Abstract
Neurologic complications of symptomatic COVID-19 are common. Brain tissues from 13 autopsies of people who died of COVID-19 were examined. Cultured endothelial and neuronal cells were incubated with and wild type mice were injected IV with different spike subunits. In situ analyses were used to detect SARS-CoV-2 proteins and the host response. In 13/13 brains from fatal COVID-19, pseudovirions (spike, envelope, and membrane proteins without viral RNA) were present in the endothelia of microvessels ranging from 0 to 14 positive cells/200× field (mean 4.3). The pseudovirions strongly co-localized with caspase-3, ACE2, IL6, TNFα, and C5b-9. The surrounding neurons demonstrated increased NMDAR2 and neuronal NOS plus decreased MFSD2a and SHIP1 proteins. Tail vein injection of the full length S1 spike subunit in mice led to neurologic signs (increased thirst, stressed behavior) not evident in those injected with the S2 subunit. The S1 subunit localized to the endothelia of microvessels in the mice brain and showed co-localization with caspase-3, ACE2, IL6, TNFα, and C5b-9. The surrounding neurons showed increased neuronal NOS and decreased MFSD2a. It is concluded that ACE2+ endothelial damage is a central part of SARS-CoV2 pathology and may be induced by the spike protein alone. Thus, the diagnostic pathologist can use either hematoxylin and eosin stain or immunohistochemistry for caspase 3 and ACE2 to document the endothelial cell damage of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Spike protein, S1 subunit
1. Introduction
Neurologic complications are now well recognized in symptomatic COVID-19 [[1], [2], [3], [4], [5]]. Estimates of neurologic findings in COVID-19 range from 7% in a study of hospitalized patients in Wuhan China to 69% among ICU patients in France [5]. The neurologic symptoms and signs include lethargy, delirium, confusion, irritability, and difficulty concentrating which may persist for months [[1], [2], [3], [4], [5]]. Possible etiologies for the neurologic manifestations include micro- and macro ischemic strokes associated with the pro-thrombotic state of severe COVID-19 [1,6,7], the viral-associated cytokine storm [2], direct neuronal invasion [3,4], and so-called bystander injury to neurons [3,4]. Although direct infection of SARS-CoV2 in the brain and neurons has been reported, most studies have documented that viral RNA is rarely detected in the brain or CSF of people with neurologic manifestations of COVID-19 [3,4].
Our group has reported that the viral pneumonia that is the essential event behind moderate/severe COVID-19 is associated with a large amount of infectious virus in the lung that infects primarily alveolar macrophages and endothelial cells and, to a lesser degree, alveolar pneumocytes [[6], [7], [8]]. This infection induces the alveolar capillaries to form thromboses due to complement activation, which is called microangiopathy [[6], [7], [8]]. It has been postulated that the subsequent death of the intrapulmonary virus releases pseudovirions (spike, envelope, and membrane proteins) disassociated from the viral RNA. The circulating pseudovirions can then attach to endothelia with the ACE2 receptor via the spike protein. It has been documented that the microvascular beds with the greatest number of ACE2 receptors in endothelia include, besides the lung, the skin's deeper dermal vessels/subcutaneous fat, brain, and liver [[6], [7], [8]]. Various investigators have shown that the endocytosed spike glycoprotein, even in the absence of viral RNA, can induce a caspase-3 mediated cell death, complement activation which could lead to a hypercoagulable state, and the increase of many cytokines/proteins associated with severe COVID-19, including TNFα, IL6, IL8, IL1β, and p38 [[6], [7], [8], [9]]. It is possible that the neurologic manifestations of moderate/severe COVID-19 may reflect a microencephalitis induced by complement and cytokine mediated endothelial damage triggered by circulating pseudovirions. This study examined the effects of various subunits of the spike protein alone (without infectious virus) on human endothelial cells in culture and after intravenous injection into mice as well as performing a comprehensive analysis of thirteen brains obtained from people with neurologic manifestations who died of COVID-19. The key finding of this study for the diagnostic anatomic pathologist is that endothelial cell damage, which can be visible on hematoxylin and eosin stains or with immunohistochemistry for caspase 3, is highly correlated with the pathophysiology of COVID-19.
2. Methods
2.1. Formalin-fixed, paraffin embedded human brain samples
Autopsy brain tissues were available from 13 patients who died of COVID-19. They ranged in age from 28 to 92 (mean 69; 7 men and 6 women) and had neurologic symptoms/signs that included lethargy, confusion, and irritability. The formalin fixed, paraffin embedded tissues were from the frontal or temporal lobe, the hippocampus, midbrain, or the pons. Ten aged matched cases from patients who had died of non-neurologic diseases prior to 2019 served as negative controls; CNS findings were described as unremarkable.
2.2. Immunohistochemistry
Immunohistochemistry was done as per a previously published protocol [8,10]. In brief, optimal conditions for each antibody were determined by testing various dilutions and pretreatment conditions. The antibodies used follow; ABCAM, Cambridge MA (IL6, TNFα, NMDAR2, SHIP1, caspase 3, and C5b-9), Enzo Life Sciences (neuronal NOS, MFSD2a), ACE2 (PROSCI, Poway CA) (cat # 3215) and optimal pretreatment in each case was 30 min with the Leica EDTA antigen retrieval solution.
Detection of the SARS-CoV-2 spike glycoprotein, membrane and/or envelope proteins was as previously described [8,10]. In brief, the Leica Bond Max automated platform was used with the primary antibodies (PROSCI) (membrane, cat # 3527), (spike; cat # 3525) and (envelope; cat # 3521) after antigen retrieval for 30 min. The specific PROSCI antibodies used to detect the S1 subunit, the truncated S1 subunit, and the S2 subunit were: 9083, 9087, and 9123, respectively. The HRP conjugate from Enzo Life Sciences (Farmingdale, New York, USA) was used in place of the equivalent reagent from Leica as this has been shown to substantially reduce background [10].
2.3. In situ hybridization
Detection of SARS-CoV-2 RNA was done using the ACD RNAscope (Newark, California, USA) probe (Cat No. 848561-C3) through a previously published protocol [8,10].
2.4. Co-expression and statistical analyses
Co-expression analyses were done using the Nuance/InForm system whereby each chromogenic signal is separated, converted to a fluorescence-based signal, then mixed to determine what percentage of cells were expressing the two proteins of interest as previously described [8,10].
The number of positive cells/200× field was counted with the InForm software or manually in 10 fields/tissue. Manual and InForm computer-based counts were equivalent. Statistical analysis was done using the InStat Statistical Analysis Software (version 3.36) and a paired t-test (also referred to as a “repeated measure t-test”). The null hypothesis was rejected if the significance level was below 5%.
2.5. Recombinant spike proteins
The recombinant spike proteins, all from PROSCI, were: Val16-Arg685 (cat #10-300) = full length S1 subunit; Arg319-Phe541 (10-303) = truncated S1 subunit (contains only the receptor binding domain) and full length S2 subunit = (Ser686–Pro1273 (10-426).
2.6. Cell culture
HUVEC (human umbilical vein/vascular endothelium) (CRL-1730) and RAW 264.7 (ATCC® TIB-71™) murine macrophage cells, was purchased from the ATCC. Motor Neuron-Like Cell line MN1 (also known as NSC-34) is a hybrid cell line produced by the fusion of motor neurons from the spinal cords of mouse embryos with mouse neuroblastoma cells N18TG2 [11]. The cells were grown in RPMI-1640 Medium, 5% heat inactivated FBS, and Penicillin/Streptomycin used at 1×.
2.7. Mouse IV injection studies
Nine months old female C57BL/6 mice (purchased from Jackson laboratory, Maine) were tail vein injected with different spike peptides and euthanized using CO2 after five days. Ten μg/mouse of Spike S1 and ten μg/mouse of S1peptide that contains only the Receptor binding domain (RBD) were injected in 3 mice per group, except for the controls (n = 4). Spike S2 was used at 3 μg /mouse. The peptides were diluted in PBS; the total volume of injection was 150 μl. Mice were monitored every 12 h. Blood smears were prepared right after euthanasia and fixed in formalin overnight; the organs were fixed in 10% buffered formalin for 3 days. All procedures were approved by and performed in accordance with The Ohio State University's Institutional Lab Animal Care and Use Committee (Columbus, Ohio).
3. Results
3.1. Human COVID-19 brain findings
The molecular and histologic changes were analyzed in the 26 COVID-19 brain tissues from 13 patients (2 tissues/person) and 10 age-matched controls in a blinded fashion. The hematoxylin and eosin findings revealed two differences: 1) Microvessels of the brain of COVID-19 patients showed perivascular edema as defined by a zone of edema at least 50 μm in size around a capillary. 2) Less commonly, microvessels in the COVID-19 cases had endothelial cells that were degenerated, mummified, and detached, showed focal basement membrane duplication, and/or microthrombi. The perivascular edema was evident in between 5.3% and 25.9% of the microvessels in the COVID-19 brains (Fig. 1 ).
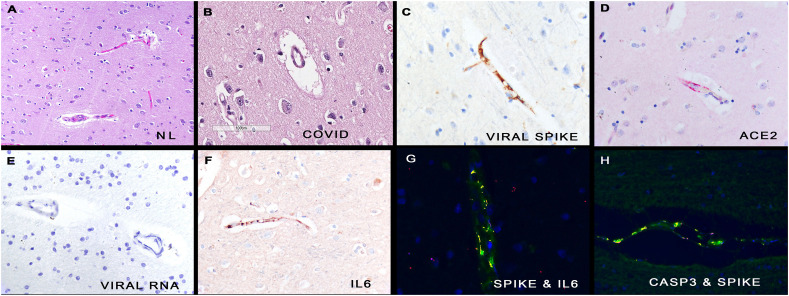
Fig. 1.
Histologic and molecular correlates of COVID-19 in human brains.
Panel A shows the microvessels in normal brain. In comparison, many of the capillaries in COVID-19 brain tissues show marked perivascular edema (panel B). Serial section analyses of the COVID-19 brain shows that the endothelial cells of the microvessels contained the spike glycoprotein (panel C), the ACE2 receptor (panel D) and IL 6 (panel F), but not viral RNA (panel E). The fluorescent yellow signal marks co-localization of the spike protein with IL6 (panel G) and caspase 3 (panel H), respectively, in these endothelial cells. Each magnification is 800× with DAB (brown) signal (panels C, E, F) or Fast Red (red) (panel D). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, the distribution of the infectious virus versus the pseudovirion (defined as spike protein with other capsid proteins without viral RNA) was analyzed. In eight cases, autopsy lung tissue was also available and, in each case, both viral RNA and viral capsid proteins were detected. Viral RNA was detected in 2/26 brain tissues in the COVID-19 patients and, in each case, was seen in less than 3 cells/cm; the cells had the cytology of microglia (data not shown). Viral spike protein was detected in 26/26 brain tissues and none of the controls. Over 95% of the cells with spike protein were the endothelial cells of microvessels (Fig. 1) (Table 1 ). Scattered cells directly outside the affected microvessels had detectable spike protein (Fig. 1). Serial section analyses (tissues four microns apart) of spike protein and ACE2 showed the same distribution (Fig. 1). Co-expression data confirmed that spike and ACE2 strongly co-localized. Viral spike, envelope, and membrane proteins showed the same distribution and co-localized (data not shown).
Table 1.
Quantification of biomarkers associated with endothelialopathy of the brains in people who died of COVID-19 and in the mice with IV injection of different spike protein subunits.
| Protein | Human normal brains mean with SD | COVID-19 brains mean with SD | Mouse normal brains mean with SD | S1 subunit brains mean with SD | Truncated S1 subunit brains mean with SD | S2 subunit brains mean with SD |
|---|---|---|---|---|---|---|
| Spike protein SARS-CoV-2 | 0a | 4.3 (0.9) | 0 | 1.3 (0.4) | 0 | 0 |
| Caspase 3 | 0.4 (0.1) | 4.8 (1.5) | 0 | 1.7 (0.5) | 0.3 (0.1) | 0 |
| IL6 | 0 | 5.1 (1.3) | 0 | 1.2 (0.2) | 0 | 0 |
| TNFα | 0 | 5.9 (1.5) | 0 | 1.8 (0.6) | 0 | 0 |
| MFSD2a | 94.7 (5.1)b | 19.1 (7.3) | 76.9 (6.1) | 30.2 (7.9) | 56.8 (9.0) | 73.9 (8.2) |
| SHIP1 | 95.2 (6.0) | 17.2 (2.9) | 57.4 (9.6) | 39.9 (10.3) | 40.2 (13.3) | 33.9 (11.4) |
| NMDAR2 | 0 | 9.9 (3.9) | 0 | 2.3 (0.8) | 1.9 (1.0) | 2.0 (0.7) |
| Neuronal NOS | 5.5 (1.8) | 69.2 (10.1) | 3.8 (1.3) | 55.2 (7.1) | 9.1 (3.3) | 10.3 (2.9) |
Counts for spike, caspase 3, IL6 and TNFα are the number of + endothelial cells/200× field with a minimum of 10 fields counted/case. The controls and cases were read in a blinded fashion.
Counts for MFSD2a, NMDAR2, neuronal NOS, and SHIP1 are the % of positive neurons/200× field with a minimum of 10 fields counted/case. The controls and cases were read in a blinded fashion.
The histologic findings suggested that endocytosis of the spike protein could induce endothelial cell damage. The cases and controls were each tested for activated caspase-3 and read in a blinded fashion (Table 1). Note the highly significant correlation between caspase-3 expression and SARS-CoV2 capsid protein within the brain microvessels. Co-expression confirmed that caspase-3 and spike glycoprotein co-localized in endothelial cells (Fig. 1).
Cytokine expression was examined in the 26 tissues/13 cases and 10 controls and focused on IL6 and TNFα, since these two cytokines are increased in severe COVID-19 [7,8]. The data presented in Table 1 highlights the significant association between each cytokine and SARS-CoV-2 protein detection in the microvessels (each p < 0.001). Scattered extravascular cells did show signal for each cytokine, but over 95% of the positive cells were endothelia and the cytokines co-localized with the spike glycoprotein (Fig. 1).
Next examined was if there was an effect on neuronal function presumably as a sequela to the SARS-CoV-2 associated microangiopathy. MFSD2A plays an essential role in the blood brain barrier (BBB), and supply of omega-3 fatty acids to neurons [[12], [13], [14]]. The normal brain cases showed strong expression of MFSD2A in over 90% of neurons (Table 1). In comparison, the percentage of neurons with detectable MFSD2A decreased significantly (p < 0.001) in the COVID-19 brains (Table 1) (Fig. 2 ). SHIP1 (Src-homology-2 containing inositol phosphatase) is another key brain protein that has a strong anti-inflammatory effect [15]. It was strongly expressed in the normal brains in over 85% of neurons. As seen in Table 1, there was a significant decreased expression of SHIP1 in the COVID-19 brain tissues (p value <0.001) (Fig. 2).
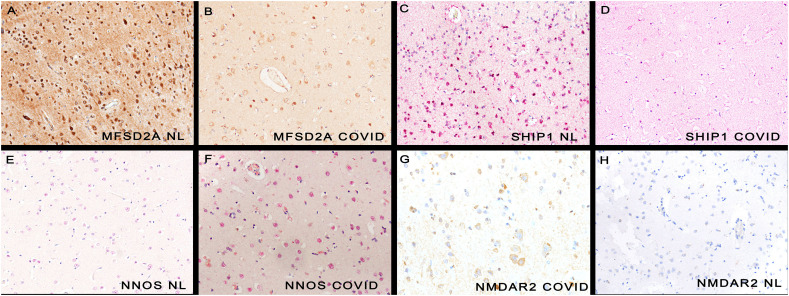
Fig. 2.
Molecular changes in neurons in COVID-19 human brains.
Panels A and C shows the strong neuronal expression of MFSD2A and SHIP1 in normal brain, respectively. The signal is much decreased for each in the COVID-19 brain (panels B/D). Neither nNOS nor NMDAR2 (panels E/H) were strongly expressed in normal brains, respectively. Each protein was markedly increased in COVID-19 brains (panels F, G, respectively; note the nuclear localization of nNOS). Each magnification is 600× with DAB (brown) signal (panels A,B,G,H) or Fast Red (red) (panels C,D,E,F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition, Glutamate NMDA receptor 2 (NMDAR2) and neuronal nitric oxide synthetase (nNOS) have each been implicated as markers of neuronal dysfunction [[15], [16], [17], [18], [19], [20]]. NMDAR2 was not detected in the normal brain tissues. It was expressed in the COVID-19 brains where it was evident in neurons adjacent to SARS-CoV-2 positive microvessels (Table 1 and Fig. 2). Similarly, the percentage of neurons with nNOS was significantly greater (p < 0.001) in the COVID-19 brain tissues compared to the controls (Table 1 and Fig. 2).
3.2. Spike subunit incubation in cell lines
Three different cell lines were incubated with the different subunits of the SARS-CoV-2 spike protein for two days: RAW264.7 (murine, macrophage), MN1 (murine, motorneuron), and HUVEC (human, endothelial). Macroscopic changes included increased adhesion and degeneration of the HUVEC cells and degeneration in the MN1 cell lines. The immunohistochemistry data is presented in Table 2 . Note that the RAW264.7 cells were negative for ACE2 expression and showed no evidence of the spike protein internalization nor caspase-3 induction. The HUVEC and MN1 cell lines each had strong ACE2 expression. The S1 subunit was detected in 11.1% (HUVEC) and 5.1% (MN1) of the cells. Co-expression analyses showed that the MN and HUVEC cells positive for S1 subunit also expressed caspase-3 (Fig. 3 ).
Table 2.
Quantification of cell line data after incubation with different subunits of the SARS-CoV2 spike protein.
| Protein tested | RAW | RAW 70 ng/ml spike S1 | HUVEC | HUVEC 70 ng/ml spike S1 | HUVEC 350 ng/ml spike S1 | HUVEC 70 ng/ml spike S1 truncated | HUVEC 70 ng/ml spike S2 | Motor neurons | Motor neurons 70 ng/ml spike S1 |
|---|---|---|---|---|---|---|---|---|---|
| ACE2a | 2.2 (0.5) | 2.7 (0.7) | 97.3 (0.4) | 96.2 (0.8) | 75,6 (2.0) | 97.2 (0.4) | 95.1 (0.8) | 95.0 (1.0) | 94.5 (0.4) |
| Caspase 3 | 0 | 0 | 0.2 (0.1) | 4.1 (0.9) | 14.2 (3.8) | 0.6 (0.1) | 0.9 (0.3) | 0 | 3.5 (1.2) |
| Spike protein | 0 | 0 | 0 | 11.1 (1.9) | 26.7 (4.2) | 1.2 (0.6) | 0.2 (0/1) | 0 | 5.1 (1.4) |
Counts for ACE2, caspase 3, and the spike protein are the % of positive cells/200× field with a minimum of 10 fields; several thousands of cells were scored per each data point with n = 3.
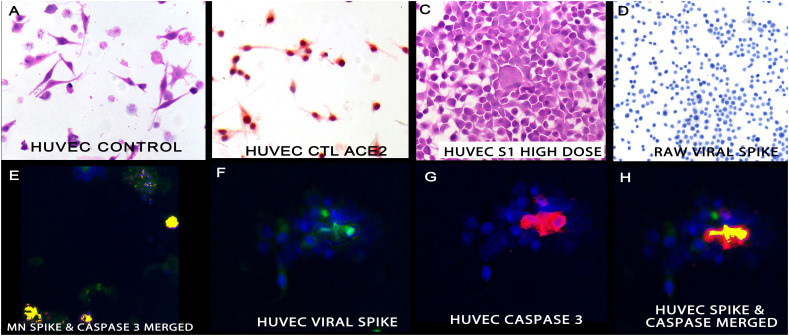
Fig. 3.
Cytologic and molecular correlates of the S1 subunit of the spike protein in cell lines.
Panel A shows the cytology of untreated HUVEC cells; these endothelial cells strongly express ACE2 (panel B). Treatment with the S1 subunit of spike protein induced cell aggregation and degeneration (panel C). The S1 subunit was not endocytosed by the RAW cells (panel D) and was evident in the HUVEC cells where it co-localized with caspase 3 as seen in panels F-H. Panel F is the isolated spike data (fluorescent green), panel G the isolated caspase 3 data (fluorescent red) and panel H the merged image with fluorescent yellow marking the co-localization of the two protein. The MN cells likewise showed co-localization of the spike protein and caspase 3 (panel E). The magnifications are 600× (panels A-D) and 1000× (panels E-F) with DAB (brown) signal (panel D). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Increasing the concentration of S1 spike protein from 70 to 350 ng/ml resulted in increased HUVEC cell degeneration and adhesion, as well as a significant increase in the % of cells with the spike subunit and caspase-3 (Table 2 and Fig. 3).
The HUVEC cells were then treated with 70 ng/ml of the truncated S1 subunit and the full length S2 subunit. Scattered degenerative changes were seen with the latter. As seen in Table 2, the % of cells with identifiable spike subunit or caspase-3 was significantly less than for the full length S1 subunit (p < 0.001).
3.3. Spike subunit injection into mouse
Mice were tail vein injected with the different spike peptides (S1 full length subunit, truncated S1 subunit, and full length S2 subunit) and euthanized using CO2 after five days. The mice injected with the S2 subunit showed no behavioral changes; the mice injected with the S1 subunit had significantly increased thirst (consumed 11 g of hydrogel/mouse at day 1 versus 3 g/mouse for sham injected) and showed stress-related behavior that included eating the plastic support in their cages. The mice injected with the truncated S1 subunit showed less thirst than the mice who received the full S1 subunit (4.5 g hydrogel/mouse at day 1) and no stressed behavior.
At autopsy, no gross changes were evident. The major organs were analyzed for the spike subunit as well as IL6, TNFα, C5b-9, and caspase 3 blinded to the experimental conditions. The mice treated with the S2 subunit showed no evidence of the spike protein internalization and no increased expression of IL6, TNFα, C5b-9, or caspase 3 (Table 1 and Fig. 4 ). The mice treated with the full length S1 subunit showed the viral protein primarily in the brain, subcutaneous fat of the skin, and liver. Over 90% of S1 spike positive cells were endothelia in microvessels (Fig. 4). ACE2 analyses by immunohistochemistry showed that the three organs with the strongest expression in the microvessels were the skin/subcutaneous fat, brain, and liver. Co-expression analyses confirmed that S1 spike subunit co-localized with ACE2 (data not shown). Endothelial cell damage, with increased caspase-3, as well as TNFα, IL6, and C5b-9 expression, was seen in the microvessels of the skin and brain in the mouse model with co-localization with the S1 spike protein (Fig. 4).
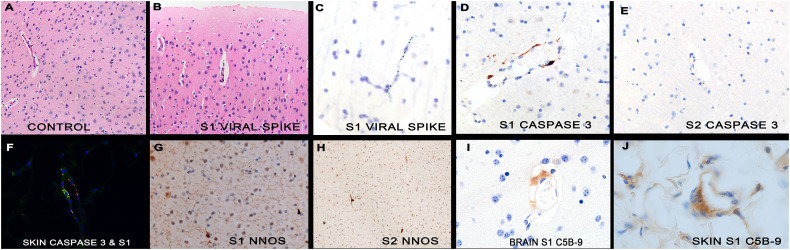
Fig. 4.
Histologic and molecular results in mice after tail vein injection of different spike subunits.
Panel A shows the capillaries of normal mouse brain in which no perivascular space is evident (box). The microvessels of the mice brains after spike S1 subunit injection do show edema (panel B, box). The spike S1 subunit was evident in the capillaries of the brain (panel C) and deep fat of the skin (panel F) as was activated caspase 3 (panel D). However, mice injected with the spike S2 subunit showed no endocytosis in the endothelia in the brain nor caspase-3 activation (panel E). The fluorescent yellow in panel F indicates that the S1 spike subunit did co-localize with caspase-3. As in the human COVID-19 brains, the mouse brains showed increase nNOS after spike S1 subunit injection (panel G) but not S2 subunit injection (panel H). Complement cascade activated, documented by C5b-9 expression, was seen in the capillaries of the brain (panel I) and deep fat of the skin (panel J) after S1 injection. Magnifications are at 600× (panels A, B, G, H) or at 1000× (other panels) with DAB (brown) signal. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The mice treated with the truncated S1 subunit showed scattered cells positive for this spike protein but not in the endothelia. Rather, the viral protein was seen in cells in the brain and liver that had the morphology of macrophages and microglia, respectively. In these mice no increased caspase-3, IL6, or TNFα expression was noted compared to the controls (Table 1); rare C5b-9 positive endothelial cells were evident but far fewer compared to the mice treated with the full S1 spike subunit (data not shown).
The mouse brain data is presented in Table 1 in parallel with the human brain data. Only the mice treated with the full length S1 subunit showed a significant increase in caspase-3, IL6, C5b-9, and TNFα in the brain. Given the marked changes in MFSD2a, SHIP1, NMDAR2, and nNOS in the human COVID-19 brains, the same proteins were analyzed in the mice brains. Note the significant decrease in MFSD2a and increase in nNOS in the mice brains exposed to the full length S1 subunit, but not in the other mice (Table 1 and Fig. 4) (p < 0.001). However, the significant change in the human COVID-19 brain tissues with regard to NMDAR2 or SHIP1 expression were not evident in the mice brains treated with the full length S1 subunit.
4. Discussion
The two main findings of this study were: 1) human COVID-19 cases show diffuse pauci-inflammatory microvessel endothelial damage in the brain and other organs including the skin [[6], [7], [8]] from the endocytosis of circulating viral spike protein that induces C5b-9, caspase-3 and cytokine production that is associated with a microencephalopathy. This encephalopathy is marked, in part, by neuronal dysfunction, evidenced by increased nNOS and NMDAR2 plus the reduction of key neuronal proteins that include MFSD2a and SHIP1. 2) Injection of the S1 full length spike subunit into the tail vein of mice, but not the S2 subunit or truncated S1 subunit, induces an equivalent microvascular encephalopathy that shares with the human COVID-19 brain disease the endocytosis of the S1 subunit in ACE2+ endothelia, caspase 3, C5b-9, TNFα, and IL6 activation, and the over-expression of nNOS and much reduced expression of MFSD2a. The much milder clinical effects of the truncated S1 subunit suggests that the N-terminal region plays a role in spike S1-induced CNS injury.
The human and murine CNS data are consistent with the term “neurovascular unit” that has been used to reflect the intimate association between neuronal circuits and their associated microvessels [[21], [22], [23]]. MFSD2A plays an essential role in this unit in the blood brain barrier (BBB) and in the transport of the fatty acid docosahexaenoic acid to neurons [[12], [13], [14]]. NMDAR2 signaling is a marker of neuronal dysfunction in this unit, indicative of an increased ratio of excitatory versus inhibitory impulses [16,17]. nNOS plays a more complex role in the unit where increased nuclear expression, as evident in the COVID-19 brain tissues, can facilitate mitochondrial biogenesis to compensate for reduced energy production [[18], [19], [20]]. In mice, increased nNOS can signal increased stress-related behavior [20], as seen in this study. We had previously reported that the SUR1 and TRPM4, which are markers of blood brain barrier dysfunction, were increased in the endothelial cells in the COVID-19 brains and that they co-localized with the spike protein [8]. These findings were similar to that described in acute spinal cord injury associated with either open repair or endovascular stent repair of an aortic aneurysm model, where microvascular injury is also postulated to play a role in pathogenesis [24].
It is well documented that the spike protein of SARS-CoV-2 is composed of two subunits, S1 and S2. The S1 subunit contains a receptor-binding domain that recognizes and binds to ACE 2, while the S2 subunit, after cleavage from the S1 subunit, mediates viral cell membrane fusion and RNA entry by forming a six-helical bundle via the two-heptad repeat domain [25,26]. Wang et al. [9] did demonstrate that the truncated form of the S1 subunit, containing the ACE2 receptor domain (RBD), could be internalized in embyronic kidney cells in vitro without the N-glycosylation that occurs in the intact virus. Their study showed that the HEK293 cells would internalize the RBD spike protein with ACE2. Although we showed similar findings, our data suggested that the full length S1 subunit was more effective in this regard both in vitro and in vivo. This may reflect the different cell lines used: embryonic kidney cells versus endothelial cells. It has also been shown that binding of the spike protein to endogenous ACE2 in mice resulted in down-regulation of ACE2 surface expression [9]. We noted the same observation in vitro with the HUVEC cells but only at the higher dosage of the S1 full length subunit, suggesting that in vivo it may be difficult to verify this phenomenon due to relatively lower concentrations of the circulating pseudovirions and the relatively high ACE2 expression in the endothelia of microvessels, especially in the brain and skin.
The mouse and human data in this paper both strongly suggest to the diagnostic anatomic pathologist that the key finding of COVID-19 is degeneration/necrosis of ACE2+ endothelial cells. This can be diagnosed on hematoxylin/eosin stains in conjunction with immunohistochemistry for ACE2 and caspase 3 in serial sections. Although this study focused on the brain, it should be stressed that there are other sites where there is a rich bed of microvessels with the ACE2 receptor, including the skin/subcutaneous fat and the liver. As has been documented in human patients, microvessels at these sites can also display an endothelialitis that, in the skin/fat can induce complement activation/hypercoagulable state and the so-called cytokine storm typical of fatal COVID-19 [[7], [8], [9]].
The mechanism of the observed endothelialopathy is likely complex after S1 spike glycoprotein endocytosis. Given that complement activation is a key component of both the pulmonary microangiopathy associated with the infectious virus, and the pseudovirion induced diseased, C5b-9 mediated endothelial cell necrosis evidenced by caspase 3 expression may play an important role. Endothelial based cytokine production may be a sequela of mannan binding lectin pathway activation and could also contribute to vascular thrombosis [6,7]. The resultant ischemic sequelae of this complement and cytokine driven endotheliopathy could, in turn, lead to the neuronal dysfunction evident in both the human and murine diseases. The data strongly suggests that ACE2+ endothelia is a biomarker of the endothelialopathy since organs with the highest concentration of ACE2+ microvessels (brain, skin, liver) show the most disease in both humans and mice. It is possible to use this mouse model as a simple assay to test the safety of vaccines in regards to any potential adverse neurologic sequela related to its administration and also allows one to examine the efficacy of other treatment regimens that target the pseudovirion induced microangiopathy and thrombosis of severe COVID-19.
In sum, the data presented indicates that the full-length S1 subunit of the spike protein of SARS-CoV-2 alone is capable, without the infectious virus, of inducing systemic microendothelial cell damage in mice with a cognate pattern of complement activation and increased cytokine expression and the concomitant thromboses/hypercoagulable state. This disease pattern strongly parallels the extra-pulmonary manifestations of severe human COVID-19 and suggests that the latter may not represent systemic infectious virus. Thus, prevention of the CNS disease so common in severe COVID-19 may require neutralization/removal of the circulating pseudovirions. The key point for the diagnostic anatomic pathologist is to focus their studies in patients with SARS-CoV-2 infection on degeneration of ACE2+ endothelia using routine H&E stains in conjunction with immunohistochemistry for ACE2, caspase 3 and, when possible, the spike protein.
CRediT authorship contribution statement
Acquisition of data: GJN, CM, TS, HA, DS, SM, BY, BL, ET.
Verification of data: GJN, CM, BY, ET.
Writing/review of manuscript: GJN, CM, ET, JJM.
The authors thank Dr. Margaret Nuovo for the excellent work with the photomicrographs.
Footnotes
No external funding.
The authors have no conflicts of interest to report.
References
- 1.Paterson R.W., Brown R.L., Benjamin L., et al. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar A., Pareek V., Prasoon P., et al. Possible routes of SARS-CoV-2 invasion in brain: in context of neurological symptoms in COVID-19 patients. J Neurosci Res. 2020 doi: 10.1002/jnr.24717. [DOI] [PubMed] [Google Scholar]
- 3.Azizi S.A., Azizi S.A. Neurological injuries in COVID-19 patients: direct viral invasion or a bystander injury after infection of epithelial/endothelial cells. J Neurovirol. 2020;26:631–641. doi: 10.1007/s13365-020-00903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall M. How Covid-19 can damage the brain. Nature. 2020;585:343. doi: 10.1038/d41586-020-02599-5. [DOI] [PubMed] [Google Scholar]
- 5.Ellul M.A., Benjamin L., Singh B., et al. Neurological associations of COVID-19. The Lancet Neurology. 2020;10:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magro C., Mulvey J.J., Nuovo G.J., et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magro C., Mulvey J.J., Laurence J., et al. The differing pathophysiologies that underlie COVID-19 associated perniosis and thrombotic retiform purpura: a case series. Br J Dermatol. 2020 Jul 22 doi: 10.1111/bjd.19415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magro C., Mulvey J.J., Kubiak J., et al. Severe COVID-19: a multifaceted viral vasculopathy syndrome. Ann Diagn Pathol. 2020 doi: 10.1016/j.anndiagpath.2020.151645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S., Guo F., Liu K., et al. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136(1–2):8–15. doi: 10.1016/j.virusres.2008.03.004. [Epub 2008 Jun 12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuovo G.J., Magro C., Mikhail A. Cytologic and molecular correlates of SARS-CoV-2 infection of the nasopharynx. Ann Diagn Pathol. 2020;48 doi: 10.1016/j.anndiagpath.2020.151565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cashman N.R., Durham H.D., Blusztajn J.K., et al. Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Zhao Z., Zlokovic B.V. Blood-brain barrier: a dual life of MFSD2A? Neuron. 2014;82(4):728–730. doi: 10.1016/j.neuron.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Zvi A., Lacoste B., Kur E., et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509(7501):507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L.N., Ma D., Shui G., et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 2014;509(7501):503–506. doi: 10.1038/nature13241. [Epub 2014 May 14. PMID: 24828044] [DOI] [PubMed] [Google Scholar]
- 15.Pauls S.D., Marshall A.J. Regulation of immune cell signaling by SHIP1: a phosphatase, scaffold protein, and potential therapeutic target. Eur J Immunol. 2017;47(6):932–945. doi: 10.1002/eji.201646795. Jun. Epub 2017 May 26. PMID: 28480512. [DOI] [PubMed] [Google Scholar]
- 16.Eugenin Eliseo A., King Jessie E., Nath Avindra, Calderon Tina M., Zukin R. Suzanne, Bennett Michael V.L., et al. HIV-tat induces formation of an LRP–PSD-95–NMDAR–nNOS complex that promotes apoptosis in neurons and astrocytes. Proc Natl Acad Sci. 2007;104(9):3438–3443. doi: 10.1073/pnas.0611699104. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frega M., Linda K., Keller J.M., et al. Neuronal network dysfunction in a model for Kleefstra syndrome mediated by enhanced NMDAR signaling. Nat Commun. 2019;10:4928–4933. doi: 10.1038/s41467-019-12947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tricoire L., Vitalis T. Neuronal nitric oxide synthase expressing neurons: a journey from birth to neuronal circuits. Front Neural Circuits. 2012;6:82. doi: 10.3389/fncir.2012.00082. Published 2012 Dec 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aquilano K., Baldelli S., Ciriolo M.R. Nuclear recruitment of neuronal nitric-oxide synthase by α-syntrophin is crucial for the induction of mitochondrial biogenesis. J Biol Chem. 2014;289(1):365–378. doi: 10.1074/jbc.M113.506733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang H., Chen Z., Xiao H., et al. nNOS-expressing neurons in the vmPFC transform pPVT-derived chronic pain signals into anxiety behaviors. Nat Commun. 2020;11:2501–2511. doi: 10.1038/s41467-020-16198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moskowitz M.A., Lo E.H., Iadecola C. The science of stroke: mechanisms in search of treatments. Neuron. 2010;67(2):181–198. doi: 10.1016/j.neuron.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zlokovic B. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat Rev Neurosci. 2011;12:723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iadecola C. The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron. 2017;96:17–42. doi: 10.1016/j.neuron.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Awad H., Bratasz A., Nuovo G., Tili E., et al. MiR-155 deletion reduces ischemia-induced paralysis in an aortic aneurysm repair mouse model: utility of immunohistochemistry and histopathology in understanding etiology of spinal cord paralysis. Ann Diagn Pathol. 2018;36:12–20. doi: 10.1016/j.anndiagpath.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y., Yang C., Xu X., et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1–9. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]