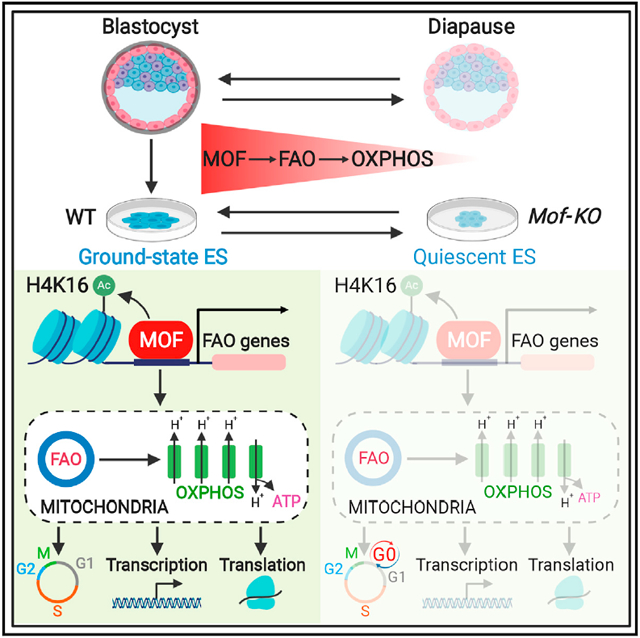
SUMMARY
Self-renewing embryonic stem cells (ESCs) respond to environmental cues by exiting pluripotency or entering a quiescent state. The molecular basis underlying this fate choice remains unclear. Here, we show that histone acetyltransferase MOF plays a critical role in this process through directly activating fatty acid oxidation (FAO) in the ground-state ESCs. We further show that the ground-state ESCs particularly rely on elevated FAO for oxidative phosphorylation (OXPHOS) and energy production. Mof deletion or FAO inhibition induces bona fide quiescent ground-state ESCs with an intact core pluripotency network and transcriptome signatures akin to the diapaused epiblasts in vivo. Mechanistically, MOF/FAO inhibition acts through reducing mitochondrial respiration (i.e., OXPHOS), which in turn triggers reversible pluripotent quiescence specifically in the ground-state ESCs. The inhibition of FAO/OXPHOS also induces quiescence in naive human ESCs. Our study suggests a general function of the MOF/FAO/OXPHOS axis in regulating cell fate determination in stem cells.
Graphical Abstract
In Brief
Khoa et al. find that ground-state embryonic stem cells (ESCs) readily use fatty acid oxidation (FAO) as the carbon source for mitochondrial oxidative phosphorylation (OXPHOS), which is directly regulated by histone acetyltransferase MOF. ESCs lacking the MOF/FAO/OXPHOS axis reside in a reversible pluripotent quiescence, mimicking embryonic diapause in mammals.
INTRODUCTION
Pluripotency is a transient state in vivo that first arises in the inner cell mass cells within the blastocyst and is defined as the ability of a single cell to give rise to all somatic cell lineages and the germline (Hackett and Surani, 2014; Nichols and Smith, 2012). Mouse embryonic stem cells (mESCs) are the first pluripotent stem cells directly derived from the blastocyst (Evans and Kaufman, 1981; Martin, 1981). Conventional embryonic stem cells (ESCs) grown in serum and leukemia inhibitory factor (LIF) (i.e., S/L ESCs) exhibit the heterogeneous expression of pluripotency markers, and only a small population of these cells exhibits a transcriptional landscape of pre-implantation epiblast (Chambers et al., 2007; Marks et al., 2012). The combined inhibition of mitogen-activated protein kinase/extracellular-signal-regulated kinase (MAPK/ERK) and glycogen synthase kinase 3 (GSK3) (hereafter called 2i) plus LIF promotes a robust pluripotent state (i.e., ground-state ESCs) that displays the molecular signatures of epiblast cells in embryonic day 4.5 (E4.5) blastocysts (Boroviak et al., 2014; Ying et al., 2008). As such, the ground-state ESCs have become a foundation for studying how pluripotency is established, maintained, and progressed during development.
A delicate balance of ESC self-renewal and differentiation is tightly regulated by the interconnected networks of transcription factors, environmental cues, and epigenetic regulators (Li and Belmonte, 2017; Niwa, 2007). ESC self-renewal is associated with high levels of histone acetylation, high chromatin accessibility (Atlasi et al., 2019; Finley et al., 2018), and the hyperactive core pluripotency transcription network (Kim et al., 2015; Li et al., 2012; Moris et al., 2018). Consistent with the importance of histone acetylation in ESC self-renewal, the reduction of glycolysis and glycolysis-derived acetyl coenzyme A (Ac-CoA) in ESCs leads to pluripotency exit and lineage differentiation (Moussaieff et al., 2015; Shyh-Chang and Ng, 2017). It is generally believed that reduction of the intermediate metabolite Ac-CoA is able to influence histone acetylation, which in turn alters chromatin dynamics and epigenomics in an instructive manner. In support, the deletion of histone acetyltransferases (HATs) often leads to the loss of self-renewal in S/L ESCs (Chen et al., 2008; Fazzio et al., 2008; Li et al., 2012; Lin et al., 2007; Zhong and Jin, 2009).
HAT MOF (males absent on the first, also known as KAT8 or MYST1) is a member of the highly conserved MYST family HATs. MOF acetylates histone H4 lysine 16 (H4K16ac) on chromatin and non-histone substrates (Li et al., 2009b; Luo et al., 2016; Morales et al., 2004). MOF plays vital roles in DNA damage repair, autophagy, lamin organization, and female fertility (Dou et al., 2005; Füllgrabe et al., 2013; Karoutas et al., 2019; Sharma et al., 2010; Yin et al., 2017). Mice with Mof deficiency die at the peri-implantation stage, with severe disruption of chromatin architecture and widespread apoptosis (Gupta et al., 2008; Thomas et al., 2008). Tissue-specific deletion of Mof in hematopoietic stem cells and cardiomyocyte leads to hematopoietic failure and cardiomyopathy, respectively (Chatterjee et al., 2016; Valerio et al., 2017). We and others previously showed that Mof deletion in S/L ESCs results in rapid ESC differentiation, followed by apoptosis of the differentiated cells (Chelmicki et al., 2014; Li et al., 2012). However, the function of MOF in ground-state ESCs, which have distinct glucose and glutamine metabolism (Carey et al., 2015; Hwang et al., 2016; Schvartzman et al., 2018; Vardhana et al., 2019), has not been tested.
Fatty acid oxidation (FAO) is an essential energy source to fuel the tricarboxylic acid (TCA) cycle in energy-demanding tissues (e.g., heart, liver), producing Ac-CoA and reducing equivalents (nicotinamide adenine dinucleotide [NADH], flavin adenine dinucleotide [FADH2]) for ATP generation (Carracedo et al., 2013; Qu et al., 2016). A high level of FAO activity is required for sustaining the self-renewal of quiescent adult stem cells (Ito et al., 2012; Knobloch et al., 2017; Mihaylova et al., 2018; Stoll et al., 2015). It also plays a role in the self-renewal of breast cancer stem cells (Wang et al., 2018) and supports the survival of a variety of tumors under metabolic stress conditions (Carracedo et al., 2013). Recently, it has been reported that the proliferating naive human ESCs (hESCs) have elevated FAO levels, along with other metabolic differences (e.g., high oxidative phosphorylation [OXPHOS], glycolysis, and amino acid metabolism), as compared to the primed hESCs (Gu et al., 2016; Sperber et al., 2015; Zhang et al., 2016). It remains unclear whether high FAO activity is a conserved feature in ground-state mESCs. The causal function of FAO metabolism in naive hESCs and ground-state mESCs, especially its role as the main carbon fuel source for mitochondrial respiration, is unknown. The upstream transcriptional regulator for FAO/OXPHOS axis in ground-state ESCs is also unclear.
Here, we report that the deletion of Mof in ground-state ESCs leads to pluripotent quiescence with an intact core transcription network. This is due to direct activation of the FAO pathway by MOF and specific reliance of ground-state ESCs on FAO activity as the major carbon fuel source for OXPHOS and cell proliferation. The role of the FAO/OXPHOS axis in restricting quiescence is conserved in naive hESCs. Inhibiting FAO or OXPHOS also induces pluripotent quiescence in naive and ground-state ESCs, distinct from their previously characterized roles in adult or cancer stem cells.
RESULTS
Deletion of Mof Induces a Pluripotent Quiescence in Ground-State ESCs
Consistent with our previous study (Li et al., 2012), 4-hydroxyta-moxifen (4-OHT)-induced deletion of Mof in Moffl/fl; Cre-ERTM S/L ESCs resulted in pluripotency exit, including loss of self-renewal, decreased expression of ESC core transcription factors, and increased expression of lineage-specific markers (Figures S1A, S1B, S1D, and S1E). It had no effects on Moffl/fl ESCs that expressed no Cre recombinase (Figure S1C). Mof deletion led to an increase in MAPK/ERK signaling and a concurrent decrease in WNT signaling in ESCs grown under S/L conditions (Figures S1F and S1G) (Li et al., 2012; Taylor et al., 2013). We were interested to find that 2i treatment blocked the differentiation of Mof null S/L ESCs (Figure S1H). Despite similar levels of MOF and H4K16ac in S/L and ground-state ESCs (Figure S1A; data not shown), Mof deletion in the ground-state Moffl/fl; Cre-ERTM ESCs (hereafter referred to as Mof null 2i ESCs) did not alter the expression of pluripotency markers such as KLF4, TFCP2L1, and OCT3/4 (Figure 1A). Quantitative mass spectrometry revealed that among >200 histone post-translational modifications (PTMs), H4K16ac was the only histone mark significantly decreased upon Mof deletion (Figure 1B, left; Table S1), which was validated by immunoblot (Figure 1B, right). Mof null 2i ESCs formed much smaller but typical dome-shaped colonies that had homogeneous expression of OCT3/4 and NANOG (Figure 1C). Mof null 2i ESCs showed slow proliferation (Figure 1D), a significant reduction of cells in G1 (16%) and S (12%) phases (Figure 1E), and a concomitant increase (~4-fold) in quiescent cells in G0 phase (Figure 1E). Notably, the increase in G0 cells in Mof-KO (knockout) cells quantitatively corresponded to a decrease in G1 cells (Figure 1E, bottom). ESCs with Mof deletion underwent a sharp reduction in total RNA amounts per cell (Figure 1F), a characteristic of quiescent adult stem cells (Cheung and Rando, 2013). It also resulted in a significant reduction in new DNA and protein synthesis (Figures 1G and 1H). These results indicated that the deletion of Mof in ground-state ESCs induces a bona fide quiescent state without affecting the core pluripotency network.
Figure 1. Deletion of Mof Induces a Pluripotent Quiescence in Ground-State ESCs.
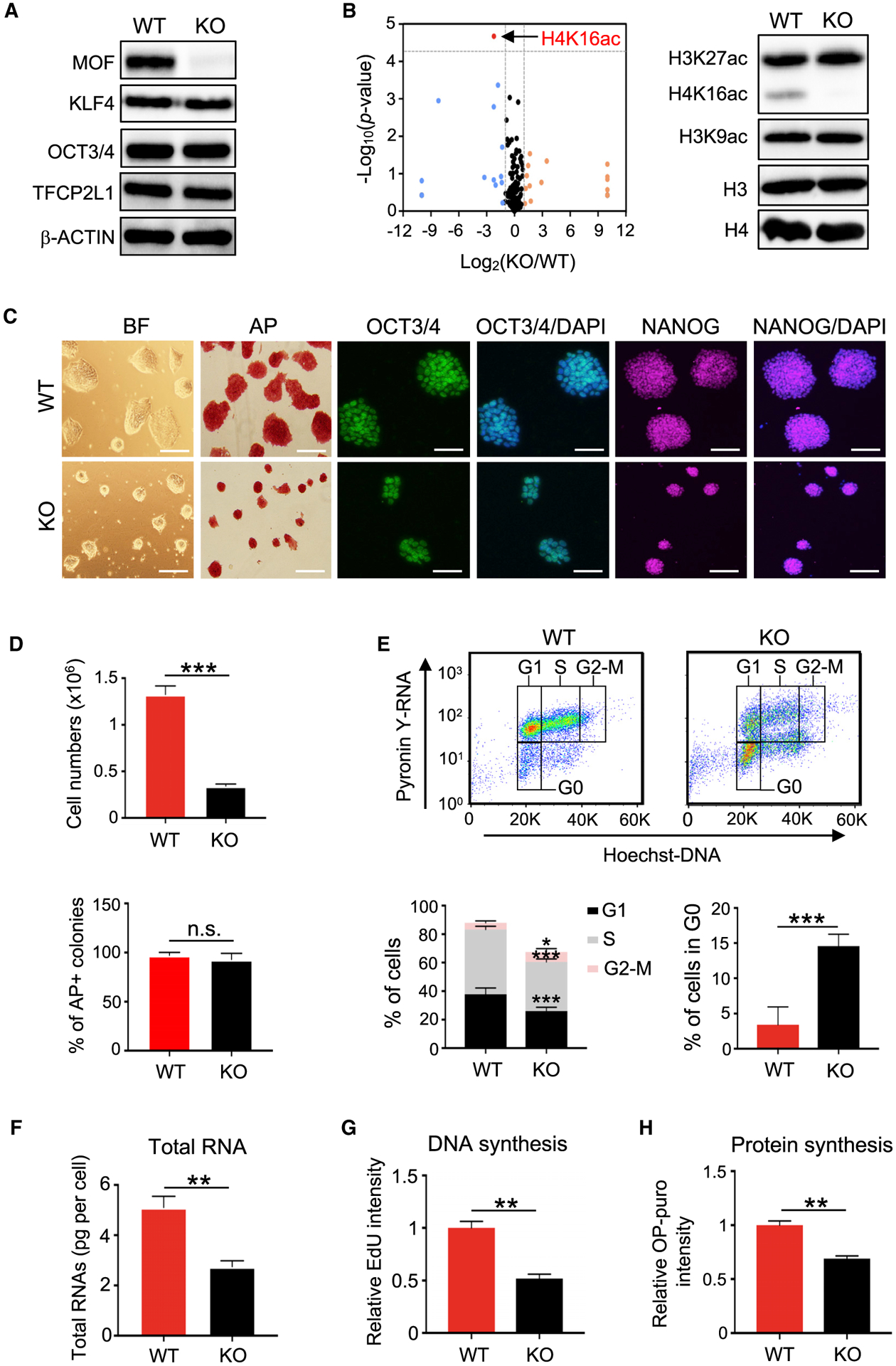
(A) Western blot analysis in wild-type (WT) and Mof null 2i ESCs. β-Actin was included as a loading control.
(B) Left, volcano plot of mass spectrometry for 231 histone PTMs and unmodified peptides ranked by log2 fold change (KO/WT) (x axis) and the (−) log10 p value (y axis). Right, western blot validation of selected histone PTMs.
(C) Representative images of bright-field (BF), alkaline phosphatase (AP) staining, or immunofluorescence (IF) of OCT3/4 and NANOG for WT and Mof null 2i ESCs. Nuclei were co-stained with DAPI. Scale bar, 100 μm.
(D) Qualifications of cell numbers (top) and AP-positive colonies (bottom) from (C). Cells were plated at the same numbers and scored after 4 days. Data are shown as means ± SEMs from six biological replicates.
(E) Cell-cycle analysis. FACS plots (top) and quantifications (bottom) of six biological replicates are shown as means ± SEMs.
(F) Total RNA amounts per cell for WT and Mof null 2i ESCs. Data are presented as means ± SEMs from three independent experiments.
(G) Measurement of DNA synthesis. Quantifications of three biological replicates are shown as means ± SEMs.
(H) Measurement of new protein synthesis. Data are presented as means ± SEMs from three biological replicates.
For (D)-(H), *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant.
MOF Represses the Gene Program Reflective of Pluripotent Quiescence
Illumina-based RNA sequencing (RNA-seq) showed that Mof-KO caused a significant downregulation of 775 genes and an upregulation of 1,245 genes (false discovery rate [FDR] < 0.05) (Figure S2A; Table S2). There were no significant changes in the transcripts of pluripotency genes such as Pou5f1 (Oct3/4), Klf4, Nanog, Tfcp2l1, Sox2, and Esrrb upon Mof deletion (Figure S2B). Likewise, the expression levels of lineage-specific genes associated with ectoderm (e.g., Krt18, Nes, Tcf15), mesoderm (e.g., Srf, Runx1, Bmp4) and endoderm (e.g., Gata4, Gata6, Sox17) differentiation were also unaltered (Figure S2C), confirming that the quiescent Mof null 2i ESCs exhibited no precocious differentiation. Gene set enrichment analysis (GSEA) showed that genes upregulated upon Mof deletion were significantly enriched in the negative regulation of multiple pathways such as cell proliferation, cell-cycle process, transcription, and translation (Figures 2A and S2D; Table S2). Conversely, mitochondrion and lipid metabolism processes were overrepresented among genes downregulated in Mof null 2i ESCs (Figure 2B).
Figure 2. MOF Directly Activates the FAO Pathway in Ground-State ESCs.
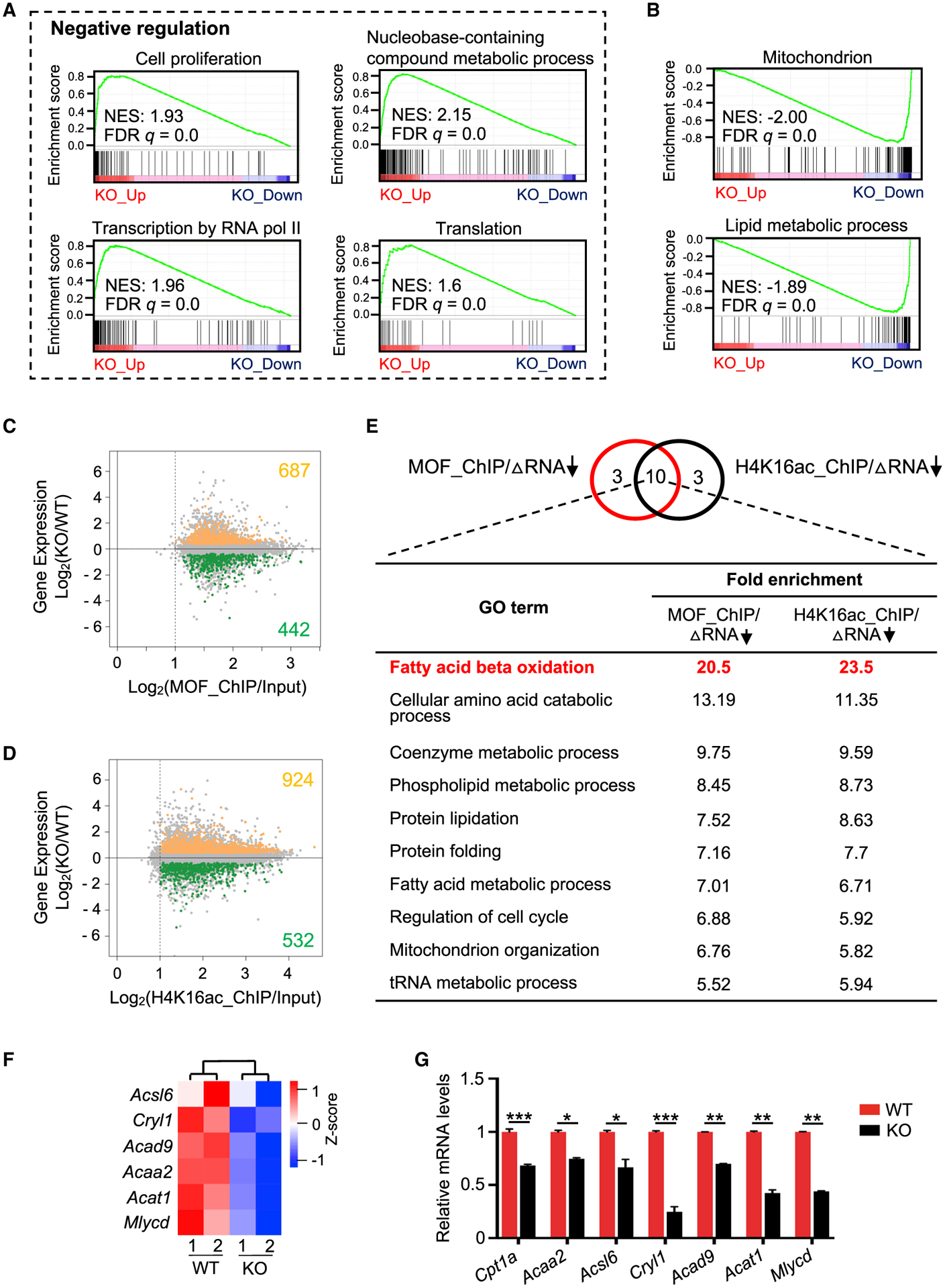
(A and B) GSEA plots for upregulated (A) and downregulated (B) genes in Mof null 2i ESCs. NES, normalized enrichment score.
(C and D) Intersection of RNA-seq (y axis) with ChIP-seq datasets (x axis) for MOF (C) and H4K16ac (D). See also Method Details.
(E) Top, Venn diagram for top 13 overrepresented GO terms for MOF or H4K16ac direct targets that were downregulated upon Mof deletion. Ten common metabolic pathways are shown at bottom.
(F) Heatmap (Z score) for the expression levels of FAO-related genes in WT and Mof null 2i ESCs. Heatmap key is shown at right.
(G) qRT-PCR validation for FAO-related genes. Relative expression from two biological replicates was normalized against the expression levels in WT ESCs, which was arbitrarily set as 1, and presented as means ± SEMs. *p < 0.05, **p < 0.01, ***p < 0.001.
MOF Directly Activates the FAO Pathway in Ground-State ESCs
To explore the mechanism by which Mof deletion induces cellular quiescence in 2i ESCs, we compared H4K16ac chromatin immunoprecipitation sequencing (ChIP-seq) data in S/L and 2i ESCs (Taylor et al., 2013; Valsecchi et al., 2018). As shown in Figure S1I, the global H4K16ac profiles were highly correlated in these two conditions. Cross-reference of the RNA-seq and ChIP-seq datasets for MOF (Li et al., 2012) and H4K16ac (Taylor et al., 2013) identified 442 and 532 downregulated genes that were directly regulated by MOF or H4K16ac, respectively (Figures 2C and 2D; Table S3). Significant overlap for the downregulated gene pathways was found for MOF- and H4K16ac-enriched targets (Figure 2E), many of which were associated with metabolic processes (Table S3). Among them, β-oxidation of fatty acids was the most enriched Gene Ontology (GO) term (Figure 2E; Table S3). The expression levels of FAO-related genes (Acsl6, Acad9, Mlycd, Acat1, Cryl1, Cpt1a, and Acaa2) were significantly reduced in Mof null 2i ESCs, as shown by RNA-seq and qRT-PCR validation (Figures 2F and 2G).
Mof Deletion Significantly Alters the Metabolic State in Ground-State ESCs
To determine metabolomic profile of the quiescent Mof null 2i ESCs, we performed liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS)-based metabolomics. Mof deletion led to 41 downregulated and 58 upregulated metabolites (Figure 3A; Table S4). In agreement with the decrease in DNA synthesis, RNA content and protein synthesis in Mof null 2i ESCs (Figures 1E–1H and 2A–2E), metabolites for purine metabolism, glycolysis, glycine and serine metabolism, glycerol phosphate shuttle, and de novo triacylglycerol biosynthesis were significantly downregulated upon Mof deletion (Figure 3B). Some metabolites in purine metabolism were also upregulated (Figures 3B and S2E). No other pathways were enriched for the elevated metabolites in Mof null 2i ESCs (Figure 3B, blue). Consistent with gene expression changes (Figure 2E), β-oxidation of long-chain fatty acids (i.e., FAO) was one of the significantly downregulated pathways in Mof null 2i ESCs (Figure 3B). The levels of metabolites in the FAO pathway (i.e., l-carnitine, O-acetyl-l-carnitine, CoA, and 5-aminoimidazole-4-carboxamide-1-β-d-ribofurano-side [AICAR]) were shown in Figure 3C. The supplementation of l-carnitine was able to rescue the quiescent phenotype of Mof null 2i ESCs in a dose-dependent manner (Figure 3D). l-carnitine treatment (3 mM) led to a significant decrease in G0 quiescent cells (Figure 3E). It is worth noting that the addition of the metabolite cocktail, including l-carnitine, O-acetyl-l-carnitine, CoA, and AICAR, did not further increase cell proliferation as compared to l-carnitine alone (Figures 3D and S2F). This is probably due to l-carnitine transportation as a rate-limiting step in the FAO pathway (see Discussion). Nonetheless, the dose-dependent rescue of Mof deletion by l-carnitine suggests that Mof deletion probably induces pluripotent quiescence by downregulating FAO activity in ground-state ESCs.
Figure 3. Mof Deletion Significantly Alters the Metabolic State in Ground-State ESCs.
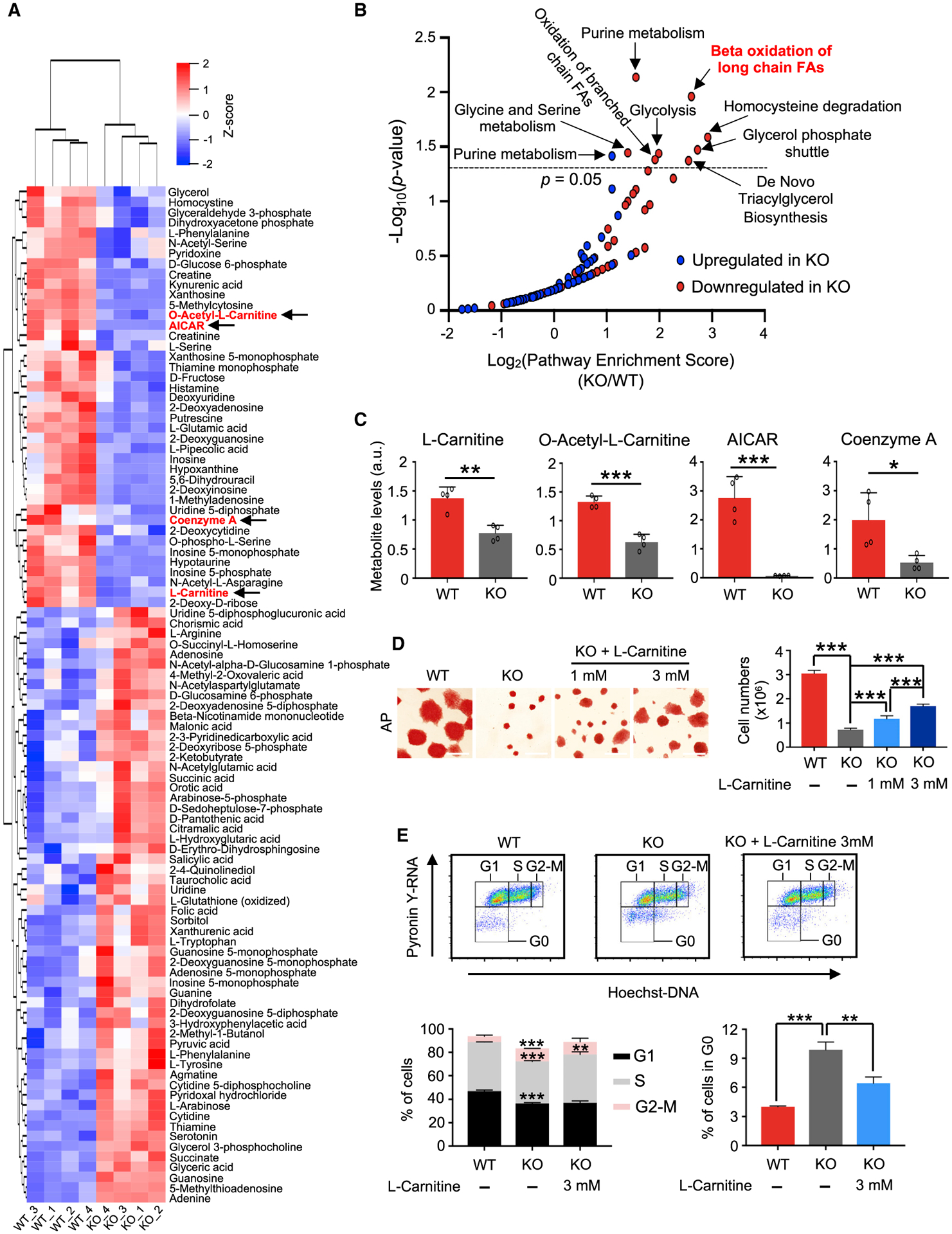
(A) Heatmap for metabolites that were significantly changed in Mof null 2i ESCs (p < 0.05). Z score values range from −2 to 2. FAO-related metabolites were highlighted in red and noted with arrows.
(B) Pathway analysis for metabolites that were changed in Mof null 2i ESCs. p < 0.05 serves as the cutoff for significantly enriched pathways. FAs, fatty acids.
(C) Levels of selected FAO metabolites. Data are shown as means ± SEMs, n = 4 biological replicates. a.u., arbitrary unit.
(D) l-Carnitine supplementation rescues proliferation defects in Mof null 2i ESCs. Left, AP staining of WT; KO and KO cells cultured with l-carnitine at a final concentration of 1 or 3 mM. Scale bar, 100 μm. Right, cell proliferation is shown as means ± SEMs, n = 4 biological replicates.
(E) Cell-cycle analysis. FACS plots (top) and quantifications (bottom) of three biological replicates are shown as mean ± SEM.
For (C)-(E), *p < 0.05, **p < 0.01, ***p < 0.001.
Mouse Ground-State ESCs Have Elevated FAO Activity
Because Mof deletion-induced quiescence was only observed in 2i ESCs, we hypothesized that 2i ESCs may have distinct FAO activity as compared to S/L ESCs. In support, FAO-related genes were expressed at higher levels in 2i ESCs (Figures 4A and 4B). Moreover, principal-component analysis (PCA) showed distinct metabolic states for 2i and S/L ESCs. There were 106 metabolites that were significantly increased (51) or decreased (55) in ground-state ESCs as compared S/L ESCs (Figures 4C and 4D; Table S4). Metabolites for glutamate metabolism were significantly upregulated in 2i ESCs (Figure 4E), consistent with a previous study (Carey et al., 2015). In contrast, metabolites in pathways such as glycine and serine metabolism, arginine and proline metabolism, and methionine metabolism were significantly downregulated in 2i ESCs (Figure 4E; Table S4). Notably, metabolites associated with carnitine synthesis were significantly upregulated in 2i ESCs (Figures 4E and 4F). Moreover, AICAR, a stimulator of FAO activity (Herms et al., 2015; Valsangkar and Downs, 2013), was also dramatically elevated in 2i ESCs (Figure 4G). Consistent with elevated FAO activity, palmitate oxidation was significantly higher in 2i ESCs (Figure 4H).
Figure 4. Inhibition of FAO Induces a Reversible Pluripotent Quiescence in Ground-State ESCs.
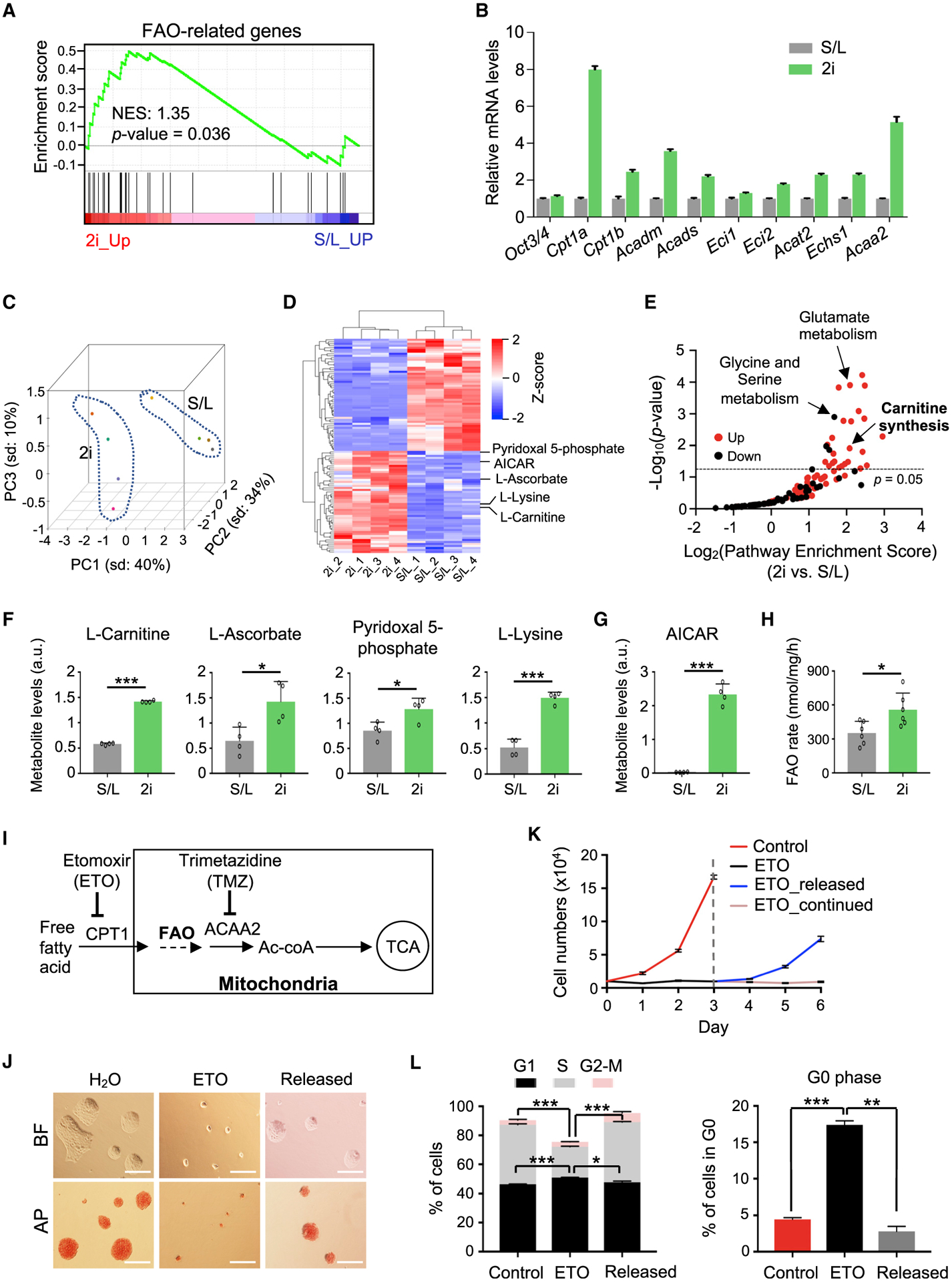
(A) GSEA for FAO genes in S/L versus 2i ESCs. RNA-seq data are from Marks et al. (2012).
(B) qRT-PCR validation for representative FAO genes. Relative expression from 3 technical replicates was normalized against the expression levels in S/L ESCs, which was arbitrarily set as 1. Data are presented as means ± SEMs.
(C) PCA plot for metabolites in S/L and 2i ESCs. Dashed lines indicate a clear separation of metabolome profiles in S/L and 2i ESCs. Colored dots indicate biological replicates.
(D) Heatmap (Z score) for significantly upregulated and downregulated metabolites in S/L and 2i ESCs. Metabolites associated with carnitine synthesis are highlighted at right.
(E) Pathway enrichment analysis for significantly up- and downregulated metabolites in 2i ESCs as shown in (D). p < 0.05 serves as the cutoff for significantly enriched pathways.
(F and G) Levels of metabolites associated with carnitine synthesis (F) and FAO (G). Data are presented as means ± SEMs, n = 4 biological replicates.
(H) Rate of FAO using [3H]-labeled palmitate. Data from two independent experiments are shown as means ± SEMs.
(I) Schematic overview of ETO and TMZ targets in the FAO pathway.
(J) Representative images of BF and AP staining for Oct4-GiP 2i ESCs with or without ETO treatment and after ETO release. Scale bar, 100 μm.
(K) Growth curves of 2i ESCs under indicated conditions. Data are presented as means ± SEMs of quadruplicate wells from a representative experiment.
(L) Quantifications of cell-cycle analysis. Data from two independent experiments are shown as means ± SEMs.
For (F)-(H) and (L), *p < 0.05, **p < 0.01, ***p < 0.001.
See also Figures S3 and S4 and Table S4.
Inhibition of FAO Induces a Reversible Pluripotent Quiescence in Ground-State ESCs
We next tested whether inactivating FAO leads to quiescence, similar to Mof deletion. To this end, we used two independent FAO inhibitors, etomoxir (ETO) and trimetazidine (TMZ) that target the first (CPT1) and the last (ACAA2) enzymes of the FAO pathway, respectively (Figure 4I) (see Method Details). We used ETO at a final concentration of 100 μM, which was commonly used in cell and mouse embryo cultures (Dunning et al., 2010; Knobloch et al., 2017; Paczkowski et al., 2014; Wang et al., 2018). At this concentration, ETO did not affect CoA homeostasis (Figures S3A and S3B; Table S4), indicating no apparent off-target effects. Remarkably, while FAO inhibition (FAOi) had moderate effects on S/L ESC proliferation (Figures S3C–S3H) or histone acetylation in general (Figure S3J), FAOi (i.e., ETO and TMZ)-treated 2i ESCs ceased proliferation and formed tiny dome-shaped colonies that maintained the expression of pluripotency markers (AP, OCT3/4, and TFCP2L1) (Figures 4J, 4K, S3K, and S3L). Switching 2i to the S/L condition in the presence of FAOi was sufficient to partially restore cell proliferation (Figure S3I). FAOi treatment led to a marked increase in cells in G0 phase (Figures 4L and S3L). The cells rapidly exited quiescence and resumed normal proliferation upon release from FAOi (referred to as released cells) (Figures 4J–4L, S4A, and S4B). The released cells were able to differentiate into primed epiblast-like cells (EpiLCs) (Brons et al., 2007; Tesar et al., 2007) with typical EpiSC characteristics (Figures S4C and S4D), as well as ectoderm (TUJ1), mesoderm (SMA), and endoderm (SOX17) lineages (Figure S4E) in embryoid body (EB) formation. These results demonstrate that FAOi is sufficient to induce a reversible pluripotent quiescence in ground-state ESCs, recapitulating Mof deletion-induced pluripotent quiescence.
Transcriptional changes in ETO-induced quiescent cells and the released cells showed that ETO treatment led to the significant downregulation of 2,720 genes (FDR < 0.05; Figure 5A; Table S5). The expression of 75% of these genes (2,044) was restored in the released cells (Figure 5A). GO term analysis revealed that these genes were mostly associated with cell proliferation (e.g., cell cycle, DNA replication), metabolism (e.g., mitochondrial respiratory chain, nucleotide synthesis), and cellular growth processes (e.g., ribosome biosynthesis, translation) (Figure 5B; Table S5). Genes upregulated by FAOi were enriched in negative regulation of cell proliferation and transcription (Figure S4F), similar to that of Mof deletion (Figures 2A and S2D).
Figure 5. Inhibition of FAO Promotes a Blastocyst Pausing Ex Vivo.
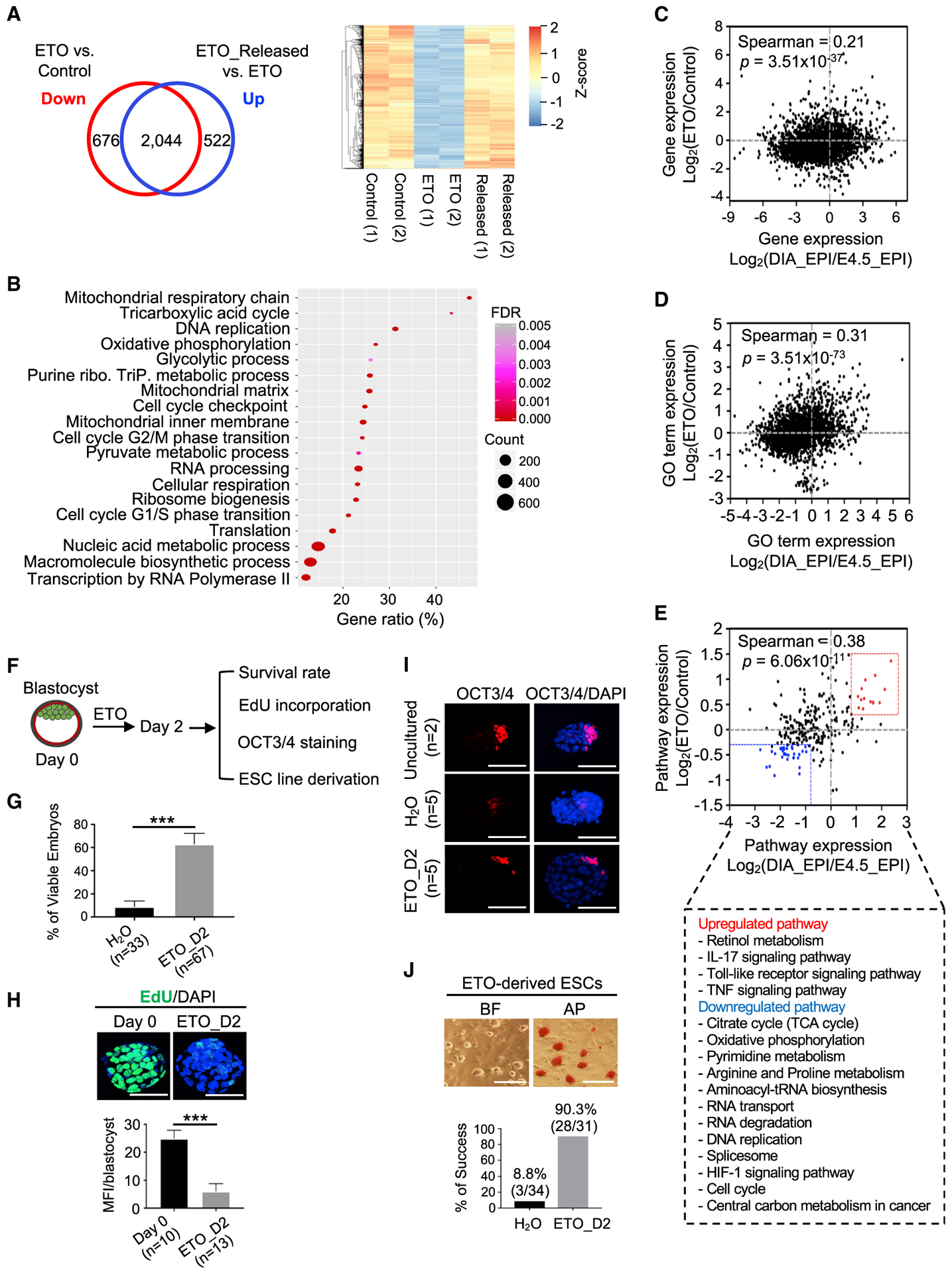
(A) Left, Venn diagram comparing ETO-downregulated genes and upregulated genes upon ETO release (FDR < 0.05). Right, heatmap (Z score) for 2,044 overlapping genes.
(B) GO term analysis for 2,044 genes identified in (A).
(C-E) Scatterplots for the correlation of gene expression (C), the GO term (D), and pathway expression (E) between the ETO-induced quiescent state (y axis) and diapaused epiblast (x axis). Selected pathways are shown at bottom (E).
(F) Experimental scheme for testing ETO in stimulating a reversible blastocyst pausing ex vivo.
(G) Survival rate of blastocysts cultured in the presence of H2O or ETO. Data are presented as means ± SEMs. n, number of blastocysts. ***p < 0.001. D2, day 2.
(H) Top, confocal images of EdU incorporation (green). Nuclei were co-stained with DAPI. Scale bar, 20 μm. Bottom, quantifications of EdU incorporation. Data are presented as means ± SEMs. n, number of blastocysts; MFI, mean fluorescence intensity. ***p < 0.001.
(I) Confocal images of OCT3/4 IF for E3.5 blastocysts, H2O control, and ETO-treated blastocysts. Nuclei were co-stained with DAPI. Scale bar, 20 μm. n, number of blastocysts.
(J) Derivation of ESC lines from H2O control and ETO-treated blastocysts. Top, BF and AP staining from 1 representative ESC line derived from an ETO-treated blastocyst. Scale bar, 50 μm. Bottom, quantifications for the efficiency of ESC derivation. Percentage of success was calculated by the number of successfully derived ESC lines from total blastocysts.
FAOi Promotes a Blastocyst Pausing Ex Vivo
The observation that FAOi induced a reversible pluripotent quiescence in ground-state ESCs is reminiscent of embryonic diapause, a naturally occurring dormant state at the blastocyst stage in many invertebrates and mammals (Fenelon et al., 2014; Renfree and Fenelon, 2017). By comparing gene expression profiles, we found that FAO pathway genes such as Acaa2, Acads, Eci2, Hadha, and Hadhb were expressed at lower levels in diapaused epiblasts relative to E4.5 epiblasts (Figure S5A). More importantly, transcriptome from ETO-induced quiescent cells and diapaused epiblasts (Boroviak et al., 2015) were correlated (Figures 5C–5E). Pathways upregulated in ETO-induced quiescent cells and diapaused epiblasts comprised Toll-like receptor, interleukin-17 (IL-17), and tumor necrosis factor (TNF) signaling pathways, whereas downregulated pathways were involved in nucleotide synthesis, cell cycle, and energy metabolism (Figure 5E). Unsupervised hierarchical clustering also confirmed the close correlation of ETO-induced quiescence and diapaused epiblasts at the transcription level (Figure S5B). FAOi-induced quiescence transcriptionally resembled that of mammalian target of rapamycin (mTOR) inhibition-mediated paused pluripotency (Figure S5C) (Bulut-Karslioglu et al., 2016). We speculated that FAOi might trigger changes in cellular energy homeostasis, which leads to the activation of AMP-activated protein kinase (AMPK) signaling, a known negative regulator of mTOR signaling (Inoki et al., 2012). To test this hypothesis, we examined AMPK phosphorylation in 2i ESCs treated with ETO, oligomycin, or mTOR inhibitor INK128. As shown in Figure S5D, while total AMPK remained unchanged across the treatments, AMPK phosphorylation was elevated in ETO-, oligomycin-, and mTOR inhibitor (mTORi)-treated cells. Consistent with AMPK activation, phosphorylation of S6K1 (mTORC1 signaling) and AKT (mTORC2 signaling) decreased in these cells. Thus, there is a potential functional interplay between FAO and mTOR inhibition in pluripotent quiescence (see Discussion).
To explore whether ETO treatment induces embryonic diapause ex vivo, E3.5 blastocysts derived from either pregnant mice or in vitro cultured one-cell embryos were used (Figure 5F). In line with previous studies (Naeslund, 1979), H2O-treated E3.5 blastocysts were collapsed after 24-48 h in culture (Figure 5G). Remarkably, blastocysts treated with ETO exhibited a significantly higher rate of viable embryos, and these blastocysts could be suspended in culture for up to 5 days (Figures 5G and S5E). ETO-treated blastocysts showed a striking decrease in DNA synthesis (Figure 5H). However, they maintained OCT3/4 expression in the intact inner cell mass (Figure 5I) and were able to give rise to stable ESC lines with high efficiency (90.3%) upon the withdrawal of ETO (Figure 5J). These results indicated that transient inhibition of FAO by ETO is sufficient to promote a reversible blastocyst dormancy ex vivo.
FAOi Induces Pluripotent Quiescence in Naive Human ESCs
Since naive hESCs have elevated FAO activity and can efficiently use fatty acid as a source of energy (Sperber et al., 2015), we tested whether FAOi induces pluripotent quiescence in naive hESCs. To this end, we used transgene-independent naive hESCs generated by the transient inhibition of histone deacetylase in combination with 2i/LIF and protein kinase C (PKC) inhibitor Gö6983 (Guo et al., 2017; Takashima et al., 2014). To eliminate the potential effects of ETO on cell attachment and viability, primed and naive hESCs were treated with ETO at day 2 or 3 after plating (Figure 6A). Inhibition of FAO by ETO in naive hESCs resulted in a rapid cessation of cell proliferation without affecting cell viability (Figures 6B and 6C). In comparison, ETO had no such effects on primed hESCs (Figure 6E). The expression of pluripotency markers (e.g., OCT3/4) was not affected by FAOi in both naive and primed hESCs (Figures 6D, 6E, S6A, and S6B). Cell-cycle analysis revealed that ETO treatment caused a modest but significant increase in cells in G0 phase for naive hESCs (Figure S6C). Cells in S, G2, and M phases were slightly decreased in ETO-treated naive hESCs (Figure S6C). ETO-treated naive hESCs exhibited a higher retention of pulse-labeled 5-ethynyl-2′-deoxyuridine (EdU) signals in comparison to untreated cells (Figure S6D). Quiescent naive hESCs maintained typical dome-shaped colony morphology and expression of naive pluripotency marker TFCP2L1 (Figures 6D and S6B). Upon release from ETO, the quiescent cells resumed their growth (Figures 6A and 6B) and underwent normal differentiation into three-germ layers in vitro (Figures 6F and 6G). Our data indicated that the inhibition of FAO by ETO induces reversible pluripotent quiescence in naive hESCs.
Figure 6. Inhibition of FAO Induces Reversible Pluripotent Quiescence in Naive Human ESCs.
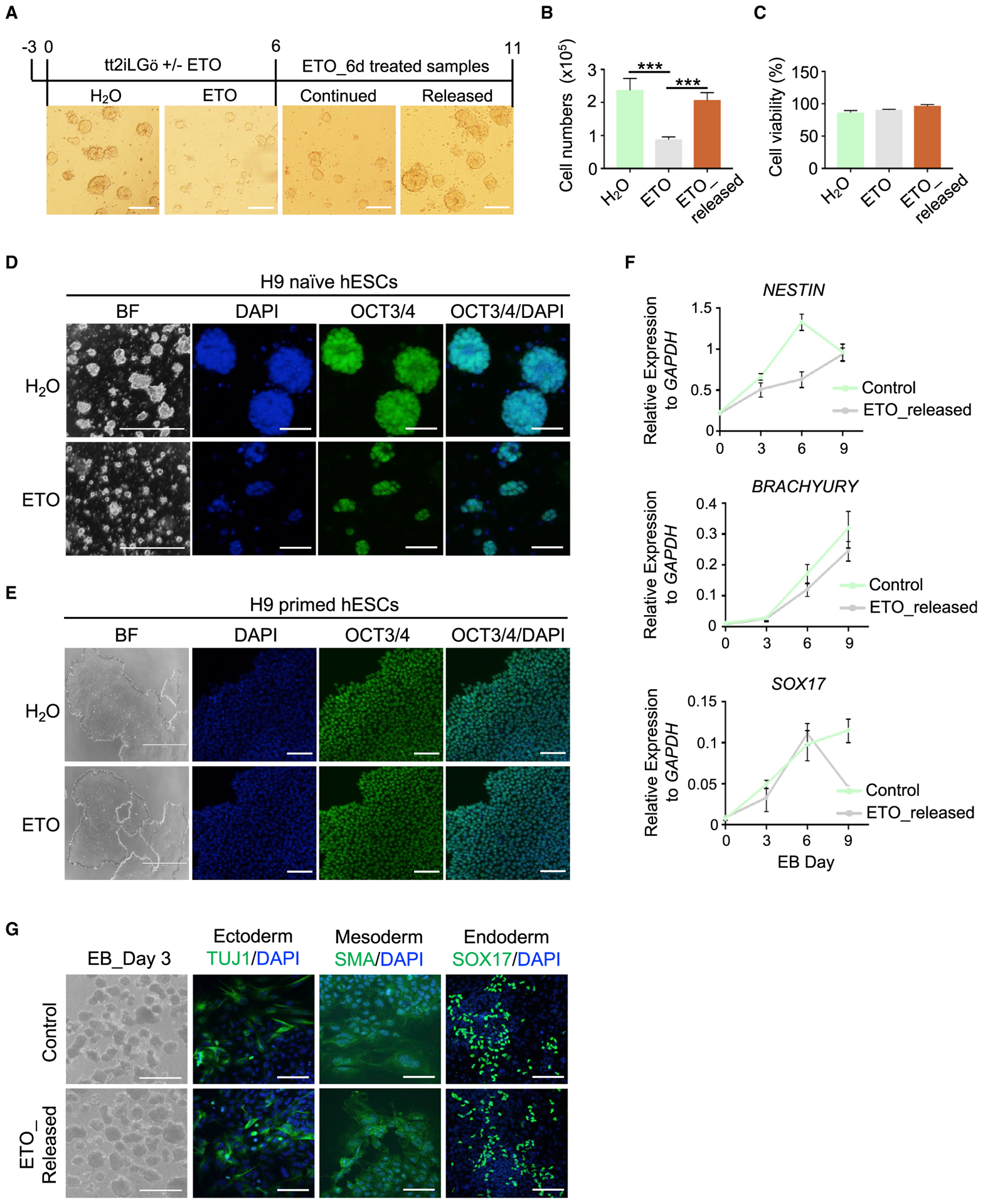
(A) BF images of naive hESCs after ETO treatment. Treatment time is shown at top. Scale bar, 100 μm.
(B) Qualifications of cell numbers at day 6 of ETO treatment and 5 days after ETO release. Data are presented as mean ± SEMs, n = 4 biological replicates. ***p < 0.001.
(C) Percentage of cell viability. Data are shown as means ± SEMs, n = 4 biological replicates.
(D and E) Representative images of BF and IF of OCT3/4 for naive (D) and primed (E) hESCs cultured with or without ETO. Nuclei were co-stained with DAPI. Scale bar, 2,000 μm for BF and 50 μm for IF in (D); 1,000 μm for BF and 100 μm for IF in (E).
(F) qRT-PCR analysis for representative genes of 3-germ layer differentiation from control and ETO-released naive hESCs. Data are presented as means ± SEMs, n = 3 biological replicates. EB, embryoid body.
(G) EB differentiation in control and ETO-released naive hESCs. Representative images of BF for EB and IF for 3-germ layer markers in EB outgrowth. Scale bar, 1,000 μm for BF and 50 μm for IF.
See also Figure S6.
Mof Deletion and FAOi Converge on Reduced Mitochondrial Respiration
Given that Mof deletion and FAOi phenocopy each other in promoting pluripotent quiescence in ground-state ESCs, we sought to investigate the major downstream effectors in this process. The intersection of transcriptome changes upon Mof deletion and FAOi identified 164 common genes that were significantly decreased in both Mof deletion and ETO treatment (Figure 7A, top). These commonly downregulated genes were mainly involved in mitochondrial function and structures (Figure 7A, bottom; Table S6). Consistently, Mof deletion resulted in a significant reduction in mitochondrial membrane potential (ΔΨm) (Figure S7A), as well as in basal and maximal respiration (Figure 7B). Mitochondrial mass in Mof null and ETO-induced quiescent cells was also strongly inhibited and rapidly recovered in the released cells (Figure 7C). The 2i ESCs treated with oligomycin, a wellcharacterized inhibitor of mitochondrial complex V (Raud et al., 2018; Zhang et al., 2011), entered pluripotent quiescence (Figures 7D, S7B, and S7C), similar to MOF/FAO-depleted 2i ESCs. Upon the withdrawal of oligomycin, the cells were able to differentiate into ectoderm, mesoderm, and endoderm lineages (Figure S7D). In contrast, S/L ESCs treated with oligomycin lost cell-to-cell contact and pluripotency marker OCT3/4 (Figure S7E). Our results support an essential role of the MOF/FAO/mitochondria axis in the regulation of ground-state quiescence (Figures 3, 4, and 7A–7D). The induction of pluripotent quiescence by oligomycin was also observed in naive hESCs (Figures S7F and S7G). In comparison, primed hESCs treated with oligomycin rapidly lost proliferation and pluripotency even at low concentrations (Figure S7H). These data indicated that reduced mitochondrial respiration is a major driving force in the induction of pluripotent quiescence in mouse ground-state and human naive pluripotency.
Figure 7. Mof Deletion and FAOi Converge on Reduced Mitochondrial Respiration.
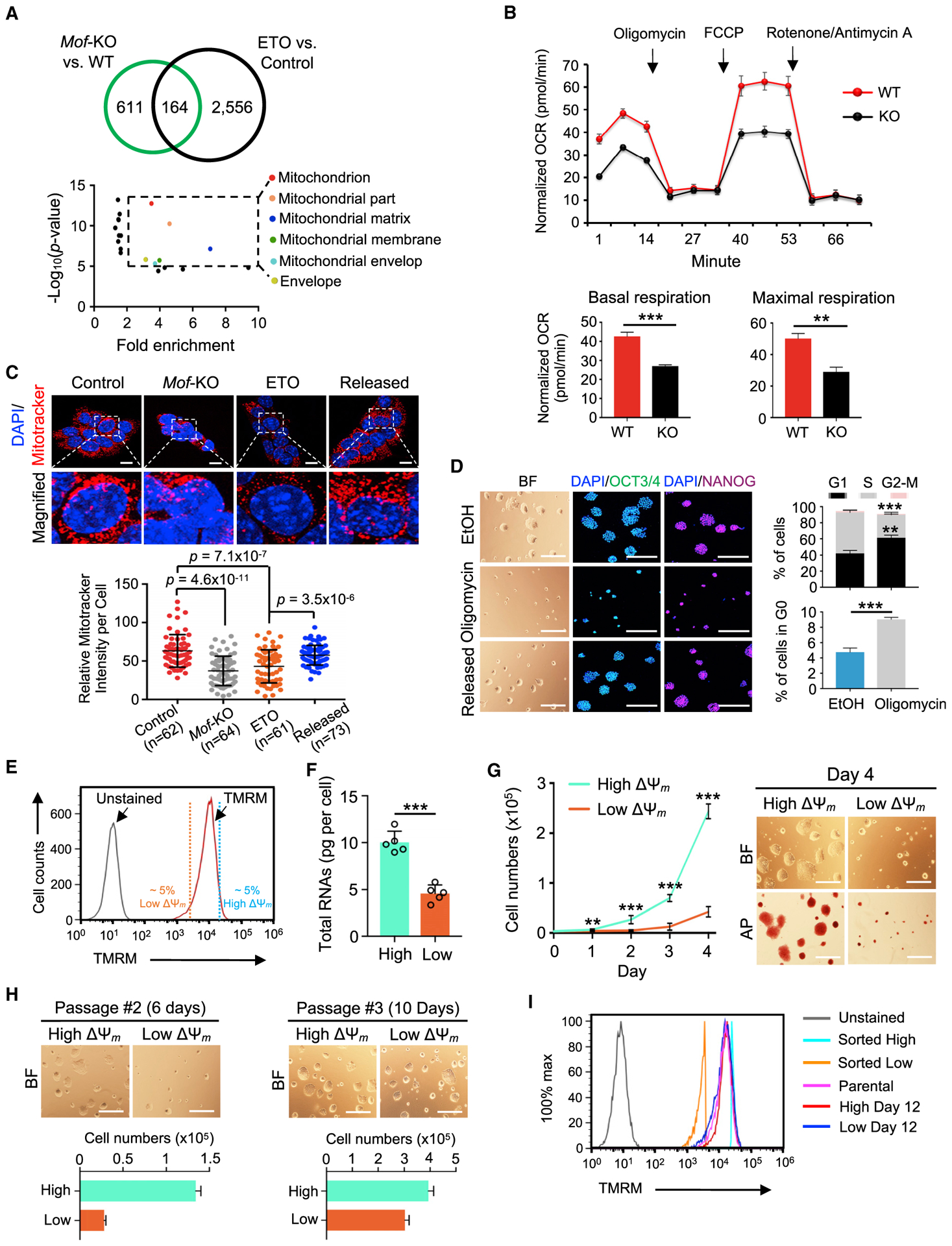
(A) Top, Venn diagram showing 164 genes commonly downregulated in Mof KO and ETO-treated 2i ESCs. Bottom, GO terms for 164 genes. Dashed box denotes GO terms with significant enrichment (fold enrichment > 2).
(B) Top, oxygen consumption rate (OCR) for WT and Mof-KO 2i ESCs. Arrows indicate MitoStress conditions. Bottom, quantifications of basal and maximal OCR. Data are presented as means ± SEMs, n = 4 biological replicates.
(C) Top, MitoTracker staining (red) at indicated conditions. Nuclei were co-stained with DAPI. Scale bar, 10 μm. Representative confocal images from three independent experiments. Bottom, quantifications of MitoTracker intensity in each cell by ImageJ.
(D) Left, BF and IF confocal images of OCT3/4 and NANOG for EtOH control, Oligomycin-treated and released 2i ESCs. Nuclei were co-stained with DAPI. Scale bar, 100 μm for BF and 50 μm for IF. Right, quantifications of cell-cycle analysis. Data are presented as means ± SEMs, n = 3 biological replicates.
(E) FACS plots showing mitochondrial labeling with TMRM (ΔΨm).
(F) Total RNA amounts per cell for high and low ΔΨm 2i ESCs. Data are shown as means ± SEMs from five independent experiments.
(G) Left, growth curves of high and low ΔΨm 2i ESCs. Data are presented as means ± SEMs, n = 6 biological replicates. Right, representative images of BF and AP staining for high and low ΔΨm cells at day 4. Scale bar, 100 μm.
(H) Representative images of BF (top) and qualifications of cell numbers (bottom) at passages #2 (left) and #3 (right) of high and low ΔΨm cells. Data are shown as means ± SEMs, n = 3 technical replicates. Scale bar, 100 μm.
(I) FACS plots showing TMRM profiles for parental, sorted high, and low ΔΨm 2i ESCs at days 0 and 12.
For (B), (D), (F), and (G), **p < 0.01, ***p < 0.001.
p values are calculated by the unpaired two-tailed Student’s t-test. Data are presented as mean ± SEMs. n, number of cells.
Low Mitochondrial Activity Is a Hallmark of Quiescence in Ground-State ESCs
To explore whether reduced mitochondrial activity, or OXPHOS, is an inherent feature of the quiescent ground-state ESCs, we isolated cells with intrinsically high ΔΨm (top 5%) or low ΔΨm (bottom 5%) levels by fluorescence-activated cell sorting (FACS) after staining cells with tetramethylrhodamine methyl ester (TMRM) (Figure 7E) (Schieke et al., 2008). As shown in Figure 7F, low ΔΨm cells had significantly lower amounts of total RNAs per cell and slower proliferation rates as compared to high ΔΨm cells (Figure 7G). These low ΔΨm cells were able to increase their growth at 10-12 days post-FACS (Figure 7H), coinciding with regaining the heterogeneity of mitochondrial activities (Figure 7I). These results suggest that reduced mitochondrial activity is a hallmark of reversible pluripotent quiescence in 2i ESCs.
DISCUSSION
Previous studies show that the loss of function of MOF or H4K16ac eventually leads to cell death via apoptosis or autophagy (Füllgrabe et al., 2013; Thomas et al., 2008). Here, we show that Mof deletion in ground-state ESCs leads to a quiescent state that maintains the pluripotent characteristics. This is in contrast to the function of MOF in the maintenance of S/L ESC self-renewal (Chelmicki et al., 2014; Li et al., 2012). The specific function of MOF in ground-state ESCs is not due to differences in the levels of MOF and global distribution of H4K16ac (Figures S1A and S1I). Instead, MOF regulation of FAO activity is the main contributor to the distinct cell fate choices. Notably, while the regulation of FAO-related genes by MOF has been observed in other contexts such as cardiomyocytes (Chatterjee et al., 2016), Mof deletion-mediated quiescence is only observed in ground-state ESCs, probably due to the ground-state-specific metabolic state.
It has been reported that MOF directly regulates the expression of respiratory genes in the mitochondria of HeLa cells (Chatterjee et al., 2016). Although we cannot completely rule out the direct role of MOF in regulating the mitochondrial genome in ESCs, our data suggest that MOF probably acts indirectly through the regulation of the nuclear FAO pathway in ground-state ESCs. The inhibition of Cpt1a or Acaa2, two nuclear FAO pathway genes, phenocopies Mof deletion (Figures 4I–4L, S3K, and S3L). Furthermore, FAO metabolite (e.g., l-carnitine) is able to partially rescue Mof deletion-induced quiescence (Figures 3D and 3E). The partial rescue by l-carnitine or the metabolite cocktail is not entirely surprising, given that MOF regulates multiple rate-limiting steps of the FAO pathway (Figure 2G). The function of MOF in regulating FAO metabolites implies that it may also be involved in balancing the content of saturated and unsaturated fatty acids, an important feature for highly proliferating cells (Zhu and Thompson, 2019), which warrants future investigations. Although FAO is one of the potential sources for Ac-CoA production (Carracedo et al., 2013), FAOi does not significantly change global histone acetylation, including H4K16ac (Figure S3J). It suggests that pluripotent quiescence is probably not induced by alterations in the nuclear pool of Ac-CoA. Whether FAOi affects protein acetylome and the epigenomic landscape remains to be determined.
Our study shows that the FAO pathway plays an essential role in cell fate decisions in ground-state ESCs and naive hESCs. Unlike FAO functions in adult stem cells (Ito et al., 2012; Knobloch et al., 2017), the inhibition of FAO does not affect self-renewal or pluripotency in ground-state or naive ESCs. Instead, these cells enter a reversible pluripotent quiescence that is differentiation competent. We find that FAO is likely a carbon fuel source for mitochondrial respiration (i.e., OXPHOS) in naive and ground-state ESCs. We show that this function is conserved in naive hESCs, which have a higher mitochondrial oxygen consumption relative to primed hESCs and require mitochondrial respiration for maximal proliferation (Carbognin et al., 2016; Sperber et al., 2015; Takashima et al., 2014; Theunissen et al., 2014). Our study shows that MOF and FAO are critical upstream regulators of OXPHOS (Figures 7A–7C). It highlights a specific role of FAO-dependent OXPHOS activities in establishing quiescence in naive and ground-state ESCs, which is consistent with the preference for mitochondrial OXPHOS in pre-implantation embryos (Zhang et al., 2018a). We find that ground-state ESCs harboring low ΔΨm exhibit defined features of quiescence without compromising the self-renewal or differentiation potential (Figures 7E–7G and S7B–S7D). These cells are able to re-enter the proliferation phase based on the OXPHOS flux (Figures 7H, 7I and S7F). In comparison, inhibiting OXPHOS in S/L ESCs or primed hESCs leads to a distinct cell fate (Figures S7E and S7H). It will be of future interest to dissect the underlying mechanism for the OXPHOS flux and unveil the role of mitochondrial retrograde signaling (Butow and Avadhani, 2004; Xu et al., 2013) in the regulation of pluripotency.
Implantation is a critical event in mammalian development and results from a functional coordination between the blastocyst and the receptive uterus (Wang and Dey, 2006). The process of implantation is temporarily suspended at the blastocyst stage under unfavorable environments (e.g., temperature change, limited nutrients), protecting the fetus from stress during pregnancy in >130 mammalian species (Fenelon et al., 2014; Renfree and Fenelon, 2017). Transcriptome analysis in mouse and human pre-implantation embryos reveals a significant upregulation of OXPHOS-related genes, which are inactivated after implantation (Zhang et al., 2018a). Furthermore, a metabolic switch from OXPHOS to glycolysis occurs right after implantation (Barbehenn et al., 1978; Brinster and Troike, 1979), suggesting that metabolic plasticity at the onset of implantation acts as a rheostat to enable the spatiotemporal control of implantation. While no significant change in Mof expression is documented in diapaused epiblasts (Boroviak et al., 2015), the downregulation of H4K16ac has been reported in rodent embryonic diapause (Bulut-Karslioglu et al., 2016; Hussein et al., 2020). Furthermore, we show that diapaused epiblasts exhibit a lower expression of FAO-related genes relative to E4.5 epiblasts (Figure S5A), and the pharmacological inhibition of FAO activity by ETO induces a blastocyst pausing ex vivo (Figures 5F–5J and S5E). FAOi-induced transcriptional changes share key features with developmental diapause (Figures 5C–5E and S5B). Future studies on whether FAOi induces embryonic diapause in vivo will be important to understand how the FAO pathway regulates blastocysts to enter and exit from embryonic diapause. The dynamic regulation of MOF, H4K16ac, and FAO during embryonic diapause also merits further investigations. In comparing our study to previous Myc deletion- and mTORi-induced pluripotent quiescence (Bulut-Karslioglu et al., 2016; Scognamiglio et al., 2016), we found that FAOi-induced transcriptome changes are more similar to those of mTOR inhibition (Figure S5C). It is likely that FAOi modulates mTOR signaling through AMPK activation (Figure S5D). However, since mTORi also induces paused pluripotency in S/L ESCs (Bulut-Karslioglu et al., 2016), the interplay between mTOR and FAO may be context dependent, which warrants future studies.
Limitations of Study
Our study provides mechanistic insights into how MOF rewires cellular metabolism for cell fate determination. However, there are limitations that need to be recognized. Due to the lack of a high-quality inhibitor of MOF, we are unable to test whether MOF inhibition induces a reversible pluripotent quiescence. In addition, it remains unclear what factors contribute to sustaining the self-renewal of Mof null 2i ESCs. Further investigations on the types of fatty acids and lipid constituents affected by MOF depletion will be important to supplement our findings. Finally, we have not yet determined whether FAOi induces embryonic diapause in human blastocysts, which have distinct developmental windows as compared to mouse blastocysts. A further understanding of this will provide potential therapeutic opportunities for reproductive health.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yali Dou (yalidou@usc.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets/code generated or analyzed during this study. The accession numbers for previously published RNA-seq and ChIP-seq datasets are provided in the “Other” section of the Key Resources Table. The accession number for the RNA-seq data files reported in this paper is NCBI GEO: GSE137138: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137138
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-MOF | Bethyl Laboratories | Cat#A300-992A; RRID:AB_805802 |
| Anti-OCT3/4 | Santa Cruz Biotechnology | Cat#sc-5279; RRID:AB_628051 |
| Anti-NANOG | Novus Biologicals | Cat#AF2729; RRID:AB_2150103 |
| Anti-OTX2 | R & D Systems | Cat#AF1979; RRID:AB_2157172 |
| Anti-SMA | SIGMA ALDRICH | Cat#A2547; RRID:AB_476701 |
| Anti-TUJ1 | Biolegend | Cat#801201; RRID:AB_2313773 |
| Anti-SOX17 | R & D Systems | Cat#AF1924; RRID:AB_355060 |
| Anti-KLF4 | Santa Cruz Biotechnology | Cat#sc-166238; RRID:AB_2130234 |
| Anti-TFCP2L1 | R & D Systems | Cat#AF5726; RRID:AB_2202564 |
| Anti-β-ACTIN | Santa Cruz Biotechnology | Cat#sc47778; RRID:AB_2714189 |
| Anti-AMPKα | Cell signaling | Cat#2532; RRID:AB_330331 |
| Anti-Phospho-AMPKα (Thr172) | Cell signaling | Cat#4188; RRID:AB_2169396 |
| Anti-S6K1 | Santa Cruz Biotechnology | Cat#sc-230; RRID:AB_632156 |
| Anti-Phospho-S6K1 (Thr389) | Cell signaling | Cat#9234; RRID:AB_2269803 |
| Anti-AKT | Cell signaling | Cat#9272; RRID:AB_329827 |
| Anti-Phospho-AKT (Ser473) | Cell signaling | Cat#4060; RRID:AB_2315049 |
| Anti-H4K16ac | Millipore | Cat#07-329; RRID:AB_310525 |
| Anti-H3K27ac | Abcam | Cat#ab4729; RRID:AB_2118291 |
| Anti-H3K9ac | Abcam | Cat#ab4441; RRID:AB_2118292 |
| Anti-H3K56ac | Abcam | Cat#ab76307; RRID:AB_1523762 |
| Anti-H4ac | Millipore | Cat# 06-866; RRID:AB_310270 |
| Anti-Histone H3 | Abcam | Cat#ab1791; RRID:AB_302613 |
| Anti-Histone H4 | Active Motif | Cat#39269; RRID:AB_2636967 |
| Goat anti-Mouse IgG Alexa FluorR 555 | Abcam | Cat#ab150114; RRID:AB_2687594 |
| Goat anti-Rabbit IgG Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat#A32733; RRID:AB_2633282 |
| Goat anti-Mouse IgG FITC-conjugated | Thermo Fisher Scientific | Cat#62-6511; RRID:AB_2533946 |
| Donkey anti-Mouse Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-31571; RRID:AB_162542 |
| Donkey anti-Goat IgG AlexaFluorR 555 | Abcam | Cat#ab150130; RRID:AB_10894526 |
| Donkey anti-Goat IgG AlexaFluor 488 | Thermo Fisher Scientific | Cat#A-11055; RRID:AB_2534102 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ESGRO® leukemia inhibitory factor (LIF) | EMD Millipore | Cat#ESG1107 |
| human LIF | SIGMA ALDRICH | Cat#L5283 |
| PD0325901 | SIGMA ALDRICH | Cat#PZ0162 |
| CHIR99021 | SIGMA ALDRICH | Cat#SML1046 |
| Geltrex | Thermo Fisher Scientific | Cat#A1413202 Cat#A1413302 |
| Ndiff227 media | Clontech | Cat#Y40002 |
| Activin A | R & D Systems | Cat#338-AC |
| bFGF | R & D Systems | Cat#233-FB |
| N2 supplement 100X | Thermo Fisher Scientific | Cat#17502001 |
| B27 supplement 50X | Thermo Fisher Scientific | Cat#17504-044 |
| mTeSR1 media | STEMCELL Technologies | Cat#85850 |
| Gö6983 | SIGMA ALDRICH | Cat#G1918 |
| L-Carnitine | SIGMA ALDRICH | Cat#C0158 |
| O-Acetyl-L-Carnitine | SIGMA ALDRICH | Cat#A6706 |
| AICAR | Cayman | Cat#10010241 |
| Coenzyme A | SIGMA ALDRICH | Cat#C4780 |
| Etomoxir | SIGMA ALDRICH | Cat#E1905 |
| Trimetazidine | SIGMA ALDRICH | Cat#653322 |
| INK128 | Medchem Express | Cat#HY-13328 |
| Oligomycin A | Cayman | Cat#11342 |
| FCCP | Cayman | Cat#15218 |
| Antimycin A | SIGMA ALDRICH | Cat#A8674 |
| Rotenone | SIGMA ALDRICH | Cat#R8875 |
| KSOMaa EvolveR medium | Zenith Biotech | Cat#ZEKS-050 |
| Bovine Serum Albumin solution | SIGMA ALDRICH | Cat#A8412 |
| MitoTracker Red CMXRos | SIGMA ALDRICH | Cat#M7512 |
| Tetramethylrhodamine methyl ester | Thermo Fisher Scientific | Cat#T668 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#62249 |
| Pyronin Y | SIGMA ALDRICH | Cat#213519 |
| Propidium Iodide | SIGMA ALDRICH | Cat#00-6990-50 |
| FITC-Annexin V | Bioscience | Cat#556419 |
| O-Propargyl Puromycin (OP-Puro) | Jena Bioscience | Cat#NU-931-05 |
| Critical Commercial Assays | ||
| VECTOR® Red Alkaline Phosphatase Kit | Vector Laboratories | Cat#SK-5100 |
| LookOut® Mycoplasma PCR Detection Kit | SIGMA ALDRICH | Cat#MP0035 |
| Click-iT™ Plus EdU Flow Cytometry Assay Kits | Thermo Fisher Scientific | Cat#C10632 Cat#C10634 |
| Click-iT Cell Reaction Buffer Kit | Thermo Fisher Scientific | Cat#C10269 |
| Qubit™ RNA HS assay kit | Thermo Fisher Scientific | Cat#Q32855 |
| Seahorse XF Base Medium | Agilent Technologies | Cat#102353-100 |
| Deposited Data | ||
| RNA-seq | This paper | GSE137138 |
| Experimental Models: Cell Lines | ||
| Mouse ES cells: E14tg2a | ATCC | Cat#CRL-1821™ |
| Mouse ES cells: Moffl/fl; Cre-ER™ | Y. Dou | Li et al., 2012 |
| Mouse ES cells: Oct4-GiP | Austin Smith | Ying et al., 2008 |
| MEFs | This paper | N/A |
| Human ES cells: Primed H9 | WiCell | Cat#WAe009-A |
| Human ES cells: Naive H9 | Austin Smith | Guo et al., 2017 Takashima et al., 2014 |
| Experimental Models: Organisms/Strains | ||
| (C57BL/6J × DBA/2J)F1 females | Jackson Laboratory | Cat#100006 |
| (C57BL/6J × DBA/2J)F1 males | Jackson Laboratory | Cat#100006 |
| C57BL/6NCrl female | Charles River Laboratory | Cat#493 |
| (C57BL/6J × SJL/J)F1 males | Jackson Laboratory | Cat#100012 |
| Oligonucleotides | ||
| Primers for RT-qPCR (see Table S7) | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Tophat2, v2.1.1 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/manual.shtml |
| Integrative Genomics Viewer (IGV), v2.3 | Thorvaldsdóttir et al., 2013 | http://software.broadinstitute.org/software/igv/ |
| Cufflinks, v2.2.1 | Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cuffiinks/ |
| CummeRbund, v2.1 | N/A | http://compbio.mit.edu/cummeRbund/ |
| Bowtie2, v2-2.2.4 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS, v1.4 | Zhang et al., 2008 | http://liulab.dfci.harvard.edu/MACS/ |
| HOMER v4.10 | Salk Institute | http://homer.ucsd.edu/homer/ |
| DeepTools, v3.2.1 | Ramírez et al., 2016 | https://github.com/deeptools/deepTools |
| Panther Classification System, v13.0 | N/A | http://pantherdb.org/ |
| Mouse ENCODE Consortium | Amemiya et al., 2019; ENCODE Project Consortium et al., 2012 | https://sites.google.com/site/anshulkundaje/projects/blacklists |
| GraphPad Prism, v7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo, v10.0.7r2 | FlowJo LLC | https://docs.fiowjo.com:443 |
| bcl2fastq2 software (v2.17) | Illumina LLC | https://support.illumina.com/sequencing/conversion-software.html |
| Cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| FastQC | Babraham institute | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trim Galore | Babraham institute | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| EpiProfileLite | Sidoli et al., 2019 | https://github.com/zfyuan/EpiProfileLite |
| MetaboAnalyst 4.0 | Chong et al., 2018 | https://www.metaboanalyst.ca |
| Agilent MassHunter Quantitative Analysis (for QQQ) Version B.07 | Agilent Technologies | https://www.agilent.com/en/support/software-informatics/qual-b-0700-sp1 |
| Other | ||
| RNA-seq for early embryonic stages | Boroviak et al., 2015 | E-MTAB-2958 |
| RNA-seq for dormant state by Myc deletion | Scognamiglio et al., 2016 | E-MTAB-3386 |
| RNA-seq for paused state by mTOR inhibition | Bulut-Karslioglu et al., 2016 | GSE81285 |
| ChIP-seq for MOF | Li et al., 2012 | GSE37268 |
| ChIP-seq for H4K16ac | Valsecchi et al., 2018 | GSE109897 |
| ChIP-seq for H4K16ac | Taylor et al., 2013 | GSE47761 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
3-4-week-old female and 2-6-month-old male C57BL/6 mice were used for all experiments. Mice were maintained on 12 hr light/dark cycle in the Transgenic Animal Model Core, University of Michigan. All animal procedures were reviewed and approved by the University of Michigan Institutional Animal Care and Use Committee. All animal experiments were conducted in accordance with “The Guide for the Care and Use of Laboratory Animals” Revised 2011. ULAM is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC, Intl).
Mouse and human cell lines
Moffl/fl; Cre-ERTM ESCs were derived from C57BL/6 mouse strain, and maintained as previously described (Li et al., 2012). E14tg2a (E14) (ATCC, #CRL-1821™) and Oct4-GiP ESC lines (from Dr. Austin Smith, University of Cambridge, United Kingdom) were used for all pharmacological experiments. Human (h) female ESC line H9 was purchased from WiCell (#WAe009-A). Naive hESCs (from Dr. Austin Smith, University of Cambridge, United Kingdom) were derived from H9 hESCs via transient inhibition of histone deacetylase and maintained as previously described (Guo et al., 2017; Takashima et al., 2014). E13.5-derived mouse embryonic fibroblasts (MEFs) were isolated from FVB triple transgenic mice developed at the University of Michigan Transgenic Animal Model Core, carrying resistance cassettes for Neomycin, Hygromycin and Puromycin. Mitomycin-C-treated MEFs were maintained in KnockOut DMEM medium (Thermo Fisher Scientific, #10829018) containing 15% Fetal Bovine Serum (FBS, Atlas Biologicals, #F-0500-D), 2 mM Glutamine (Thermo Fisher Scientific, #25030-024), 1X Non-essential amino acids (Thermo Fisher Scientific, #11140050) and 0.055 mM 2-mercaptoethanol (Thermo Fisher Scientific, #21985023). Unless otherwise indicated, the sexes of commercial and requested cell lines have not originally been reported.
METHOD DETAILS
ESC culture and drug treatment
All mouse ESC lines were maintained on dishes coated with Geltrex (Thermo Fisher Scientific, #A1413202, 1:100) at 37°C and 5%CO2 in 2i/LIF culture condition. 2i/LIF medium contains N2B27 or Ndiff227 (Clontech, #Y40002) supplemented with PD0325901 (SIGMA ALDRICH, #PZ0162, 1 μM), CHIR99021 (SIGMA ALDRICH, #SML1046, 3 μM) and LIF (Millipore, #ESG1107, 1000 U/ml). N2B27 basal medium consists of a 1:1 mix of DMEM/F12 (Thermo Fisher Scientific, #11320-033) and Neurobasal (Thermo Fisher Scientific, #21103-049), N2 (Thermo Fisher Scientific, #17502001, 1:100), B27 (Thermo Fisher Scientific, #17504-044, 1:100), 2 mM Glutamine and 0.055 mM 2-mercaptoethanol. Medium change was performed every other day, and cells were passaged every two days by dissociation with StemPro Accutase (GIBCO, #A1110501). To delete Mof, the Moffl/fl; Cre-ERTM ESCs were treated with 4-OHT (SIGMA ALDRICH, #H7904, 0.1 μM) for 3 days. Deletion of Mof was confirmed by genotyping and western blotting. Mouse S/L ESC lines were grown in the KnockOut DMEM medium containing 15% FBS or 15% KnockOut Serum Replacement (Thermo Fisher Scientific, #10828028) plus 5% FBS, 2 mM Glutamine, 1X Non-essential amino acids, 0.055 mM 2-mercaptoethanol and 1000 U/ml LIF. E14 and Moffl/fl; Cre-ERTM ESC lines were routinely tested for negative mycoplasma contamination using the LookOut® Mycoplasma PCR Detection Kit (SIGMA ALDRICH, #MP0035) according to the manufacturer’s instructions.
Primed H9 hESCs (WiCell) were grown on Geltrex (Thermo Fisher Scientific, #A1413302) coated tissue culture dishes according to manufacturer’s recommendation, and maintained in mTeSR1 media (StemCell Technologies, Inc., #85850) at 37°C and 5% CO2. Medium change was performed daily. Differentiating cells were cleaned daily and healthy cells were passaged every 4-5 days using the chemical dissociation method with Versene (Thermo Fisher Scientific, #15040066). To promote cell survival, 1 μM of ROCK inhibitor Y-27632 (Tocris, #1254) was added in the media for the first day after each passaging. Transgene-independent naive hESCs derived from H9 hESCs were generously provided by the Austin Smith lab, and maintained as described (Guo et al., 2017; Takashima et al., 2014). Briefly, naive hESCs were routinely propagated on top of MEF feeders in N2B27+t2iLGö media consisting of 1 μM PD0325901, 1 μM CHIR99021, 10 ng/ml human LIF (SIGMA ALDRICH, #L5283) and 2 μM Gö6983 (SIGMA ALDRICH, #G1918). Naive hESCs were adapted to feeder-free condition for at least 2 passages prior to experiments and maintained in media described above except that CHIR99021 was used at a final concentration of 0.3 μM (also known as N2B27+tt2iLGö). Geltrex was added at the time of plating. Naive hESCs were maintained at 37°C, 5% CO2 and 5% O2, and passaged every 3-5 days by dissociation with StemPro Accutase at 37°C for 5 min. ROCKi was added at the time of re-plating and removed after 24 hr. No mycoplasma contamination test was conducted for hESC lines.
To inhibit fatty acid oxidation (FAO), Etomoxir (SIGMA ALDRICH, #E1905, 100 μM) or Trimetazidine (SIGMA ALDRICH, #653322, 200 μM) was added at the time of plating (for mouse ESCs) or 2-3 days after plating (for hESCs). To rule out potential off-target effects of Etomoxir (Divakaruni et al., 2018; Raud et al., 2018), we used Etomoxir at a final concentration of 100 μM. This concentration has also been used for mouse embryo culture (Dunning et al., 2010; Paczkowski et al., 2014). Oligomycin A (Cayman, #11342) was used to inhibit mitochondrial respiration at indicated concentrations. INK128 (Medchem Express, HY-13328) was used at a final concentration of 200 nM to inhibit mTOR activity (Bulut-Karslioglu et al., 2016). To rescue the quiescent phenotypes of Mof null 2i ESCs, the following FAO-related metabolites were used: L-Carnitine (SIGMA ALDRICH, #C0158, 1 or 3 mM), O-Acetyl-L-Carnitine (SIGMA ALDRICH, #A6706, 0.5 mM), AICAR (Cayman, #10010241, 0.1 mM) and Coenzyme A (SIGMA ALDRICH, #C4780, 0.1 mM). Cells were subjected to alkaline phosphatase (AP) staining using VectorR Red Alkaline Phosphatase Substrate Kit (VECTOR Laboratories, #SK-5100) according to the manufacturer’s instructions. Images were captured by the IX73 microscope system (Olympus).
Induction of epiblast-like cells (EpiLCs)
ESC-derived EpiLCs were performed as described (Buecker et al., 2014) with minor modifications. Specifically, 2×103 ground-state ESCs were seeded on top of MEF in 24-well plates. 24 hr later, cells were washed once with PBS and maintained in EpiLC medium containing Ndiff227, 1% KnockOut Serum Replacement, 20 ng/ml Activin A (R & D Systems, #338-AC) and 10 ng/ml bFGF (R & D Systems, #233-FB) for 5 days. Cells were then subjected to AP staining and immunofluorescence as described below.
ES cell derivation
Derivation of ESC lines was performed using N2B27+2i/LIF media as previously described (Mulas et al., 2019) with minor modifications. Briefly, E3.5 blastocysts cultured in the presence of H2O or Etomoxir were gently washed in M2 medium for 5 times, and each blastocyst was immediately placed on a layer of MEF feeders in pre-equilibrated N2B27+2i/LIF media. After 5-6 days in culture, each inner cell mass (ICM) was picked under a dissection microscope, dissociated into single cells with 20 μl drop of 0.25% Trypsin-EDTA, and seeded onto a new culture dish containing MEF feeders. ESCs were routinely cultured in N2B27+2i/LIF till establishing the cell lines.
Embryo culture
The blastocysts were obtained from (C57BL/6J × DBA/2J) F1 females (Jackson Laboratory, #100006) mated with (C57BL/6J × DBA/2J) F1 males (Jackson Laboratory, #100006). E3.5 blastocysts were directly flushed in M2 medium from super-ovulated pregnant females. Alternatively, super-ovulated C57BL/6NCrl female (Charles River Laboratory, #493) treated with PMSG and HCG (Van Keuren et al., 2009) were mated with (C57BL/6J × SJL/J) F1 males (Jackson Laboratory, #100012). One-cell embryos were collected approximately 12 hr after fertilization by flushing oviducts in M2 medium supplemented with penicillin and streptomycin (SIGMA ALDRICH). Embryos were subsequently cultured in KSOMaa EvolveR medium (Zenith Biotech, #ZEKS-050) supplemented with 4 mg/ml bovine serum albumin (BSA) in a 37°C, 5% CO2 humidified incubator until they reached to blastocyst stage. To inhibit FAO, the blastocysts were incubated with Etomoxir at a final concentration of 100 μM. The same volume of H2O was used as a vehicle. mTOR inhibitor INK128 (200 nM) was used for embryo culture as described above.
Immunofluorescence
Embryos were fixed in 4% paraformaldehyde (PFA, Ted Pella Inc. #18505)/PBS for 2 hr at 4°C. Fixed embryos were then washed with 0.1% Triton X-100 (SIGMA ALDRICH, #93443)/PBS twice, permeabilized in 0.25% Triton X-100/PBS (PBST) for 30 min at 4°C and blocked in blocking solution (10% Goat serum (Cell Signaling Technology, #5425) or 10% Donkey serum (SIGMA ALDRICH, #D9663), 0.1% BSA and 0.01% Tween20 in PBS) for 1 hr at 4°C. Incubation of primary antibodies diluted in blocking solution was performed at 4°C overnight. The next day, embryos were washed with 0.25% PBST three times and incubated with appropriate secondary antibodies at room temperature for 1 hr. Embryos were then washed with 0.25% PBST three times, and nuclei were stained with DAPI (Thermo Fisher Scientific, #62248) diluted in 0.25% PBST at room temperature for 15 min. After washing with 0.25% PBST three times, embryos were mounted on glass slides in Vectashield solution (Vector Laboratories, #H-1200) for imaging. Images were captured by a Leica SP5 upright 2-photon confocal microscope (Leica Microsystems). Fluorescence was excited with a 405 nm UV laser (DAPI), a 488 nm laser (Alexa Fluor488), a 561 nm laser (Alexa Fluor555) and a 633 nm laser (Alexa Fluor647). All confocal images were acquired using HC PL APO CS2 40X/1.25 oil immersion objective lens, with an optical section thickness of 0.5 μm. Image data were further processed by either Leica software or ImageJ software (Schneider et al., 2012). Immunofluorescence for ESCs was performed as described above, except that the fixation with 4% PFA/PBS was performed at 4°C for 30 min. Images were captured by a Leica SP5 upright 2-photon confocal microscope or the IX73 microscope system (Olympus). The primary antibodies and dilution were: Mouse anti-OCT3/4 (Santa Cruz Biotechnology, #sc-5279, 1:50), Goat anti-NANOG (Novus Biologicals, #AF2729, 1:100), Goat anti-OTX2 (R & D Systems, #AF1979, 1:50), Goat anti-TFCP2L1 (R & D Systems, #AF5726, 1:100), Mouse anti-SMA (SIGMA ALDRICH, #A2547, 1:200), Mouse anti-TUJ1 (Biolegend, #801201, 1:200), Goat anti-SOX17 (R & D Systems, #AF1924, 1:200). The secondary antibodies used were: Goat anti-Rabbit IgG Alexa Fluor Plus 647 (Thermo Fisher Scientific, #A32733, 1:1000), Goat anti-Mouse IgG FITC-conjugated (Thermo Fisher Scientific, #62-6511, 1:50), Goat anti-Mouse IgG Alexa FluorR 555 (Abcam, #ab150114, 1:1000), Donkey anti-Goat IgG AlexaFluor 488 (Thermo Fisher Scientific, #A-11055, 1:1000), Donkey anti-Goat IgG AlexaFluorR 555 (Abcam, #ab150130, 1:1000) and Donkey anti-Mouse Alexa Fluor 647 (Thermo Fisher Scientific, #A-31571, 1:1000).
Cell proliferation assay
1×104 cells were plated on 12-well tissue culture dishes with or without Etomoxir. At day 3, Etomoxir-treated cells with or without releasing from Etomoxir were cultured for another 3 days. Cells were then dissociated by StemPro Accutase, stained with Trypan Blue (Thermo Fisher Scientific, #15250061) and counted every 24 hr for 6 consecutive days. For mitochondrial respiration inhibition, 2×104 cells were seeded in a 6-well tissue culture dish with or without Oligomycin A, and counted every 24 hr for 3 consecutive days.
Three-germ layer differentiation
Embryoid body (EB) formation was performed by seeding 2×106 mouse ESCs onto 10 cm Petri-dish containing 12 mL of EB medium (the KnockOut DMEM, 15% FBS, 2 mM Glutamine, 1X Non-essential amino acids and 0.055 mM 2-mercaptoethanol) for 5 days. EBs were transferred to gelatin-coated dishes and cultured for 5 days prior to immunofluorescence. EB formation from naive hESCs was conducted as follows: Feeder-free undifferentiated naive hESC colonies were mechanically scrapped and cultured in suspension for 10 days in a modified FGF2-free hESC media (Thomson et al., 1998) containing 20% KnockOut Serum Replacement, 1X GlutaMAX (Thermo Fisher Scientific, #35050061) and 1X Non-essential amino acid in DMEM/F12. EBs were then plated and further differentiated on gelatin-coated dishes for one week. Differentiated cells were fixed with 4% PFA/PBS and stained for ectoderm (TUJ1), mesoderm (SMA) and endoderm (SOX17) markers as described above. EB suspension from naive hESCs at day 3, 6 and 9 were harvested for RT-qPCR analyses.
Mitotracker staining
Cells were grown on Lab-Tek II Chamber Coverglass (Thermo Fisher Scientific, #155382) and incubated with 250 nM MitoTracker Red CMXRos (Thermo Fisher Scientific, # M7512) under routine culture condition for 30 min. Cells were then washed twice with PBS, fixed in 4% PFA/PBS and stained with DAPI at room temperature. Mitochondrial morphology was visualized by the Leica SP5 upright 2-photon confocal microscope. Relative intensities of Mitotracker were quantified in each cell using ImageJ as previously described (Ma et al., 2019). Briefly, confocal images were converted into binary images, and threshold was set to remove the cytosolic or background signals. Fluorescence intensity per cell was then corrected by subtracting the background signals. Unpaired two-tailed Student’s t test was used for statistical analyses.
Mitochondrial membrane potential (ΔΨm) staining and FACS sorting
ΔΨm was stained using the dye tetramethylrhodamine methyl ester (TMRM) (Thermo Fisher Scientific, #T668) as described (Schieke et al., 2008). Briefly, cells were dissociated into single cells by StemPro Accutase, resuspended in FACS buffer (Phenol red-free HBSS 1X, 10 mM HEPES and 2% FBS) containing 25 nM TMRM and incubated at 37°C, 5% CO2 for 15 min. Cells were then washed and resuspended in FACS buffer with SYTOX™ Blue (Thermo Fisher Scientific, #S34857). Cells of top and bottom 5% of TMRM fluorescence intensity were sorted on the MoFlo Astrios cell Sorter (BECKMAN COULTER Life Sciences).
Cell cycle analysis
To distinguish resting cells at G0 phase from other phases of the cell cycle, we measured DNA and RNA contents by double staining live cells with Hoechst 33342 (Thermo Fisher Scientific, #62249) and Pyronin Y (SIGMA ALDRICH, #213519), respectively (Eddaoudi et al., 2018). Staining solution containing culture media and 25 mM HEPES (Thermo Fisher Scientific, #15630080) was pre-warmed at 37°C, and used to resuspend single cells at a final concentration of 1×106 cells/ml. Cells were stained with 5 μg Hoechst 33342 at 37°C for 30 min in dark, followed by an incubation with 1 μg Pyronin Y at 37°C for 10 min in dark. Cells were then collected by centrifugation at 2500 rpm at 4°C for 10 min and resuspended in ice-cold staining solution. Labeled cells were subjected to flow cytometry analysis within 1 hr using the MoFlo Astrios cell Sorter.
Analysis of apoptosis
Single cell suspension was washed once with cold PBS, and resuspended in 100 μl of 1X Binding Buffer diluted from 10X Binding Buffer (0.1 M HEPES pH 7.4, 1.4 M NaCl and 25 mM CaCl2). 5 μl of FITC-Annexin V (Bioscience, #556419) was added and cells were incubated at room temperature for 15 min in dark. Cells were then resuspended in 400 μl of 1X Binding Buffer with Propidium Iodide (Thermo Fisher Scientific, #00-6990-50). Cells were left on ice and subjected to flow cytometry analysis within 1 hr on the MoFlo Astrios cell Sorter.
EdU incorporation assay
ESCs or embryos were incubated with 10 μM EdU under routine culture condition for 90 min. For EdU tracing experiments, naive hESCs were first incubated with 10 μM EdU for 2 hr (day 0). After washing once with PBS, cells were cultured and harvested at day 2 and 4 for subsequent analyses. EdU-labeled samples were fixed and stained using the Click-iT™ Plus EdU Flow Cytometry Assay Kits (Thermo Fisher Scientific, #C10632 or #C10634) according to manufacturer’s instructions. Samples were analyzed by flow cytometry analysis (for cells) or a Leica SP5 upright 2-photon confocal microscope (for embryos). EdU incorporation was quantified by median fluorescence intensity using FlowJo v10.0.7r2 software or mean fluorescence intensity using ImageJ.
Measurement of protein synthesis
Newly synthesized proteins in cells were determined by O-Propargyl Puromycin labeling (OP-Puro) (Jena Bioscience, #NU-931-05), a cell-permeable alkyne analog of Puromycin capable of incorporating into the C terminus of translating polypeptide chains. ESCs were incubated with 50 μM OP-Puro for 1 hr. Cells were then dissociated into single cells, fixed for 15 min at room temperature with 4% PFA/PBS, and permeabilized with PBS containing 1% BSA and 0.1% Saponin for 10 min at room temperature. Alexa 647-azide was then added together with Click-iT Cell Reaction Buffer Kit (Thermo Fisher Scientific, #C10269) according to the manufacturer’s instructions. Data were analyzed by flow cytometry using the MoFlo Astrios cell Sorter. Median OP-Puro fluorescence intensity was determined using FlowJo v10.0.7r2 software.
RNA sequencing
Total RNAs were extracted using the RNeasy mini kit (QIAGEN) in accordance to manufacturer’s instructions. Any residual genomic DNAs were removed on-column by treating with RNA-free DNase I (Thermo Fisher Scientific) for 15 min at room temperature. Ten nanogram of total extracted RNAs were used for library preparation. In all samples, duplicates were sequenced on Illumina Hi-Seq2500 High-Output with v4 single end 50 cycle at the University of Michigan core facility.
Quantitative reverse transcription PCR (RT-qPCR)
Total RNAs were extracted by the RNeasy mini kit (QIAGEN). cDNA was synthesized using Superscript III (Thermo Fisher Scientific) and oligoT primer (Thermo Fisher Scientific). qPCR was performed with Radiant Green Lo-ROX qPCR Kit (Alkali Scientific Inc.) in triplicate wells per sample. Results were analyzed by the comparative cycling threshold (ΔΔt) method and normalized against β-actin or GAPDH genes. Primers were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) or designed by Primer3 (http://primer3.ut.ee), and included in Table S7.
Determination of total RNAs per cell
Single cell suspension was filtered through 40 μm sterile cell strainer and incubated with either propidium iodide (Thermo Fisher Scientific, #P3566) or SYTOX™ Blue for 10 min on ice to exclude dead cells. Equal cell numbers from different conditions were sorted with MoFlo Astrios cell Sorter. Total RNAs were extracted by the RNeasy mini kit (QIAGEN) as described above. Total RNA amounts were quantified using Qubit™ RNA HS assay kit (Thermo Fisher Scientific, #Q32855) and normalized against the cell numbers.
Western blotting
Cells were washed twice in ice-cold PBS with protease inhibitor cocktail (SIGMA ALDRICH, #11836153001), lysed in RIPA buffer (SIGMA ALDRICH, #R0278) and chilled on ice for 5 min. The extracts were then centrifuged at 15000 rpm for 5 min at 4°C. The suspension was collected and resuspended in SDS-sample buffer. 10-20 μg of proteins or histones were separated on a 10%−15% Protogel (National Diagnostics, #EC-890) and transferred onto polyvinylidene difluoride membrane (Millipore). The membrane was blocked in blocking solution (5% milk/PBS), followed by incubation at 4°C overnight with primary antibody in blocking solution. Membranes were washed 3 times in 0.1% tween20/TBS 1X (TBST) and incubated with the HRP-conjugated anti-Mouse/Rabbit/Goat secondary antibodies at room temperature for 1 hr. The membrane was developed using by Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, #32106), and images were captured by ChemiDoc™ Touch Imaging System (Bio-rad). The primary and secondary antibodies included: Anti-β-ACTIN (Santa Cruz Biotechnology, #sc47778, 1:2000), Mouse anti-OCT3/4 (Santa Cruz Biotechnology, #sc-5279, 1:100), Mouse anti-KLF4 (Santa Cruz Biotechnology, #sc-166238, 1:100), Goat anti-TFCP2L1 (R & D Systems, #AF5726, 1:100), Rabbit anti-MOF (Bethyl Laboratories, #A300-992A, 1:1000), Rabbit anti-H4K16ac (Millipore, #07-329, 1:2000), Rabbit anti-H3K27ac (Abcam, #ab4729, 1:1000), Rabbit anti-H3K9ac (Abcam, #ab4441, 1:1000), Rabbit anti-Histone H3 (Abcam, #ab1791, 1:2000), Rabbit anti-Histone H4 (Active Motif, #39269, 1:2000), Rabbit anti-H4ac (Millipore, #06-866, 1:2000), Rabbit anti-H3K56ac (Abcam, #ab76307, 1:2000), anti-mouse IgG Horseradish Peroxidase-linked whole antibody from Sheep (GE Healthcare, #NX931V, 1:3000), anti-Rabbit IgG Horseradish Peroxidase-linked whole antibody (GE Healthcare, #NA934, 1:3000) and Donkey anti-Goat IgG H&L HRP (Abcam, #ab6885, 1:2000).
Detection of phosphorylated proteins was performed as previously described (Kazyken et al., 2019). 2% BSA diluted in PBS were used for blocking reaction. Antibodies included: Rabbit-anti-AMPKα (Cell signaling, #2532, 1:3000), Rabbit anti-Phospho-AMP-Kα-Thr172 (Cell signaling, #4188, 1:5000), Rabbit anti-S6K1 (Santa Cruz Biotechnology, #sc-230, 1:7000), Rabbit anti-Phospho-S6K1-Thr389 (Cell signaling, #9234, 1:3000), Rabbit anti-AKT (Cell signaling, #9272, 1:2000) and Rabbit anti-Phospho-AKT-Ser473 (Cell signaling, #4060, 1:5000).
Histone extraction
Acid extraction of histones was performed as previously described (Shechter et al., 2007; Sidoli et al., 2016) with modifications. Briefly, approximately 7×106 cells were washed twice in ice-cold PBS followed by centrifugation at 700 rpm for 10 min at 4°C. Cell pellets were then resuspended in the hypotonic lysis buffer (10 mM Tris-Cl pH 8.0, 1 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 200 mM AEBSF and 2.5 μM Microcystin) and incubated for 30 min at 4°C. The intact nuclei were collected by centrifugation at 10,000 rpm for 10 min at 4°C, followed by incubation with 0.4 N H2SO4 for 2 hr. Acid-extracted proteins were precipitated overnight with Trichloroacetic acid at a final concentration of 33%. Pellets were collected by spinning in cooled tabletop centrifuge at 15,000 rpm for 10 min at 4°C, washed with ice-cold acetone, air-dried and resuspended in double distilled H2O.
Quantitative mass spectrometry analysis of histone
To prepare the histones for mass spectrometry (MS), three biological replicates of acid-extracted histones were chemically derivatized at the unmodified lysine residues and digested with trypsin, as described (Sidoli et al., 2016). These peptides were desalted and eluted by 70% acetonitrile/0.1% formic acid and injected directly for mass spectrometry with liquid chromatography. Briefly, samples were queued in a TriVersa NanoMate (Advion) and acquired by a contact closure in the Orbitrap Fusion Tribrid (Thermo Fisher Scientific) with a spray voltage of 1.7 kV and a gas pressure of 0.5 psi in the Nanomate. All scans were acquired in the orbitrap, at 240,000 resolution for the full MS and at 120,000 resolution for MS/MS. The AGC target for the tSIM-MSX scans was set to 10E6. The full description of the DI-MS acquisition method was previously described (Sidoli et al., 2019). For histone peptide quantification, raw files were searched using software EpiProfileLite (Sidoli et al., 2019). Intensities of histone peptides were analyzed in MS scans, and those of isobaric peptides were collected in targeted pre-set MS/MS scans. Intensities from MS scans were used to calculate the percentage of all peptides with the same amino acid sequence. Unique fragment ions in the MS/MS scans were extracted to discriminate isobaric peptide intensities from the MS scans.
Unlabeled targeted metabolomics
Metabolomics analyses were performed as previously described (Lee et al., 2019). For all metabolomics experiments, cells were seeded in 10 cm plates under the indicated conditions. The medium was aspirated, and cells were lysed using 80% cold methanol. The extracts were incubated at −80°C for 10 min and centrifugated at 14,000 rpm for 10 min at 4°C. Protein concentration was determined by processing a parallel 10 cm dish for each sample and used to normalize metabolite fractions across samples. Aliquots of the supernatants were then transferred to a fresh microcentrifuge tube and dried. Metabolite extracts were then re-suspended in 35 μl 50:50 MeOH:H2O mixture for LC-MS analysis. LC-MS analysis was performed using an Agilent 1290 UHPLC-6490 Triple Quadrupole MS system ran in negative and/or positive ion acquisition modes. For negative ion acquisition, a Waters Acquity UPLC BEH amide column (2.1 × 100 mm, 1.7 μm) with the mobile phase (A) consisting of 20 mM ammonium acetate, pH 9.6 in water, and mobile phase (B) consisting of acetonitrile was used. Gradient program was: mobile phase (B) was held at 85% for 1 min, increased to 65% in 12 min, then to 40% in 15 min and held for 5 min before going to initial condition and held for 10 min. For positive ion acquisition, a Waters Acquity UPLC BEH TSS C18 column (2.1 × 100 mm, 1.7 μm) was used with mobile phase (A) consisting of 0.5 mM NH4F and 0.1% formic acid in water and a mobile phase (B) consisting of 0.1% formic acid in acetonitrile. Gradient program was: mobile phase (B) was held at 1% for 1.5 min, increased to 80% in 15 min, then to 99% in 17 min and held for 2 min before going back to initial condition for 10 min. The column was kept at 40°C and 3 μL of sample were injected into the LC-MS/MS with a flow rate of 0.2 ml/min. Key parameters utilized by positive acquisition modes of AJS ESI were: Gas temp 250°C, Gas Flow 14 l/min, Nebulizer at 20 psi, Sheath Gas Heater 250°C, Sheath Gas Flow 11 L/min, Capillary 3000 V, Delta EMV was 350 V. For negative mode, Gas temp 250°C, Gas Flow 14 l/min, Nebulizer at 20 psi, Sheath Gas Heater 250°C, Sheath Gas Flow 11 L/min, Capillary 3000 V MS, Delta EMV was 350 V, Cell accelerator voltage was 4 V. Metabolomics data analysis was performed as follows: all targeted compounds were optimized with authentic standards for optimal detection by MS/MS conditions including collision energies and greatest abundant fragment ion, either manually or by Agilent optimization software tool. Retention times of listed compounds were measured by the LC-MRMs methods, and the final LC-dynamic MRMs methods were created with delta retention time windows of 1.4 to 3.4 min. Initial data processing was performed by Agilent MassHunter Quantitative Analysis (for QQQ) Version B.07. Integration of peak areas of detected compounds was performed by software analysis and validated by manually inspection. Metabolite counts were then normalized by the total intensity of all metabolites to reflect equal sample loading. Finally, each metabolite abundance in the sample was divided by the median of all abundance levels across all samples for statistical analyses and visualizations. The significance test was done by the two-tailed Student’s t test with a significant threshold level of 0.05. The p values were not adjusted in favor of downstream visual inspection of the data and a more flexible biological interpretation. Additional statistical analyses were carried out for further quality control in program R. Metabolite pathway enrichment analysis was performed using MetaboAnalyst 4.0 (Chong et al., 2018). The software is available at: https://www.metaboanalyst.ca
Determination of fatty acid oxidation (FAO) rate
FAO rate determination was measured as described (Zhang et al., 2018b). Briefly, cells with 70%−80% confluency were incubated in 250 μL of FAO reaction buffer (0.1 μL of 9,10-3H palmitate (1.0 mCi/ml), 3 μL of 10 mM cold palmitate in 10% BSA and 247 μL of serum-free Williams Medium E) at 37°C for 3 hr. FAO reaction buffer was collected and mixed with 96 μL of 1.3 M perchloric acid for protein precipitation. Total of 62.4 μL of 2 M KOH/0.6 M MOPS was added to neutralize pH. Cells left in each well were lysed using 200 μL of 0.1 M HCl. 2 μL of each lysate were used for protein concentration measurement using BCA method (BioRad). The FAO reaction buffer was loaded into equilibrated anion-exchange Dower 1×8-200 resin column and the effluent was collected. The column was washed with 2 mL of ddH2O and collected in vials. 500 μL of the mixed effluent were added into a scintillation vial for radioactivity measurement. Water (500 μl) alone or with 10 μL of FAO reaction buffer were used as the negative control and standard, respectively. FAO rate (nmol/mg/h) was calculated after normalization against total protein level for each sample.
Determination of oxygen consumption rate (OCR)
OCR was determined using the XFe96 extracellular flux analyzer (Seahorse Bioscience) in accordance with manufacturer’s instructions. WT and Mof null ESCs were seeded at 4×104 and 5×104 cells per well, respectively, onto Geltrex-coated XF96 cell culture microplates (Agilent Technologies, #102601-100). The plate was incubated overnight at 37°C, 5% CO2. Culture medium was exchanged to Seahorse XF base medium (Agilent Technologies, #102353-100) supplemented with 25 mM glucose and 1 mM sodium pyruvate for 1 hr prior to the assay. Mitochondrial stress was induced by sequential injection of 1 μM Oligomycin A, 0.25 μM FCCP (Cayman, #15218) and 2.5 μM Antimycin A (SIGMA ALDRICH, #A8674) plus 2.5 μM Rotenone (SIGMA ALDRICH, #R8875). OCR (pmol/min) values were normalized against protein concentration in each well. Average values of the first three measurements (min 1-14) were presented as the basal OCR, whereas the maximal OCR was determined by differences in OCR values after FCCP injection (min 40-53) and Antimycin A/Rotenone injection (min 60-73).
RNA-seq data analysis
For base calling and data cleaning, Fastq generation was performed using Illumina’s bcl2fastq2 software (v2.17). The cutadapt (https://cutadapt.readthedocs.io/en/stable/) and FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) tools wrapped in Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) were used to trim low quality bases (Q < 20) and adaptor for raw sequences. For RNA-seq read mapping and signal normalization, RNA-seq data were mapped to the mm10 genome of mouse using Tophat2 (2.1.1) (Kim et al., 2013). Duplicated reads for paired-end data from public datasets were removed, but not for single-end data by SAMtools (v1.5) (Li et al., 2009a). The bigwig files for visualization in Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al., 2013) were generated from BAM files by using “bamCoverage” from deepTools (v3.2.1) (Ramírez et al., 2016) with parameters “-normalizeUsingRPKM-minMappingQuality 30-binSize 50.” To perform peak calling and gene annotation, BAM files of mapping results were merged for the same sample using SAMtools and converted to BED format using BEDTools (Quinlan and Hall, 2010). Peaks of regulatory regions were called for each sample using MACS (v 1.4) (Zhang et al., 2008) with parameters “-w -S -p 0.00001.” The input signal was used as the control to call peaks for the ChIP-seq datasets (see also below). The heatmap plot of signals centered on peaks and gene promoters was performed by deepTools2 (v3.2.1) (Ramírez et al., 2016). Gene annotation of peaks was performed by using HOMER (v4.10) (http://homer.ucsd.edu/homer/) with default parameters (Heinz et al., 2010). For RNA-seq quantification and differential expression analysis, aligned reads were assembled using Cufflinks (v2.2.1) (Trapnell et al., 2012) and assembled transcriptome catalog was used as input for Cuffdiff2 to determine expression levels (FPKM, Fragments Per Kilobase per Million mapped reads) and differential expression between conditions using default options. CummeR-bund v2.1 (http://compbio.mit.edu/cummeRbund/) was used to process, index and visualize the output of the Cuffdiff2 analyses. Genes with FDR < 0.05 were considered to be significant. For Pearson correlation coefficients, Log10 transformed FPKM values of all the selected genes were used as input, and Pearson correlation coefficients between each two samples were calculated using function cor() in R. The matrix of Pearson correlation coefficients was used to generate heatmap using R package pheatmap.
For suppressing batch effects and hierarchical clustering, we followed the “batch mean-centering” approach for batch effect removal (Lazar et al., 2013) in order to compare our RNA-seq data with public RNA-seq datasets. Briefly, we have separately mean-centered the Log2(FPKM+1) value of each gene by subtracting the mean Log2(FPKM+1) across all samples and across each RNA-seq samples. We used “1 - Spearman” correlation coefficient as the distance in the hierarchical clustering since numerical values of the mean-centered expression may have different dynamic ranges in different batches (Bulut-Karslioglu et al., 2016). We computed the dissimilarity values with “as.dist” function in R and then fed these values into hclust with “average” algorithm to do the agglomeration. Finally, we plotted the dendrogram by R.
ChIP-seq data analysis
ChIP-seq data were mapped to the mm10 genome of mouse by using Bowtie2 (v2-2.2.4) (Langmead and Salzberg, 2012). Any duplicated reads were removed using SAMtools (v1.5) (Li et al., 2009a). The fold enrichments of IP to input for IGV visualization were implemented by deepTools2 subcommands bamCompare with parameters “-ignoreDuplicates-minMappingQuality 30-normali-zeUsingRPKM-binSize 1-ratio ratio” (Thorvaldsdóttir et al., 2013).
H4K16ac ChIP-seq data of S/L and 2i ESCs were downloaded from GEO accession number GSE47761 and GSE109897, respectively. Raw reads were cleaned and mapped to mouse mm10 genome using Bowtie2. Genome-wide RPKM value with 20 bp resolution was calculated from mapping results by using deepTools subcommand “bamCoverage” with parameters “-outFileFormat bigwig-normalizeUsing RPKM-minMappingQuality 30-binSize 20-smoothLength 60-ignoreDuplicates-ignoreForNormalization chrM chrX chrY-blackListFileName blacklist.” We filtered artifact regions that tend to show artificially high signals by using the annotations from Mouse ENCODE Consortium (Amemiya et al., 2019; ENCODE Project Consortium et al., 2012) (https://sites.google.com/site/anshulkundaje/projects/blacklists). DeepTools subcommand “multiBigwigSummary” was used to compute the average RPKM scores for each sample in 100 kb resolution genomic windows. Spearman correlation and scatterplot were performed by ggplot2.
Graphical abstract generation
The graphical abstract of this study was generated using the web-based tool BioRender (https://biorender.com).
QUANTIFICIATION AND STATISTICAL ANALYSIS
Statistical analyses were performed by the two-tailed Student’s t test using GraphPad Prism 7.0 software. Data were presented as standard error of the mean (SEM). Details of individual tests including number and type of replication were highlighted within each figure legend. p values of less than 0.05 were considered statistically significant. All statistics are *p < 0.05, **p < 0.01, ***p < 0.001.
GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis was performed using Cytoscape (version 3.7.1) (Shannon et al., 2003) plugin ClueGO (version 2.5.0) (Bindea et al., 2009). p values were calculated by hypergeometric test and corrected to q values by Benjamini-Hochberg procedure that reduces the false discovery rate of multiple hypothesis testing. Dot plots of GO enrichment were implemented by R package clusterProfiler (version 1.4.0) (Yu et al., 2012). Alternatively, GO analysis was conducted using Panther Classification System Version 13.1 (http://pantherdb.org/). Statistical overrepresentation test was performed with “default settings.” We employed “Fisher’s Exact with FDR multiple test correction” for further analyses. GO terms with FDR < 0.05 or 0.01 and fold change > 2 were considered as significant enrichment.
Gene set enrichment analysis of certain GO terms was performed by GSEA (version 3.0) (Subramanian et al., 2005). A detailed description of GSEA methodology and interpretation is available at http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. Significance of gene sets from the GSEA was based on normalized enrichment score (NES) and p value or false discovery rate q value (FDR q-val), which determines the probability that a gene set with a given NES represents a false-positive finding.
For cross-reference of RNA-seq with ChIP-seq data, the fold enrichment (FE) of MOF and H4K16Ac peaks was calculated by map function in BEDTools (Quinlan and Hall, 2010). Starburst plot was performed as previously described (Wang et al., 2014) to show association between FE (Log2[FC] > 1) of H4K16Ac or MOF ChIP-seq peaks and expression changes ([FC] > 1.2 and [FDR] < 0.05) of corresponding genes after Mof deletion.
Supplementary Material
Highlights.
MOF blocks acquisition of the quiescent state in ground-state ESCs
MOF directly activates the fatty acid oxidation (FAO) pathway
MOF/FAO axis provides the carbon source for fueling mitochondrial respiration
FAO/OXPHOS axis-mediated blocking of quiescence is conserved in naive human ESCs
ACKNOWLEDGMENTS
We are grateful to Ms. Elizabeth Hughes, Wanda Filipiak, Galina Gavrilina, and Laura Keller for help with the embryo work; to Drs. David Lombard and Surinder Kumar for help with the Seahorse assay; to Dr. Diane C. Fingar for providing valuable reagents; to Dr. Hui Zhang for valuable advice; and to Dr. Austin Smith for providing Oct4-GiP ESCs and naïve hESCs. T.L.S. thanks the Rogel Cancer Center for research support. This work is supported by NIH grants GM082856 and NS101597 to Y.D. and NIHgrant NS101597 to S.B. C.A.L. is supported by the American Association for Cancer Research (AACR) NextGen Grant for Transformative Cancer Research (17-20-01-LYSS), the American Cancer Society (ACS) Research Scholar Grant (RSG-18-186-01), and the University of Michigan Comprehensive Cancer Center (UMCCC) Core Grant (P30 CA046592). The Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (grant agreement NNF14CC0001 to C.C.). Metabolomics studies performed at the University of Michigan are supported by NIH grant DK097153, the Charles Woodson Research Fund, and the UM Pediatric Brain Tumor Initiative.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2020.06.005.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Amemiya HM, Kundaje A, and Boyle AP (2019). The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep 9, 9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Megchelenbrink W, Peng T, Habibi E, Joshi O, Wang SY, Wang C, Logie C, Poser I, Marks H, and Stunnenberg HG (2019). Epigenetic modulation of a hardwired 3D chromatin landscape in two naive states of pluripotency. Nat. Cell Biol 21, 568–578. [DOI] [PubMed] [Google Scholar]
- Barbehenn EK, Wales RG, and Lowry OH (1978). Measurement of metabolites in single preimplantation embryos; a new means to study metabolic control in early embryos. J. Embryol. Exp. Morphol 43, 29–46. [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, and Galon J (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, and Nichols J (2014). The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol 16, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A, and Bertone P (2015). Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev. Cell 35, 366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, and Troike DE (1979). Requirements for blastocyst development in vitro. J. Anim. Sci 49 (Suppl 2), 26–34. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, and Vallier L (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, and Wysocka J (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Biechele S, Jin H, Macrae TA, Hejna M, Gertsenstein M, Song JS, and Ramalho-Santos M (2016). Inhibition of mTOR induces a paused pluripotent state. Nature 540, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, and Avadhani NG (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1–15. [DOI] [PubMed] [Google Scholar]
- Carbognin E, Betto RM, Soriano ME, Smith AG, and Martello G (2016). Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 35, 618–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, and Thompson CB (2015). Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, and Pandolfi PP (2013). Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, and Smith A (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Seyfferth J, Lucci J, Gilsbach R, Preissl S, Bottinger L, Martensson CU, Panhale A, Stehle T, Kretz O, et al. (2016). MOF Acetyl Transferase Regulates Transcription and Respiration in Mitochondria. Cell 167, 722–738.e23. [DOI] [PubMed] [Google Scholar]
- Chelmicki T, Dündar F, Turley MJ, Khanam T, Aktas T, Ramírez F, Gendrel AV, Wright PR, Videm P, Backofen R, et al. (2014). MOF-associated complexes ensure stem cell identity and Xist repression. eLife 3, e02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117. [DOI] [PubMed] [Google Scholar]
- Cheung TH, and Rando TA (2013). Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, and Xia J (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46 (W1), W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Hsieh WY, Minarrieta L, Duong TN, Kim KKO, Desousa BR, Andreyev AY, Bowman CE, Caradonna K, Dranka BP, et al. (2018). Etomoxir Inhibits Macrophage Polarization by Disrupting CoA Homeostasis. Cell Metab. 28, 490–503.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, and Roeder RG (2005). Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885. [DOI] [PubMed] [Google Scholar]
- Dunning KR, Cashman K, Russell DL, Thompson JG, Norman RJ, and Robker RL (2010). Beta-oxidation is essential for mouse oocyte developmental competence and early embryo development. Biol. Reprod 83, 909–918. [DOI] [PubMed] [Google Scholar]
- Eddaoudi A, Canning SL, and Kato I (2018). Flow Cytometric Detection of G0 in Live Cells by Hoechst 33342 and Pyronin Y Staining. Methods Mol. Biol 1686, 49–57. [DOI] [PubMed] [Google Scholar]
- Evans MJ, and Kaufman MH (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, and Panning B (2008). An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenelon JC, Banerjee A, and Murphy BD (2014). Embryonic diapause: development on hold. Int. J. Dev. Biol 58, 163–174. [DOI] [PubMed] [Google Scholar]
- Finley LWS, Vardhana SA, Carey BW, Alonso-Curbelo D, Koche R, Chen Y, Wen D, King B, Radler MR, Rafii S, et al. (2018). Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat. Cell Biol 20, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, and Joseph B (2013). The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Gaeta X, Sahakyan A, Chan AB, Hong CS, Kim R, Braas D, Plath K, Lowry WE, and Christofk HR (2016). Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 19, 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Rostovskaya M, Clarke J, Dietmann S, Baker D, Sahakyan A, Myers S, Bertone P, Reik W, et al. (2017). Epigenetic resetting of human pluripotency. Development 144, 2748–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. (2008). The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol. Cell. Biol 28, 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, and Surani MA (2014). Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15, 416–430. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A, Bosch M, Reddy BJ, Schieber NL, Fajardo A, Rupérez C, Fernández-Vidal A, Ferguson C, Rentero C, Tebar F, et al. (2015). AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun 6, 7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein AM, Wang Y, Mathieu J, Margaretha L, Song C, Jones DC, Cavanaugh C, Miklas JW, Mahen E, Showalter MR, et al. (2020). Metabolic Control over mTOR-Dependent Diapause-like State. Dev. Cell 52, 236–250.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IY, Kwak S, Lee S, Kim H, Lee SE, Kim JH, Kim YA, Jeon YK, Chung DH, Jin X, et al. (2016). Psat1-Dependent Fluctuations in α-Ketoglutarate Affect the Timing of ESC Differentiation. Cell Metab 24, 494–501. [DOI] [PubMed] [Google Scholar]
- Inoki K, Kim J, and Guan KL (2012). AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol 52, 381–400. [DOI] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, and Pandolfi PP (2012). A PML-PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med 18, 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoutas A, Szymanski W, Rausch T, Guhathakurta S, Rog-Zielinska EA, Peyronnet R, Seyfferth J, Chen HR, de Leeuw R, Herquel B, et al. (2019). The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell Biol 21, 1248–1260. [DOI] [PubMed] [Google Scholar]
- Kazyken D, Magnuson B, Bodur C, Acosta-Jaquez HA, Zhang D, Tong X, Barnes TM, Steinl GK, Patterson NE, Altheim CH, et al. (2019). AMPK directly activates mTORC2 to promote cell survival during acute energetic stress. Sci. Signal 12, eaav3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Cho HI, Park SH, Kim JH, Chai YG, and Jang YK (2015). The histone acetyltransferase Myst2 regulates Nanog expression, and is involved in maintaining pluripotency and self-renewal of embryonic stem cells. FEBS Lett. 589, 941–950. [DOI] [PubMed] [Google Scholar]
- Knobloch M, Pilz GA, Ghesquière B, Kovacs WJ, Wegleiter T, Moore DL, Hruzova M, Zamboni N, Carmeliet P, and Jessberger S (2017). A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep. 20, 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar C, Meganck S, Taminau J, Steenhoff D, Coletta A, Molter C, Weiss-Solís DY, Duque R, Bersini H, and Nowé A (2013). Batch effect removal methods for microarray gene expression data integration: a survey. Brief. Bioinform 14, 469–490. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kremer DM, Sajjakulnukit P, Zhang L, and Lyssiotis CA (2019). A large-scale analysis of targeted metabolomics data from heterogeneous biological samples provides insights into metabolite dynamics. Metabolomics 15, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, and Belmonte JC (2017). Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet 18, 180–191. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009a). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, and Dou Y (2009b). Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol. Cell 36, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, and Dou Y (2012). The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 11, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, and Dent SY (2007). Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev. Dyn 236, 1547–1557. [DOI] [PubMed] [Google Scholar]
- Luo H, Shenoy AK, Li X, Jin Y, Jin L, Cai Q, Tang M, Liu Y, Chen H, Reisman D, et al. (2016). MOF Acetylates the Histone Demethylase LSD1 to Suppress Epithelial-to-Mesenchymal Transition. Cell Rep. 15, 2665–2678. [DOI] [PubMed] [Google Scholar]
- Ma Y, Ma M, Sun J, Li W, Li Y, Guo X, and Zhang H (2019). CHIR-99021 regulates mitochondrial remodelling via b-catenin signalling and miRNA expression during endodermal differentiation. J. Cell Sci 132, jcs229948. [DOI] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, and Stunnenberg HG (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, Abu-Remaileh M, Clavain L, Erdemir A, Lewis CA, et al. (2018). Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell 22, 769–778.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, and Becker PB (2004). Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 23, 2258–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris N, Edri S, Seyres D, Kulkarni R, Domingues AF, Balayo T, Frontini M, and Pina C (2018). Histone Acetyltransferase KAT2A Stabilizes Pluripotency with Control of Transcriptional Heterogeneity. Stem Cells 36, 1828–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21, 392–402. [DOI] [PubMed] [Google Scholar]
- Mulas C, Kalkan T, von Meyenn F, Leitch HG, Nichols J, and Smith A (2019). Defined conditions for propagation and manipulation of mouse embryonic stem cells. Development 146, dev173146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeslund G (1979). The effect of glucose-, arginine- and leucine-deprivation on mouse blastocyst outgrowth in vitro. Ups. J. Med. Sci 84, 9–20. [DOI] [PubMed] [Google Scholar]
- Nichols J, and Smith A (2012). Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol 4, a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H (2007). How is pluripotency determined and maintained? Development 134, 635–646. [DOI] [PubMed] [Google Scholar]
- Paczkowski M, Schoolcraft WB, and Krisher RL (2014). Fatty acid metabolism during maturation affects glucose uptake and is essential to oocyte competence. Reproduction 148, 429–439. [DOI] [PubMed] [Google Scholar]
- Qu Q, Zeng F, Liu X, Wang QJ, and Deng F (2016). Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis. 7, e2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raud B, Roy DG, Divakaruni AS, Tarasenko TN, Franke R, Ma EH, Samborska B, Hsieh WY, Wong AH, Stüve P, et al. (2018). Etomoxir Actions on Regulatory and Memory T Cells Are Independent of Cpt1a-Mediated Fatty Acid Oxidation. Cell Metab. 28, 504–515.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, and Fenelon JC (2017). The enigma of embryonic diapause. Development 144, 3199–3210. [DOI] [PubMed] [Google Scholar]
- Schieke SM, Ma M, Cao L, McCoy JP Jr., Liu C, Hensel NF, Barrett AJ, Boehm M, and Finkel T (2008). Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J. Biol. Chem 283, 28506–28512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Thompson CB, and Finley LWS (2018). Metabolic regulation of chromatin modifications and gene expression. J. Cell Biol 217, 2247–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, Baumgärtner D, Carnevalli LS, Atzberger A, Haas S, et al. (2016). Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 164, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. (2010). MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell. Biol 30, 3582–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, and Hake SB (2007). Extraction, purification and analysis of histones. Nat. Protoc 2, 1445–1457. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, and Ng HH (2017). The metabolic programming of stem cells. Genes Dev. 31, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Bhanu NV, Karch KR, Wang X, and Garcia BA (2016). Complete Workflow for Analysis of Histone Post-translational Modifications Using Bottom-up Mass Spectrometry: From Histone Extraction to Data Analysis. J. Vis. Exp (111), 54112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Kori Y, Lopes M, Yuan ZF, Kim HJ, Kulej K, Janssen KA, Agosto LM, Cunha JPCD, Andrews AJ, and Garcia BA (2019). One minute analysis of 200 histone posttranslational modifications by direct injection mass spectrometry. Genome Res. 29, 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. (2015). The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol 17, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll EA, Makin R, Sweet IR, Trevelyan AJ, Miwa S, Horner PJ, and Turnbull DM (2015). Neural Stem Cells in the Adult Subventricular Zone Oxidize Fatty Acids to Produce Energy and Support Neurogenic Activity. Stem Cells 33, 2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. (2014). Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GC, Eskeland R, Hekimoglu-Balkan B, Pradeepa MM, and Bickmore WA (2013). H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 23, 2053–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, and McKay RD (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. (2014). Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Dixon MP, Kueh AJ, and Voss AK (2008). Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol. Cell. Biol 28, 5093–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio DG, Xu H, Eisold ME, Woolthuis CM, Pandita TK, and Armstrong SA (2017). Histone acetyltransferase activity of MOF is required for adult but not early fetal hematopoiesis in mice. Blood 129, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsangkar D, and Downs SM (2013). A requirement for fatty acid oxidation in the hormone-induced meiotic maturation of mouse oocytes. Biol. Reprod 89, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi CIK, Basilicata MF, Semplicio G, Georgiev P, Gutierrez NM, and Akhtar A (2018). Facultative dosage compensation of developmental genes on autosomes in Drosophila and mouse embryonic stem cells. Nat. Commun 9, 3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keuren ML, Gavrilina GB, Filipiak WE, Zeidler MG, and Saunders TL (2009). Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 18, 769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana SA, Arnold PK, Rosen BP, Chen Y, Carey BW, Huangfu D, Carmona Fontaine C, Thompson CB, and Finley LWS (2019). Glutamine independence is a selectable feature of pluripotent stem cells. Nat. Metab 1, 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, and Dey SK (2006). Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet 7, 185–199. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li G, Mao F, Li X, Liu Q, Chen L, Lv L, Wang X, Wu J, Dai W, et al. (2014). Ras-induced epigenetic inactivation of the RRAD (Ras-related associated with diabetes) gene promotes glucose uptake in a human ovarian cancer model. J. Biol. Chem 289, 14225–14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Fahrmann JF, Lee H, Li YJ, Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, et al. (2018). JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 27, 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Duan S, Yi F, Ocampo A, Liu GH, and Izpisua Belmonte JC (2013). Mitochondrial regulation in pluripotent stem cells. Cell Metab. 18, 325–332. [DOI] [PubMed] [Google Scholar]
- Yin S, Jiang X, Jiang H, Gao Q, Wang F, Fan S, Khan T, Jabeen N, Khan M, Ali A, et al. (2017). Histone acetyltransferase KAT8 is essential for mouse oocyte development by regulating reactive oxygen species levels. Development 144, 2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, and Smith A (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. (2011). UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 30, 4860–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Badur MG, Divakaruni AS, Parker SJ, Jäger C, Hiller K, Murphy AN, and Metallo CM (2016). Distinct Metabolic States Can Support Self-Renewal and Lipogenesis in Human Pluripotent Stem Cells under Different Culture Conditions. Cell Rep. 16, 1536–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Dahan P, Lu V, Zhang C, Li H, and Teitell MA (2018a). Metabolism in Pluripotent Stem Cells and Early Mammalian Development. Cell Metab. 27, 332–338. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kim H, Fu Z, Qiu Y, Yang Z, Wang J, Zhang D, Tong X, Yin L, Li J, et al. (2018b). Deficiency of the Mitochondrial NAD Kinase Causes Stress-Induced Hepatic Steatosis in Mice. Gastroenterology 154, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, and Jin Y (2009). Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J. Biol. Chem 284, 9168–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, and Thompson CB (2019). Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol 20, 436–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets/code generated or analyzed during this study. The accession numbers for previously published RNA-seq and ChIP-seq datasets are provided in the “Other” section of the Key Resources Table. The accession number for the RNA-seq data files reported in this paper is NCBI GEO: GSE137138: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137138
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-MOF | Bethyl Laboratories | Cat#A300-992A; RRID:AB_805802 |
| Anti-OCT3/4 | Santa Cruz Biotechnology | Cat#sc-5279; RRID:AB_628051 |
| Anti-NANOG | Novus Biologicals | Cat#AF2729; RRID:AB_2150103 |
| Anti-OTX2 | R & D Systems | Cat#AF1979; RRID:AB_2157172 |
| Anti-SMA | SIGMA ALDRICH | Cat#A2547; RRID:AB_476701 |
| Anti-TUJ1 | Biolegend | Cat#801201; RRID:AB_2313773 |
| Anti-SOX17 | R & D Systems | Cat#AF1924; RRID:AB_355060 |
| Anti-KLF4 | Santa Cruz Biotechnology | Cat#sc-166238; RRID:AB_2130234 |
| Anti-TFCP2L1 | R & D Systems | Cat#AF5726; RRID:AB_2202564 |
| Anti-β-ACTIN | Santa Cruz Biotechnology | Cat#sc47778; RRID:AB_2714189 |
| Anti-AMPKα | Cell signaling | Cat#2532; RRID:AB_330331 |
| Anti-Phospho-AMPKα (Thr172) | Cell signaling | Cat#4188; RRID:AB_2169396 |
| Anti-S6K1 | Santa Cruz Biotechnology | Cat#sc-230; RRID:AB_632156 |
| Anti-Phospho-S6K1 (Thr389) | Cell signaling | Cat#9234; RRID:AB_2269803 |
| Anti-AKT | Cell signaling | Cat#9272; RRID:AB_329827 |
| Anti-Phospho-AKT (Ser473) | Cell signaling | Cat#4060; RRID:AB_2315049 |
| Anti-H4K16ac | Millipore | Cat#07-329; RRID:AB_310525 |
| Anti-H3K27ac | Abcam | Cat#ab4729; RRID:AB_2118291 |
| Anti-H3K9ac | Abcam | Cat#ab4441; RRID:AB_2118292 |
| Anti-H3K56ac | Abcam | Cat#ab76307; RRID:AB_1523762 |
| Anti-H4ac | Millipore | Cat# 06-866; RRID:AB_310270 |
| Anti-Histone H3 | Abcam | Cat#ab1791; RRID:AB_302613 |
| Anti-Histone H4 | Active Motif | Cat#39269; RRID:AB_2636967 |
| Goat anti-Mouse IgG Alexa FluorR 555 | Abcam | Cat#ab150114; RRID:AB_2687594 |
| Goat anti-Rabbit IgG Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat#A32733; RRID:AB_2633282 |
| Goat anti-Mouse IgG FITC-conjugated | Thermo Fisher Scientific | Cat#62-6511; RRID:AB_2533946 |
| Donkey anti-Mouse Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-31571; RRID:AB_162542 |
| Donkey anti-Goat IgG AlexaFluorR 555 | Abcam | Cat#ab150130; RRID:AB_10894526 |
| Donkey anti-Goat IgG AlexaFluor 488 | Thermo Fisher Scientific | Cat#A-11055; RRID:AB_2534102 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ESGRO® leukemia inhibitory factor (LIF) | EMD Millipore | Cat#ESG1107 |
| human LIF | SIGMA ALDRICH | Cat#L5283 |
| PD0325901 | SIGMA ALDRICH | Cat#PZ0162 |
| CHIR99021 | SIGMA ALDRICH | Cat#SML1046 |
| Geltrex | Thermo Fisher Scientific | Cat#A1413202 Cat#A1413302 |
| Ndiff227 media | Clontech | Cat#Y40002 |
| Activin A | R & D Systems | Cat#338-AC |
| bFGF | R & D Systems | Cat#233-FB |
| N2 supplement 100X | Thermo Fisher Scientific | Cat#17502001 |
| B27 supplement 50X | Thermo Fisher Scientific | Cat#17504-044 |
| mTeSR1 media | STEMCELL Technologies | Cat#85850 |
| Gö6983 | SIGMA ALDRICH | Cat#G1918 |
| L-Carnitine | SIGMA ALDRICH | Cat#C0158 |
| O-Acetyl-L-Carnitine | SIGMA ALDRICH | Cat#A6706 |
| AICAR | Cayman | Cat#10010241 |
| Coenzyme A | SIGMA ALDRICH | Cat#C4780 |
| Etomoxir | SIGMA ALDRICH | Cat#E1905 |
| Trimetazidine | SIGMA ALDRICH | Cat#653322 |
| INK128 | Medchem Express | Cat#HY-13328 |
| Oligomycin A | Cayman | Cat#11342 |
| FCCP | Cayman | Cat#15218 |
| Antimycin A | SIGMA ALDRICH | Cat#A8674 |
| Rotenone | SIGMA ALDRICH | Cat#R8875 |
| KSOMaa EvolveR medium | Zenith Biotech | Cat#ZEKS-050 |
| Bovine Serum Albumin solution | SIGMA ALDRICH | Cat#A8412 |
| MitoTracker Red CMXRos | SIGMA ALDRICH | Cat#M7512 |
| Tetramethylrhodamine methyl ester | Thermo Fisher Scientific | Cat#T668 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#62249 |
| Pyronin Y | SIGMA ALDRICH | Cat#213519 |
| Propidium Iodide | SIGMA ALDRICH | Cat#00-6990-50 |
| FITC-Annexin V | Bioscience | Cat#556419 |
| O-Propargyl Puromycin (OP-Puro) | Jena Bioscience | Cat#NU-931-05 |
| Critical Commercial Assays | ||
| VECTOR® Red Alkaline Phosphatase Kit | Vector Laboratories | Cat#SK-5100 |
| LookOut® Mycoplasma PCR Detection Kit | SIGMA ALDRICH | Cat#MP0035 |
| Click-iT™ Plus EdU Flow Cytometry Assay Kits | Thermo Fisher Scientific | Cat#C10632 Cat#C10634 |
| Click-iT Cell Reaction Buffer Kit | Thermo Fisher Scientific | Cat#C10269 |
| Qubit™ RNA HS assay kit | Thermo Fisher Scientific | Cat#Q32855 |
| Seahorse XF Base Medium | Agilent Technologies | Cat#102353-100 |
| Deposited Data | ||
| RNA-seq | This paper | GSE137138 |
| Experimental Models: Cell Lines | ||
| Mouse ES cells: E14tg2a | ATCC | Cat#CRL-1821™ |
| Mouse ES cells: Moffl/fl; Cre-ER™ | Y. Dou | Li et al., 2012 |
| Mouse ES cells: Oct4-GiP | Austin Smith | Ying et al., 2008 |
| MEFs | This paper | N/A |
| Human ES cells: Primed H9 | WiCell | Cat#WAe009-A |
| Human ES cells: Naive H9 | Austin Smith | Guo et al., 2017 Takashima et al., 2014 |
| Experimental Models: Organisms/Strains | ||
| (C57BL/6J × DBA/2J)F1 females | Jackson Laboratory | Cat#100006 |
| (C57BL/6J × DBA/2J)F1 males | Jackson Laboratory | Cat#100006 |
| C57BL/6NCrl female | Charles River Laboratory | Cat#493 |
| (C57BL/6J × SJL/J)F1 males | Jackson Laboratory | Cat#100012 |
| Oligonucleotides | ||
| Primers for RT-qPCR (see Table S7) | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Tophat2, v2.1.1 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/manual.shtml |
| Integrative Genomics Viewer (IGV), v2.3 | Thorvaldsdóttir et al., 2013 | http://software.broadinstitute.org/software/igv/ |
| Cufflinks, v2.2.1 | Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cuffiinks/ |
| CummeRbund, v2.1 | N/A | http://compbio.mit.edu/cummeRbund/ |
| Bowtie2, v2-2.2.4 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS, v1.4 | Zhang et al., 2008 | http://liulab.dfci.harvard.edu/MACS/ |
| HOMER v4.10 | Salk Institute | http://homer.ucsd.edu/homer/ |
| DeepTools, v3.2.1 | Ramírez et al., 2016 | https://github.com/deeptools/deepTools |
| Panther Classification System, v13.0 | N/A | http://pantherdb.org/ |
| Mouse ENCODE Consortium | Amemiya et al., 2019; ENCODE Project Consortium et al., 2012 | https://sites.google.com/site/anshulkundaje/projects/blacklists |
| GraphPad Prism, v7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo, v10.0.7r2 | FlowJo LLC | https://docs.fiowjo.com:443 |
| bcl2fastq2 software (v2.17) | Illumina LLC | https://support.illumina.com/sequencing/conversion-software.html |
| Cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| FastQC | Babraham institute | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trim Galore | Babraham institute | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| EpiProfileLite | Sidoli et al., 2019 | https://github.com/zfyuan/EpiProfileLite |
| MetaboAnalyst 4.0 | Chong et al., 2018 | https://www.metaboanalyst.ca |
| Agilent MassHunter Quantitative Analysis (for QQQ) Version B.07 | Agilent Technologies | https://www.agilent.com/en/support/software-informatics/qual-b-0700-sp1 |
| Other | ||
| RNA-seq for early embryonic stages | Boroviak et al., 2015 | E-MTAB-2958 |
| RNA-seq for dormant state by Myc deletion | Scognamiglio et al., 2016 | E-MTAB-3386 |
| RNA-seq for paused state by mTOR inhibition | Bulut-Karslioglu et al., 2016 | GSE81285 |
| ChIP-seq for MOF | Li et al., 2012 | GSE37268 |
| ChIP-seq for H4K16ac | Valsecchi et al., 2018 | GSE109897 |
| ChIP-seq for H4K16ac | Taylor et al., 2013 | GSE47761 |


