CalREDIE to better capture parent-child elements and ensure that the data collected can be accessed by the Messaging module for seamless message creation for CDC. NYSDOH, on the other hand, did not identify any major areas for system improvements.
Discussion
No standard guideline for reporting molecular DST results exists within the United States. Reporting of molecular DST results is often unique to the public health laboratory performing the test. Different laboratories are using different LOINC codes to report the same battery of tests. For example, results of Cepheid Xpert® MTB/RIF are currently being reported with five different LOINC codes (i.e., 38379-4; 48176-2; 46244-0; 33634-7; and 48174-7) to CalREDIE by laboratory facilities in the state of California. The common molecular resistance LOINC panel harmonizes across all five different LOINC codes for the Cepheid Xpert® MTB/RIF results to support efficient reporting of molecular DST. The panel is also extended to include all molecular DST results generated by DNA sequencing services similar to those provided by CDC and NYSDOH.
Promoting broad adoption of the common molecular resistance LOINC panel remains a challenge. The testing infrastructure services and certification programs developed by the National Institute of Science and Technology (NIST) for sending nationally notifiable laboratory test results to public health agencies does not address the multiple competing approaches that many healthcare providers and health information technology (HIT) vendors are using to represent laboratory tests and their associated results. Integrating these fragmented, heterogeneous, and voluminous data into a single common format for use by the public health surveillance community is proving to be difficult.
NIST has a suite of HL7 v2 ELR testing tools [23] that test conformance of implementations and compliance with the HL7 v2.5.1 ELR R1 message specifications. The NIST ELR testing tools validate ELR messages created by healthcare providers and HIT vendors for the purposes of satisfying the 2014 and 2015 edition certification testing criteria for EHR technology mandated by the Office of the National Coordinator for HIT [23]. The NIST suite of validation tools also includes test cases to test the implementation guide entitled “HL7 Version 2.5.1 Implementation Guide: S&I Framework Lab Results Interface, Release 1, DSTU Release 2 – US Realm” (referred to as LRI IG), which provides guidance on how to communicate laboratory test results from a Laboratory Information System to a recipient system (e.g., EHR, public health, other laboratory) [12]. However, since the LRI IG was not named in the MU standards, it is not widely implemented, thus contributing to many variations of existing HL7 interfaces between laboratories and healthcare providers.
Efforts are underway to ensure that the common molecular resistance LOINC panel is widely understood by leading public health and clinical laboratories across the nation. In order to create awareness, representatives from APHL are providing education and technical assistance to industry stakeholders, such as FDA and manufacturers of laboratory instruments/toolkits, regarding the availability, flexibility and usability of the panel. For example, if this panel were to be included in the FDA’s SHIELD (Systemic Harmonization and Interoperability Enhancement for Lab Data) program, the LOINC codes could be harmonized across manufacturers and across coded results [24].
The study is limited to the data elements and formats of molecular DST results received from CDPH and NYSDOH. Majority of molecular DST results are currently reported by public health and clinical laboratories in pure narrative text formats with no computer accessible coding and processing of the results [9]. The common molecular resistance LOINC panel is, however, designed to provide a unified template for reporting machine-processable molecular DST results. Also, considering the fragmented and heterogeneous nature of voluminous data coming from the various laboratories and public health agencies, one cannot ignore the problems associated with the practicality of integrating them into a single common format for use by the public health surveillance community. To alleviate these burdens, early adopters of the panel may acquire and maintain functional interface/integration engine (i.e., Rhapsody, Mirth Connect, Cloverleaf, etc.) to support seamless data mapping routines and health information exchanges.
Conclusion
This study demonstrates that it is possible to exchange molecular DST data across the continuum of disparate healthcare information systems in integrated public health environments using the common molecular resistance LOINC panel. Even though this paper has focused on the usage of the panel in the TB domain, the common molecular resistance LOINC panel is designed to be generalizable across other areas of antimicrobial resistance (e.g., Staphylococcus aureus resistance to Vancomycin) and ideally also applicable to other disease domains. Moreover, the common molecular resistance LOINC panel is also intended to be more sustainable over time compared to creating new LOINC codes for every microorganism + gene + antibiotic combination.
Supplementary Material
Illustration A: Sample Supported Segments for Cepheid Xpert® MTB/RIF.
Illustration B: Sample Supported Segments for DNA Sequencing Results.
Acknowledgments
This work was supported in part by an appointment to the Public Health Informatics Fellowship Program at the Centers for Disease Control and Prevention (CDC) with funding from CDC’s Combating Antimicrobial Resistant Bacteria Initiative.
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC). References in this manuscript to any specific commercial products, process, service, manufacturer, or company does not constitute its endorsement or recommendation by the U.S. Government or CDC.
Footnotes
Financial Disclosure: No financial disclosures.
Competing Interests: No competing interests.
References
- 1.HL7 International. HL7 Messaging Standard Version 2.5.1: An application protocol for electronic data exchange in healthcare environments. 2007 [cited 2020 March 12]. Available from: https://www.hl7.org/implement/standards/product_brief.cfm?product_id=144.
- 2.HL7 International. HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 1 (US Realm). 2010 [cited 2020 March 12]. Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=98.
- 3.U.S. Department of Health and Human Services. Health Information Technology: Initial Set of Standards, Implementation Specifications, and Certification Criteria for Electronic Health Record Technology; Final Rule. In: Department of Health and Human Services, editor.: Federal Register; 2010. p. 44590-654. [PubMed] [Google Scholar]
- 4.U.S. Centers for Disease Control and Prevention. Meaningful Use Fact Sheet. [cited 2020 January 27]. Available from: https://www.cdc.gov/ehrmeaningfuluse/docs/factsheets/FS_MU_RLR.pdf.
- 5.Oemig F, Snelick R. Healthcare Interoperability Standards Compliance Handbook: Conformance and Testing of Healthcare Data Exchange Standards. Cham, Switzerland: Springer; 2016. [Google Scholar]
- 6.Bonney W, Price SF, Miramontes R. Improving the Quality of Data Exchange Formats in the U.S. National Tuberculosis Surveillance System. Online J Public Health Inform. 2018 2018-05-17;10(1):e48. Epub 2018-05-17.
- 7.U.S. Centers for Disease Control and Prevention. Reported Tuberculosis in the United States, 2018. 2018 [cited 2020 March 24]. Available from: https://www.cdc.gov/tb/statistics/reports/2018/default.htm.
- 8.Tornheim JA, Starks AM, Rodwell TC, Gardy JL, Walker TM, et al. 2019. Building the Framework for Standardized Clinical Laboratory Reporting of Next-generation Sequencing Data for Resistance-associated Mutations in Mycobacterium tuberculosis Complex. Clin Infect Dis. 69(9), 1631-33. 10.1093/cid/ciz219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonney W, Price SF, Miramontes R. 2019. Design and Implementation of Data Exchange Formats for Molecular Detection of Drug-Resistant Tuberculosis. AMIA Jt Summits Transl Sci Proc. 2019, 686-95. Epub 07 2019. [PMC free article] [PubMed] [Google Scholar]
- 10.HL7 International. HL7 Version 2 Implementation Guide: Clinical Genomics; Fully LOINC-Qualified Genetic Variation Model (US Realm). 2013 [cited 2020 March 12]. Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=23.
- 11.HL7 International. HL7 Version 2 Implementation Guide: Clinical Genomics Coded Reporting - “Lite”, Release 1 (1st DSTU Ballot) (US Realm). 2016 [cited 2020 March 11]. Available from: http://www.hl7.org/Special/committees/clingenomics/index.cfm.
- 12.HL7 International. HL7 Version 2.5.1 Implementation Guide: Laboratory Results Interface, Release 1 STU Release 3 - US Realm. 2017 [cited 2020 March 12]. Available from: http://hl7.org/implement/standards/product_brief.cfm?product_id=279.
- 13.HL7 International. HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 2 (US Realm). 2014 [cited 2020 March 11]. Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=329.
- 14.U.S. National Library of Medicine. API for NCBI Genes. [cited 2019 October 12]. Available from: https://clinicaltables.nlm.nih.gov/apidoc/ncbi_genes/v3/doc.html.
- 15.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, et al. 2016. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 37(6), 564-69. Epub 03 2016. 10.1002/humu.22981 [DOI] [PubMed] [Google Scholar]
- 16.Bodenreider O, Cornet R, Vreeman DJ. 2018. Recent Developments in Clinical Terminologies - SNOMED CT, LOINC, and RxNorm. Yearb Med Inform. 27(1), 129-39. Epub 08 2018. 10.1055/s-0038-1667077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vreeman DJ, Abhyankar S. Introduction to LOINC. 2018 [cited 2020 March 11]. Available from: https://danielvreeman.com/amia2018.
- 18.Regenstrief Institute Inc. 92254-2: Microorganism identification and resistance pattern determination panel by Molecular genetics method. 2019 [cited 2019 October 1]. Available from: https://loinc.org/92254-2/.
- 19.Lin SY, Desmond EP. 2014. Molecular diagnosis of tuberculosis and drug resistance. Clin Lab Med. 34(2), 297-314. Epub 05 2014. 10.1016/j.cll.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 20.Orion Health™. Rhapsody: Intelligent integration for healthcare systems. 2020 [cited 2020 January 17]. Available from: https://orionhealth.com/global/strategic-partners/rhapsody/.
- 21.Curry International Tuberculosis Center and California Department of Public Health. Drug-Resistant Tuberculosis: A Survival Guide for Clinicians. 3rd ed. USA 2016. [Google Scholar]
- 22.Association of Public Health Laboratories (APHL). Laboratory Considerations for Use of Cepheid GeneXpert MTB/RIF Assay. 2013 [cited 2020 March 12]. Available from: https://www.aphl.org/AboutAPHL/publications/Documents/ID_2013Nov_Cepheid-Xpert-Fact-Sheet.pdf.
- 23.National Institute of Science and Technology (NIST). Electronic Laboratory Reporting (ELR) Validation Suite. 2016 [cited 2020 January 22]. Available from: https://hl7v2-elr-testing.nist.gov/mu-elr/.
- 24.Paxton A. 2019. FDA nudges standards adoption in electronic reporting. CAP Today. (March), 2019. [Google Scholar]