Abstract
Single-cell analysis to investigate cellular heterogeneity and cell-to-cell interactions is a crucial compartment to answer key questions in important biological mechanisms. Droplet-based microfluidics appears to be the ideal platform for such a purpose because the compartmentalization of single cells into microdroplets offers unique advantages of enhancing assay sensitivity, protecting cells against external stresses, allowing versatile and precise manipulations over tested samples, and providing a stable microenvironment for long-term cell proliferation and observation. The present Review aims to give a preliminary guidance for researchers from different backgrounds to explore the field of single-cell encapsulation and analysis. A comprehensive and introductory overview of the droplet formation mechanism, fabrication methods of microchips, and a myriad of passive and active encapsulation techniques to enhance single-cell encapsulation efficiency were presented. Meanwhile, common methods for single-cell analysis, especially for long-term cell proliferation, differentiation, and observation inside microcapsules, are briefly introduced. Finally, the major challenges faced in the field are illustrated, and potential prospects for future work are discussed.
I. INTRODUCTION
The central dogma of biology states the transformation from genetic material DNA to RNA to protein through the processes of translation and transcription. Genomic, transcriptomic, and proteomic profiles can accurately reveal the states of the cells in question. Current molecular biology technologies, such as qPCR, RNA-seq, and Western Blotting, enable the investigation of the average expression of genome, transcriptome, and proteome of a certain cell population. However, the molecular status of each individual cell as well as cell-to-cell interactions remains mostly unrevealed. In fact, recent progress in biology demands cellular screening technologies with higher resolution. For instance, using single-cell ChiP-seq, Rotem et al. combined microfluidics, DNA barcoding, and next-generation sequencing technologies to de-convolute the low-coverage chromatin state of thousands of single embryonic stem (ES) cells.1 They identified a spectrum of three subpopulations distinguished by their signals over loci either bound by pluripotency-associated transcription factors or targeted by epigenetic repressors. The genotyping at single-cell resolution instead of bulk population offered a very low level of background noise and, consequently, enhanced the assay sensitivity. Thus, the existence of cellular heterogeneity within a cell population urges the development of single-cell analysis.
Recently, single-cell analysis has shown its applicability and effectiveness in early disease diagnosis (e.g., single-cell analysis of circulating tumor cells), drug development, and many other biological applications.2–5 Shembekar et al. co-encapsulated OKT9 hybridoma cells and leukemic K562 cells along with fluorescently labeled goat anti-mouse IgG Alexa 488 antibody into microcapsules. They investigated the cell-to-cell interactions by detecting the appearance of fluorescent peaks inside individual droplets when the OKT9-secreted antibodies bound to the transferrin receptors on leukemic K562 cells and subsequently activated the fluorescent IgG Alexa 488.6 The high throughput and resolution of this system greatly enhanced the speed and efficiency of drug screening. Witte et al. also recently co-encapsulated prokaryotic L. lactis and eukaryotic bone marrow-derived mesenchymal stem cells (BM-MSCs) into alginate microcapsules and reported the symbiotic relationship between these two cell types within the microgels. They observed that the differentiation of MSCs was enhanced by the proteins and cell factors secreted by L. lactis.7 On the other hand, single-cell analysis is favored in the investigation of scarce samples as it reduces the reagent/substrate consumption because of its minimal reaction volume.8 Weitz et al. reported a general screening platform based on droplet microfluidic for directed evolution that is 103 times faster and consumes 107 times less volume of reagent compared to the state-of-the-art robotic screening method. The platform can single out new mutants of the enzyme horseradish peroxidase (HRP) from approximately 108 individual enzymatic reactions within 10 h at the cost of merely 150 μl of total reagents or less.9 Using this microdroplet-based platform, they successfully discovered variants of HRP that are 10-fold faster than the parental strain. Alternatively, Martino et al. achieved in situ immunoassay in the droplet-based microfluidic system with high sensitivity. They investigated the intracellular proteins in MCF-7 and HEK-293 cells at concentrations between 10−12 M and 10−6 M, enabling protein analysis of scarce samples.10
The first step of single-cell analysis is to isolate individual cells from their population and remove undesired impurities from the mixture if needed. Various cell isolation technologies, including microwells, micropattern, application of external forces, hydrodynamic trapping, fluorescent-activated cell sorter (FACS), and droplet microfluidics, have been developed in the past few years.11–16 Microwells achieve single-cell isolation comparable in size via cell sedimentation while allowing the free passage of solution and other impurities.13 Negative pressure is often applied to facilitate the sedimentation of individual cells. In general, the microwell-based single-cell isolation method is characterized for its high capturing throughput, low sample requirement, and robust operation, as cells can hardly be washed away due to the limited fluid impact above microwells. Nevertheless, undesired cell loss due to the missed sedimentation between microwells and the mandatory cell transfer for downstream analysis limit the employment of such a method for scarce samples, not to mention cell death caused by the application of negative pressure. Moreover, lack of compartmentalization can result in difficult sample manipulation and a higher risk of contamination. Micropattern implies the printing of cell-adhesive ink on cell-repulsive platform, which guides the selective cell landing on those printed patterns. Unfortunately, cell seeding is largely unpredictable and the efficiency of obtaining one cell per pattern is very limited. Consequently, it requires further cell manipulation to isolate single cells from the printed pattern.15 The application of external forces such as optical, electrophoretic, and optoelectrophoretic tweezers can manipulate the cell movement by exerting external force fields to subject samples that are responsive to these stimulations.17–19 Despite its prominent cell capture efficiency, this type of cell isolation and manipulation techniques suffers from its low capturing rate and intensive operation, making it not suitable for the high-throughput single-cell analysis system. Currently, commercialized FACS systems offer an efficient state-of-the-art cell manipulation and analysis platform.16,20,21 It utilizes the effect of hydrodynamic focusing to line up cells in suspension via a sheath flow liquid and passes the cell stream by a laser beam to acquire an optical signal from targeted cells at a rate of the order of 102–103 cells per second. Nonetheless, FACS requires sample cells to be in suspension, rendering it unable to investigate cell–cell interaction and tissue architecture. In addition, it has difficulties in differentiating subpopulation with similar marker expressions, which leads to a high level of background noise and the incapability of detecting low-concentration samples. The exerted shear stress on the cell sample could also result in non-negligible cell damage.
Droplet microfluidics is one of the most promising and applicable methods to isolate cells in single-cell analysis. It has attracted great attention from many research groups around the world because of its high throughput, biocompatibility, and simple operability.22 Passive generation of microdroplets with microfluidic devices can generate over thousands of highly monodisperse microdroplets per second.23 Such a monodispersity of generated microcapsules contributes to the authenticity of the results of downstream single-cell analysis. In addition, the encapsulating shell provides a physical barrier of protection for encapsulated cells, reducing the risk for cross-contamination and avoiding any undesired cell damage during the sample manipulations and downstream analysis. Here, the reaction volume is confined within the microdroplet, which increases the sample and reagent concentration for a better test result. Also, microfluidic system enables precise and relatively simple manipulation of droplet size, shape, and motion by altering flow conditions or microchannel geometries, facilitating the downstream analysis process.24
In sum, droplet microfluidic technology offers incomparable advantages over other methods of cell manipulation for single-cell analysis. Thanks to the recent progress in micromachining, more sophisticated microchannel structures in a wider range of potential working material can be manufactured with superior precision. Hence, greater control of cell manipulation can be achieved. This review aims to give preliminary guidance for researchers from different backgrounds on the single-cell encapsulation and analysis with droplet-based microfluidic systems. The mechanism of droplet generation will first be introduced, followed by the basic principle of cell encapsulation in microfluidic systems. Then, basic methods to fabricate microfluidic system, especially those used for single-cell encapsulation, will be presented. Various passive and active cell encapsulation methods will be discussed and compared in detail. Finally, commonly used methods for single-cell analysis will be briefly presented with a focus on long-term monitoring of live cells.
II. DROPLET GENERATION MECHANISM AND DEVICE MANUFACTURING
A. Physics of droplet generation
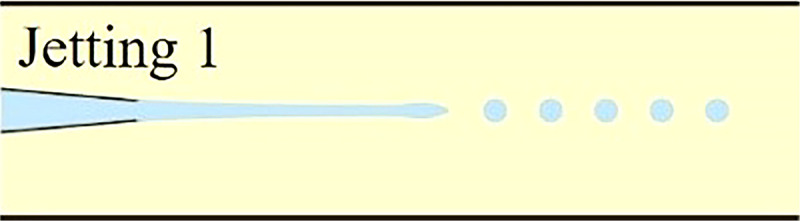
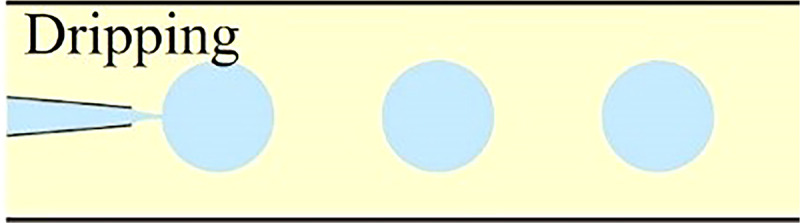
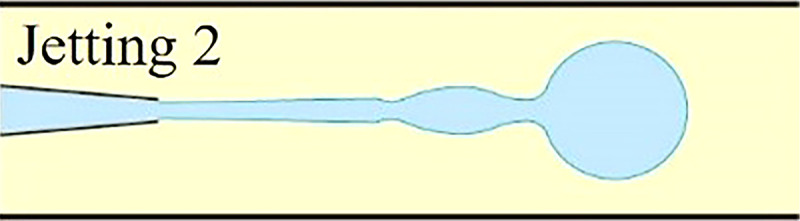
Understanding the physics behind droplet generation in microfluidic systems is crucial for proper single-cell encapsulation. It provides a theoretical foundation to tune both hydrodynamic profiles within microchannel and morphologies of generated droplets in terms of droplet volume, shape, and monodispersity. In microfluidic devices, the breakup of the stream into droplets is achieved through the introduction of the dispersed stream into a continuous stream. In fact, the droplet generation in microfluidic devices relies on fluid instability, notably at the interface of two immiscible phases. Here, viscous and inertial forces that tend to deform the liquid interface counteract the interfacial tension resisting such a deformation. The competition among all these three forces determines the breakup regime of streams into discrete droplets. Depending on the force interaction at the interface, the droplet generation for cell encapsulation occurs in either squeezing, dripping, or jetting mode.25–29 These hydrodynamic modes are often realized via three typical microfluidic geometries: cross-flow, co-flow, and flow-focusing microchannel. Logically, different droplet generation modes would form droplets of different morphologies, which are crucial for proper cell encapsulation as well as downstream cell analysis. In this section, the droplet formation mechanisms in squeezing, dripping, and jetting modes were briefly described. Moreover, some common microfluidic geometries to generate cell-laden microcapsules were summarized along with some examples.
B. Microfluidic motion within microchannel
The hydrodynamic mode of droplet generation is determined by the force balance inside microchannel. Such a force balance can be described by different dimensionless numbers that compare the relative predominance of various physical properties such as characteristic length, inertial stress, viscous stress, gravity, and capillary pressure. Due to the miniature of the microchannel, the interfacial tension and viscous force play a much more dominant role than other forces such as buoyancy. Consequently, commonly used dimensionless numbers for characterizing droplet generation consist of Reynolds number, Capillary number, Bond number, and Weber number.30The computational formulas and the physical significance of these dimensionless numbers are summarized in Table I. Among them, capillary number (Ca) illustrating the relative importance of viscous force and interfacial tension is the most crucial parameter in characterizing droplet formation modes.
TABLE I.
Dimensionless numbers describing hydrodynamic regime of droplet generation inside microchannel; μ is the fluid viscosity, u is the average fluid velocity, γ is the interfacial tension between two phases, dh is the characteristic length of microchannel, ρ is the fluid density, and g is the gravitational acceleration.
| Dimensionless number | Formula | Physical significance |
|---|---|---|
| Capillary number (Ca) | ||
| Reynolds number (Re) | ||
| Weber number (We) | ||
| Bond number (Bo) |
C. Geometries of microfluidic devices for droplet generation
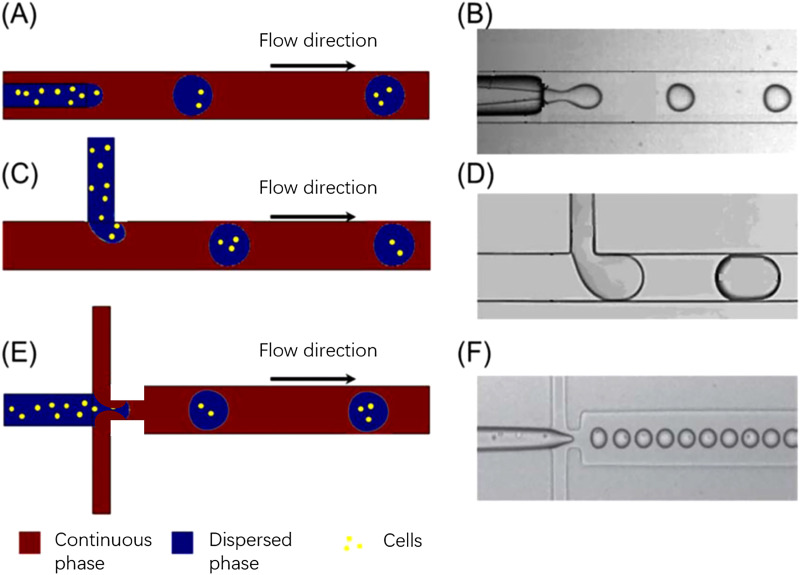
The three most common microfluidic geometries used in droplet generation for cell encapsulation are cross-flow, co-flow, and flow-focusing. The schematic illustrations of droplet generations with different microchannel geometries are presented in Fig. 1.
FIG. 1.
The schematic illustrations of droplet generations at microchannel geometries as well as the experimental results: (a), (c), and (e) are the schematic illustration of droplet generation and cell encapsulation of co-flow, T-junction (cross-flow), and flow-focusing, respectively. (b), (d), and (f) are experimental images. Reproduced and modified with permission from Huang et al., Lab Chip 17, 1913–1932 (2017). Copyright 2017 Royal Society of Chemistry.31
In cross-flow geometries, the continuous and dispersed phases meet at an angle between 0° and 180°. The T-junction where the continuous and dispersed phases orthogonally intersect with each other is the most common. It is also the first cross-flow microfluidic device manufactured for droplet formation. It applies a perpendicular shear stress to introduce non-linearity and instability to the immiscible interface. This geometry is widely employed because of its simplicity and ability to produce a highly monodisperse droplet with a coefficient of variation (CV, the ratio of the standard deviation to the mean of the droplet radius) as low as 2%.30–32 In addition, the droplet production rate of cross-flow microfluidics can reach as high as 7400 droplets per second at the cost of polydispersity of generated droplets. This showcases the potential of cross-flow geometries in high-throughput single-cell analysis in conditions of further improvement in droplet monodispersity.32 Xu et al. reported that the hydrodynamic regime at the T-junction is determined by the interfacial instability, wetting properties, and viscous forces of the continuous phase, as well as the interfacial tension between the two immiscible flows.33 An empirical equation was developed to estimate the droplet volume
| (1) |
where is the droplet volume, is the water flowrate, and is the oil flowrate. Meanwhile, the droplet diameter can also be estimated as where is the hydraulic diameter at the T-junction. Hence, the droplet size is positively correlated to the as well as the continuous phase viscosity (i.e., Ca). However, the utilization of T-junction for cell encapsulation requires extra consideration as the relatively high shear stress may harm the integrity and activity of biological samples.
The idea of employing co-flow geometries in microfluidic devices for droplet generation was first reported by Umbanhowar et al. Such a configuration can either be a quasi-two-dimensional planar device or a three-dimensional coaxial device. In general, it aligns dispersed and continuous phase fluids in parallel streams to generate an immiscible interface.34 Once found in the dripping mode, generated droplets are highly monodisperse with CV less than 3% and diameter greater than that of the dispersed channel. Nonetheless, significant polydispersity is found in droplets generated in jetting mode.26,34 Definition and significance of different fluid modes in droplet generation would be discussed in Sec. II D. The droplet generation rate can vary from hundreds of droplets per second to tens of thousands of droplets per second depending on the specific flow and/or microchannel parameters.33
Alternatively, in flow-focusing microfluidic devices, dispersed and continuous phase fluids are fed at a cross junction such that the dispersed phase is focused by the continuous stream into elongating flows. Here, droplets are generated with size distribution significantly narrower than those generated in other microchannel geometries. The droplet size is also smaller than the microchannel diameter, i.e., often used to overcome dimensional limitations of microchannels. Similar to the co-flow geometries, flow-focusing microchannel system can be either two-dimensional quasi-planar or three-dimensional axis symmetric. Three-dimensional device is generally preferred as it confines droplets in the central axis of the microchannel and protects them from shear stress, damage from wall adhesion and wetting instability at the microchannel wall.35 For instance, Takeuchi et al. managed to generate monodisperse microgel at a rate of approximately 500 droplets per minute with a CV of 2.5% prior to polymerization and of 4.1% postpolymerization. The utilization of three-dimensional flow-focusing geometry avoided issues such as the wetting of microchannel walls by the aqueous dispersed phase, ensuring the generation of highly monodisperse microdroplets.35 In contrast, despite the ease in fabrication and visualization of quasi-2D flow-focusing device, the selection of its continuous fluid is often limited by the potential wall wetting. Inappropriate continuous fluid would disturb the capillary instability, lead to unstable droplet pinch-off, and result in polydisperse droplets. In the case of high flowrate and high pressure drop, leaking may also occur between the layers of quasi-2D flow-focusing devices.36
Other microchannel parameters could also contribute in altering the morphologies of generated droplets. For instance, the microchannel wettability, which is quantified by the contact angle between fluid and wall, plays a significant role in the flow pattern during droplet generation. The increase in the contact angle via the addition of surfactant or surface treatment of the microchannel wall would adversely affect the droplet volume. Many research studies were conducted to study the dynamics of surfactant effects on droplet generation and to develop empirical models that correlate the droplet morphologies to the presence of surfactant.37–40
D. Different fluid modes for droplet generation
As previously described, the droplet formation for cell encapsulation within microfluidic devices occurs in either squeezing, jetting, or dripping mode, which is determined by the stress profile within the microchannel, notably at the immiscible interface. The transition from one mode to another can happen by manipulating different flow parameters such as flow rate, fluid density, and fluid viscosity, as summarized in Table II. It can also occur by altering microchannel parameters such as diameter and wall wettability. Since each of the three modes of droplet generation would generate droplets of different morphologies at different rates, one should carefully choose the appropriate flow and microchannel parameters depending on the requirement of the downstream processes.
TABLE II.
The transition among different droplet generation modes in co-flowing and cross-flowing microchannel configuration; Cac is the capillary number of the outer fluid, and Wed is the Weber number of the inner fluid. Reproduced and modified with permission from Geng et al., Small 16, 1906357 (2020).41 Copyright 2020 Wiley.
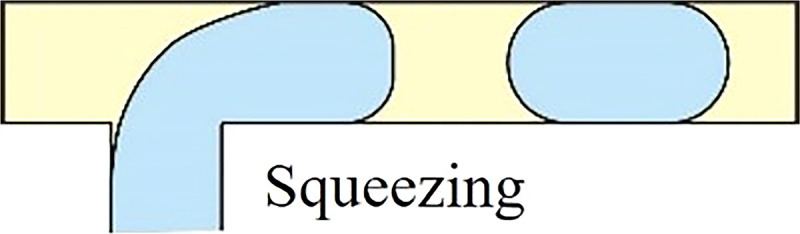
Squeezing mode is observed at low capillary number, where the shear stress from the continuous fluid is much lower than the interfacial stress and insufficient to rupture the dispersed phase to discrete droplets. Hence, the viscous stress is dominated by the confinement of microchannel walls, i.e., high interfacial tension between fluid and wall. Here, droplet pinch-off occurs when the pressure difference between dispersed and continuous phases is large enough to overcome the pressure inside the droplet. Then, the liquid interface is squeezed, deformed, and eventually necked into a separated droplet. Droplets generated in squeezing mode are often larger than the channel dimension, highly monodisperse, and plug-like due to the significance of wall confinement. The pressure instability inside the dispersed phase in the squeezing mode can greatly harm the viability of suspended cells, thereby limiting the applicability of the squeezing regime for cell encapsulation. Moreover, the inherent plug-shaped droplet also causes inconvenience for downstream cell analysis.
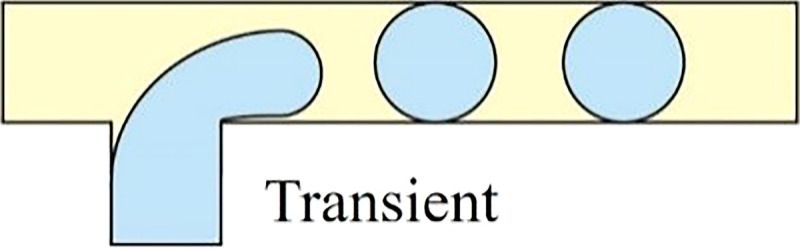
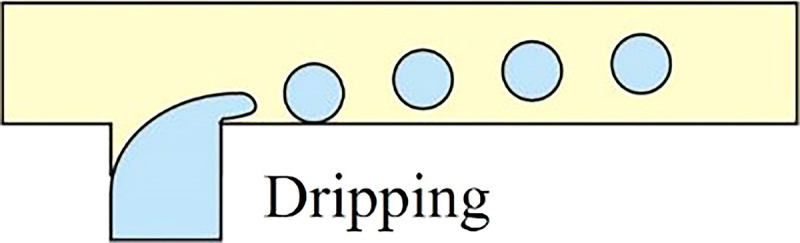
As demonstrated by Xu et al., the flow regime transforms from squeezing (Cac < 0.002) through transient (0.002 < Cac < 0.01) to dripping mode (0.01 < Cac < 0.03) when increasing the Ca value in the continuous phase.41,42 In the dripping mode, the viscous force that ruptures the fluid interface overcomes the interfacial tension stabilizing the emerging droplet against pinch-off. This large viscous force would neck the growing dispersed droplet before it obstructs the microchannel, resulting in a highly monodisperse spherical droplet smaller than the microchannel dimension. At the transient regime between dripping and squeezing, the dynamic of droplet pinch-off is highly unpredictable because it is dominated by both mechanisms mentioned above.
When increasing either the flowrate of the continuous or dispersed phase, the flow regime transits from dripping to jetting. In co-flow microfluidic device, such a transition happens either when the flowrate of the outer continuous fluid increases, so its Ca value overcomes a certain critical value or when the flowrate of inner dispersed phase increases until its We value surpasses a certain critical level.24 Overall, in the jetting regime, the dispersed liquid jets from the channel and ultimately breaks up into droplet downstream the junction by following the Rayleigh-Plateau instability. The diameter of generated droplets is defined by the dimension of the jet and not limited by that of the microchannel. Therefore, the jetting regime is often utilized to overcome the manufacturing limitation as it can produce microdroplets smaller than the dimension of the microchannel. However, due to the existence of capillary perturbations, the resulted droplets are spherical and polydisperse. The highly polydisperse morphologies of droplets limit the usage of jetting for single-cell encapsulation because it requires highly monodisperse cell-laden microcapsules, not only to facilitate downstream cell analysis but also to ensure cell viability and biological activity in desired droplet geometries. Irregular microcapsule geometries may induce ineffective oxygenation or nutrient transfer across the encapsulating shell, which would harm the cellular viability inside microdroplets. Recent efforts were made to adjust the polydispersity of droplets generated in the jetting regime. Li et al. built a new microfluidic device extending capillaries into a step microchannel to generate monodisperse droplets in a stable narrowing jetting regime.43 By carefully manipulating Ca of the continuous phase as well as We of the dispersed phase, they were able to generate droplets whose diameter ranges from 20 μm to 40 μm (two orders of magnitude smaller than the microchannel size) while maintaining a CV below 5%.
E. Manufacture of microfluidic devices for cell encapsulation
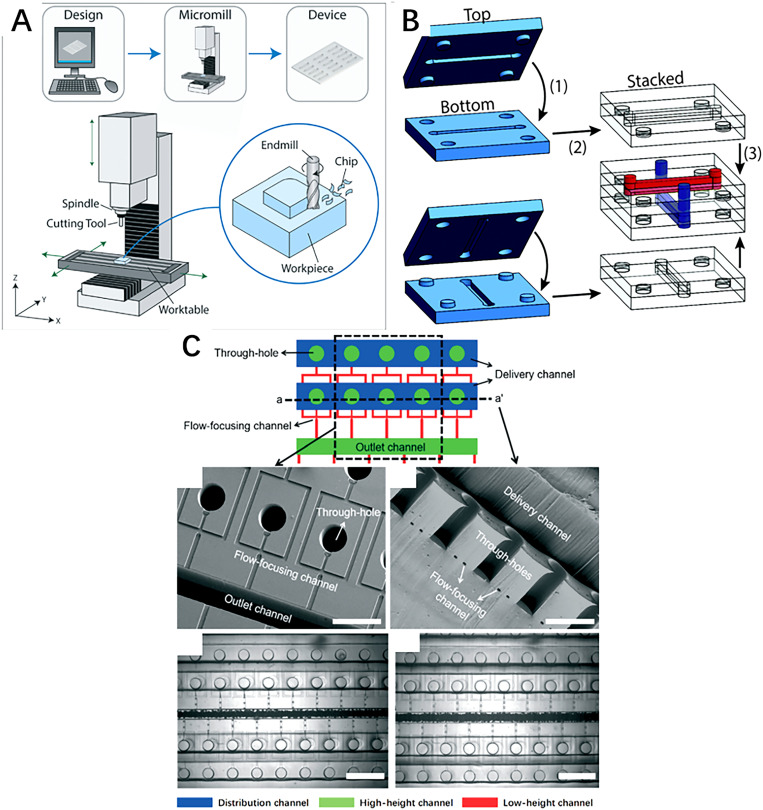
Recent advances in micromachining and material science have broadened the selection of both the manufacturing techniques and working material for the fabrication of microfluidic devices. This not only improves the mechanical, chemical, and biological properties of the microfluidic system but also allows the design of a more delicate and complex microstructure to achieve sophisticated flow manipulations inside the microchannel. Currently, the most common methods for the fabrication of microfluidic chips include subtractive manufacturing, additive manufacturing (also known as 3D printing), and photolithography.44–47 All these three methods would be introduced in the present section. More specially, photolithographed polydimethylsiloxane (PDMS) microfluidic devices were described with greater detail because of their excellent biocompatibility, mechanical flexibility, and extensive application in the field of bioengineering, notably single-cell encapsulation. The basic principles and examples of these three techniques are illustrated in Fig. 2.
FIG. 2.
(a) The schematic illustration of basic principles of micromilling technology. Reproduced with permission from Guckenberger et al., Lab Chip 15, 2364–2378 (2015).48 Copyright 2015 Royal Society of Chemistry. (b) The schematic illustration of microchannel built with light-directed LEGO-like 3D-printed building block. Reproduced with permission from Valentin et al., Polym. Chem. 10, 2015–2028 (2019).49 Copyright 2019 Royal Society of Chemistry. (c) Schematic diagram and experimental images of a 3D PDMS microchannel fabricated by PDMS micromolding. Reproduced with permission from Jeong et al., Lab Chip 15, 4387–4392 (2015).50 Copyright 2015 Royal Society of Chemistry.
F. Subtractive manufacturing
Subtractive manufacturing techniques selectively removes a portion of the substrate material, leaving the desired structure in place. Micromilling is one of the most common methods for the fabrication of microfluidic devices. With the aid of computational numerical control, it uses a rotating microscale milling cutter to remove the material from the working piece. The dimensional limit and surface quality of the fabricated microchannel largely depend on the working conditions of the micromilling machine such as the cut of depth, feed rate, spindle speed, chosen coolant, and properties of the milling cutter.51 Common working substrate of the micromilling is restricted to polymeric material such as PMMA. A metallic material is mostly avoided due to the frequent deformation burrs and high surface roughness, which may disturb the flow regime within the microchannel. The overall long-time cost, dimensional limits, and surface quality of final products are the main challenges for the wider application of micromilling in the manufacturing of single-cell encapsulating microfluidic devices.41
G. Additive manufacturing
Additive manufacturing, namely 3D printing, enables the direct transformation from the digital model into the concrete product by selective depositing, consolidating, and curing substrate material in successive layers.52 Therefore, the option of freely designing complex microstructure and ensuring mass customization becomes possible with this technology as a minimal amount of waste is generated during the manufacturing process.52 With the rapid development of 3D printing in the past few years, the selection for working materials has been greatly broadened from mainly thermoplastic and photosensitive polymer to glass and metal.53–55 Latest research dedicates to the manufacturing of LEGO-like building blocks containing microchannel, connector, and other microstructures with the goal of assembling complex 3D microfluidic devices from a selection of simple and potentially standardized units.56,57 Although additive manufacturing is often used in the fabrication of co-flow and flow-focusing microstructure, the microchannel product suffers from a relatively low resolution and poor visibility for observation.
H. PDMS molding
PDMS microfluidic chip manufactured by soft lithography technique is the most extensively used microfluidic system in biomedical applications, notably cell encapsulation into droplets. It is known for its excellent biocompatibility, optical transparency, gas permeability, mechanical elasticity, high resolution, and superior electrical insulation.47 When manufacturing standard 2D microfluidic devices in PDMS, a masterpiece, often in silicon, is shaped with different microstructures, micropatterns, and functional units using photolithography. This masterpiece then serves as the mold for PDMS molding. The resulted open microchannel is then sealed with a flat film before usage. On the other hand, to fabricate 3D microfluidic devices in PDMS, researchers utilize the mechanical elasticity and self-adhesive nature of PDMS during its curing period to fabricate the real three-dimensional multi-layer microfluidic system. Such a 3D PDMS microchip is notably equipped with co-flow or flow-focusing microstructure in order to largely enhance the droplet generation rate but at the cost of long production time and intensive operations.58 Garcia et al. utilized the PDMS photolithography method and the self-adhesive nature of PDMS to fabricate a two-layer elastomer device with six parallel flow-focusing nozzles of 100 μm width. They used the microchip to encapsulate human mesenchymal stem cells into microdroplets. The device not only showcased around 75% cell viability on day 3 postencapsulation but also enhanced the droplet generation rate by six times compared to single nozzle configuration.59 Akbari et al. also reported another PDMS device for the massive generation of cell-laden microgel.60 They designed a wide junction where the dispersed and continuous liquid phases met and streamed downward into a wide microchannel with uniformly embedded micropillars. The first few rows of micropillars served as an array of flow-focusing junctions for droplet generation, and the rest of micropillars broke incoming droplets into monodisperse daughter droplets until they reached a critical dimension. Using this device, the generation of cell-laden polyethylene glycol (PEG) microgels can achieve a rate of over 3.1 × 106 droplets per second and a single-cell encapsulation efficiency of around 30%.
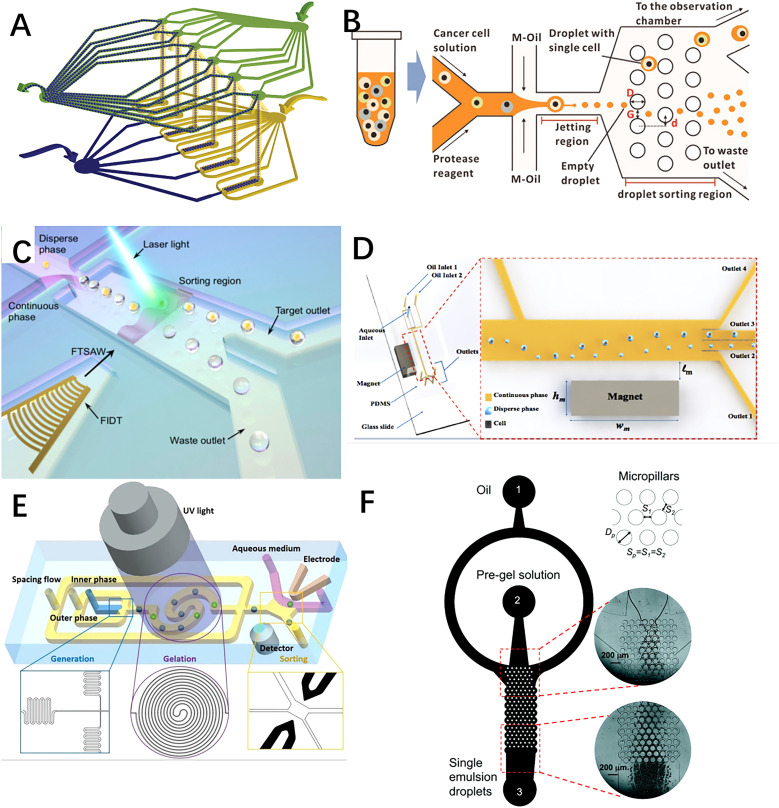
Specially designed microstructures were often integrated inside microchips using PDMS lithography techniques to enhance the single-cell encapsulation efficiency. For instance, Chen et al. fabricated a PDMS lithography flow-focusing microchip that generated cell-laden droplets in the jetting mode.61 They integrated a micropillar structure as the deterministic lateral displacement (DLD) size-sorting channel to separate lung cancer cell (PC-9)-encapsulating microdroplets from empty ones based on their difference in size, since the diameter of cell-laden droplets was on average 9 μm larger than that of the empty ones. On the other hand, external devices can also be anchored onto the PDMS microchip to amplify the single-cell encapsulation efficiency by applying external manipulations such as magnetic and pneumatic forces onto the system. Li et al. designed a detachable fluorescent-activated droplet sorting system that isolated single-cell encapsulating droplets at high accuracy and throughput using a highly focused acoustic wave.62 In this configuration, a 100 μm laser beam is alighted with the sorting region. When a droplet containing a targeted fluorescent signal is detected by the laser beam, a focused interdigitated transducer (FIDT) with 40 pairs of concentric circular electrodes would generate a focused traveling surface acoustic wave (FTSAW) to laterally translate the droplet toward the specific outlet for collection. In this case, the microchip was fabricated using standard PDMS lithography, and it consisted of two PDMS layers: a bulk PDMS layer with microcavities defining the microfluidic channel features and another thin PDMS layer with waveguide microstructures to transmit the FTSAW to the droplet sorting region. Alternatively, Tsai's group published several articles on the employment of the magnetic field to improve single-cell encapsulation efficiency.63,64 By simply putting a piece of magnet nearby their ferrofluid-flowing microchip, cell-laden microdroplets were isolated from empty ones based on the difference in their magnitude of deflection under the magnetic field. Also, employing the PDMS maskless photolithography method, Shum et al. integrated droplet generation, cell encapsulation, UV droplet gelation, and electric droplet sorting regions into a single microchip.65 Here, winding channels were added to the droplet generation region to homogenize cell density in suspension, and spiral channels were added to the UV gelation region to ensure sufficient exposure time for droplet gelation. The schematic illustrations of previously described devices are shown in Fig. 3.
FIG. 3.
Schematic illustrations of some microfluidic systems fabricated with PDMS lithography method: (c) a two-layer elastomer device with six parallel flow-focusing nozzles fabricated by Garcia et al. Reproduced with permission from Headen et al., Microsyst. Nanoeng. 4, 17076 (2018).59 Copyright 2018 Nature. (b) Jetting flow-focusing microchip using micropillar structure as the deterministic lateral displacement (DLD) size-sorting region fabricated by Chen et al. Reproduced with permission from Jing et al., Biosens. Bioelectron. 66, 19–23 (2015).61 Copyright 2015 Elsevier. (c) Detachable fluorescent-activated droplet sorting system fabricated by Li et al. Reproduced with permission from Li et al., Anal. Chem. 91, 9970–9977 (2019).62 Copyright 2019 American Chemical Society. (d) PDMS microchip using magnetic field to sort cell-encapsulating droplets fabricated by Buryk et al. Reproduced with permission from Buryk et al., AIP Adv. 9, 075106 (2019); licensed under a Creative Commons Attribution (CC BY) license. Copyright 2019 AIP Publishing LLC. (e) A microfluidic system for on-chip harvesting of single-cell-laden hydrogels in culture medium fabricated by Shum et al. Reproduced with permission from Nan et al., Adv. Biosys. 3, 1900076 (2019).65 Copyright 2019 Wiley. (f) A PDMS microfluidic system for high-throughput production of cell-laden microgels up to 3.1 × 106 droplets per second, fabricated by Akbari et al. Reproduced with permission from Akbari et al., Lab Chip 17, 2067 (2017).60 Copyright 2017 Royal Society of Chemistry.
III. PASSIVE AND ACTIVE CELL ENCAPSULATION
For cell encapsulation within microfluidic devices, the aqueous dispersed phase containing suspended cells intersects with the continuous phase, usually oil, at the microchannel junction to form cell-laden microcapsules by following mechanisms described in Sec. II. The change in dimensionless numbers of the dispersed liquid due to the presence of suspended cells was found to be negligible, allowing the application of all mechanisms previously presented. Because the gelation of microcapsule postdroplet generation is often required for the downstream process, gel precursors are usually found in the dispersed phase fluid. Depending on the requirement and nature of downstream cell analysis, different gel materials including hydrogel and phospholipid may be chosen for the formation of gel emulsion droplets.66–70 In the goal of maintaining the cell viability and its biological activity within microcapsules, the gelation condition should be relatively mild. In addition, the porosity and the pore size of the encapsulating shell should be carefully designed to ensure proper oxygenation and nutrient transport across microcapsules. Details to maintain long-term cell viability within microcapsules can be found in Sec. IV.
Typically, without the application of any external forces or manipulations, the microfluidic system can generate droplets at a rate of 1000 droplets per second or above.71 However, as a cell is randomly suspended in the dispersed phase and its encounter with the continuous phase occurs in a completely random manner, the number of cells encapsulated inside microcapsules is generally unknown.23,72,73 This raises major concerns for downstream analysis. Ideally, each microcapsule for single-cell analysis should contain one and only one cell since the number of cells per microcapsule can significantly affect the kinetics of downstream processes. For example, if a microcapsule contains two cells, the quantity of working material would double, as well as the apparent reaction kinetics. Therefore, in this section, the efficiency of single-cell encapsulation within microfluidic devices was first introduced, followed by the presentation of current passive and active cell encapsulation methods to enhance the single-cell encapsulation rate and/or efficiency.
A. Cell encapsulation efficiency
Because randomly cells suspended in the dispersed phase arrive at the oil–aqueous interface in a completely randomized manner, one cannot predict whether a microcapsule contains no cell, one cell, or multiple cells. Researchers can only assure that the quantity of cell per microcapsule in passive cell encapsulation follows the Poisson distribution, which is described by Eq. (2),
| (2) |
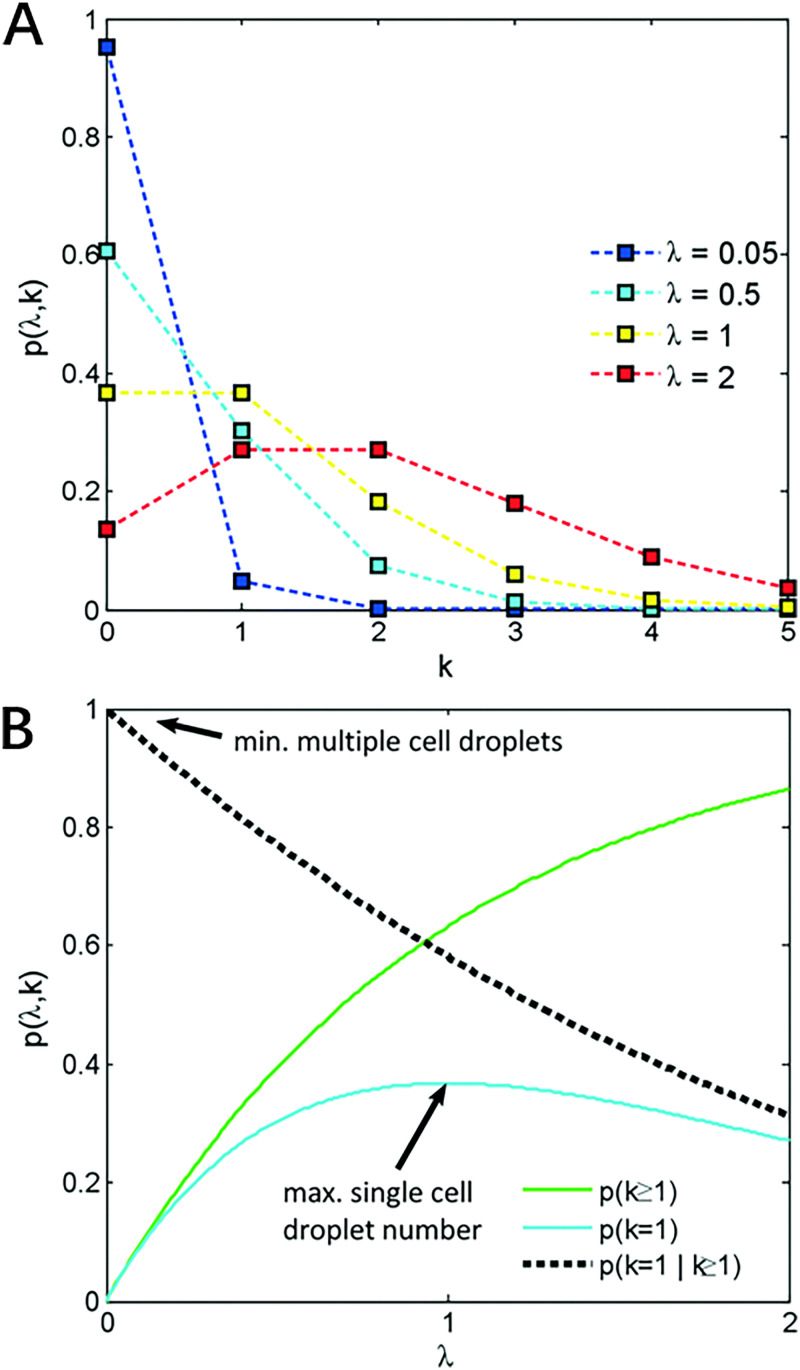
where k is the number of cells per microcapsule and is the average number of cells per microcapsule. According to statistics, when , approximately only 58% of generated droplets contain cells, and around 36% of all generated microcapsules would encapsulate only one cells, not to mention that these numbers are often unmet in real-life experiments due to the existence of experimental errors. The graphic representation of the Poisson distribution is shown in Fig. 4.
FIG. 4.
(a) The graphic representation of the Poisson distribution. (b) Proportion of droplets that contain at least one cell, p(k ≥ 1), exactly one cell, p(k = 1), and the proportion of droplets containing cells that contain exactly one cell, p(k = 1|k ≥ 1). Image reproduced with permission from Collins et al., Lab Chip 15, 3439 (2015).23 Copyright 2015 Royal Society of Chemistry.
These numbers are far below the basic requirement for authentic single-cell analysis. For this reason, many passive and active cell encapsulation methods were developed to enhance the single-cell encapsulation efficiency inside microfluidic devices.
B. Passive cell encapsulation
In the past few years, different research groups have developed numerous passive single-cell encapsulation methods in the microfluidic system to increase the efficiency of the process by exploiting the hydrodynamic profile inside the microchannel. Common methods of passive single-cell encapsulation are summarized in Table III.
TABLE III.
Common methods to passively modulate single-cell encapsulation within microfluidic systems.
| Method | Single-cell encapsulation rate (cells per second) | Single-cell encapsulation efficiency | Principle | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|
| Inertial ordering | 103–104 | Around 80% | Manipulate the hydrodynamic profile inside microchannel to localize suspended cells in an orderly and predictable manner | Very high single-cell efficiency at relatively high encapsulation rate; no need for the implementation of external devices | Requirement for meticulous manipulation of flows and high cell density (of the order of 107 cells per ml) Necessity for relatively long channel length to reach spatial equilibrium. | 77–82 |
| Hydrodynamic microvortices | Not given | Around 50% | The formation of three-dimensional hydrodynamic microvortices to physically trap suspended cells and release them to the droplet-forming junction at one-to-one manner. | Local concentration of cell density inside microvortices, enabling its applicability for low cell density (of the order of 2 × 105 cells per ml) Size-selective | Microvortices may get quickly saturated, which leads to occasional release of cells Requirement for continuous monitoring of cell density inside microvortices |
85 |
| Trapping-and-encapsulating | Not given, but the operation time per round is around 30 min | 78%–86% | Suspended cells get first hydrodynamically trapped inside microstructure, and then encapsulated into oil microcapsules. | Very high single-cell encapsulation efficiency No need for the implementation of external devices | Very long operation time, and hard to scale up | 86 |
| Deterministic encapsulation | Not given | Around 90% | Coat a layer of cross-linker over suspended cells, and then inject these cells into polymer precursor to form a layer of encapsulating polymer over their surface. | Very high single-cell encapsulation efficiency | Potential harm toward encapsulated cell from nanoparticles or processes of surface engineering; Very thin and easily degradable coating layer, may not be able to provide sufficient protection | 74, 87, 88 |
The schematic illustrations of some passive techniques to enhance single-cell encapsulation efficiency are shown in Fig. 5 and would be further discussed in this section.
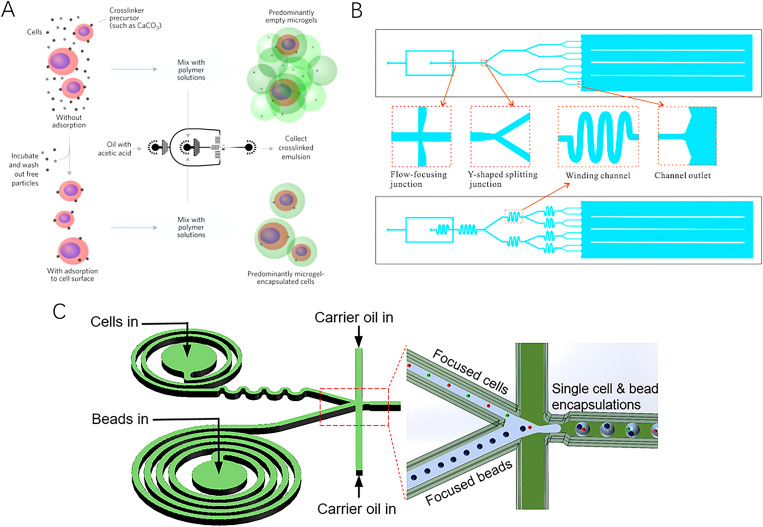
FIG. 5.
Schematic illustrations of some passive techniques to enhance single-cell encapsulation in microdroplets. (a) Strategy of deterministic encapsulation of single cells in thin alginate layer, developed by Weiz et al. Reproduced with permission from Weiz et al., Nature Mater. 16, 236–243 (2017).74 Copyright 2017 Nature. (b) Microfluidic system with winding and Y-shaped splitting channels developed by Yang et al. Reproduced with permission from Yang et al., Chin. Chem. Lett. 26, 1450–1454 (2015).75 Copyright 2015 Elsevier. (c) Spiral channel designed by Li et al. to apply extra viscous drag and centrifugal forces to cells. Reproduced with permission from Li et al., ACS Sens. 4, 1299−1305 (2019).76 Copyright 2019 ACS.
Among them, inertial ordering in either a straight or spiral channel draws particular attention because of its high single-cell encapsulation efficiency (over 80%) and rate (of the order of 103–104 droplets per second).77,78 This method hydrodynamically localizes suspended particles to a restricted region of the microchannel. Those particles then line up and reach the droplet formation/encapsulation region in an orderly manner. Consequently, the chance of multiple or empty encapsulation is greatly reduced, and the single-cell encapsulation efficiency is largely enhanced. In the case of the straight microchannel, suspended cells experience both the force resulted from the parabolic-profile shear gradient, which pushes them to the channel edges, and the wall interaction force analogous to the ground effect utilized in some aircraft.23,77 Consequently, the lateral motion of suspended cells can be stabilized by the balance of the two above-mentioned forces. Longitudinally, the hydrodynamic repulsion effect originated from inter-particle interactions, i.e., the repelling effect of reversing fluid streaming in the vicinity of a rotating particle, ordering the movement of suspended cells. It can be effectively manipulated by altering the distance between neighboring suspended cells, which is determined by either cell concentration, input flow rates, or microchannel dimensions. Because it is rather hard to control microchannel width in situ, the former two factors are more conveniently used to adjust cell ordering and spacing.
Similar to the principles of inertial ordering in straight microchannel, suspended cells in the spiral microchannel are subjected to both drag and lift forces. In fact, the inertial lift experienced by suspended cells comes from the combination of the wall effect, where an asymmetric wake of cells near the microchannel wall induces a lift force away from the wall, and the shear-gradient-induced lift that directs cells down the shear gradient and towards the wall.78 Specifically for curved microchannel, the Dean flow, namely, the secondary rotational flow originated from the imbalance in both viscosity and velocity of fluids between the channel center and the near-wall regions, applies a drag force on suspended cells. Hence, the balance between inertial lift and Dean drag forces determines the preferred and ordered motion of suspended cells inside the spiral microchannel. Theoretical studies predicted that suspended particles in curved channels can reach a single equilibrium position when the ratio of values larger than 0.07, where is the diameter of particles and Dh is the hydraulic diameter of microchannel.79–82 Also, the lateral velocity of particles can be described by Eq. (3),
| (3) |
where is the density of the fluid, is the flow velocity of the fluid, is the dynamic viscosity of the fluid, and CL is the lift coefficient, which is a function of the particle position on the channel cross-section.
Overall, implementing inertial ordering of suspended cells in either straight or spiral microchannel has an extremely high requirement in controlling the flow regime, i.e., the hydrodynamic force balance within the microchannel. In addition, one should also fabricate the channel long enough to ensure sufficient time for proper cell ordering before the occurrence of encapsulation. It was reported that the ordering channel should measure 1 and 5 cm for spiral and straight inertial ordering, respectively.77,78 To avoid extremely long spiral channel, Li et al. added winding serpentine channels post spiral channel. These serpentine channels applied extra viscous drag and centrifugal forces to suspended cells and consequently shortened the required time for cells to reach their equilibrium position.76 Interestingly, winding channels are also used in many other microfluidic single-cell encapsulation systems to homogenize cell distribution in suspension, and consequently avoid multiple or empty encapsulation. For instance, Yang et al. integrated winding channels in a microfluidic multi-step droplet splitting system and greatly enhanced the single-cell encapsulation efficiency of the device.75 Interestingly, they implemented three levels of Y-shaped splitting junction where nozzles contracted from the width of the upstream channel to the width of the downstream channel, and each mother droplet was split into two daughter droplets of equal volume. Consequently, multiple cell encapsulating droplets can be split into single-cell-laden droplets. Here, the splitting primarily depends on the effect of viscous stresses and interfacial tension, which can be described by Ca. Eventually, they achieved a generation rate of 1300 droplets per second with a standard deviation of droplet volume ranged from 3% to 6%. The single-cell encapsulation efficiency was reported to be 31%, much higher than what was reported in ordinary passive encapsulation experiments. Other methods to achieve homogenous cell distribution in suspension include mechanical stirring or rapid mixing at high flow by chaotic advection.83,84
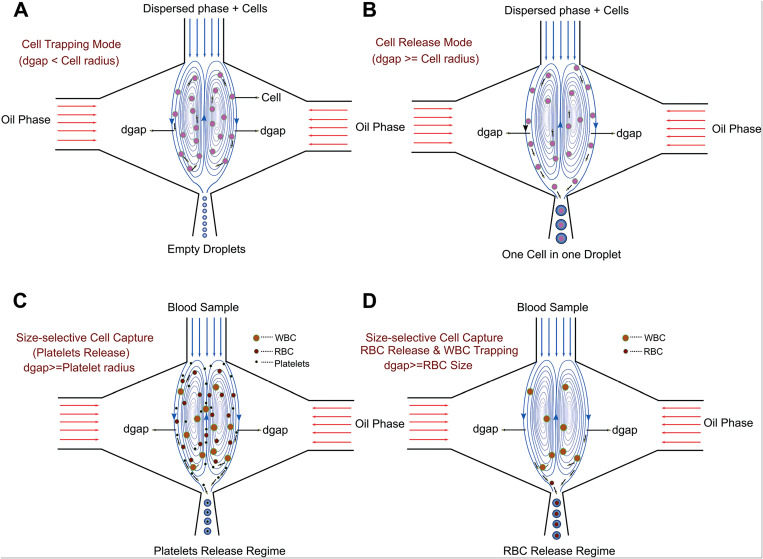
Despite the high single-cell encapsulation rate and efficiency of inertial ordering, the complexity in precisely controlling flow regime as well as the requirement for relatively high cell concentration (of the order of 107 cells per ml) limit the application of this method in some scenario of single-cell analysis, especially in the case of scarce cell samples. Kamalakshakurup and Lee developed an alternative three-dimensionally confined hydrodynamic micro-vortices method to overcome the intrinsic limitation of Poisson distribution.85 They used it as a physical tool to trap cells and then released them one-to-one into the droplet formation region, which eventually resulted in a single-cell efficiency of around 50%. Here, the dispersed phase containing suspended cells was injected into the central channel, and then focused into a narrow orifice by symmetric co-flowing continuous streams. The latter streams exert a very high viscosity shear on the dispersed stream, leading to the formation of two symmetric three-dimensionally confined hydrodynamic micro-vortices in the aqueous dispersed phase. These micro-vortices are forward-oriented at the periphery and reverse-oriented at the center. The schematic illustration of the hydrodynamic profiles is shown in Fig. 6.
FIG. 6.
Schematic illustration of three-dimensionally confined hydrodynamic micro-vortices. WBC is the white blood cell and RBC is the red blood cell. (a) Cell trapping mode where dgap is smaller than the radius of the cells and all the cells are trapped within the micro-vortices; (b) cell release mode where dgap is comparable to the radius of the cells, which allows one-to-one encapsulation; (c) dgap is comparable to the radius of blood platelets that are released whereas RBCs and WBCs are trapped within the micro-vortices; (d) by increasing dgap, platelets, RBCs and WBCs are released in ascending order of their sizes. Reproduced with permission Kamalakshakurup and Lee, Lab Chip 17, 4324 (2017).85 Copyright 2017 Royal Society of Chemistry.
The cell-containing dispersed phase recirculating within the vortex would exit through the dgap into the orifice in the form of droplets, where dgap is the width separating the closed loop vortex streamlines from the immiscible interface. If the radii of suspended cells are greater than the dgap, cells would not exit micro-vortices along with the dispersed flow. If their radii are comparable to the dgap, cells would enter droplets one-to-one and the diameters of generated droplets are determined by the dimension of encapsulated cells. Otherwise, if the cell radii are much smaller than the dgap, multiple encapsulations would occur. Therefore, this system can also isolate a single-cell type from a cell mixture based on their difference in size. One can adjust the value of dgap by carefully manipulating the pressure ratio between dispersed and continuous phases. Kamalakshakurup and Lee reported that 50% single-cell encapsulation efficiency was achieved with a cell density as low as 2 × 105 cells per ml, since cells were locally concentrated within the micro-vortices.
Overall, the deterministic methods of improving single-cell encapsulation efficiency, e.g., inertial ordering and hydrodynamic micro-vortices, can overcome the intrinsic Poisson statistic by modulating hydrodynamic profiles within microchannels. Nevertheless, other than the requirement of a meticulous flow motion, these methods merely suit the encapsulation of homogeneous cells as any change in shape, size, or other physical properties of suspended particles demands the re-adjustment of hydrodynamic profiles.
Besides the deterministic methods described above, one can integrate specially designed microstructure inside a microchip to first trap single cells and then encapsulate them into microcapsules. For instance, Sauzade and Brouzes designed a series of microstructures to hydrodynamically trap single cells, which were then engulfed/encapsulated inside oil microcapsule.86 Despite the high single-cell encapsulation efficiency of the system ranged between 78% and 86%, its total operation time was approximately 30 min, not to mention the number of cells encapsulated per round was limited by the number of traps on the microchip. Moreover, the size of the microcapsule was confined by the dimensional limitation of trapping microstructures, making the generation of smaller microcapsules impossible. All these make the “trapping-and-encapsulating” pathways unsuitable for the large-scale single-cell encapsulation under current technical constraints.
Other passive methods to encapsulate a single cell that circumvent the involvement of multi-phase microfluidics were also reported in the past few years. For instance, Weitz et al. incubated cells with cross-linker precursors such as CaCO3 and washed off free particles with buffer solution, leaving a layer of cross-linker precursor particles at the cell surface.74,87 Then, by injecting those cells into hydrogel solution, a micrometer-scale encapsulating gel layer was grown at the cell surface, with an encapsulation efficiency of over 90%. Similarly, Oh et al. combined click-chemistry and glycoengineering to encapsulate single neural progenitor cells (NPCs) with a layer of the polymer at different stiffnesses.88 By treating NPCs with Ac4ManNAz, azide modified cells were generated. Then, a layer of PEG with varying molecular weights was coated over engineered cells via a click crosslinking reaction between the azide group and the alkyne group of DBCO modified PEGs. Nevertheless, the potential effects of the modification of cellular membrane on cellular behavior render this method questionable for the purpose of single-cell analysis. The relatively long incubation time required for the formation of the encapsulating layer (around 60 min) is another obstacle preventing the wider application of such a method for single-cell encapsulation at a commercial scale. Overall, the minimal thickness of the encapsulating layer and the incapability to co-encapsulate other substances, e.g., culture medium and fluorescent signal molecules, along with encapsulated cells in both above-mentioned methods limit the application of this pathway for single-cell encapsulation and downstream analysis.
C. Active cell encapsulation
There are two pathways to actively increase the single-cell encapsulation efficiency within the microchannel: (1) manipulating cell displacement and/or rate for droplet generation to increase the number of microcapsules containing only one cell; (2) sorting out microcapsules containing only one cell post passive cell encapsulation. In this section, cell encapsulation technologies developed based on the first pathways are discussed and summarized in Table IV. The latter pathway was dismissed as it does not directly involve droplet generation and cell encapsulation processes.
TABLE IV.
Common methods used to actively modulate cell encapsulation within microfluidic devices.
| Methods | Order of magnitude of rate (Hz, or droplet per second) | Advantages | Disadvantages | Principles | Reference |
|---|---|---|---|---|---|
| Extrinsic | |||||
| Electronic | 101–102 | Well-studied fundamental theories/ the application of electrical field may not be compatible with the integrity or viability of biological samples. | Low production rate and potential harm of magnetic nanoparticles toward biological sample. | The application of electrical fields can manipulate the displacement of water–oil interface, and thereby accelerate the droplet pinch-off. | 89 and 91–99 |
| Magnetic | 102 | Easy to implement and manipulate | Converting the fluid into ferrofluid, by the addition of magnetic nanoparticles for example, can be used to control the fluid motion. | 63, 64, 101 | |
| Optical | |||||
| Laser | 103–104 | High production rate, precise control | potential harm of laser towards biological samples and requirement of integrating complex laser equipment into the system | Laser-induced cavitation is used to disturb immiscible interphase. | 103 |
| Off-chip affiliation | 102 | Non-invasive manipulation of fluid hydraulic pressure | The application of off-chip pressure source to induce hydraulic pressure cycles, and thereby manipulate droplet generation. | ||
| Microvibrator | very low production rate of droplets | 90 | |||
| Mechanical valve | 108 and 109 | ||||
| On-chip affiliation | 103 | Control over the production rate and droplet morphologies | very low production rate | On-chip actuators or valves to induce mechanical pulses can be used to control the droplet generation. | |
| Piezoelectric actuator | 113 | ||||
| SAW actuator | 111 | ||||
| Pneumatic valves | 116 and 119 | ||||
| Intrinsic | |||||
| Electronic | Not given | Able to achieve on-demand droplet generation as the response time is within the order of millisecond. | Potential harm of electrical field to biological samples | The ordering of suspended non-conductive but electrically active particles under the application of electrical field can change the fluid viscosity. | 94 |
| Thermal | Not given | The application of heat may not be compatible with the integrity or activity of biological samples. | Potential harm of heat to biological samples | The change in fluid viscosity and volume along with the change in temperature can be used to modulate the viscous and interfacial tension force profile within microchannel, and thereby manipulate droplet generation. | 121 and 122 |
| Optical/dielectrophoresis tweezers | <1 | Extremely precise control over cell displacement, near 100% encapsulation efficiency | extremely low encapsulation rate | Application of optical or electrical field to control the cell displacement. | 17–19 |
Active cell encapsulation prior droplet generation relies on the implementation of additional forces such as electricity, heat, acoustic waves, and light to manipulate the rate of droplet production. In this manner, droplet formation coincides with the arrival of suspended cells, and on-demand encapsulation, i.e., the single-cell encapsulation efficiency of nearly 100%, can be achieved. Therefore, a detection module, e.g., fluorescence detection section, can be integrated within the microfluidic system to predict the arrival of single cells. Once the target cell arrives at the droplet-forming junction, the external force field can be applied onto the system and actively triggers the generation of single-cell-laden microcapsules. On-demand encapsulation can also be realized by actively changing the intrinsic properties of the microfluidic system, e.g., inertial, viscous, and capillary force profiles. In that manner, one can manipulate not only the rate of droplet generation but also droplet morphologies. Moreover, the introduction of additional forces can shorten the time required for system stabilization compared to the simple passive cell encapsulation process. The schematic illustrations of some active single-cell encapsulations are shown in Fig. 7.
FIG. 7.
Schematic illustrations of some active techniques to enhance single-cell encapsulation in microdroplets. (a) Active manipulation of droplet generation using electrowetting, developed by Gu et al. Reproduced with permission from Gu et al., Biomicrofluidics 5, 11101 (2011).89 Copyright 2017 AIP Publishing LLC. (b) Integration of mechanical vibrator to facilitate droplet generation with liquids of ultra-low interfacial tension, developed by Sauret et al. Reproduced with permission from Sauret et al., Lab Chip 12, 3380–3386 (2012).90 Copyright 2012 Royal Society of Chemistry. (c) Nanoscale dieletrophoretic tweezers designed by Nadappuram et al. to capture single cells or subcellular constituents. Reproduced with permission from Nadappuram et al., Nat. Nanotechnol. 14, 80–88 (2019).17 Copyright 2019 Nature.
D. Active single-cell encapsulation by modulating extrinsic parameters
1. Electrical field
The application of the electrical field is one of the most common and mature methods to actively enhance single-cell encapsulation efficiency by utilizing tools such as dielectrophoresis, electrowetting, and electrorheological effects.89,91–94 It is usually achieved by directly embedding electrodes into a microchip, connecting them to high voltage, and grounding the oil continuous phase as an insulator.95 Under the effect of the electrical field, the charge at the water–oil interface will migrate and accumulate. By finely tuning the strength, direction, and distribution of the applied electrical field, one can direct the movement of the water–oil interface and control droplet generation. For instance, the application of high AC voltage can direct the edge of the immiscible interface along the electric potential gradient, thereby manipulating droplet generation in an even on-demand manner.92 Such a phenomenon was first described by Taylor et al. They reported that the formation of the conical interface resulted from a static balance between electric stress and surface tension.96 The electrical potential gradient varies according to the local radius of the vertex as , and the conical interface erupts because the tangential and normal stresses exert enough electric pressure to overcome surface tension. Consequently, a jet is formed and accelerated away from the interface. This jet formation from a Taylor cone is subjected to interfacial stress conditions (i.e., the combination of tangential and normal electric stresses), interfacial tension, and velocity pressure. It can be theoretically predicted by simultaneously solving the Navier–Stokes equation, the continuity equation, and the Laplace equation.92 If the electric field is sufficiently strong, the conical interface would pulsate and eventually break the jet into droplets as a result of the Rayleigh-Plateau instability. Otherwise, if the applied high AV voltage (>600 V) is at a sufficiently low frequency (around 3 Hz), the jet on the water–oil interface in a flow-focusing device could be elongated and destabilized to generate droplets when the electrical potential at the jet tip is below a certain value.92,97–99 Alternatively, when choosing an appropriate substrate for the microchannel, applying an electrical field on the microchannel can lower the wettability of the microchannel wall and consequently manipulate droplet generation in a manner similar to that of surfactant effect.89 For example, Gu et al. applied a continuous AC voltage to a PDMS flow-focusing microchip mounted on a Teflon insulator. Under the effect of the electrical field, the aqueous wettability of the Teflon layer increases. This effect facilitated the neck formation and consequently accelerated the droplet generation.89
Despite numerous studies reported the applicability of the electrical field to actively produce cell-laden microcapsules, the wider application of electrical manipulation of droplet generation at a commercial scale remains limited due to its relatively low on-demand droplet generation rate ranged from 101 to 102 droplets per second and the potential damage to encapsulated cells originated from the introduction of high voltage. Recent progress in utilizing electrical fields to aid cell encapsulation is rarely reported.
2. Magnetic field
Another common method to manipulate the fluid displacement inside the microchannel is the application of the magnetic field. In this case, nanometer-scale magnetic particles are often added in either stream, which can be easily magnetized and demagnetized with the application and removal of the magnetic field, respectively. To control the droplet generation, the dispersed phase fluid is usually converted into ferrofluid and manipulated with an external magnetic field.100 Alternatively, the direct usage of ferrofluid as the dispersed phase is also possible and already discussed in Sec. II.102,103 However, it is not desired for the purpose of cell encapsulation as most commercially available ferrofluids are proven to have high cytotoxicity for mammalian cells. In fact, various aspects of the implemented magnetic fields such as the type of magnet, the location of implementation, and uniformity, direction as well as the polarity of the magnetic fields, can affect the morphologies of the generated droplet and its rate of production. Nevertheless, only a few studies have been reported to employ the magnetic field into droplet generation for single-cell encapsulation due to their low rate of production far from the requirement of downstream single-cell analysis as well as the potential harm of magnetic nanoparticles toward suspended cells.63,64,101,102
3. Optical field
Besides the application of external forces to directly manipulate the fluid movement, one can achieve active droplet generation and cell encapsulation by altering the extrinsic and intrinsic properties of fluids flowing inside the microchannel. For instance, the fluid velocity of either dispersed or continuous fluids can be modified by modulating the hydraulic pressure and flow resistance.
An intense laser pulse can be implemented on the microfluidic system, specifically at the junction where a stable interface between the aqueous dispersed phase and the oil continuous phase is located. When an intense laser pulse is focused on the aqueous phase that is usually capable of absorbing optical energy, molecules get quickly heated up and transform into the plasma state. In order to dissipate heat induced by the laser pulse, laser-induced cavitation bubbles are rapidly arisen and act like a micropump pushing the aqueous dispersed phase into the oil continuous phase. Consequently, the immiscible interface is destabilized, leading to droplet generation. Because of the great controllability and stability of laser pulse, on-demand monodisperse droplet generation could be achieved with the rate of production as high as 10 000 droplets per second and a wide volume range for generated droplets from 1 to 150 pl.103 On the other hand, the focused laser beam can also induce a local thermal gradient, resulting in a spatial imbalance of surface tension. Such an imbalance would induce a flow inside and around the droplet, which in turn serves as a contactless optical valve to block the oil–water interface, delay the pinch-off time and enlarge the droplet volume.104,105 However, one should be careful when employing laser force for single-cell encapsulation due to the potential harm of laser-induced heat toward encapsulated cells, as well as the possible interference of laser pulse to pre-mixed fluorescent tag for downstream analysis.
4. Extra pressure source
The hydraulic pressure of fluids can also be controlled by connecting microfluidic chips to external pressure sources. This method is often employed when using fluids with more complex theological and interfacial properties, e.g., non-Newtonian behaviors and fluids with low interfacial tensions. Notably, aqueous two-phase systems (ATPSs) are favored for biomedical applications because of their excellent biocompatibility. However, they usually exhibit an interfacial tension over 500 times lower than the conventional oil–water system, which urges the involvement of external force to facilitate droplet pinch-off.106 For example, a mechanical vibrator can be integrated into the microfluidic system to facilitate droplet generation by perturbing the microtubings of the aqueous dispersed phase. Then, the pressure of the dispersed fluid would pulsate and an oscillatory vibration could be found at the nozzles.90 Interestingly, similar methods can be employed to the oil–water systems in the presence of a significant amount of surfactants.107 In the absence of vibration, a stable jet of constant radius is found at the junction since disturbance cannot grow along the jet due to low interfacial tension. Under the effect of a mechanical vibrator, the dispersed-fluid inertia would be enhanced and counteract against the viscous stress. This accelerates the pinch-off of the aqueous droplet and reduces the size of generated microcapsules. For example, Sauret et al. reported the droplet generation of PEG/K3PO4 ATPS in the co-flowing device using the mechanical vibrator.90 When introducing vibration at a low frequency of 3 Hz, the immiscible interface oscillated and the growth rate of the jet remained relatively slow, resulting in a longer droplet pinch-off time. If increasing the vibration frequency to the optimal range of 4–5 Hz, the growth rate reached its maximum, and microdroplets are quickly generated because of the Rayleigh-Plateau instability. When the vibration frequency is slightly above its optimal range, generated droplets become monodisperse but not spherical. Finally, if the vibration frequency is further increased, the formation of a corrugated interface instead of droplets is observed. Moreover, research reported that within a certain critical frequency range, when the vibrator frequency synchronizes with that of droplet generation, the droplet size is controllable regardless of properties of continuous phases, microchannel, and fluid dynamics.108 In fact, the droplet generation under the effect of external pressure pulsation can be categorized into three distinct modes: (1) non-synchronization mode 1 where the vibration frequency is lower than the natural frequency of droplet generation; here, the droplet generation relies on the superposition of several sources of instability including the intrinsic Rayleigh-Plateau instability and the perturbation caused by the mechanical vibrator, which results in polydisperse droplets; (2) synchronization mode where external vibration dominates the hydrodynamic profile and monodisperse droplets are generated with weak dependence on other parameters such as flowrate and microchannel geometries; (3) non-synchronization mode 2 where the vibration frequency has little impact over both generation rate and droplet volume, and the droplet can be either polydisperse or monodisperse depending on the vibration amplitude.
Alternatively, another off-chip affiliation device to alter the hydraulic pressure of the fluid is the mechanical valve. It is capable of achieving on-demand droplet generation by inducing a square-wave pressure pulse to the dispersed phase fluid when the continuous phase is driven by some constant pressure source.109,110 By implementing different pressure cycles onto the dispersed phase, droplets with different volumes can be generated at a controllable rate, but usually ranged in the order of 102 droplets per second, far below the standard requirement for single-cell encapsulation and downstream applications.
Besides off-chip controller, on-chip actuator can also pulsate the hydraulic pressure of either dispersed or continuous fluids. In general, there are two common categories of on-chip actuators: one using the pulse of piezoelectric force to deform the geometries of the microchannel, and one exploiting surface acoustic wave (SAW).111,112 In the former case, the microfluidic system needs to be fabricated with a soft material like PDMS, and the piezoelectric actuator should be placed atop the dispersed phase feedstock chamber and separated by a flexible membrane. Such a configuration can control both the droplet morphologies and the generation frequency independently. A research study demonstrated that the droplet volume is linearly correlated with the pulse amplitude and the pulse duration of the actuator before reaching the plateau, whereas the pulse shape can significantly affect the monodispersity of droplets.113 In addition, when carefully designing the microfluidic system, researchers reported the possibility to achieve on-demand droplet generation, where no droplet is generated in the absence of actuation.113 On the other hand, on-chip SAW actuator utilizes acoustic radiation force (ARF) arisen from the gradient of fluid density at the immiscible interphase between the aqueous dispersed phase and the oil continuous phase. Generally, the SAW actuator consists of a series of gold-focused interdigital transducers arrayed on a lithium niobite substrate. Its resonant frequency is determined by the distance between each successive transducer. When an AC signal is applied across the actuator, the electrochemical displacement would induce a SAW radiating acoustic energy at the Rayleigh angle.114 The ARF can deform the immiscible interface by pushing one phase into the other, thereby accelerating the droplet generation. Both the droplet morphologies, generation rate, and even on-demand droplet generation can be modulated by steering SAW. In principle, the generation frequency is affected by the pulse duration whereas droplet size is determined by the applied power, pulse duration, channel geometry, and continuous phase flowrate. Research showed that when applying SAW directly to the oil–water interface or to the oil continuous phase, a decrease in the droplet size is observed due to a SAW-induced pressure raise at the inlet channel of the continuous phase.111 In addition, on-demand microcapsule generation can be achieved by carefully manipulating the applied power and pulse duration.
Alternatively, on-chip valves that are usually pneumatically driven by compressed air or hydraulically by the pressurized liquid in the actuation channel can also be used to control the droplet generation and droplet morphologies by deforming or even blocking the fluidic microchannel.115,116 The actuation channel can be placed upstream, downstream, or at the junction, and it is generally separated from the fluidic microchannel by a flexible membrane. When located upstream of the junction, on-chip actuation valves can deform or even block the dispersed channel, thereby modulating the velocity of the flowing fluid. Both the underlying mechanism and the computational equation were developed to correlate the relation between the resultant droplet size and the pressure actuation.117 On the other hand, valves placed at the junction directly alter the immiscible interface by deforming or blocking the fluidic microchannel, which destabilizes the shear stress profile of the continuous phase at the junction and tunes the droplet size.32,118 When placing downstream the junction, microvalves are often used to break the already generated droplets into smaller daughter droplets, which is not often used for the purpose of single-cell encapsulation.119 The implementation of actuation valves underneath the entire microfluidic channel was also reported. Similar to previously mentioned on- and off-chip actuator, on-chip actuation microvalve also suffers from its low droplet production rate usually ranged from 100 to 101 droplets per second, making it inappropriate for most of the applications of single-cell encapsulation despite its great biocompatibility.120
On the other hand, optical or dielectrophoresis tweezers can be employed to bring the cell to the oil–water interface at the moment of droplet generation or stimulate on-demand droplet generation when cell is brought to the junction. Many excellent reviews describe the application of optical and/or electrical tweezers in the field of cell manipulation and encapsulation with much detail.17–19 Recently, Nadappuram et al. reported nanoscale dielectrophoresis tweezers even capable of capturing and moving sub-cellular components, giving the new possibility for single-cell analysis.17 Despite the perfect single-cell encapsulation efficiency using various types of tweezers, its droplet generation, and cell encapsulation rate are usually found to be lower than 1 droplet per second, which makes its practical application in single-cell encapsulation nearly impossible for the moment.
E. Active single-cell encapsulation by modulating intrinsic parameters
Aside from adjusting extrinsic parameters of fluids such as velocity and flow regime, intrinsic properties of fluid such as viscosity can also be modulated to actively generate droplets in the microfluidic system. For instance, the application of an electrical field on electrorheological (ER) fluid containing non-conducting but electrically active particles would polarize and aggregate those suspended particles into chain/column in the direction of the electrical field, which restricts the fluid flow perpendicular to the direction of the field and enhances the apparent viscosity of the fluid.94 Such a viscosity change of ER fluid is reversible and its response time is as short as milliseconds. Hence, on-demand droplet generation is attainable by altering the viscosity of ER fluid, i.e., its Ca value. Alternatively, fluid viscosity can also be thermally modulated via micro-heaters, micro-heat-exchangers, or laser beams. Usually, the viscosity of most liquids as well as their interfacial tension would decrease with the rise in temperature. Nonetheless, the decrease in viscosity is often faster than that in interfacial tension. This would lead to the increase in droplet volume up to almost two orders of magnitude when selecting continuous phase fluid that has a much larger variation in viscosity than that in interfacial tension. For instance, Yeh et al. showed that when heating the gelatin continuous phase over 25°, the fluid transforms from a gel state into liquid, which consequently fastens the release of gelatin droplet, reduces the continuous phase supply at the droplet generating junction, and enlarges the volume of microcapsules.121 In addition, an enhancement to the temperature dependence of the droplet size is observed for the microchannel with smaller dimensions. On the other hand, thermal control can be directly applied at the immiscible interface with the goal of exploiting the Marangoni effect, i.e., tangential stress along the liquid–liquid interface.122 In fact, when heating the liquid–liquid interface in a non-uniform manner and creating a thermal gradient, a gradient of interfacial tension could be observed at the interface, and the droplet generation is affected.
In general, on-demand single-cell encapsulation can be achieved via the application of the external field. Despite the high single-cell encapsulation efficiency, the throughput of active methods usually proves to be of the order of magnitude lower than that of passive encapsulation. Furthermore, active encapsulation assisted by the application of external force not only requires a more complex experimental setup, e.g., the implementation of fluorescence detection modules, SAW module, etc., but may also deteriorate cell growth and living conditions by exposing target cells under external stresses. This could alter the cellular behaviors and lead to false observation. Nevertheless, while passive single-cell encapsulation methods merely utilize hydrodynamic profiles within microchannels to differ cells based on their physical properties such as size, shape, and stiffness, active encapsulation methods can label specific biomarkers within cells. These biomarkers later serve as the reference for selective encapsulation. This is especially useful for the capture of rare cell samples like in the case of circulating tumor cells (CTCs) that need to be singled out of millions of normal blood cells. From our perspective, though passive encapsulation methods outcompete active methods because of their relatively high encapsulation efficiency (around 80%) and high rate, their inherent incapability to handle heterogeneous cell groups and necessity for meticulous modulation of hydrodynamic profiles within microchannels all constraint their wider applicability in more complicated real-life scenarios. For this reason, in view of the application perspective, more studies are suggested to further enhance the encapsulation rate of active methods, which involve the collaboration of diverse fields of science and engineering such as electrical, chemical, material engineering, etc.
IV. SINGLE-CELL ANALYSIS WITHIN DROPLET-BASED MICROFLUIDICS
As previously mentioned, many biological assays at the single-cell resolution were recently developed because they reveal valuable information such as cellular heterogeneities and cell-to-cell interactions that are often missed in the measurement of the bulk population. A droplet-based microfluidic device offers a unique platform for the development of a single-cell assay as it does not only enable precise manipulations over tested samples but also provides a physical barrier to avoid cross-contamination, confines reaction volume, reduces the reagent consumption, and enhances the sensitivity of assays. In addition, the compartmentalization of single cells into microdroplets favors their long-term culture and the measurement of their dynamic cellular behaviors when comparing to other single-cell analysis assays. Therefore, in this section, common single-cell analysis methods were presented in brief. Recent advances in the studies of long-term cellular behaviors as well as factors influencing cellular proliferation within microdroplets were also introduced. Some examples of single-cell analysis methods are summarized in Table V.
TABLE V.
Some examples of single-cell analysis methods.
| Method | Principle | Purpose | Reference |
|---|---|---|---|
| Optical analysis | |||
| Lab-tag methods | |||
| Fluorescence resonance energy transfer | Detect fluorescent signal originated from the energy transfer from an excited donor fluorophore to an acceptor molecule. | Usually used to detect membrane proteins or secreted proteins at single cell levels. | 123 and 124 |
| Radiofluorescence | Detect positron emission through a scintillator substrate at single-cell level | Used for the detection of the presence or uptake of small biomolecules | 125 |
| Immunofluorescence | Detect the fluorescent signal emitted once specific fluorescent antibody attached to corresponding biomolecules | Usually used to detect single-cell proteomics, and often required signal amplification or concentration process to enhance the test sensitivity | 126–128 |
| Single copy genetic amplification | Co-encapsulation of single cells along with PCR mixture and an agarose bead covalently labeled with reversed primers. The cell lysis and PCR are performed off-chip, and flow cytometry is used to measure the fluorescent intensity. | Used to test single-cell genomics; if the RT-PCR mix was inserted, also able to test single-cell transcriptomics. | 131 |
| Agarose-based emulsion PCR | Similar to single copy genetic amplification, but here the reversed primers are labeled on the encapsulating agarose layer, avoiding the necessity of co-encapsulating single cells along with agarose beads. | Used to test single-cell genomics; if the RT-PCR mix was inserted, also able to test single-cell transcriptomics. | 132 |
| Label-free method | |||
| Raman spectrometry | Relies on the dispersion and detection of inelastically scattered photons. | Usually used to detect the single-cell proteomics, often required extra signal enhancers to ensure proper test sensitivity. | 134–136 |
| Electrochemical analysis | Relies on electrochemical conversion or a physical interaction of the analyte at an electrode surface generates a detectable electrical signal | Usually used to detect the secretomics at single-cell resolution. | 140–143 |
| Mass spectrometry analysis | Relies on the use of heavy metal element labels and separation of ions according to their mass-to-charge ratio, which can be detected either sequentially or quasi-simultaneously and generate a short transient signal. | Usually sued to detect the metabolomics at single-cell resolution, or the analysis of single-cell proteomics. | 144 and 145 |
A. Methods for single-cell analysis within droplet-based microfluidics
1. Optical analysis—Label tag methods
One of the most widely employed and straightforward methods in single-cell analysis is the optical analysis with or without external labels. It can reveal the temporal–spatial expression and distribution of subcellular constituents as well as dynamic cellular behaviors. Notably, the recent development of fluorescence-based assays offers a facile and straightforward pathway to observe cellular behavior at the micrometer or even the nanometer resolution. For example, the fluorescence resonance energy transfer (FRET) technique is widely utilized to study cell biology, especially cellular secretome, by detecting the fluorescent signal originated from the energy transfer from an excited donor fluorophore to an acceptor molecule.123 Utilizing a T-junction microfluidic device, Ng et al. encapsulated single cells along with multicolored FRET-based enzymatic substrates to measure the secretion rate and binding efficiency of several specific proteases at a single-cell resolution [Fig. 8(a)].124 When a specific protease was secreted by encapsulated single cells within microdroplets, it would cleave the fluorescence quencher unit off the modified FRET substrate and activate the emission of the fluorescence signal, which could then be detected via fluorescence microscopy. The platform was tested effective on both single-cell lysate and live PC-9 cancer cells as well as leukemia K-562 cells.
FIG. 8.
Schematic illustrations of some single-cell analysis methods in droplet-based microfluidics. (a) Analysis of cellular secretome at single-cell level suing FRET, developed by Ng et al. Reproduced with permission from Biosens. Bioelectron. Ng et al., 81, 408–414 (2016).124 Copyright 2016 Elsevier. (b) Determine [18F]fluorodeoxyglucose uptake by droplet radiofluidics at single-cell resolution, developed by Türkcan et al. Reproduced with permission from Türkcan et al., Anal. Chem. 87, 6667–6673 (2015). Copyright 2015 American Chemical Society.125 (c) DropMap system to measure dynamic phenotyping of single-cell, developed by Baudry et al. Reproduced with permission from Nat. Biotechnol. 35, 977–982 (2017).128 Copyright 2017 Nature. (d) SERS system for single-cell analysis, developed by Willner et al. Reproduced with permission from Willner et al., Anal. Chem. 90, 12004–12010 (2018).135 Copyright 2018 American Chemical Society.
Alternatively, some small biomolecules such as ions, amino acids, metabolites, and drugs cannot be easily tagged by fluorescent dye. Radioluminescence microscopy is a prominent alternative to overcome this obstacle as it can detect positron emission through a scintillator substrate at a single-cell level. Türkcan et al. reported the combination of radioluminescence microscopy and droplet microfluidics for the single-cell analysis of [18F]fluorodexyglucose (FDG) uptake [Fig. 8(b)].125 After the incubation with radionuclide probe FDG, individual cells were encapsulated into microdroplets and underwent radioluminescence microscopic analysis. They observed cellular heterogeneity in FDG uptake rate with the coefficient of variation of 50% for the long incubation and 80% for the short incubation, which was never reported in the bulk assay.
Immunofluorescence staining is another extensively employed fluorescence method for live single-cell analysis. Targeted cells can be either pre-stained prior to encapsulation, co-encapsulated with staining reagents or strained during the culture via perfusion, depending on the nature of assays and encapsulating conditions. For example, An et al. employed a flow-focusing microchannel to encapsulate Salmonella along with red fluorescent resorufin as a cellular metabolic indicator to quantitatively determine the proliferation of food pathogen.126 On the other hand, the droplet-based single-cell proteomic analysis often suffers from the intense background noise and low detection sensitivity. Many efforts were conducted to solve this issue. The group of Griffiths and Baudry developed a dynamic single-cell phenotyping system named DropMap, where individual mice cells were encapsulated along with red fluorescent antibodies, green fluorescent antigens, and paramagnetic nanoparticles functionalized for capturing secreted IgG, a common immunology marker [Fig. 8(c)].127,128 Using time-resolved fluorescent microscopy, both the secretion rate of IgG and its affinity toward antigen were determined simultaneously in a quantitative and dynamic fashion. Under the effect of a magnetic field, co-encapsulated paramagnetic nanoparticles formed a detection rod, which concentrated the analyte signals, reduced the background noise, and enhanced the detection limit. Similarly, Wei et al. utilized antibody-conjugated gold nanorods (AuNR) as the plasmonic sensors that were co-encapsulated with single cells inside microcapsules and bound to target analytes.129 The usage of AuNR increased the detection limit up to 6–7 ng/ml. Details on long-term cell culture within microcapsules are presented in a later section.
Finally, the genetic and transcriptomic profiles at the single-cell resolution can be measured using the state-of-the-art digital PCR and RT-PCR. Generally, the analyte from a single cell is first randomly distributed into many small volumes. The number of discretized volumes is sufficiently high so that some volumes do not contain any analyte. Then, after the PCR or RT-PCR is finished within each volume, endpoint detection is performed to quantify initial analytes by assuming a Poisson distribution of analytes into discretized assay volumes. Since the commercialization of these two techniques is relatively mature, they were not presented in further detail in the present review. One can refer to the review written by Thompson et al. if interested.130 Alternatively, targeted DNA amplification of single cells can also be performed within microcapsules. For instance, with the assistance of microfluidic devices, single copy genetic amplification (SCGA) first co-encapsulates single cells along with an agarose bead covalently labeled with reverse primers, and PCR mixture containing dye-labeled forward primers and enzymes. Then, the microcapsules are collected off-chip for cell lysis and PCR thermocycles. The resulted dye-labeled double-stranded genetic materials are accumulated on the agarose bead surface, and the fluorescent intensity is measured via flow cytometry. To further enhance the throughput of SCGA method, Zeng et al. employed a microfabricated emulsion array (MEGA) with 96 channels, which enabled the generation of approximately 940 fluorescent droplets per second readily testable by flow cytometry.131 Nevertheless, the PCR efficiency remained low (around 40%) compared to standard PCR operated in the bulk environment, not to mention the Poisson statistic limitation of co-encapsulating single cells along with single agarose beads. All these circumstances constraint the efficiency and throughput of SCGA in large-scale applications. On the other hand, in order to circumvent the Poisson statistic for co-encapsulation, Yang et al. developed agarose-based emulsion PCR (ePCR).132 Similar to SCGA, single cells are encapsulated inside an agarose microcapsule along with the PCR mixture containing fluorescently labeled forward primers and enzymes. Here, the reverse primers are covalently attached to the agarose layer, which avoids the necessity to co-encapsulate agarose beads with single cells. The resulted PCR amplicons are attached to the encapsulating agarose matrix, ready to be measured under flow cytometry. Compared to SCGA, ePCR showcases one order of magnitude higher in efficiency, which increases the test throughput while reducing the duration and costs of the experiments. In general, analysis methods on single-cell genomics and transcriptomics rely on the co-encapsulation of single cells along with reagent mixtures. Some barcoding beads are often added to concentrate test products with the goal of increasing the detection limit of downstream analysis. One can refer to a recent review written by Huck et al. if interested.133 After all, there are several major technical concerns that droplet-based single-cell genomic/transcriptomic analysis needs to overcome: (1) the PCR efficiency remains relatively low within microcapsules compared to the reaction performed in the bulk environment, which leads to a longer experimental duration and a higher cost; (2) the PCR amplification as well as the fluorescent analysis with flow cytometry needs to be performed off-chip; such a lack of integration enhances the laboring, cost, and duration of the experiments; (3) product analysis is often realized in a serial manner (e.g., like in the case of flow cytometry), which limits the absolute throughput of the test; more studies are suggested to increase the efficiency and throughput of the testing protocol.
2. Optical analysis—Label-free methods
Despite the rapid development of single-cell analysis using fluorescent tags, label-free methods for single-cell analysis also attract a lot of attention as they offer direct readings on cellular profiles without potential complications such as label affinity, specificity, etc. Raman spectroscopy, which is initially used to observe vibrational, rotational, and other low-frequency movement of a system, has been recently reported as a potential tool for single-cell analysis. In fact, Raman spectroscopy relies on the dispersion and detection of inelastically scattered photons. When a monochromatic laser hits the sample object, most of the interacting photons experience elastic scattering, whereas a small fraction of photons undergo inelastic scattering, resulting in exchanges of energy between photons and the vibrational energy level of the molecules. The loss of energy in photons, i.e., the Stokes-shifted Raman scattering, would result in a spectrum of discrete peaks, which reveals not only the molecular bond of the sample object but also its chemical micro-environment.134 Nevertheless, the signal of ordinary Raman spectroscopy is relatively weak, and many methods were developed to enhance the acquired signal. For example, Willner et al. employed surface-enhanced Raman scattering (SERS) technique to determine glycan expression on the surface of prostate cancer cells by binding wheat germ agglutin (WGA)-modified metallic nanoparticles onto cellular membrane as a signal reporter and amplifier [Fig. 8(d)].135 They encapsulated functionalized single PC3 cancer prostate cells into droplets using a T-junction device. Under the effect of laser excitation, WGA functionalized nanoprobes on the cellular membrane, which are specific to the glycan N-acetyl neuraminic, can reveal glycan surface coverage at single-cell resolution. Similarly, using 5-bromo-4-chloro-3indolyl phosphate as the signal reporter and enhancer, Sun et al. identified the cellular heterogeneity in alkaline phosphatase (ALP) activities via a SERS droplet microfluidic platform at a single-cell level.136 More information on Raman spectroscopy for single-cell analysis can be found in the review written by Schie et al.134
After all, optical analysis of single-cell at high throughput relies on techniques for rapid image capture and auto-image analysis. While state-of-the-art microscopy platforms for auto-screening and image capture are already very mature, many automated image analysis techniques were developed to study behaviors of single cells in microdroplets, especially for time-resolved dynamic studies with a massive amount of data to be analyzed.137–139
3. Electrochemical analysis
Electrochemical analysis utilizes the electrochemical conversion or the physical interaction of the analyte at an electrode surface that leads to the generation of a detectable electrical signal such as current, charge, or potential. In recent years, the electrochemical techniques are widely applied in single-cell analysis, especially in the studies on exocytosis. For example, Li et al. described the capability of carbon microdisk electrodes to adsorb nanoscale mammalian vesicles onto their surface and subsequently rupture and expel the contents within these vesicles under the application of the electrical field.140 This rupture elicited an oxidation current that can be used to quantify the catecholamine content inside vesicles. Furthermore, they flame-etched carbon-fiber microelectrodes so that these microelectrodes could be inserted inside single PC12 cells and determined the quantal neurotransmitter content by following the same pathway.141 Similarly, these electrodes can be inserted inside cell-encapsulating microdroplets for the detection of the cellular secretome. Alternatively, electrochemical techniques can also be utilized in enzymatic effects. Safaei et al. reported the detection of circulating tumor cells (CTC) using electrochemical effects.142 By tagging captured cells with functionalized antibodies specific to the universal epithelial cancer marker cytokeratin (anti-CK18), alkaline phosphatase (ALP) labeled onto anti-CK18 would activate an electrochemical reporter once bound to the cancer marker and produce a change of current. Such a signal would be acquired for the capture of CTC. Recent advances in the usage of electrochemical techniques for single-cell analysis can be found in the review written by Zhang et al.143
4. Mass spectrometry analysis
Mass spectrometry (MS) is recently used to detect biomolecules in single cells because of its high detection limits, high sensitivity, high selectivity, wide applicability, and label-free nature. This technique uses a heavy metal element as detection labels and separates ions according to their mass-to-charge ratio, which can be detected either sequentially or quasi-simultaneously. Besides its wide application in single-cell proteomic analysis, the mass spectrometry technology is often used to detect metabolomic at single-cell resolution. In fact, the study of cellular metabolomic involves compounds with a molecular weight smaller than 1500 Da, i.e., a relatively weak signal. The inability of amplifying metabolites further enhances the detection difficulties. Therefore, mass spectrometry with very high sensitivity, great selectivity, and wide applicability is the ideal tool for such a purpose. For instance, Wang et al. built a time-resolved MS platform for the detection of zinc distribution as well as the uptake/adsorption of ZnO nanoparticles in single HepG2 cells encapsulated in droplets using a simple flow-focusing design.144 More recent applications of MS for single-cell analysis can refer to the review written by Yin et al.145
B. Long-term live cell analysis
Microdroplets generated through droplet-based microfluidic can provide a three-dimensional niche for the long-term culture and real-time analysis of single cells. It can be utilized in studying dynamic cellular behavior, notably during single-cell proliferation and differentiation. Logically, maintaining the cell viability within microdroplets over the studying period is crucial for such a purpose. This requires the efficient exchange of oxygen and nutrient into microdroplets while avoiding the accumulation of cytotoxic substances within the confinement.
1. Ionic polymerization
Recently, hydrogels and other microgels have been widely used as encapsulating material for long-term culture and single-cell analysis within microcapsules because they allow the exchange of a fresh culture medium and the doping of chemicals. Among all types of hydrogels employed in cell encapsulation, alginate is the most commonly used material in research. In fact, alginate macromer consists of b-D-mannuronic acid (M block) and a-L-guluronic acid (G block) containing a sodium ion, which are arranged in a block-wise pattern.146,147 The gelation of alginate can occur via an ionic crosslinking mechanism, where a divalent ion, usually calcium (Ca2+), substitutes sodium ions and binds to the free carboxyl group of the G block, forming an egg-box structure.147,148 The mechanical size and the porosity of alginate microgels are determined by their degree of crosslinking, which can be tuned by adjusting the ratio of M and G blocks as well as the chain's molecular weight. There are two distinct gelation pathways for alginate: internal and external gelation. In external gelation, a stream of cell-containing alginate simply co-flows with a CaCl2 containing oil stream, resulting in heterogeneous, non-spherical, and core-shell microbeads in the case of rapid polymerization.146,147 Li et al. fabricated a microfluidic device with two flow-focusing geometries to generate cell-laden disk-shaped microgels using external gelation on-chip. The post-encapsulation cell viability was reported to be only 50%.149 Alternatively, generated droplets can be collected in a bath of CaCl2 and complete the external gelation off-chip, which results in relatively high cell viability and cell mitosis rate.150 In contrast, during the internal gelation process, the alginate dispersed phase that contains suspended cells and insoluble CaCO3 is pinched off by the continuous phase containing acidic oil.151–153 The acidic conditions facilitate the gradual release of Ca2+ into the dispersed phase, allowing ionic polymerization of alginate. Despite the homogeneity, monodispersity, and controllability of droplets generated via internal gelation, the exposure to the acidic environment can lower the cell viability, which is unfavorable for long-term cell culture in microdroplets. Many research studies work on solving such an issue. For instance, higher cell viability was achieved when streaming flow with a greater concentration of CaCO3 as CO32– can act as a buffer to increase the pH of the system.154 On the other hand, Akbari et al. proposed the usage of a stoichiometric equivalent of acetic acid with respect to CaCO3 in order to reduce the cell exposure to acidic conditions.152 Kim et al. proposed to wash off acid from alginate microgels with a high flow of biocompatible oil and achieved 95% of cell viability immediately postencapsulation.155 However, in both internal and external gelation, either the usage of insoluble calcium salt or the delivery of calcium ions via the oil phase risks a heterogenous distribution of calcium ions, i.e., heterogeneous polymerization in cell-laden droplets. This may cause local stress concentration and harm cell viability within microcapsules. For this reason, Weitz et al. proposed the dissolution of water-soluble Ca-ethylenediaminetetraacetic acid (EDTA) complex directly in the aqueous phase along with alginate macromers and suspended cells.87 Upon the diffusion of acetic acid from the oil phase into the droplets, calcium ions dissociate from the complex and induce the alginate gelation. The post-encapsulation cell viability was reported to be 83%.
However, studies showcase several drawbacks of ionic crosslinking pathways in generating cell-laden microgels. For example, ionically crosslinked alginate is characterized by a slow, uncontrollable, and unpredictable degradation rate.156 This nature limits its application in tissue engineering that requires the degradation of the cell-laden scaffold to be at a similar rate as new tissue formation. In addition, the difficulties of these hydrogels to form a non-spherical structure further constraint their usages in biomedical applications.157
2. Photopolymerization
Photo-crosslinking polymerization, where a gel polymerizes through exposing monomers or pre-polymer substrates to photoinitiators and light source (usually UV light), offers an alternative to produce cell-laden microgels. Wang et al. investigated the gelling conditions (monomer concentration, photoinitiator concentration, UV intensity, and UV exposure time), mechanical properties (monodispersity, degradation rate, elastic modulus, etc.), and cytocompatibility of cell-laden oxidized methacrylated alginate (OMA). They optimized the photo-crosslinking polymerization of OMA and reported over 90% cell viability postencapsulation.158 Interestingly, Etter et al. combined the ionic and photo-crosslinking mechanisms to generate homogeneous alginate microgel.159 Cell-laden -cyclodextrin-methacrylated alginate first underwent an ionic polymerization of low-concentration CaCl2 to prevent cell escape and then a photopolymerization of green light to finish the gelation with minimal cell damage.
Alternatively, PEG-based photopolymerized hydrogel is another common photo-crosslinked microgel in biomedical applications. It has attracted researchers’ attention for cell encapsulation because of its excellent cytocompatibility for many cell types such as mesenchymal stem cells and beta-cells.160,161 It can be generally formed via either acrylate or thiol-ene chemistry by following chain growth [poly(ethylene glycol) diacrylate (PEGDA)] or step-growth [poly(ethylene glycol) norbornene (PEGNB)] polymerization mechanism.24 However, at the presence of oxygen, chain growth polymerization is inhibited since the rate of radical quenching by oxygen is much faster than the rate of chain growth.162,163 This radical scavenging would lead to the accumulation of reactive oxygen species (ROS) and raise intracellular oxidative stress that can damage biomolecules.164 Therefore, step-growth PEGNB exhibits lower stress onto encapsulated cells and showcases higher post-encapsulation cell viability than PEGDA, especially for cells types sensitive to the presence of ROS. Jiang et al. described the encapsulation of single cells in PEGNB hydrogel microspheres and performed in situ photopolymerization, which can greatly shorten the required polymerization time.165 By simply staining cell-laden microgel with LIVE/DEAD assay, they reported over 50% cell viability after 30 days of incubation. Similarly, Akbari reported the encapsulation of single cells into step-growing Arg-Gly-Asp (RDG)-functionalized 8-arm poly(ethylene glycol) hydrogel microspheres and observed around 80% cell viability after 13 days of incubation.60 Alternatively, to solve the problem of ROS in PEGDA microgel, Xia et al. recently presented a nitrogen-jacketed microfluidic device that encapsulated cells into in situ polymerized PEGDA droplets [Fig. 9(c)].166 By deprived oxygen diffusion into the PDMS microfluidic device, they reported high post-encapsulation viability of 95.6%, much higher than that of standard PEGD microgels. However, the long-time culture of cells under an oxygen-deprived environment could reversely harm cellular viability, and cell-laden microgels should be quickly transferred into a suitable cultivation environment once the polymerization process is finished.158 Overall, the morphologies of photo-cross-linked microgels are related to many operating parameters including macromer and photoinitiator concentrations, UV intensity, and UV exposure time. One should carefully adjust these parameters to create sufficiently mild gelling conditions in order to maximize cellular viability within microgels.
FIG. 9.
Schematic illustrations of some techniques for long-term cell culture in droplet-based microfluidics. (a) PEG-based microgel formed via T-MA, developed by Guerzoni et al. Reproduced with permission from Guerzoni et al., Small 15, 1900692 (2019).173 Copyright 2019 Wiley. (b) W/O/W double emulsions for the encapsulation of non-adherent cells, developed by Cai et al. IA is the inner aqueous phase, OA is the outer aqueous phase and OP is the oil phase. Reproduced with permission from Cai et al., Lab Chip 19, 422–431 (2019).177 Copyright 2019 Royal Society of Chemistry. (c) Nitrogen-jacketed microfluidic system to produce cell-laden PEGDA microgels, developed by Jiang et al. Reproduced with permission from Jiang et al., J. Mater. Chem. B 5, 173–180 (2017).165 Copyright 2017 Royal Society of Chemistry.
3. Thiol-Michael addition polymerization
Another common method to generate a PEG-based microgel is the thiol-Michael addition (T-MA), which is a click reaction between a thiol-containing species (Michael donor) and an electron-deficient olefin (Michael acceptor).167–170 Compared to the photo-crosslinked PEG-based microgel, T-MA offers greater versatility in terms of the nature of the precursors, integration of functional groups, as well as milder reaction conditions while avoiding the potential cytotoxicity of UV exposure.171,172 It is believed to be very biocompatible because of its UV-free and radical-free polymerization reaction. Guerzoni et al. recently reported the generation of cell-laden PEG-based microgel via base-catalyzed T-MA. In their study, star-PEG-VS and star-PEG-SH were used as polymer precursors, and N,N,N′,N′-tetramethylethylenediamine (TEMED) as base catalysis [Fig. 9(a)].173 Particularly, GRGDSPC peptide chains were bound to the star-PEG-VS as the adhesion sites for encapsulated primary normal dermal human fibroblasts. Here, two aqueous streams, each containing one of the precursors, flowed parallelly to each other until the introduction of a first continuous oil stream that pinched off the precursor stream into microdroplets. Further downstream, a secondary oil stream containing TEMED was injected and triggered the T-MA polymerization. The resulted cell-laden microgel showcased 90% cell viability at day 2 postencapsulation. Similarly, Siltanen et al. utilized T-MA to generate a bioactive co-polymer microgel system composed of heparin and PEG. In fact, heparins are highly sulfated glycosaminoglycans that can regulate cell signaling through reversible sequestration of numerous growth factors expressing heparin-binding domains. For this reason, the minimal dosage of growth factors co-encapsulated along with stem cells inside microcapsules was required to induce a high level of differentiation. Here, the heparin/PEG microgels encapsulated mice embryonic stem cells (mESCs) with growth factors Nodal and FGF-2, and were cultivated over 5 days of differentiation. Results demonstrated that not only high expressions of endoderm marker Sox17/FoxA2 were found in encapsulated mESCs, this strategy also reduced total Nodal/FGF-2 consumption by approximately 90-fold compared to traditional 2D or 3D differentiation protocols.174
4. Catalytic polymerization
As previously described, cell encapsulation within ATPS microgels has drawn considerable attention because of its low cytotoxicity, despite its low interfacial tension that inhibits the easy formation of microdroplets. Liu et al. described the encapsulation of human liver carcinoma HepG2 cells in the aqueous two-phase dextran (DEX)/PEG microgels.175 With the assistance of a cyclic pressure change, a DEX-based solution containing suspended HepG2 cells, alginate-Ph, and HRP was first introduced into pure PEG solution to form droplets. The generated microdroplets were then injected into PEG solution containing 2 mH H2O2, which triggered the catalytic activities of HRP to crosslink the Ph moieties of alginate gel. Using Calcein AM/PI double staining, the cell viability within microcapsules was measured to be over 90% after 6 days of culture. Mastiani et al. also reported the cell encapsulation in ATPS droplets.176 They compared the standard DEX/PEG system with the PEG/MgSO4 system and concluded that PEG/MgSO4 system can generate droplets with higher monodispersity while maintaining a high cell viability. However, the salt concentration should be carefully controlled as only 50% of cell viability was observed when it exceeded 15% w/v.
5. Other factors influencing long-term cellular viability inside microcapsules
The microenvironment of microgel is drastically different from the aqueous surrounding where non-adherent cells naturally suspend. To solve this problem, Cai et al. utilized anchored water-in-oil-in-water (W/O/W) double emulsion as a niche to realize in situ real-time treatment and monitor non-adherent cells at single-cell resolution [Fig. 9(b)].177 Generated W/O/W double emulsion provides an aqueous core for the suspension of non-adherent cells and allows the mass transfer of small molecules across the silicone oil/PDMS shell to maintain cell viability. They used fluorescein diacetate (FDA) perfusion culture to monitor cellular FDA uptake and subsequent metabolism, which was an indication of cell viability within microcapsules. Cell viability and proliferation were reported to be maintained during 24 h post-encapsulation under continuous perfusion.
On top of the material for single-cell encapsulation, droplet size and cell location are also essential to ensure proper cell proliferation and/or differentiation in the long-term. Joensson's group investigated the influence of droplet size on Chinese Hamster Ovary (CHO) cell division and observed a clear correlation between droplet size and cell division/viability over a period of 3 days, demonstrating the importance of selecting appropriate droplet size with corresponding cell density for long-term cell screening.178 On the other hand, Kamperman et al. highlighted the importance of centering a single cell within microgels to ensure long-term 3D culture and prevent cell escaping.179 As cells prefer to locate at the immiscible interface during the droplet formation, they delayed the polymerization of encapsulating tyramine conjugated dextran (Dex-TRA) microgels by slowly diffusing the crosslinking catalysis hydroperoxide into the system. In this manner, they allowed enough time for cells to migrate back to the center of microgels and observed merely 5% of cell escape on day 7 of incubation and over 90% cell viability on day 28 of incubation. Additional cell adhesion sites can also be added to the polymer as previously described. Other key factors to ensure long-term cell viability inside microgels are the selection of oil/surfactant phase and how to quickly transfer cell-laden droplets from the oil phase (often cytotoxic toward cells) to an aqueous environment for a long-term culture. For instance, fluorocarbon oil is an excellent biocompatible oil phase for the generation of cell-laden microgels in microfluidic.127,128,180 With around 20 times oxygen and carbon dioxide dissolution than water and immiscibility with organic as well as aqueous solvents, fluorocarbon oil offers an effective barrier to protect cell-laden microgel from external stress while alleviating oxygen stress within microcapsules.181,182 Baudry et al. utilized the combination of fluorinated oil and fluorosurfactants in their DropMap system to ensure relatively high cell viability up to 12 h without the addition of fresh culture medium.127,128 However, a certain combination of fluorinated oils and fluorosurfactants can lead to nonspecific protein adsorption onto the oil–water interface, which could potentially affect downstream proteomic analysis.183 Hence, one should carefully choose the oil phase based on the encapsulated content and downstream applications in order to ensure proper transport of desired molecules in-and-out the cell-laden droplets.
On the other hand, rapid extraction of cell-laden microgels from the cytotoxic oil phase into the aqueous phase is crucial for the long-term cell culture in microcapsules. The traditional method for this purpose involves strong centrifugation and cyclic washing with an aqueous buffer off-chip. Nevertheless, the relatively slow extraction speed, i.e., prolonged exposure to the oil phase, and the requirement for high centrifugation speed reduce the cell viability. Hence, on-chip extraction methods with shorter time and milder conditions are preferred. Current common on-chip extraction methods include a fluid exchange with the use of filter blocks, integration of an extraction chamber with the use of filter blocks, the employment of oil sacrificial layer, and the utilization of interfacial tension between oil and microdroplets.149,184–186 For example, Liao et al. reported a W/O/W double emulsion system for cell encapsulation in droplet microfluidic.187 Generated ionically crosslinked alginate microspheres were covered by a layer of mineral oil, which would detach from microbeads spontaneously upon the contact with the outer aqueous layer because of hydrodynamic force and density difference. Alternatively, Mohamed et al. designed a hydrodynamic filter postextraction chamber as the filtration unit to wash the oil phase away with an aqueous washing buffer. They achieved for the first time the integration of all steps of cell-laden microgel fabrication on a chip: from droplet generation to in situ photopolymerization of microgels to microgel extraction from the oil phase.188
V. CONCLUSIONS AND OUTLOOK
Recent progress in biomedicine and biology urges the development of single-cell encapsulation and analysis. Because of its simplicity, biocompatibility, and monodispersity of generated microcapsules, droplet-based microfluidics is the most promising platform for this purpose. The present Review systematically summarized the basic principles of droplet generation mechanism inside microfluidic devices, basic methods to fabricate microfluidic devices, and various passive as well as active techniques to enhance single-cell encapsulation efficiency. Moreover, common techniques of single-cell analysis based on droplet microfluidics were briefly introduced, with a focus on the long-term proliferation, differentiation, and observation of encapsulated cells within microcapsules. In principle, passive droplet generation depends on the flow regime within the microchannel, which is determined by the stress competition among capillary stress, viscous stress, and interfacial tensions. By carefully manipulating the flow conditions and channel parameters, one can modulate the generation rate as well as morphologies of droplets. Various passive and active methods were developed in the past few years to manipulate either extrinsic parameters or intrinsic features of the microfluidic system with the goal of enhancing single-cell encapsulation efficiency.
Compared with other methods of single-cell isolation, droplet-based microfluidics offers the unique advantage of compartmentalizing single-cell into a physically confined microenvironment. This can not only avoid cross-contamination between samples, increase assay sensitivity, lower background noise signal, and protect encapsulated cells against external stresses, but also most importantly provides the possibility of observing cell behaviors in a dynamic and real-time manner while allowing more precise and diverse manipulations over tested samples. Considering that most of the current molecular biology techniques still rely on endpoint measurement, monitoring long-term single-cell dynamic status, e.g., dynamic cellular profiles during stem cell differentiation, will certainly bring revolutionary progress to the modern biomedicine and biology.
With an increasing number of researchers and companies working in this novel technology, a massive number of fundamental research studies and potential applications have been reported in recent years. Despite the rapid advances in this field of research, plenty of novel technologies stay at the bench level and wait for commercialization since many technical issues still need to be addressed for their application on a larger scale. Most importantly, current passive and active single-cell encapsulation technologies suffer from the problems of either extremely low cell encapsulating rate, the potential harm of external forces onto biological samples, or the rigorous requirement in flow manipulation (i.e., lack of robustness). Various multi-layer 3D microfluidic devices for cell encapsulation were manufactured to enhance the production rate of the single-cell microcapsule, but the improvement is still limited by the low encapsulation efficiency, long production period, intense operation, and high cost.59,60,189,190
In the future, researchers in the field of single-cell encapsulation and analysis should collaborate with biologists, chemists, electrical engineers, software engineers, and experts from other related fields to conquer challenges in both fundamental research and practical application. For example, factors and mechanisms affecting single-cell proliferation and differentiation within microdroplets need to be further unveiled.191 Several major technical concerns in droplet-based single-cell analysis need to be resolved. First, the confined environment of microcapsules is vastly different from the bulk environment in which cells are naturally found. This may alter the cellular behaviors and lead to false observation. In fact, how to mimic in vivo conditions has attracted great attention of the scientific community recently, especially with the advancement in constructing 3D environment in vitro to simulate the biomechanical and biochemical profile in real-life. More efforts are suggested to create an in vivo like microenvironment within microcapsules. Second, existing microfluidic modules merely include simple functionalities such as cell culture, cell encapsulation, flow mixing, and signal detection. Microfluidic features with more complex functionalities need to be further investigated and developed. For example, current common methods for single-cell genomic analysis still require off-chip PCR amplification and flow cytometry analysis. This not only burdens the experimental laboring and cost but also disturbs the continuity of the entire analysis process. On the other hand, while providing a physical barrier for encapsulated single cells, microcapsules inherently prevent the application of multiple-step assays, which may require the addition of reagents or other manipulations. Despite the appearance of the electro-coalescence technique to merge two distinct droplets together, the potential harm of applying electrical field onto biological samples makes the method questionable, not to mention the inevitable limitation of Poisson statistics in pairing two single droplets. Therefore, more studies are required to develop microfluidic modules with more complicated functionalities in order to achieve the complete integration of single-cell analysis onto one single microchip. Third, due to the difficulties of calibration within microcapsules, the absolute quantification is rather hard for droplet-based single-cell analysis, limiting the analysis to qualitative level rather than quantitative level. More effective calibration methods are necessary in the future. All these require collaboration between experts of different fields to investigate the problem from all perspectives including material science, biomechanics, developmental biology, etc. On the other hand, as current single-cell encapsulation and analysis remain an expensive, time-consuming, and heavy laboring technique, researchers should work on the engineering of automated on-chip single-cell culture, encapsulation, and analysis platform while integrating newly emerging technologies like MEMS and machine learning to enhance the efficiency, versatility, applicability, and throughput of the system.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support by the National Natural Science Foundation of China (NNSFC) (Nos. 22025801 and 21991101).
Contributor Information
Yanan Du, Email: .
Jianhong Xu, Email: .
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Rotem A. et al. , “Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state,” Nat. Biotechnol. 33, 1165–1172 (2015). 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu X., Wu N., Chen F., Wei J., and Zhao Y., “Engineering microfluidic chip for circulating tumor cells: From enrichment, release to single cell analysis,” Trends Anal. Chem. 117, 27–38 (2019). 10.1016/j.trac.2019.03.027 [DOI] [Google Scholar]
- 3.Tavakoli H. et al. , “Recent advances in microfluidic platforms for single-cell analysis in cancer biology, diagnosis and therapy,” Trends Anal. Chem. 117, 13–26 (2019). 10.1016/j.trac.2019.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang L., Chung B. G., Langer R., and Khademhosseini A., “Microfluidics for drug discovery and development: From target selection to product lifecycle management,” Drug Discovery Today 13, 1–13 (2008). 10.1016/j.drudis.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein A. M. et al. , “Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells,” Cell 161, 1187–1201 (2015). 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shembekar N., Hu H., Eustace D., and Merten C. A., “Single-cell droplet microfluidic screening for antibodies specifically binding to target cells,” Cell Rep. 22, 2206–2215 (2018). 10.1016/j.celrep.2018.01.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witte K., Rodrigo-Navarro A., and Salmeron-Sanchez M., “Bacteria-laden microgels as autonomous three-dimensional environments for stem cell engineering,” Mater. Today Bio 2, 100011 (2019). 10.1016/j.mtbio.2019.100011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P., Chen D., Li S., Ou X., and Liu B.-F., “Microfluidics towards single cell resolution protein analysis,” Trends Anal. Chem. 117, 2–12 (2019). 10.1016/j.trac.2019.06.022 [DOI] [Google Scholar]
- 9.Agresti J. J. et al. , “Ultrahigh-throughput screening in drop-based microfluidics for directed evolution,” Proc. Natl. Acad. Sci. U.S.A. 107, 4004 (2010). 10.1073/pnas.0910781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martino C. et al. , “Intracellular protein determination using droplet-based immunoassays,” Anal. Chem. 83, 5361–5368 (2011). 10.1021/ac200876q [DOI] [PubMed] [Google Scholar]
- 11.Fallah-Araghi A., Baret J. C., Ryckelynck M., and Griffiths A. D., “A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution,” Lab Chip 12, 882–891 (2012). 10.1039/c2lc21035e [DOI] [PubMed] [Google Scholar]
- 12.Zare R. N. and Kim S., “Microfluidic platforms for single-cell analysis,” Annu. Rev. Biomed. Eng. 12, 187–201 (2010). 10.1146/annurev-bioeng-070909-105238 [DOI] [PubMed] [Google Scholar]
- 13.Swennenhuis J. F. et al. , “Self-seeding microwell chip for the isolation and characterization of single cells,” Lab Chip 15, 3039–3046 (2015). 10.1039/C5LC00304K [DOI] [PubMed] [Google Scholar]
- 14.Xu X. et al. , “Microfluidic single-cell omics analysis,” Small 16, 1903905 (2020). 10.1002/smll.201903905 [DOI] [PubMed] [Google Scholar]
- 15.Lin L., Chu Y.-S., Thiery J. P., Lim C. T., and Rodriguez I., “Microfluidic cell trap array for controlled positioning of single cells on adhesive micropatterns,” Lab Chip 13, 714–721 (2013). 10.1039/C2LC41070B [DOI] [PubMed] [Google Scholar]
- 16.Gross A. et al. , “Technologies for single-cell isolation,” Int. J. Mol. Sci. 16, 16897–16919 (2015). 10.3390/ijms160816897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadappuram B. P. et al. , “Nanoscale tweezers for single-cell biopsies,” Nat. Nanotechnol. 14, 80–88 (2019). 10.1038/s41565-018-0315-8 [DOI] [PubMed] [Google Scholar]
- 18.Hunt T. P. and Westervelt R. M., “Dielectrophoresis tweezers for single cell manipulation,” Biomed. Microdevices 8, 227–230 (2006). 10.1007/s10544-006-8170-z [DOI] [PubMed] [Google Scholar]
- 19.Zhang H. and Liu K.-K., “Optical tweezers for single cells,” J. R. Soc. Interface 5, 671–690 (2008). 10.1098/rsif.2008.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shenkman R. M., Godoy-Silva R., Papas K. K., and Chalmers J. J., “Effects of energy dissipation rate on islets of Langerhans: Implications for isolation and transplantation,” Biotechnol. Bioeng. 103, 413–423 (2009). 10.1002/bit.22241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L., Cheung T. H., Charville G. W., and Rando T. A., “Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting,” Nat. Protoc. 10, 1612–1624 (2015). 10.1038/nprot.2015.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao D., Jin F., Zhou M., and Jiang Y., “Recent advances in single cell manipulation and biochemical analysis on microfluidics,” Analyst 144, 766–781 (2019). 10.1039/C8AN01186A [DOI] [PubMed] [Google Scholar]
- 23.Collins D. J., Neild A., deMello A., Liu A.-Q., and Ai Y., “The Poisson distribution and beyond: Methods for microfluidic droplet production and single cell encapsulation,” Lab Chip 15, 3439–3459 (2015). 10.1039/C5LC00614G [DOI] [PubMed] [Google Scholar]
- 24.Alkayyali T., Cameron T., Haltli B., Kerr R. G., and Ahmadi A., “Microfluidic and cross-linking methods for encapsulation of living cells and bacteria—A review,” Anal. Chim. Acta 1053, 1–21 (2019). 10.1016/j.aca.2018.12.056 [DOI] [PubMed] [Google Scholar]
- 25.De Menech M., Garstecki P., Jousse F., and Stone H. A., “Transition from squeezing to dripping in a microfluidic T-shaped junction,” J. Fluid Mech. 595, 141–161 (2008). 10.1017/S002211200700910X [DOI] [Google Scholar]
- 26.Utada A. S., Fernandez-Nieves A., Stone H. A., and Weitz D. A., “Dripping to jetting transitions in coflowing liquid streams,” Phys. Rev. Lett. 99, 094502 (2007). 10.1103/PhysRevLett.99.094502 [DOI] [PubMed] [Google Scholar]
- 27.Marín A. G., Campo-Cortés F., and Gordillo J. M., “Generation of micron-sized drops and bubbles through viscous coflows,” Colloids Surf. A 344, 2–7 (2009). 10.1016/j.colsurfa.2008.09.033 [DOI] [Google Scholar]
- 28.Castro-Hernández E., Gundabala V., Fernández-Nieves A., and Gordillo J. M., “Scaling the drop size in coflow experiments,” New J. Phys. 11, 075021 (2009). 10.1088/1367-2630/11/7/075021 [DOI] [Google Scholar]
- 29.Zhu P., Kong T., Kang Z., Tian X., and Wang L., “Tip-multi-breaking in capillary microfluidic devices,” Sci. Rep. 5, 11102–11102 (2015). 10.1038/srep11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cech J., Přibyl M., and Snita D., “Three-phase slug flow in microchips can provide beneficial reaction conditions for enzyme liquid–liquid reactions,” Biomicrofluidics 7, 54103–54103 (2013). 10.1063/1.4821168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H. S. et al. , “Generation and manipulation of hydrogel microcapsules by droplet-based microfluidics for mammalian cell culture,” Lab Chip 17, 1913–1932 (2017). 10.1039/c7lc00262a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abate A. R. and Weitz D. A., “Air-bubble-triggered drop formation in microfluidics,” Lab Chip 11, 1713–1716 (2011). 10.1039/C1LC20108E [DOI] [PubMed] [Google Scholar]
- 33.Xu J. H., Luo G. S., Li S. W., and Chen G. G., “Shear force induced monodisperse droplet formation in a microfluidic device by controlling wetting properties,” Lab Chip 6, 131–136 (2006). 10.1039/B509939K [DOI] [PubMed] [Google Scholar]
- 34.Umbanhowar P. B., Prasad V., and Weitz D. A., “Monodisperse emulsion generation via drop break Off in a coflowing stream,” Langmuir 16, 347–351 (2000). 10.1021/la990101e [DOI] [Google Scholar]
- 35.Takeuchi S., Garstecki P., Weibel D. B., and Whitesides G. M., “An axisymmetric flow-focusing microfluidic device,” Adv. Mater. 17, 1067–1072 (2005). 10.1002/adma.200401738 [DOI] [Google Scholar]
- 36.McDonald J. C. et al. , “Fabrication of microfluidic systems in poly(dimethylsiloxane),” Electrophoresis 21, 27–40 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Xu J.-H., and Luo G.-S., “The dynamic adsorption of different surfactants on droplet formation in coaxial microfluidic devices,” Chem. Eng. Sci. 138, 655–662 (2015). 10.1016/j.ces.2015.08.048 [DOI] [Google Scholar]
- 38.Xu J. H., Dong P. F., Zhao H., Tostado C. P., and Luo G. S., “The dynamic effects of surfactants on droplet formation in coaxial microfluidic devices,” Langmuir 28, 9250–9258 (2012). 10.1021/la301363d [DOI] [PubMed] [Google Scholar]
- 39.Baret J.-C., “Surfactants in droplet-based microfluidics,” Lab Chip 12, 422–433 (2012). 10.1039/C1LC20582J [DOI] [PubMed] [Google Scholar]
- 40.Wang K., Lu Y. C., Xu J. H., and Luo G. S., “Droplet generation in micro-sieve dispersion device,” Microfluid. Nanofluid. 10, 1087–1095 (2011). 10.1007/s10404-010-0737-6 [DOI] [Google Scholar]
- 41.Geng Y., Ling S., Huang J., and Xu J., “Multiphase microfluidics fundamentals, fabrication, and functions,” Small 16, 1906357 (2020). 10.1002/smll.201906357 [DOI] [PubMed] [Google Scholar]
- 42.Xu J. H., Li S. W., Tan J., and Luo G. S., “Correlations of droplet formation in T-junction microfluidic devices: From squeezing to dripping,” Microfluid. Nanofluid. 5, 711–717 (2008). 10.1007/s10404-008-0306-4 [DOI] [Google Scholar]
- 43.Li Y. K., Liu G. T., Xu J. H., Wang K., and Luo G. S., “A microdevice for producing monodispersed droplets under a jetting flow,” RSC Adv. 5, 27356–27364 (2015). 10.1039/C5RA02397A [DOI] [Google Scholar]
- 44.Raju L. and Hiremath S. S., “A state-of-the-art review on micro electro-discharge machining,” Proc. Technol. 25, 1281–1288 (2016). 10.1016/j.protcy.2016.08.222 [DOI] [Google Scholar]
- 45.Wang X., Jiang M., Zhou Z., Gou J., and Hui D., “3D printing of polymer matrix composites: A review and prospective,” Compos. Pt. B 110, 442–458 (2017). 10.1016/j.compositesb.2016.11.034 [DOI] [Google Scholar]
- 46.Gal-Or E. et al. , “Chemical analysis using 3D printed glass microfluidics,” Anal. Methods 11, 1802–1810 (2019). 10.1039/C8AY01934G [DOI] [Google Scholar]
- 47.Chen W., Lam R. H., and Fu J., “Photolithographic surface micromachining of polydimethylsiloxane (PDMS),” Lab Chip 12, 391–395 (2012). 10.1039/c1lc20721k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guckenberger D. J., de Groot T. E., Wan A. M. D., Beebe D. J., and Young E. W. K., “Micromilling: A method for ultra-rapid prototyping of plastic microfluidic devices,” Lab Chip 15, 2364–2378 (2015). 10.1039/C5LC00234F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valentin T. et al. , “3D printed self-adhesive PEGDA-PAA hydrogels as modular components for soft actuators and microfluidics,” Polym. Chem. 10, 2015 (2019). 10.1039/C9PY00211A [DOI] [Google Scholar]
- 50.Jeong H.-H., Yelleswarapu V. R., Yadavali S., Issadore D., and Lee D., “Kilo-scale droplet generation in three-dimensional monolithic elastomer device (3D MED),” Lab Chip 15, 4387–4392 (2015). 10.1039/C5LC01025J [DOI] [PubMed] [Google Scholar]
- 51.Chen P.-C., Pan C.-W., Lee W.-C., and Li K.-M., “An experimental study of micromilling parameters to manufacture microchannels on a PMMA substrate,” Int. J. Adv. Manuf. Technol. 71, 1623–1630 (2014). 10.1007/s00170-013-5555-z [DOI] [Google Scholar]
- 52.Horn T. J. and Harrysson O. L. A., “Overview of current additive manufacturing technologies and selected applications,” Sci. Prog. 95, 255–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhattacharjee N., Urrios A., Kang S., and Folch A., “The upcoming 3D-printing revolution in microfluidics,” Lab Chip 16, 1720–1742 (2016). 10.1039/c6lc00163g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielson R., Kaehr B., and Shear J. B., “Microreplication and design of biological architectures using dynamic-mask multiphoton lithography,” Small 5, 120–125 (2009). 10.1002/smll.200801084 [DOI] [PubMed] [Google Scholar]
- 55.Ladd C., So J.-H., Muth J., and Dickey M. D., “3D printing of free standing liquid metal microstructures,” Adv. Mater. 25, 5081–5085 (2013). 10.1002/adma.201301400 [DOI] [PubMed] [Google Scholar]
- 56.Hsieh Y.-F. et al. , “A lego®-like swappable fluidic module for bio-chem applications,” Sens. Actuators B 204, 489–496 (2014). 10.1016/j.snb.2014.07.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee M. and Burns M. A., “Microfluidic assembly blocks,” Lab Chip 8, 1365–1373 (2008). 10.1039/B805137B [DOI] [PubMed] [Google Scholar]
- 58.Chan H. N. et al. , “Direct, one-step molding of 3D-printed structures for convenient fabrication of truly 3D PDMS microfluidic chips,” Microfluid. Nanofluid. 19, 9–18 (2015). 10.1007/s10404-014-1542-4 [DOI] [Google Scholar]
- 59.Headen D. M., García J. R., and García A. J., “Parallel droplet microfluidics for high throughput cell encapsulation and synthetic microgel generation,” Microsyst. Nanoeng. 4, 17076 (2018). 10.1038/micronano.2017.76 [DOI] [Google Scholar]
- 60.Akbari S., Pirbodaghi T., Kamm R. D., and Hammond P. T., “A versatile microfluidic device for high throughput production of microparticles and cell microencapsulation,” Lab Chip 17, 2067–2075 (2017). 10.1039/C6LC01568A [DOI] [PubMed] [Google Scholar]
- 61.Jing T. et al. , “Jetting microfluidics with size-sorting capability for single-cell protease detection,” Biosens. Bioelectron. 66, 19–23 (2015). 10.1016/j.bios.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 62.Li P. et al. , “Detachable acoustophoretic system for fluorescence-activated sorting at the single-droplet level,” Anal. Chem. 91, 9970–9977 (2019). 10.1021/acs.analchem.9b01708 [DOI] [PubMed] [Google Scholar]
- 63.Buryk S., Kieda J., and Tsai S., “Diamagnetic droplet microfluidics applied to single-cell sorting,” AIP Adv. 9, 075106 (2019). 10.1063/1.5095884 [DOI] [Google Scholar]
- 64.Navi M., Abbasi N., Jeyhani M., Gnyawali V., and Tsai S. S. H., “Microfluidic diamagnetic water-in-water droplets: A biocompatible cell encapsulation and manipulation platform,” Lab Chip 18, 11 (2018). 10.1039/c8lc00867a [DOI] [PubMed] [Google Scholar]
- 65.Nan L., Yang Z., Lyu H., Lau K. Y. Y., and Shum H. C., “A microfluidic system for one-chip harvesting of single-cell-laden hydrogels in culture medium,” Adv. Biosyst. 3, 1900076 (2019). 10.1002/adbi.201900076 [DOI] [PubMed] [Google Scholar]
- 66.Tan Y. C., Hettiarachchi K., Siu M., and Pan Y. P., “Controlled microfluidic encapsulation of cells, proteins, and microbeads in lipid vesicles,” J. Am. Chem. Soc. 128, 5656–5658 (2006). 10.1021/ja056641h [DOI] [PubMed] [Google Scholar]
- 67.Zhu J., “Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering,” Biomaterials 31, 4639–4656 (2010). 10.1016/j.biomaterials.2010.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puppi D., Chiellini F., Piras A. M., and Chiellini E., “Polymeric materials for bone and cartilage repair,” Prog. Polym. Sci. 35, 403–440 (2010). 10.1016/j.progpolymsci.2010.01.006 [DOI] [Google Scholar]
- 69.Topuz F., Henke A., Richtering W., and Groll J., “Magnesium ions and alginate do form hydrogels: A rheological study,” Soft Matter 8, 4877–4881 (2012). 10.1039/C2SM07465F [DOI] [Google Scholar]
- 70.Rokstad A. M. et al. , “Alginate microbeads are complement compatible, in contrast to polycation containing microcapsules, as revealed in a human whole blood model,” Acta Biomater. 7, 2566–2578 (2011). 10.1016/j.actbio.2011.03.011 [DOI] [PubMed] [Google Scholar]
- 71.Xu J. H., Li S. W., Tan J., Wang Y. J., and Luo G. S., “Preparation of highly monodisperse droplet in a T-junction microfluidic device,” AIChE J. 52, 3005–3010 (2006). 10.1002/aic.10924 [DOI] [Google Scholar]
- 72.Yu J. Q. et al. , “Droplet optofluidic imaging for λ-bacteriophage detection via co-culture with host cell Escherichia coli,” Lab Chip 14, 3519–3524 (2014). 10.1039/C4LC00042K [DOI] [PubMed] [Google Scholar]
- 73.Najah M., Griffiths A. D., and Ryckelynck M., “Teaching single-cell digital analysis using droplet-based microfluidics,” Anal. Chem. 84, 1202–1209 (2012). 10.1021/ac202645m [DOI] [PubMed] [Google Scholar]
- 74.Mao A. S. et al. , “Deterministic encapsulation of single cells in thin tunable microgels for niche modelling and therapeutic delivery,” Nat. Mater. 16, 236–243 (2017). 10.1038/nmat4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang C.-G., Pan R.-Y., and Xu Z.-R., “A single-cell encapsulation method based on a microfluidic multi-step droplet splitting system,” Chin. Chem. Lett. 26, 1450–1454 (2015). 10.1016/j.cclet.2015.10.016 [DOI] [Google Scholar]
- 76.Li L. et al. , “Dean flow assisted single cell and bead encapsulation for high performance single cell expression profiling,” ACS Sens. 4, 1299–1305 (2019). 10.1021/acssensors.9b00171 [DOI] [PubMed] [Google Scholar]
- 77.Edd J. F. et al. , “Controlled encapsulation of single-cells into monodisperse picolitre drops,” Lab Chip 8, 1262–1264 (2008). 10.1039/B805456H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Di Carlo D., Irimia D., Tompkins R. G., and Toner M., “Continuous inertial focusing, ordering, and separation of particles in microchannels,” Proc. Natl. Acad. Sci. U.S.A. 104, 18892–18897 (2007). 10.1073/pnas.0704958104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramachandraiah H. et al. , “Dean flow-coupled inertial focusing in curved channels,” Biomicrofluidics 8, 034117–034117 (2014). 10.1063/1.4884306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kemna E. W. M. et al. , “High-yield cell ordering and deterministic cell-in-droplet encapsulation using dean flow in a curved microchannel,” Lab Chip 12, 2881–2887 (2012). 10.1039/C2LC00013J [DOI] [PubMed] [Google Scholar]
- 81.Nivedita N., Ligrani P., and Papautsky I., “Dean flow dynamics in low-aspect ratio spiral microchannels,” Sci. Rep. 7, 44072–44072 (2017). 10.1038/srep44072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiang N., Ni Z., and Yi H., “Concentration-controlled particle focusing in spiral elasto-inertial microfluidic devices,” Electrophoresis 39, 417–424 (2018). 10.1002/elps.201700150 [DOI] [PubMed] [Google Scholar]
- 83.Köster S. et al. , “Drop-based microfluidic devices for encapsulation of single cells,” Lab Chip 8, 1110–1115 (2008). 10.1039/b802941e [DOI] [PubMed] [Google Scholar]
- 84.Song H., Tice J. D., and Ismagilov R. F., “A microfluidic system for controlling reaction networks in time,” Angew. Chem. Int. Ed. Engl. 42, 768–772 (2003). 10.1002/anie.200390203 [DOI] [PubMed] [Google Scholar]
- 85.Kamalakshakurup G. and Lee A. P., “High-efficiency single cell encapsulation and size selective capture of cells in picoliter droplets based on hydrodynamic micro-vortices,” Lab Chip 17, 4324–4333 (2017). 10.1039/c7lc00972k [DOI] [PubMed] [Google Scholar]
- 86.Sauzade M. and Brouzes E., “Deterministic trapping, encapsulation and retrieval of single-cells,” Lab Chip 17, 2186–2192 (2017). 10.1039/C7LC00283A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Utech S. et al. , “Microfluidic generation of monodisperse, structurally homogeneous alginate microgels for cell encapsulation and 3D cell culture,” Adv. Healthcare Mater. 4, 1628–1633 (2015). 10.1002/adhm.201500021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oh B. et al. , “Single-cell encapsulation via click-chemistry alters production of paracrine factors from neural progenitor cells,” Adv. Sci. 7, 1902573 (2020). 10.1002/advs.201902573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gu H., Murade C. U., Duits M. H. G., and Mugele F., “A microfluidic platform for on-demand formation and merging of microdroplets using electric control,” Biomicrofluidics 5, 11101–11101 (2011). 10.1063/1.3570666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sauret A., Spandagos C., and Shum H. C., “Fluctuation-induced dynamics of multiphase liquid jets with ultra-low interfacial tension,” Lab Chip 12, 3380–3386 (2012). 10.1039/C2LC40524E [DOI] [PubMed] [Google Scholar]
- 91.Tang S.-Y. et al. , “Liquid-Metal microdroplets formed dynamically with electrical control of size and rate,” Adv. Mater. 28, 604–609 (2016). 10.1002/adma.201503875 [DOI] [PubMed] [Google Scholar]
- 92.He M., Kuo J. S., and Chiu D. T., “Electro-generation of single femtoliter- and picoliter-volume aqueous droplets in microfluidic systems,” Appl. Phys. Lett. 87, 031916 (2005). 10.1063/1.1997280 [DOI] [Google Scholar]
- 93.He P., Kim H., Luo D., Marquez M., and Cheng Z., “Low-frequency ac electro-flow-focusing microfluidic emulsification,” Appl. Phys. Lett. 96, 174103 (2010). 10.1063/1.3424791 [DOI] [Google Scholar]
- 94.Zhang M., Wu J., Niu X., Wen W., and Sheng P., “Manipulations of microfluidic droplets using electrorheological carrier fluid,” Phys. Rev. E 78, 066305 (2008). 10.1103/PhysRevE.78.066305 [DOI] [PubMed] [Google Scholar]
- 95.Link D. R. et al. , “Electric control of droplets in microfluidic devices,” Angew. Chem. Int. Ed. 45, 2556–2560 (2006). 10.1002/anie.200503540 [DOI] [PubMed] [Google Scholar]
- 96.Taylor G. I., “Disintegration of water drops in an electric field,” Proc. R. Soc. A 280, 383–397 (1964). 10.1098/rspa.1964.0151 [DOI] [Google Scholar]
- 97.Tan S. H., Maes F., Semin B., Vrignon J., and Baret J.-C., “The microfluidic jukebox,” Sci. Rep. 4, 4787 (2014). 10.1038/srep04787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Castro-Hernández E. et al. , “AC electrified jets in a flow-focusing device: Jet length scaling,” Biomicrofluidics 10, 043504 (2016). 10.1063/1.4954194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Castro-Hernández E. et al. , “Breakup length of AC electrified jets in a microfluidic flow-focusing junction,” Microfluid. Nanofluid. 19, 787–794 (2015). 10.1007/s10404-015-1603-3 [DOI] [Google Scholar]
- 100.Chen A. et al. , “On-chip magnetic separation and encapsulation of cells in droplets,” Lab Chip 13, 1172–1181 (2013). 10.1039/C2LC41201B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yan Q., Xuan S., Ruan X., Wu J., and Gong X., “Magnetically controllable generation of ferrofluid droplets,” Microfluid. Nanofluid. 19, 1377–1384 (2015). 10.1007/s10404-015-1652-7 [DOI] [Google Scholar]
- 102.Lee C.-P., Lan T.-S., and Lai M.-F., “Fabrication of two-dimensional ferrofluid microdroplet lattices in a microfluidic channel,” J. Appl. Phys. 115, 17B527 (2014). 10.1063/1.4867964 [DOI] [Google Scholar]
- 103.Park S.-Y., Wu T.-H., Chen Y., Teitell M. A., and Chiou P.-Y., “High-speed droplet generation on demand driven by pulse laser-induced cavitation,” Lab Chip 11, 1010–1012 (2011). 10.1039/C0LC00555J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baroud C. N., Delville J.-P., Gallaire F., and Wunenburger R., “Thermocapillary valve for droplet production and sorting,” Phys. Rev. E 75, 046302 (2007). 10.1103/PhysRevE.75.046302 [DOI] [PubMed] [Google Scholar]
- 105.Baroud C. N., Robert de Saint Vincent M., and Delville J.-P., “An optical toolbox for total control of droplet microfluidics,” Lab Chip 7, 1029–1033 (2007). 10.1039/B702472J [DOI] [PubMed] [Google Scholar]
- 106.Hardt S. and Hahn T., “Microfluidics with aqueous two-phase systems,” Lab Chip 12, 434–442 (2012). 10.1039/C1LC20569B [DOI] [PubMed] [Google Scholar]
- 107.Shum H. C., Sauret A., Fernandez-Nieves A., Stone H. A., and Weitz D. A., “Corrugated interfaces in multiphase core-annular flow,” Phys. Fluids 22, 082002 (2010). 10.1063/1.3480561 [DOI] [Google Scholar]
- 108.Zhu P., Tang X., Tian Y., and Wang L., “Pinch-off of microfluidic droplets with oscillatory velocity of inner phase flow,” Sci. Rep. 6, 31436–31436 (2016). 10.1038/srep31436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou H. and Yao S., “A facile on-demand droplet microfluidic system for lab-on-a-chip applications,” Microfluid. Nanofluid. 16, 667–675 (2014). 10.1007/s10404-013-1268-8 [DOI] [Google Scholar]
- 110.Jakiela S., Debski P., Dabrowski B., and Garstecki P., “Generation of nanoliter droplets on demand at hundred-Hz frequencies,” Micromachines 5, 1002–1011 (2014). 10.3390/mi5041002 [DOI] [Google Scholar]
- 111.Collins D. J., Alan T., Helmerson K., and Neild A., “Surface acoustic waves for on-demand production of picoliter droplets and particle encapsulation,” Lab Chip 13, 3225–3231 (2013). 10.1039/C3LC50372K [DOI] [PubMed] [Google Scholar]
- 112.Xu J. and Attinger D., “Drop on demand in a microfluidic chip,” J. Micromech. Microeng. 18, 065020 (2008). 10.1088/0960-1317/18/6/065020 [DOI] [Google Scholar]
- 113.Bransky A., Korin N., Khoury M., and Levenberg S., “A microfluidic droplet generator based on a piezoelectric actuator,” Lab Chip 9, 516–520 (2009). 10.1039/B814810D [DOI] [PubMed] [Google Scholar]
- 114.Friend J. and Yeo L. Y., “Microscale acoustofluidics: Microfluidics driven via acoustics and ultrasonics,” Rev. Mod. Phys. 83, 647–704 (2011). 10.1103/RevModPhys.83.647 [DOI] [Google Scholar]
- 115.Christopher G. F. and Anna S. L., “Microfluidic methods for generating continuous droplet streams,” J. Phys. D Appl. Phys. 40, R319–R336 (2007). 10.1088/0022-3727/40/19/r01 [DOI] [Google Scholar]
- 116.Willaime H., Barbier V., Kloul L., Maine S., and Tabeling P., “Arnold tongues in a microfluidic drop emitter,” Phys. Rev. Lett. 96, 054501 (2006). 10.1103/PhysRevLett.96.054501 [DOI] [PubMed] [Google Scholar]
- 117.Raj A., Halder R., Sajeesh P., and Sen A. K., “Droplet generation in a microchannel with a controllable deformable wall,” Microfluid. Nanofluid. 20, 102 (2016). 10.1007/s10404-016-1768-4 [DOI] [Google Scholar]
- 118.Abate A. R., Agresti J. J., and Weitz D. A., “Microfluidic sorting with high-speed single-layer membrane valves,” Appl. Phys. Lett. 96, 203509 (2010). 10.1063/1.3431281 [DOI] [Google Scholar]
- 119.Choi J.-H., Lee S.-K., Lim J.-M., Yang S.-M., and Yi G.-R., “Designed pneumatic valve actuators for controlled droplet breakup and generation,” Lab Chip 10, 456–461 (2010). 10.1039/B915596A [DOI] [PubMed] [Google Scholar]
- 120.Lee C.-H., Hsiung S.-K., and Lee G.-B., “A tunable microflow focusing device utilizing controllable moving walls and its applications for formation of micro-droplets in liquids,” J. Micromech. Microeng. 17, 1121 (2007). 10.1088/0960-1317/17/6/004 [DOI] [Google Scholar]
- 121.Yeh C.-H., Chen K.-R., and Lin Y.-C., “Developing heatable microfluidic chip to generate gelatin emulsions and microcapsules,” Microfluid. Nanofluid. 15, 775–784 (2013). 10.1007/s10404-013-1193-x [DOI] [Google Scholar]
- 122.Suryo R. and Basaran O., “Dripping of a liquid from a tube in the absence of gravity,” Phys. Rev. Lett. 96, 034504 (2006). 10.1103/PhysRevLett.96.034504 [DOI] [PubMed] [Google Scholar]
- 123.Hohng S., Lee S., Lee J., and Jo M. H., “Maximizing information content of single-molecule FRET experiments: Multi-color FRET and FRET combined with force or torque,” Chem. Soc. Rev. 43, 1007–1013 (2014). 10.1039/C3CS60184F [DOI] [PubMed] [Google Scholar]
- 124.Ng E. X., Miller M. A., Jing T., and Chen C.-H., “Single cell multiplexed assay for proteolytic activity using droplet microfluidics,” Biosens. Bioelectron. 81, 408–414 (2016). 10.1016/j.bios.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 125.Türkcan S. et al. , “Single-cell analysis of [18F]fluorodeoxyglucose uptake by droplet radiofluidics,” Anal. Chem. 87, 6667–6673 (2015). 10.1021/acs.analchem.5b00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.An X., Zuo P., and Ye B.-C., “A single cell droplet microfluidic system for quantitative determination of food-borne pathogens,” Talanta 209, 120571 (2020). 10.1016/j.talanta.2019.120571 [DOI] [PubMed] [Google Scholar]
- 127.Bounab Y. et al. , “Dynamic single-cell phenotyping of immune cells using the microfluidic platform DropMap,” Nat. Protoc. 15, 2920–2955 (2020). 10.1038/s41596-020-0354-0 [DOI] [PubMed] [Google Scholar]
- 128.Eyer K. et al. , “Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring,” Nat. Biotechnol. 35, 977–982 (2017). 10.1038/nbt.3964 [DOI] [PubMed] [Google Scholar]
- 129.Wei S.-C., Hsu M. N., and Chen C.-H., “Plasmonic droplet screen for single-cell secretion analysis,” Biosens. Bioelectron. 144, 111639 (2019). 10.1016/j.bios.2019.111639 [DOI] [PubMed] [Google Scholar]
- 130.Thompson A. M., Paguirigan A. L., Kreutz J. E., Radich J. P., and Chiu D. T., “Microfluidics for single-cell genetic analysis,” Lab Chip 14, 3135–3142 (2014). 10.1039/c4lc00175c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zeng Y., Novak R., Shuga J., Smith M. T., and Mathies R. A., “High-performance single cell genetic analysis using microfluidic emulsion generator arrays,” Anal. Chem. 82, 3183–3190 (2010). 10.1021/ac902683t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu Z. et al. , “Highly sensitive and quantitative detection of rare pathogens through agarose droplet microfluidic emulsion PCR at the single-cell level,” Lab Chip 12, 3907–3913 (2012). 10.1039/C2LC40461C [DOI] [PubMed] [Google Scholar]
- 133.Matuła K., Rivello F., and Huck W. T. S., “Single-cell analysis using droplet microfluidics,” Adv. Biosyst. 4, 1900188 (2020). 10.1002/adbi.201900188 [DOI] [PubMed] [Google Scholar]
- 134.Schie I. W. and Huser T., “Methods and applications of Raman microspectroscopy to single-cell analysis,” Appl. Spectrosc. 67, 813–828 (2013). 10.1366/12-06971 [DOI] [PubMed] [Google Scholar]
- 135.Willner M. R., McMillan K. S., Graham D., Vikesland P. J., and Zagnoni M., “Surface-enhanced Raman scattering based microfluidics for single-cell analysis,” Anal. Chem. 90, 12004–12010 (2018). 10.1021/acs.analchem.8b02636 [DOI] [PubMed] [Google Scholar]
- 136.Sun D. et al. , “Cellular heterogeneity identified by single-cell alkaline phosphatase (ALP) via a SERRS-microfluidic droplet platform,” Lab Chip 19, 335–342 (2019). 10.1039/C8LC01006D [DOI] [PubMed] [Google Scholar]
- 137.Khorshidi M. A., Rajeswari P. K. P., Wählby C., Joensson H. N., and Andersson Svahn H., “Automated analysis of dynamic behavior of single cells in picoliter droplets,” Lab Chip 14, 931–937 (2014). 10.1039/C3LC51136G [DOI] [PubMed] [Google Scholar]
- 138.Lu H. et al. , “High throughput single cell counting in droplet-based microfluidics,” Sci. Rep. 7, 1366 (2017). 10.1038/s41598-017-01454-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brouzes E. et al. , “Droplet microfluidic technology for single-cell high-throughput screening,” Proc. Natl. Acad. Sci. U.S.A. 106, 14195–14200 (2009). 10.1073/pnas.0903542106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dunevall J. et al. , “Characterizing the catecholamine content of single mammalian vesicles by collision–adsorption events at an electrode,” J. Am. Chem. Soc. 137, 4344–4346 (2015). 10.1021/ja512972f [DOI] [PubMed] [Google Scholar]
- 141.Li X., Majdi S., Dunevall J., Fathali H., and Ewing A. G., “Quantitative measurement of transmitters in individual vesicles in the cytoplasm of single cells with nanotip electrodes,” Angew. Chem. Int. Ed. 54, 11978–11982 (2015). 10.1002/anie.201504839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Safaei T. S., Mohamadi R. M., Sargent E. H., and Kelley S. O., “In situ electrochemical ELISA for specific identification of captured cancer cells,” ACS Appl. Mater. Interfaces 7, 14165–14169 (2015). 10.1021/acsami.5b02404 [DOI] [PubMed] [Google Scholar]
- 143.Zhang J. et al. , “New frontiers and challenges for single-cell electrochemical analysis,” ACS Sens. 3, 242–250 (2018). 10.1021/acssensors.7b00711 [DOI] [PubMed] [Google Scholar]
- 144.Wang H., Chen B., He M., and Hu B., “A facile droplet-chip-time-resolved inductively coupled plasma mass spectrometry online system for determination of zinc in single cell,” Anal. Chem. 89, 4931–4938 (2017). 10.1021/acs.analchem.7b00134 [DOI] [PubMed] [Google Scholar]
- 145.Yin L., Zhang Z., Liu Y., Gao Y., and Gu J., “Recent advances in single-cell analysis by mass spectrometry,” Analyst 144, 824–845 (2019). 10.1039/C8AN01190G [DOI] [PubMed] [Google Scholar]
- 146.Eral H. B. et al. , “Biocompatible alginate microgel particles as heteronucleants and encapsulating vehicles for hydrophilic and hydrophobic drugs,” Cryst. Growth Des. 14, 2073–2082 (2014). 10.1021/cg500250e [DOI] [Google Scholar]
- 147.Visted T., Bjerkvig R., and Enger P. O., “Cell encapsulation technology as a therapeutic strategy for CNS malignancies,” Neuro-Oncol. 3, 201–210 (2001). 10.1093/neuonc/3.3.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li P., Müller M., Chang M. W., Frettlöh M., and Schönherr H., “Encapsulation of autoinducer sensing reporter bacteria in reinforced alginate-based microbeads,” ACS Appl. Mater. Interfaces 9, 22321–22331 (2017). 10.1021/acsami.7b07166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Deng Y. et al. , “Rapid purification of cell encapsulated hydrogel beads from oil phase to aqueous phase in a microfluidic device,” Lab Chip 11, 4117–4121 (2011). 10.1039/c1lc20494g [DOI] [PubMed] [Google Scholar]
- 150.Hong J., deMello A. J., and Jayasinghe S. N., “Bio-electrospraying and droplet-based microfluidics: Control of cell numbers within living residues,” Biomed. Mater. 5, 21001 (2010). 10.1088/1748-6041/5/2/021001 [DOI] [PubMed] [Google Scholar]
- 151.Teh S. Y., Lin R., Hung L. H., and Lee A. P., “Droplet microfluidics,” Lab Chip 8, 198–220 (2008). 10.1039/b715524g [DOI] [PubMed] [Google Scholar]
- 152.Akbari S. and Pirbodaghi T., “Microfluidic encapsulation of cells in alginate particles via an improved internal gelation approach,” Microfluid. Nanofluid. 16, 773–777 (2014). 10.1007/s10404-013-1264-z [DOI] [Google Scholar]
- 153.Dendukuri D. and Doyle P. S., “The synthesis and assembly of polymeric microparticles using microfluidics,” Adv. Mater. 21, 4071–4086 (2009). 10.1002/adma.200803386 [DOI] [Google Scholar]
- 154.Velasco D., Tumarkin E., and Kumacheva E., “Microfluidic encapsulation of cells in polymer microgels,” Small 8, 1633–1642 (2012). 10.1002/smll.201102464 [DOI] [PubMed] [Google Scholar]
- 155.Kim C. et al. , “Rapid exchange of oil-phase in microencapsulation chip to enhance cell viability,” Lab Chip 9, 1294–1297 (2009). 10.1039/b819044e [DOI] [PubMed] [Google Scholar]
- 156.Shoichet M. S., Li R. H., White M. L., and Winn S. R., “Stability of hydrogels used in cell encapsulation: An in vitro comparison of alginate and agarose,” Biotechnol. Bioeng. 50, 374–381 (1996). [DOI] [PubMed] [Google Scholar]
- 157.Liu K., Ding H. J., Liu J., Chen Y., and Zhao X. Z., “Shape-controlled production of biodegradable calcium alginate gel microparticles using a novel microfluidic device,” Langmuir 22, 9453–9457 (2006). 10.1021/la061729+ [DOI] [PubMed] [Google Scholar]
- 158.Wang S. et al. , “An in situ photocrosslinking microfluidic technique to generate non-spherical, cytocompatible, degradable, monodisperse alginate microgels for chondrocyte encapsulation,” Biomicrofluidics 12, 014106 (2018). 10.1063/1.5017644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Etter J. N., Karasinski M., Ware J., and Oldinski R. A., “Dual-crosslinked homogeneous alginate microspheres for mesenchymal stem cell encapsulation,” J. Mater. Sci. Mater. Med. 29, 143 (2018). 10.1007/s10856-018-6151-4 [DOI] [PubMed] [Google Scholar]
- 160.Benoit D. S., Schwartz M. P., Durney A. R., and Anseth K. S., “Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells,” Nat. Mater. 7, 816–823 (2008). 10.1038/nmat2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Weber L. M., Hayda K. N., Haskins K., and Anseth K. S., “The effects of cell-matrix interactions on encapsulated beta-cell function within hydrogels functionalized with matrix-derived adhesive peptides,” Biomaterials 28, 3004–3011 (2007). 10.1016/j.biomaterials.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 162.O'Brien A. K. and Bowman C. N., “Modeling the effect of oxygen on photopolymerization kinetics,” Macromol. Theory Simul. 15, 176–182 (2006). 10.1002/mats.200500056 [DOI] [Google Scholar]
- 163.Dendukuri D. et al. , “Modeling of oxygen-inhibited free radical photopolymerization in a PDMS microfluidic device,” Macromolecules 41, 8547–8556 (2008). 10.1021/ma801219w [DOI] [Google Scholar]
- 164.Cabiscol E., Tamarit J., and Ros J., “Oxidative stress in bacteria and protein damage by reactive oxygen species,” Int. Microbiol. 3, 3–8 (2000). [PubMed] [Google Scholar]
- 165.Jiang Z., Xia B., McBride R., and Oakey J., “A microfluidic-based cell encapsulation platform to achieve high long-term cell viability in photopolymerized PEGNB hydrogel microspheres,” J. Mater. Chem. B 5, 173–180 (2017). 10.1039/c6tb02551j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xia B., Krutkramelis K., and Oakey J., “Oxygen-Purged microfluidic device to enhance cell viability in photopolymerized PEG hydrogel microparticles,” Biomacromolecules 17, 2459–2465 (2016). 10.1021/acs.biomac.6b00597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rossow T. et al. , “Controlled synthesis of cell-laden microgels by radical-free gelation in droplet microfluidics,” J. Am. Chem. Soc. 134, 4983–4989 (2012). 10.1021/ja300460p [DOI] [PubMed] [Google Scholar]
- 168.Nair D. P. et al. , “The thiol-Michael addition click reaction: A powerful and widely used tool in materials chemistry,” Chem. Mater. 26, 724–744 (2014). 10.1021/cm402180t [DOI] [Google Scholar]
- 169.Ma Y., Neubauer M. P., Thiele J., Fery A., and Huck W. T. S., “Artificial microniches for probing mesenchymal stem cell fate in 3D,” Biomater. Sci. 2, 1661–1671 (2014). 10.1039/C4BM00104D [DOI] [PubMed] [Google Scholar]
- 170.Headen D. M., Aubry G., Lu H., and Garcia A. J., “Microfluidic-Based generation of size-controlled, biofunctionalized synthetic polymer microgels for cell encapsulation,” Adv. Mater. 26, 3003–3008 (2014). 10.1002/adma.201304880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rossow T., Lienemann P. S., and Mooney D. J., “Cell microencapsulation by droplet microfluidic templating,” Macromol. Chem. Phys. 218, 14 (2017). 10.1002/macp.201600380 [DOI] [Google Scholar]
- 172.Jiang Y., Chen J., Deng C., Suuronen E. J., and Zhong Z., “Click hydrogels, microgels and nanogels: Emerging platforms for drug delivery and tissue engineering,” Biomaterials 35, 4969–4985 (2014). 10.1016/j.biomaterials.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 173.Guerzoni L. P. B. et al. , “Cell encapsulation in soft, anisometric poly(ethylene) glycol microgels using a novel radical-free microfluidic system,” Small 15, 8 (2019). 10.1002/smll.201900692 [DOI] [PubMed] [Google Scholar]
- 174.Siltanen C. et al. , “Microfluidic fabrication of bioactive microgels for rapid formation and enhanced differentiation of stem cell spheroids,” Acta Biomater. 34, 125–132 (2016). 10.1016/j.actbio.2016.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu Y., Nambu N. O., and Taya M., “Cell-laden microgel prepared using a biocompatible aqueous two-phase strategy,” Biomed. Microdevices 19, 55 (2017). 10.1007/s10544-017-0198-8 [DOI] [PubMed] [Google Scholar]
- 176.Mastiani M., Firoozi N., Petrozzi N., Seo S., and Kim M., “Polymer-salt aqueous two-phase system (ATPS) micro-droplets for cell encapsulation,” Sci. Rep. 9, 15561 (2019). 10.1038/s41598-019-51958-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cai B. et al. , “A microfluidic platform utilizing anchored water-in-oil-in-water double emulsions to create a niche for analyzing single non-adherent cells,” Lab Chip 19, 422–431 (2019). 10.1039/C8LC01130C [DOI] [PubMed] [Google Scholar]
- 178.Periyannan Rajeswari P. K., Joensson H. N., and Andersson-Svahn H., “Droplet size influences division of mammalian cell factories in droplet microfluidic cultivation,” Electrophoresis 38, 305–310 (2017). 10.1002/elps.201600316 [DOI] [PubMed] [Google Scholar]
- 179.Kamperman T., Henke S., Visser C. W., Karperien M., and Leijten J., “Centering single cells in microgels via delayed crosslinking supports long-term 3D culture by preventing cell escape,” Small 13, 1603711 (2017). 10.1002/smll.201603711 [DOI] [PubMed] [Google Scholar]
- 180.Joensson H. N. and Andersson Svahn H., “Droplet microfluidics—A tool for single-cell analysis,” Angew. Chem. Int. Ed. Engl. 51, 12176–12192 (2012). 10.1002/anie.201200460 [DOI] [PubMed] [Google Scholar]
- 181.Curran D. P., “Strategy-level separations in organic synthesis: From planning to practice,” Angew. Chem. Int. Ed. 37, 1174–1196 (1998). [DOI] [PubMed] [Google Scholar]
- 182.Song H., Chen D. L., and Ismagilov R. F., “Reactions in droplets in microfluidic channels,” Angew. Chem. Int. Ed. Engl. 45, 7336–7356 (2006). 10.1002/anie.200601554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Roach L. S., Song H., and Ismagilov R. F., “Controlling nonspecific protein adsorption in a plug-based microfluidic system by controlling interfacial chemistry using fluorous-phase surfactants,” Anal. Chem. 77, 785–796 (2005). 10.1021/ac049061w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hong S., Hsu H. J., Kaunas R., and Kameoka J., “Collagen microsphere production on a chip,” Lab Chip 12, 3277–3280 (2012). 10.1039/c2lc40558j [DOI] [PubMed] [Google Scholar]
- 185.Choi C.-H. et al. , “One-step generation of cell-laden microgels using double emulsion drops with a sacrificial ultra-thin oil shell,” Lab Chip 16, 1549–1555 (2016). 10.1039/c6lc00261g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang H. and He X., “Interfacial tension based on-chip extraction of microparticles confined in microfluidic stokes flows,” Appl. Phys. Lett. 105, 143704 (2014). 10.1063/1.4898040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liao Q.-Q. et al. , “Biocompatible fabrication of cell-laden calcium alginate microbeads using microfluidic double flow-focusing device,” Sens. Actuators A 279, 313–320 (2018). 10.1016/j.sna.2018.06.006 [DOI] [Google Scholar]
- 188.Mohamed M. G. A. et al. , “An integrated microfluidic flow-focusing platform for on-chip fabrication and filtration of cell-laden microgels,” Lab Chip 19, 1621–1632 (2019). 10.1039/C9LC00073A [DOI] [PubMed] [Google Scholar]
- 189.Valley J. K. et al. , “Parallel single-cell light-induced electroporation and dielectrophoretic manipulation,” Lab Chip 9, 1714–1720 (2009). 10.1039/b821678a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chiou P. Y., Ohta A. T., and Wu M. C., “Massively parallel manipulation of single cells and microparticles using optical images,” Nature 436, 370–372 (2005). 10.1038/nature03831 [DOI] [PubMed] [Google Scholar]
- 191.Hidalgo San Jose L., Stephens P., Song B., and Barrow D., “Microfluidic encapsulation supports stem cell viability, proliferation, and neuronal differentiation,” Tissue Eng. Part C 24, 158–170 (2018). 10.1089/ten.TEC.2017.0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.