Abstract
Cucumber mosaic virus (CMV) is a prevalent virus affecting the quality and yield of pepper, resulting in yield losses of greater than 80% during severe local epidemics. Cultural practices and the heavy use of agrochemicals are the most common control measures for CMV. Sources of resistance provide a practical reference and a basis for breeding for CMV resistance. Genetic factors underlying CMV resistance have been studied and advanced breeding lines and cultivars with improved resistance have been developed by traditional breeding methods. Additionally, QTLs or genes for CMV resistance have been identified and can be utilized for marker-assisted resistance breeding. This review focuses on status and prospect of CMV against different virus strains, host resistance, and its applied genetics. With the advent of novel technologies, more useful markers and precise approaches can facilitate the progress for improving CMV resistance in Capsicum.
Keywords: Capsicum, cucumber mosaic virus, resistance, marker-assisted breeding, quantitative trait loci
Introduction
Pepper (Capsicum spp.) is an important vegetable and spice crop worldwide owing to its color, taste, pungency, flavor, and aroma (Rohini and Lakshmanan, 2017). In 2018, approximately 36.7 million tons of fresh pepper fruit and 4.16 million tons of dried pods were harvested from 3.76 million hectares worldwide (FAOSTAT, 2018)1. Among vegetable crops in China, pepper has the largest cultivated area, with an annual planting area of 2.13 million hectares and output of more than 25 million tons (Wang et al., 2019). Pepper production is frequently threatened by many biotic factors, such as diseases, weeds, and pests, including bacterial wilt, phytophthora root rot, cucumber mosaic virus (CMV), root-knot nematodes, aphids, and thrips (Ridzuan et al., 2018). Among these threats, CMV is the most significant constraint to pepper production and it has been found in most of pepper cultivating countries cross the worldwide (Doolittle, 1916; Jagger, 1916; Palukaitis et al., 1992). CMV is a seed-borne disease in pepper resulting in yield losses of over 80% in years of severe local epidemics (Joshi and Dubey, 1973; Florini and Zitter, 1987; Green and Kim, 1991; Avilla et al., 1997). In China, CMV results in marketable yield losses of 20 to 40% in most of the pepper-growing regions (Wu et al., 2006; Zhao et al., 2009). The host range of CMV is very broad, affecting more than 1200 plant species belonging to 100 families, including other important solanaceous crops (e.g., potato, tobacco, and tomato) (Watterson, 1993; Jacquemond, 2012). Additionally, over 80 species of aphids are known to transmit the virus in a non-persistent manner (Palukaitis and Garcíaarenal, 2003).
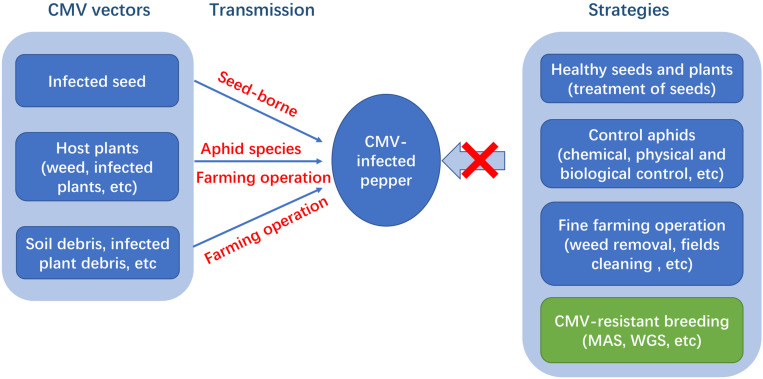
Several strategies have been implemented to control CMV, including cultural measures (e.g., weed removal, the eradication of infected plants, and disease-free seed use) and chemical and biological pesticides, such as pyrethroids, abamectin, and carbamate, to control aphids (insect vectors) (Figure 1). The broad host range and large number of insect vectors make disease control very difficult (Palukaitis et al., 1992). Additionally, chemical insecticides are considered uneconomical and environmentally unfriendly. Thus, the most effective, sustainable, and long-lasting strategy for the control of CMV is the development of disease-resistant pepper cultivars or hybrids (Yao et al., 2013). Some sources of resistance to CMV in Capsicum spp. have been identified and utilized for breeding (Green and Kim, 1994). However, the levels of CMV resistance in commercial lines are still insufficient (Pochard and Daubèze, 1989). Hence, it is of primary importance to screen new sources of CMV resistance. Additionally, the genetic basis of resistance to CMV in Capsicum is complex, unclear, and even controversial in some cases. Most studies have found partial resistance mediated by multiple loci, and a few studies reported dominance or recessive monogenic. Therefore, it requires the clarification the genetic basis of resistance as well as resistance mechanisms and the development of molecular markers linked to resistance loci. This information will contribute to breeding for resistant pepper cultivars. The current study provides an overview of CMV diseases in pepper, focusing on the characteristics of virus strains, mechanisms underlying host resistance, genetics, and efforts to breed pepper lines and cultivars with improved resistance.
FIGURE 1.
CMV vectors, virus transmission and strategies for managing CMV in pepper.
Disease Symptoms and Management
Cucumber mosaic virus was first discovered in cucumbers and muskmelons in Michigan, United States and in cucumbers in New York, United States in 1916 (Doolittle, 1916). It is now known to occur worldwide in both temperate and tropical climates, affecting many agricultural and horticultural crops, as one of the most prevalent viruses. Common symptoms of CMV infection in peppers are mottle, mosaic, vein clearing, yellow discoloration, narrowing, or shoe-stringing (Conti et al., 1979; Green and Kim, 1991; Zitter and Murphy, 2009). The symptoms of CMV infection in peppers are highly dependent on plant age; young plants tend to show more severe symptoms, while plants at later stages of development may be asymptomatic (Garcia-Ruiz and Murphy, 2011; Kenyon et al., 2014). When plants are young at the time of infection, early leaves are slightly wrinkled or bumpy and pale green. During growth, the foliage may develop oak-leaf patterns or ringspot. As new leaves emerge, they develop a chlorotic mosaic pattern that tends to encompass the entire leaf (Zitter and Murphy, 2009). CMV-infected pepper plants also tend to show severe stunting and reduced flower formation and set. The fruits are small, malformed, bumpy, patchily discolored, and may show depressed spots or necrotic lesions, leading to significantly reduced fruit yield and quality (Sugiura et al., 1975; Lockhart and Fischer, 1976; Green and Kim, 1994; Zhao et al., 2009). Furthermore, CMV composite infections with other viruses are commonly observed in the field, and the symptoms are usually difficult to distinguish from those of single CMV infection. Several studies have also reported severe synergy in some CMV strains coinfected other viruses (e.g., CMV and PepMov) in pepper plants (Tien et al., 1987; Palukaitis et al., 1992; Zhao et al., 2004; Gao et al., 2016).
Cucumber mosaic virus is transmitted by the mechanical inoculation of plant sap and aphids are the most important means of natural transmission (Edwardson and Christie, 1986). More than 80 aphid species have been described as CMV vectors and transmit the virus in a non-persistent, stylet-borne manner. CMV does not reproduce in its aphid vector and is not transmitted to progeny aphids. For vegetables, Myzus persicae and Aphis gossypii are the most efficient vectors for virus transmission (Conti et al., 1979). A major characteristic of non-persistent transmission is that aphids acquire the virus for short periods of time (normally a few seconds) and lose the virus after normal feeding on plants. CMV can also be transmitted via infected seed, infected crop debris, non-vectored soil debris, pollen, and other routes (Pares and Gunn, 1989; Zhou et al., 1994; Chen et al., 2000).
The main biological strategies for managing CMV is the eradication of infected plants from field plot. CMV has an extremely wide alternative host range, including many species of weeds, such as Carex vulpine, Solanum nigru, and Datura stramonium (Kazinczi et al., 2004; Juan et al., 2006). These weed hosts in nearby field plots are important potential virus sources for aphid transmission to pepper. When pepper crops are absent, some weed species are perennial depositories for non-persistently transmitted viruses by viruliferous vectors. Additionally, the virus can remain viable in plant debris in the soil for several months. Thus, CMV infection can occur from infected soil debris via non-vectored soil transmission (Pares and Gunn, 1989). Second, owing to the seed-borne nature, the use of healthy seeds and plants for production is necessary. The control of CMV should start by only using clean and disease-free seeds. The treatment of seeds with 15% trisodium phosphate solution is an effective method to reduce the incidence of viral disease (Rast and Stijger, 1987). Third, the application of chemical insecticides and biological pesticides to control insect vectors can be effective. However, it is not a sustainable strategy, since populations of beneficial insects (such as coccinellidae) may be adversely affected by insecticides (Wang and Uchida, 2014). Moreover, the use of insecticides can result in the rapid development of insecticide-resistant populations. High costs and environmental consequences are additional issues. Thus, the development of disease-resistant pepper cultivars is an important approach to overcoming the threat of CMV.
Overview of the Virus
Viral Structure
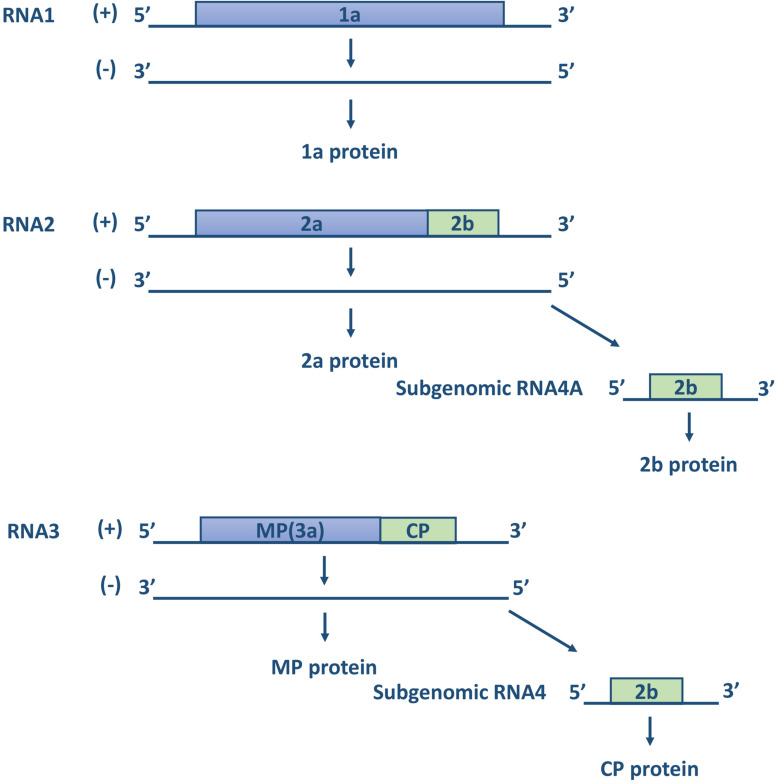
Cucumber mosaic virus is the type member of the genus Cucumovirus in the family Bromoviridae and the superfamily of alpha-like viruses (Goldbach, 1987; Kenyon et al., 2014). It is a multicomponent virus with a single-stranded positive-sense RNA genome, consisting of three genomic RNAs, designated RNA1, RNA2, and RNA3, and subgenomic RNA4 and RNA4A (Sivakumaran et al., 2000, 2002; Figure 2). RNA1 encodes a single protein, referred to as 1a, which is related to the replication of the viral genome. The sequence of protein 1a is highly conserved, with two functional domains. The N-terminal domain has methyltransferase activity for the addition of a cap structure to the 5′-terminus of genomic and subgenomic RNAs, while the C-terminal domain of the 1a protein is a putative helicase, which functions to “unwind” the double-stranded RNA that develops during viral replication (Zitter and Murphy, 2009). RNA2 encodes two proteins, referred to as 2a and 2b. The 2a protein has RNA-dependent RNA polymerase (RdRp) activity and participates in the viral replication process together with the 1a protein. The 2b protein is translated from a separate (subgenomic) RNA strand, referred to as RNA4A, within the 3′ portion of RNA2. The CMV 2b protein is involved in long-distance virus movement, expression of systemic symptoms, and the suppression of gene silencing (Ding et al., 1994; Brigneti et al., 1998). The CMV 2b protein inhibits the plant initiation of gene silencing in distant tissues, thereby allowing CMV to invade plant tissues (Zitter and Murphy, 2009). RNA3 encodes the 3a protein and the coat protein (CP) expressed from subgenomic RNA4. The 3a protein is associated with intercellular movement and long-distance transport of the virus in the host; accordingly, it is also called movement protein (MP). CP is the only protein associated with virus particles and is the sole determinant of transmission by aphid vectors. Minor changes in the sequence of the viral coat protein can affect the ability of CMV strains to be transmitted by aphids (Palukaitis et al., 1992). Moreover, according to Mochizuki and Ohki (2005), certain amino acid positions of CP have important functions, such as residue 129, whose mutation affect host symptoms and the transmission efficiency of the virus (Table 1).
FIGURE 2.
The structure of genomic RNAs of CMV. The genomic plus-strand and minus-strands are indicated with a “+” or “–” (modified according to Zitter and Murphy, 2009).
TABLE 1.
The function of protein encoded by CMV RNAs.
| RNAs | Proteins | Function |
| RNA1 | 1a | Replication of the viral genome |
| RNA2 | 2a | RdRp activity and participates in the viral replication process |
| Subgenomic RNA4A | 2b | Influence the systemic infection of the virus and inhibit virus-mediated gene silencing |
| RNA3 | MP (3a) | Virus cell-to-cell movement, intraplant long distance movement |
| Subgenomic RNA4 | CP | The sole determinant for transmission by aphid vectors |
CMV Strains
The traditional classification of CMV strains was based on pathotype and morphological symptoms in different host species (Kaper and Waterworth, 1981). For example, six different CMV isolates were identified by observed varying symptoms on host plants (Fulton, 1950), and they were named Fulton’s strain (accession from FA-CMV to FE-CMV). Representatively, Yasuo classified CMV strains in tobacco into five strain-groups using 10 different host species (Yasuo, 1963; Li, 1995). Hollings et al. (1967) identified the CMV isolates from UK as PY/Y and II subgroups according to indicator plants. However, there are many strains of CMV. Kaper and Waterworth (1981) also list false identifications of new viruses that have turned out to be strains of CMV. Therefore, according to serology and phylogenetic analyses of CP genes, CMV isolates have recently been characterized into two subgroups, I and II. Nucleotide sequences of CP genes in CMV isolates within each subgroup can share up to 95% nucleotide sequence identity, whereas isolates in different subgroups share 69–77% sequence identity (Palukaitis and Garcíaarenal, 2003). Normally, high-temperature tolerance is greater in subgroup I than in subgroup II, while symptoms in subgroup II are less severe. Peppers are more likely to be infected with subgroup I isolates, since these are more prevalent in the warm climates where peppers are grown (Moury and Verdin, 2012). Subgroup I can be further divided into subgroups IA and IB, and populations of subgroup IB is very common in East Asia (Palukaitis et al., 1992; Roossinck et al., 1999).
Although accurate classification of serology and phylogenetic, the relationship between pathogenic genes in the virus and resistance genes in hosts was still unknown. Development a set of differential hosts of CMV strains have implications for breeding and reflect the relationship between resistance phenotypes and genotypes. Yang et al. (1992) identified a set of C. annuum lines with different levels of resistance as differential hosts for distinct CMV strains in pepper (Table 2). As a result, five strain-groups in pepper were identified in China, including CMV-P0, CMV-P1, CMV-P2, CMV-P3, CMV-P4, and CMV-P5. Moreover, several studies about strain specific resistance to CMV was also reported in pepper (Kang et al., 2010; Min et al., 2014; Choi et al., 2018).
TABLE 2.
Differential hosts of CMV strains in pepper.
| Name of strains | Resistant phenotype of differential hosts of C. annuum |
Symptom of pepper | ||||
| LS-8501 | LS-8502 | LS-8503 | LS-8504 | LS-8505 | ||
| CMV-P0 | R | R | R | T | S | Moderate mosaic |
| CMV-P1 | R | T | T | S | S | Ring spot |
| CMV-P2 | R | S | S | S | S | Ferny leaf to linear leaf |
| CMV-P3 | T | S | S | S | S | Stem necrosis |
| CMV-P4 | S | S | S | S | S | Yellow mosaic |
R, resistant; T, tolerance; S, susceptible.
Disease Screening Methods
Most purification methods for CMV strains used in practice are modifications of previously described methods (Scott, 1963; Lot et al., 1972). To prepare an inoculum that can infect pepper, purified CMV strains are propagated in leaves of tobacco (Nicotiana benthamiana). The virus inoculum is prepared by grinding 1 g of CMV-infected tobacco leaves in 5 ml of 0.03 M phosphate buffer (pH 7.2), containing 0.2% ascorbic acid or sodium diethyldithiocarbamate (Feng et al., 1986). The homogenate is filtered through gauze and mixed with 600-mesh carborundum as the inoculum.
The optimal time point for the identification of resistance to CMV in pepper is at an early stage of plant growth (Agrios et al., 1985; Dogimont et al., 1994). Generally, seedlings are inoculated with inoculum at the cotyledon stage to 6-leaf stage (cotyledon stage, 1–2 leaf stage, 3–4 leaf stage, and 5–6 leaf stage) by manual rubbing (Caranta et al., 1997; Chaim et al., 2001a; Yao, 2013; Min et al., 2014). According to Tian et al. (1989), the 3–6 leaf stage is the best inoculation period for assessing the resistance level of pepper resources because young seedlings can show a higher disease index at the cotyledon stage and 1–2 leaf stage, while old seedlings show lower values at the 7–8 leaf stage. Seedlings are rinsed 2 min after inoculation to remove excess inoculum (Dogimont et al., 1994). Following inoculation, the plans are maintained at 22–28°C under a 12-h day/12-h night cycle in a growth chamber (Feng et al., 1986; Dogimont et al., 1994).
Normally, the inoculated seedlings are observed for systematic symptom development after 2–4 weeks. Resistance is assessed according to the severity of mosaic and leaf distortion on a scale (Feng et al., 1986; Monma and Sakata, 1996; Chaim et al., 2001a; Zou, 2005; Yao et al., 2013; Rahman et al., 2016), number of local lesions (Caranta et al., 1997) or ELISA absorbance values (Reddy et al., 2001; Kang et al., 2010). The most commonly used method for disease evaluation is examining visually and assigning with a disease grade. Zou (2005) described the standard of disease grade, a scale ranging from 0 to 9, as following. 0 = no symptoms, 1 = mild mosaic on inoculated leaf, no leaf distortion, 3 = mild mosaic, stem streak and leaf distortion, 5 = strong mosaic, mild leaf distortion and stem necrotizing, 7 = severe mosaic and distortion, and 9 = severely stunted and systemic necrotizing. Disease indices of a population were calculated from disease grades of individual plants of that population, according to the formula disease indices (DIs) = (Σ (numbers of plants in the disease grade × disease grade)/9 × total numbers of plants) × 100 (Zou, 2005; Yao et al., 2013, Guo et al., 2017). Besides, Chaim et al. (2001a) rated the severity of the mosaic and leaf distortion of plants using a scale of 0–3, Monma and Sakata (1996) defined a score of 1 to 4. Although there are differences in the standard of disease grade, all of them can accurately reflect the resistance level of plants. Caranta et al. (1997) assessed the resistance and defined a disease score of 1–4 using the number of local lesions. “1” = 0 to 5 lesions, “2” = 6 to 20 lesions, “3” = 21 to 50 lesions, and “4” = more than 50 lesions, and resistance was assessed 4 days after inoculation (Caranta et al., 1997). However, the disease evaluation of number of local lesions depends on the CMV strain and mode of host resistance. The Fulton CMV/N strain could induce necrotic local lesions on the inoculated leaves, that gave an estimate of resistance by directly and quantitatively method of number of local lesions (Troutman and Fulton, 1958; Caranta et al., 1997). Kang et al. (2010) tested systemic infection using ELISA and monitored the symptoms of plants simultaneously. ELISA values could be more precise programs by providing a quantitative evaluation of viral resistance.
Pepper Genetic Resources for CMV Resistance
The identification of sources of CMV resistance is a prerequisite for strategic breeding aimed at developing stable resistant varieties and hybrids. An accession of the wild pepper C. baccatum, “Pen 3-4,” was identified as resistant to CMV in greenhouse and field experiments (Pochard, 1977). Subsequently, various resistant or tolerant hybrids and breeding lines have been developed from the source, such as “Vania,” “Gadir,” “Pochard’s androgenic dihaploids” (ADH), and “Nicklow’s Emerald Bell” (Pochard and Daubèze, 1989; Caranta et al., 2002). The small-fruited Indian chili “Perennial,” which is the most well-characterized source of resistance to a broad range of viruses, shows tolerance or partial resistance to CMV (Pochard, 1977; Nono-Womdim et al., 1993; Lapidot et al., 1997). Another small-fruited pungent pepper accession, C. frutescens “BG2814-6,” possesses alleles conferring more complete resistance than that in Perennial (Grube et al., 2000). BG2814-6 crossed with Perennial has been used for the development of lines with improved CMV resistance by Mazourek et al. (2009). Suzuki et al. (2003) identified five resistant lines, “Sapporo-oonaga,” “Nanbu-oonaga,” “LS 1839-2-4,” “PI 439381-1-3,” and “Tabasco,” from 37 cultivated and wild Capsicum species. Previous study identified a CMV resistance source, Likeumjo, which was later used to develop commercial varieties, such as “Bukang” (Kang et al., 2010). Yao (2013) evaluated several breeding lines and accessions of C. annuum and found a resistant accession “BJ0747-1-3-1-1.” Screening of 199 genetic pepper resources by Shin et al. (2013) resulted in the identification of 32 resistant or moderately resistant Capsicum accessions to CMVP1. Min et al. (2014) reported three accessions, “I7339,” “ICPN-18-8,” and “Daedlbo,” with resistance to CMVP1. More recently, several accessions with resistance to CMV were identified from 50 pepper resources (Naresh et al., 2016). Rahman et al. (2016) identified two C. annuum genotypes, CA23 (Noakhali) and CA12 (Comilla-2) possess genes conferring resistance and moderate resistance phenotype to CMV, respectively. Choi et al. (2018) identified an Indian C. annuum cultivar, Lam32, with broad-spectrum resistance to CMVKorean, CMVFNY, and CMVP1. In China, at least 317 sources of resistance or moderate resistance to CMV have been identified in Capsicum spp. by breeders in different groups, such as the Chinese local variety “erfutou,” “shifangjiao,” and “Hanchuan sweet pepper” (Li and He, 2000; Wang et al., 2001; Mao et al., 2004; Sun et al., 2008). Of note, the World Vegetable Center (AVRDC, Asian Vegetable Research and Development Center) has published a catalog of sources of resistance to viruses for pepper (Capsicum spp.), listing 165 Capsicum accessions with resistance and 46 accessions with tolerance to CMV (Green and Kim, 1994). These sources of resistance to CMV provide a practical reference and a basis for CMV resistant breeding (Table 3); however, most of the sources display only partial resistance to CMV, limiting resistance breeding in pepper.
TABLE 3.
Primary genetic sources of resistance to CMV.
| Capsicum species | Original source | Developed source | Resistance reaction | References |
| C. annuum | Perennial (IHR 384) | HAD 249, HAD 260, HAD 268, HAD 293 | Resistance or tolerance to several CMV isolates | Singh and Thakur, 1977; Pochard et al., 1983 |
| C. annuum | Wild type from Nicaragua | Rama | Resistance to CMV | Pochard, 1982 |
| C. annuum | Milord | Partially resistant (reduced virus migration), resistance is suppressed at low temperatures (12–25oC) | Pochard and Daubèze, 1989; Nono-Womdim et al., 1991 | |
| C. annuum | Pusa Sadabahar, RHRC-clustering erect, Punjab Lal | Resistance to CMV | Pinaki, 1999 | |
| C. annuum | MRCH | Tolerance to CMV | Shifriss and Cohen, 1987 | |
| C. annuum | Likeumjo | Bukang | Resistance to CMVKorean and CMVFNY strains | Kang et al., 2010 |
| C. annuum | Changyang difangzhong 4 | BJ0747-1-3-1-1 | Resistance to Hubei CMV isolates | Yao et al., 2013; Li N. et al., 2018 |
| C. annuum | I7339, ICPN-18-8, Daedlbo, Lam3 | Resistance to CMVP1 strain | Min et al., 2014; Choi et al., 2018 | |
| C. annuum | CA23 (Noakhali), CA12 (Comilla-2) | Resistance to CMV subgroup-II isolate | Rahman et al., 2016 | |
| C. baccatum | pendulum 3-4 (Pen 3-4) | Vania, Philomèle 1 | Resistance to CMV-N and CMV-MES, partial resistance Resistant to CMV migration | Pochard, 1977; Lapidot et al., 1997 |
| C. baccatum | PI 439381-1-3 | Resistance to CMV-Y | Suzuki et al., 2003 | |
| C. frutescens | LP-1 | Resistance to CMV | Barrios et al., 1971 | |
| C. frutescens | BG2814-6 | Resistance to several CMV strains | Grube et al., 2000 | |
| C. frutescens | PBC688 | Resistance to CMVFNY | Guo et al., 2017 | |
| C. frutescens | LS 1839-2-4 | Resistance to CMV-Y | Suzuki et al., 2003 | |
| C. frutescens | Tabasco | Resistance to CMV-Y | Suzuki et al., 2003 |
Inheritance Studies and Genetic Mapping
The genetic basis of resistance to CMV in pepper is complex, and most studies have found partial resistance mediated by multiple loci (Grube et al., 2000; Suzuki et al., 2003; Yao et al., 2013; Wang, 2016). Table 4, displays classical genetic studies of resistance to CMV in different Capsicum accessions. Resistance in “Perennial” exhibited different inheritance patterns: monogenic recessive, partially dominant, or polygenic recessive (Singh and Thakur, 1977; Rusco and Csillery, 1980; Pochard and Daubèze, 1989; Bansal et al., 1992; Nono-Womdim et al., 1993; Lapidot et al., 1997; Wang, 2016). The line “Vania,” derived from C. baccatum “Pen 3-4,” showed dominant partial resistance to CMV (Caranta et al., 2002). The inheritance of CMV resistance in “BG2814-6” was incomplete and was controlled by at least two major recessive genes (Grube et al., 2000). The application of a mixed inheritance model revealed two additive-dominant major genes with additive-dominant polygenic traits in the CMV-resistance source “BJ0747-1-3-1-1” (Yao et al., 2013). Polygenic recessive resistance has been reported in “IHR 2451” and “IHR 4503” by Naresh et al. (2016). Single-gene control of resistance against CMV has also been reported. Kang et al. found that resistance in “Bukang” is under the control of a single dominant gene (Kang et al., 2010). Min et al. (2014) reported two different recessive genes conferring resistance to CMV in C. annuum “I7339.” More recently, Choi et al. (2018) showed that resistance in C. annuum “Lam32” is controlled by a single recessive gene, CMV resistance gene 2 (cmr2). Zhang et al. (2016) also described a single recessive gene cr underlying resistance to CMV.
TABLE 4.
Classical genetic studies of resistance to CMV in Capsicum.
| Capsicum species | Resistant parent | Genetic control and mechanisms of resistance | References |
| C. annuum | Perennial | Monogenic recessive, cm | Singh and Thakur, 1977 |
| Recessive genes | Pochard, 1982 | ||
| Polygenetic recessive inheritance, resistant to virus multiplication | Pochard and Daubèze, 1989 | ||
| Monogenic recessive and partially dominant | Rusco and Csillery, 1980 | ||
| Two or three incompletely dominant genes | Lapidot et al., 1997 | ||
| Two additive-dominant major genes with epistatic polygene or two additive-dominant major genes with additive-dominant polygene | Wang, 2016 | ||
| C. annuum | Rama | A single dominant gene Riv | Pochard, 1982 |
| C. annuum | MRCH | Polygenetic inheritance and linked to small fruit size | Shifriss and Cohen, 1987 |
| C. annuum | Vania | Polygenetic inheritance, resistant to CMV migration | Caranta et al., 2002 |
| C. annuum | Bukang | A single dominant gene Cmr1 | Kang et al., 2010 |
| C. annuum | BJ0747-1-3-1-1 | Two additive-dominant major genes with additive-dominant polygene | Yao et al., 2013 |
| C. annuum | I7339 | Two recessive, cmr3E and cmr3L | Min et al., 2014 |
| C. annuum | Q132 | Monogenic recessive, cr | Zhang et al., 2016 |
| C. annuum | Lam32 | a single recessive gene cmr2 | Choi et al., 2018 |
| C. frutescens | LP-1 | Monogenic recessive | Barrios et al., 1971 |
| C. frutescens | BG2814-6 | Incompletely dominant and at least two major recessive genes | Grube et al., 2000 |
| C. frutescens | PBC688 | Incompletely dominant and quantitatively inherited | Guo et al., 2017 |
Quantitative inheritance of resistance to CMV followed an additive-dominant genetic model, and the most important genetic effect was the additive one (Yan et al., 1996; Yu et al., 2000; Zou et al., 2004; Yao, 2013). High broad-sense heritability and narrow-sense heritability was detected for CMV resistance in different resistant lines (Yan et al., 1996; Zou et al., 2004; Yao, 2013; Herison et al., 2014). F1 plants showed intermediate in level of resistance, comparing to resistance and susceptible parents. Therefore, it is beneficial to obtain desirable resistance hybrid combinations if both parents have high resistance. However, breeding for CMV resistance totally based on phenotypic selection will take a longer time. The genetic mapping and marker-assisted breeding methods may accelerate the transfer of resistant traits.
The earliest QTL analysis of CMV-resistance in pepper was conducted by Caranta et al. (1997), that identified three resistance QTLs using a DH population of a cross between “Perennial” and “Yolo Wonder.” The three QTLs were located on chromosomes Noir, Pourpre, and linkage group 3, and together explained 57% of the total phenotypic variation. For each QTL, the Perennial allele increased the level of resistance. A digenic epistasis between one locus (TG66 on chromosome Pourpre) that controlled significant trait variation and another locus (TG124 on chromosome Noir) that by itself had no demonstrable effect on the trait was found to have an effect on CMV resistance (Caranta et al., 1997). Several other studies have identified and analyzed QTLs for CMV resistance in “Perennial“ (Chaim et al., 2001a; Zhao et al., 2011; Wang, 2016). Chaim et al. (2001a) identified four QTLs for CMV resistance on linkage groups 4, 6, 11, and 13 using “Maor (TMV-resistant line, L1/L1 genotype) × Perennial” populations. Whereas, one major QTL, cmv11.1, was detected in all three experiments and explained the higher percentage (16%–33%) of the observed phenotypic variation. This QTL is linked to the AFLP marker E35/M48–101 and L locus which confers resistance to tobacco mosaic virus (TMV). A linkage between resistance to CMV and susceptibility to TMV was observed in “Perennial.” Moreover, interaction that involved E35/M48–101, cmv11.1, and the RFLP marker TG191 on LG2 was detected. Therefore, choosing “Maor” allele at TG191 with “Perennial” allele at E35/M48–101 in “Perennial” could have a good level of resistance to CMV in pepper breeding. Markers linked to QTL for CMV resistance were also linked to QTL for fruit weight, as observed by Chaim et al. (2001b). This study also explained the cause of the relatively slow progress in the development of CMV-tolerant large-fruited pepper genotypes (Lapidot et al., 1997; Chaim et al., 2001a). The linked molecular markers facilitate the identification of genotypes from a breeding perspective. QTLs for CMV resistance were also detected on linkage groups 1, 4, and 7 by Zhao et al. (2011). Wang (2016) identified four QTLs for CMV resistance in “Perennial” using F2 (Carolina Wonder × Perennial) and RIL (83-58 × Perennial) populations. CMV12.3, which was detected in the RIL population, was also detected in the F2 population.
QTLs for CMV resistance have also been studied in other sources (Caranta et al., 2002; Yao et al., 2013; Min et al., 2016; Guo et al., 2017). Caranta et al. (2002) identified a QTL on chromosome 12 of C. annuum line “Vania” explaining 45–63.6% of the phenotypic variation, in addition to three minor QTLs on chromosomes 5 and 11. In the inbred line “BJ0747-1-3-1-1,” six QTLs, including one major effect QTL on chromosome 11 and several minor effect QTLs, conferred CMV resistance (Yao et al., 2013). A major QTL, explaining about 20% of the phenotypic variation on chromosome 11, was identified for resistance to CMV in “BJ0747-1-3-1-1” (Li N. et al., 2018). Two QTLs were detected on chromosomes 5 and 10 for CMV resistance in “A1” by Min et al. (2016). In a recent study by Guo et al. (2017), two QTLs on chromosomes 2 and 11 were shown to be associated with CMV resistance in “PBC688” in C. frutescens. qCmr2.1, on chromosome 2, was in the region containing CA02g19570 and CA02g19600. CA02g19570 exhibited high levels of expression after inoculation and is homologous to genes encoding N-like proteins associated with TMV resistance in Solanum crops.
A few monogenic, strain-specific resistance loci have been mapped (Kang et al., 2010; Park et al., 2011; Min et al., 2014). The first single dominance resistance gene, Cmr1, conferring resistance to CMVKorean and CMVFNY strains, was detected in C. annuum “Bukang.” Cmr1 is in the region on chromosome 2 associated with the ToMV resistance locus Tm-1 in tomato (Kang et al., 2010). The single recessive gene cmr2, resistant to CMVP1, has been mapped to marker Affy4 (within 2.3 cM) on chromosome 8 (Choi et al., 2018). Two recessive genes, cmr3E and cmr3L, were located on linkage group 4, associated with the RAPD primer OPAT16 (Min et al., 2014). Zhang et al. (2016) mapped a recessive gene, cr, to a linkage group including two SSR markers GI1354 and genSSR3000.
Possible Mechanisms of Host Resistance
Various mechanisms underlying resistance to CMV have been demonstrated, including the restriction of virus entry and uncoating (installation) in host cells (Lecoq et al., 1982; Caranta et al., 1997), restriction of viral multiplication (Nono-Womdim et al., 1993), or restriction of the long-distance movement of the virus (Dufour et al., 1989; Nono-Womdim et al., 1993). Different resistance mechanisms may be related to environmental conditions, CMV isolates, or genetic backgrounds (Chaim et al., 2001a). CMV resistance in C. annuum “Vania,” “Milord,” “L57,” and “L113” is governed at the level of long-distance movement and is classified as partial resistance (Caranta et al., 2002; Mihailova et al., 2013). The Indian chili “Perennial,” which is described as tolerant or partially resistant to CMV, shows the restriction of CMV installation, reduced viral multiplication, and the prevention of long distance movement (Nono-Womdim et al., 1993; Caranta et al., 1997). C. frutescence “BG2814-6” blocks viral replication and cell-to-cell movement (Grube et al., 2000). C. annuum “Bukang” inhibits the movement of CMV from the epidermal cell layer to mesophyll cells (Kang et al., 2010).
Cucumber mosaic virus infection affects defense-related enzyme metabolism in pepper plants, causing the accumulation of superoxide dismutase (SOD), peroxidase (POD), and polyphenol oxidase (PPO) activity, decreases in phenylalanine ammonia lyase (PAL) activity, and increases in the levels of lignin and total phenols. A correlation analysis has indicated that POD, PAL, PPO, lignin, and total phenols are correlated with resistance after inoculation, where resistant sources show high POD and PPO activity (Yang et al., 2006; Zhang, 2009).
In transcript profiles, several key genes associated with plant-pathogen interactions show increased transcript abundances after inoculation, including genes encoding chitinase, pathogenesis-related (PR) protein, TMV resistance protein, WRKY transcription factor, and jasmonate ZIM-domain protein (Zhu et al., 2018). Among them, four WRKY transcription factors, WRKY 6, WRKY 33, WRKY 38, and WRKY 45, responded to CMV infection in pepper. One possible candidate gene, CA02g19570, that control of CMV resistance in C. frutescens have been identified (Guo et al., 2017). Recent studies determined that CA02g19570 was the gene of qCmr2.1 governing CMV resistance in “PBC688” (Zhang et al., 2020). The expression of CA02g19570 was significantly up-regulated after inoculation, with higher expression levels in the CMV-resistant parent “PBC688.” Silencing of CA02g19570 gene exhibited severe systemic mosaic symptoms after inoculation with CMV (Zhang et al., 2020).
Breeding Strategies for CMV Resistance
Genetic Breeding for CMV Resistance
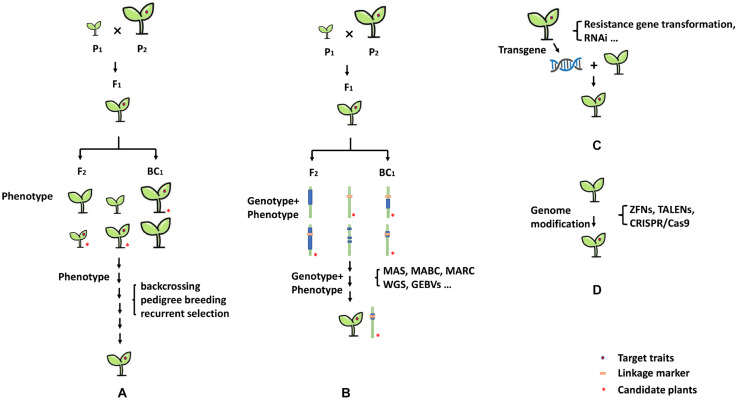
Traditional protocols for plant genetics and breeding, such as pedigree, backcrossing, and recurrent selection methods, have been applied to develop several breeding lines and cultivars with measurable levels of resistance (Figure 3A). For example, the bell-pepper inbred line “Vania” with CMV-resistant trait was developed from recurrent selection using C. baccatum “Pen 3-4” as a primary genitor and C. annuum “Antibois” and “Bastidon” as secondary genitors for resistance to CMV (Pochard and Daubèze, 1989). The inbred line “Vania” cumulated different CMV resistance genotypes and carried wide-range genetic diversity at the same time. Another example was the big-fruit sweet pepper breeding inbred line “98-42,” which was developed by Guo with CMV resistance and is widely used in China. Using the breeding line “98-42,” a series of commercial varieties, including “Zhongjiao No.4,” “Zhongjiao No.6,” and “Zhongjiao No.7,” were developed by IVF, CAAS (Institute of vegetables and Flowers, Chinese Academy of Agricultural Sciences), with good fruit yield and tolerance to CMV (Guo, 1994; Yu et al., 2019). Additionally, strategy can be elaborated doubled haploid (DH) methodology for speeding up the process of breeding lines against CMV and obtaining completely homozygous pure resistant lines. The successful applications of DH strategy for pepper resistant breeding have been described by several seed companies.
FIGURE 3.
Breeding strategies for CMV resistance. (A) Genetic breeding strategies, (B) marker-assisted breeding and genome assistance breeding strategies, (C) transgenic approaches and RNAi strategies, and (D) genome modification strategy.
Marker-Assisted Breeding for CMV Resistance
Conventional breeding involves hybridization with contrasting parental lines and the selection and evaluation of individual plants in cross and backcross populations. However, it is often very difficult and time-consuming to evaluate phenotype or pathotype resistance. The application of marker-assisted selection (MAS) has facilitated breeding for crop improvement, especially for phenotype traits that controlled by quantitative trait loci or recessive allele. Additionally, it is useful for backcross breeding, for the introgression of resistance genes from wild species while selecting against the undesirable characteristics of the wild parent (Young and Tanksley, 1989). Marker-assisted selection has been successfully used in efficient selection of many resistance genes in pepper crop improvement (Ridzuan et al., 2018). Alternatively, several modern breeding strategies, such as, marker-assisted backcrossing (MABC), marker-assisted recurrent selection (MARS), and marker-assisted pedigree selection (MAPS), have been also used for resistant breeding in pepper. These methods are essentially one type of MAS, all of which can quickly and efficiently introduce target genes while retaining the essential characteristics of the recurrent parent (Figure 3B). For CMV-resistance breeding, CMV-susceptible and -resistant genotypes can be precisely identified using molecular markers at an early stage of plant growth, without requiring field screening with artificial inoculation or any environmental influence. Reported linkage markers associated with CMV-resistant traits in Capsicum are listed in Table 5, and the utilization of the linkage markers in MAS breeding program for CMV-resistant has been reported in pepper (Zhang et al., 2016; Kim et al., 2017).
TABLE 5.
Linkage markers associated with CMV-resistant in Capsicum.
| Genetic population | Resistant loci | Chromosome/Linkage group | Marker type | Marker name | Marker size (bp) | Map location (cM) | CMV strains | References |
| Maor × Perennial | cmv11.1 | LG11/Chr. 11 | AFLP | E35/M48-101 | 101 | – | CMV-V27 | Chaim et al., 2001a |
| cmv13.1 | LG13 | RAPD | OPP11500 | 500 | – | |||
| cmv6.1 | LG6 | RAPD | OPA121700 | 1700 | – | |||
| cmv4.1 | LG4 | AFLP | E48/M49-270 | 270 | – | |||
| H3 × Vania | cmv12.1 | Chr. 12 | AFLP | E33/M48-132 | 132 | – | CMV-MES and CMV-N | Caranta et al., 2002 |
| cmv11.1 | Chr. 11 | L | – | – | CMV-N | |||
| cmv11.2 | Chr. 11 | P14/M59-137 | 137 | CMV-N | ||||
| cmv5.1 | Chr. 5 | E35/M48-475 | 475 | CMV-N | ||||
| VC16a × SS69 | - | – | ISSR | I-34450 | 450 | 27.3 | - | Wang et al., 2009 |
| Bukang × Jeju | Cmr1 | Chr.2 | SNP | CaTm-int3HRM | 158 | 2 | CMVKorean, CMVFNY and CMVP1 | Kang et al., 2010 |
| CaT1616BAC | 200 | 2 | ||||||
| 240H02sp6 | 472 | 2 | ||||||
| BJ0747-1-3-1-1 × XJ0630-2-1-2-1-1 | qcmv.hb-4.1 | LG4 | ISSR | UBC843 | – | CMV-HB | Yao et al., 2013 | |
| qcmv.hb-8.2 | LG8 | ISSR | UBC829 | – | ||||
| BJ0747-1-3-1-1 × XJ0630-2-1-2-1-1 | qcmv11.1 | Chr. 11 | SNP | Marker6201026 | – | – | CMVHB–zj | Li N. et al., 2018 |
| qcmv11.2 | Chr. 11 | Marker5409028 | – | – | ||||
| qcmv12.1 | Chr. 12 | Marker17652010 | – | – | ||||
| Q132 × FS871 | cr | – | SSR | GI1354, genSSR3000 | – – | 1.3 0.3 | – | Zhang et al., 2016 |
| Perennial × 83-58 | CMV12.3 | Chr. 12 | KASP | UN54228_1146 | – | 1.8 | – | Wang, 2016 |
| PBC688 × G29 | qCmv2.1 | Chr. 2 | Indel | Indel2-134 | 230 | 0.3 | CMVFNY | Guo et al., 2017 |
| I7339 × E1338 | cmr3E, cmr3L | – | RAPD | OPAT-16 | 22.3 20.7 | CMVP1 | Min et al., 2014 | |
| A1 × 2602 | cmvP1-5.1 | Chr. 5 | SNP | SNP13525, SNP13624 | – | – | CMVP1 | Min et al., 2016 |
| cmvP1-10.1 | Chr. 10 | SNP | SNP07415, SNP07478 | – | – | |||
| Lam32 × Jeju | cmr2 | Chr. 8 | KASP | Affy4 | – | 2.3 | CMVKorean and CMVFNY | Choi et al., 2018 |
Chr., chromosome; LG, linkage group; AFLP, amplified fragment length polymorphism; RAPD, random amplified polymorphic DNA; ISSR, inter-simple sequence repeat; SNP, single nucleotide polymorphism; CAPS, cleaved amplified polymorphic sequence; SSR, simple sequence repeat; KASP, kompetitive allele-specific PCR; Indel, insertion-deletion.
Genome Assistance Breeding for CMV Resistance
The technological advancement next-generation sequencing (NGS) and single nucleotide polymorphism (SNP) genotyping, provides powerful tools for crop improvement (Figure 3B). High-throughput SNP genotyping offers a number of advantages over previous marker systems, such as an abundance of markers, rapid processing of large populations, and low cost (Thomson, 2014). It is particularly useful for breeders to use SNP-based targeted approach to select and combine beneficial alleles at known major genes controlling traits of interest in crop breeding. Moreover, high-throughput SNP genotyping also provides the genome-wide selection (GWS) approaches based on genomic estimated breeding values (GEBVs), rather than being limited to a few known loci. Several high-throughput SNP genotyping platforms, including Affymetrix Axiom array, Fluidigm dynamic arrays, restriction-enzyme-based genotyping-by-sequencing (GBS), and Illumina Infinium iSelect HD array have been reported (Thomson, 2014). Recently, Kim et al. (2017) reported successfully development of Fluidigm SNP type genotyping assays for marker-assisted selection breeding in pepper, that could simultaneously analyze 20 SNP markers about target breeding traits. It will provide an efficient application for MAS (also including MABC, MARS, and MAPS, etc.), and easily tracks with flanking SNP markers polymorphic between the donor and recurrent parent.
Transgenic Approaches for Breeding Against CMV
The use of genetic transformation of pepper with CMV resistance gene is a strategy to obtain resistant plants, such as application of CA02g19570 of qCmr2.1 loci. Another successfully strategy against CMV infection in pepper crops is RNA-mediated virus gene silencing (Figure 3C). RNA interference (RNAi) or RNA silencing, which was first discovered in plants, is a natural defense mechanism against viruses (Hamilton and Baulcombe, 1999). It is triggered by the processing of virus-derived dsRNA by host Dicer-like (DCL) enzymes into 21- to 24-nucleotide (nt) small interfering RNAs (siRNAs) (Blevins et al., 2006; Fusaro et al., 2006; Pooggin, 2018). The expression of dsRNA from transgene plants can induce post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS). Plants expressing a copy of a viral gene in sense and/or antisense orientation have shown resistance upon infection with the virus (or other virus containing identical sequences) through RNAi (Waterhouse and Helliwell, 2003). Therefore, RNAi can be used to engineer resistance in plants against viruses. The transgenic pepper plants that express CMV-siRNA displayed delayed symptom development and significantly milder disease severity upon inoculation with CMV strains (Kim et al., 1997; Cai et al., 2003; Lee et al., 2009). In addition, it is a potential strategy to use transgenic plants for introduced defense-related genes, which are involved in the JA signaling and antiviral RNA silencing pathways that regulate against virus resistance (Yang et al., 2020).
Genome Modification for Breeding Against CMV
In the past decade, great advancements in genome or gene editing technologies, such as zinc finger nucleases (ZFNs), transcriptional activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats associated to nuclease Cas9 (CRISPR/Cas9)., has made it possible to precisely target any gene of interest (Jaganathan et al., 2018; Van Eck, 2018; Xu et al., 2019). Because of simplicity, precision and power, genome editing offers great opportunities to develop improved crop varieties with clear-cut addition of valuable traits or removal of undesirable traits. Moreover, gene editing is more precise than either conventional crop breeding methods or standard genetic engineering methods (Figure 3D). To date, these technologies are being applied in many plant species for enhancing qualities, yield, disease resistance, and other traits (Ito et al., 2015; Pyott et al., 2016; Li R. et al., 2018). Several cloned-genes conferring resistance to virus diseases have sequence variation between resistant and susceptible loci (Li et al., 2016), which might provide good sites for genome modification. Therefore, modification of the sequence of susceptible loci using genome-editing strategy has the potential to improve pepper varieties against CMV.
Conclusion and Future Prospects
Cucumber mosaic virus is one of the most serious threats to pepper production worldwide. Hence, the utilization of resistance resources for breeding resistant cultivars is an efficient method to control CMV outbreaks. The identification and characterization and introgression of new CMV resistance sources and QTLs in pepper within the previous studies have significantly contributed to the effective control of the disease. However, many CMV resistance-related issues still need to be unraveled. The levels of CMV resistance in known disease resistance resources are still insufficient and mostly partial resistance. Hence, it is requiring us to screen new and more desirable sources to diversify the genetic basis of the resistance. Recently several new sources with good levels of CMV resistance have been identified within C. annuum, which have been utilized for developing peppers with improved resistance (Rahman et al., 2016; Yu et al., 2019). It is suggesting the possibility of developing commercially accepted cultivars with broad spectrum and durable resistance. The development of markers linked to resistance loci is the most convenient means for CMV resistance breeding. Advances in NGS technologies, genome-wide association studies (GWAS), high-throughput SNP genotyping and fine mapping of resistance genes or QTLs facilities the development of reliable markers, map-based cloning and accurate transfer to elite lines via MAS or GWS in pepper. The genetic basis of resistance to CMV is quite complex, involving multiple genes, recessive or partially dominant. The mechanism of resistance to CMV in pepper still stays at observing the virus migration, viral multiplication and defense-related enzyme metabolism. There is still a lack of understanding of the resistance mechanisms at the molecular level. Thus, it is necessary to deep the study of the interaction between virus and host, clarifying the host gene-mediated virus resistance mechanisms and signaling in pepper. However, molecular and genetic studies are difficult in pepper, which hinders the identification of genes responsible for CMV resistance. So far, only CA02g19570 that control of CMV resistance in Capsicum was cloned, more resistance genes need to be identified. Remarkable technological achievements in the field of DNA sequencing will be important for identifying genes associated with CMV resistant traits. Genome editing provides great promise to improve crop productivity but relies on genetic transformation and plant regeneration, which are bottlenecks in the process (Altpeter et al., 2016; Xu et al., 2019). However, efficient protocol for transformation and regeneration from tissue culture remain arduous for pepper, successful genetic transformation of pepper is strongly dependent on the genotype. The transformation and regeneration system should be further optimized in pepper. Recently, Maher et al. (2020) described new methods to generate gene-edited dicotyledonous plants through de novo meristem induction, that could overcome the difficult of transformation and regeneration in plant gene editing.
Summarize, future efforts to identify new and more desirable sources of resistance and new resistance genes and QTLs are imperative. With the advent of novel technologies, precise markers and approaches can facilitate the progress for resistant breeding. Preliminary results from these studies have been encouraging, suggesting the possibility of developing commercially accepted cultivars with good levels of CMV resistance.
Author Contributions
NL and CY: writing and editing. YY and SG: literature collection. FW: project administration. CJ and MY: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Mogo Internet Technology Co., Ltd., for providing the language editing services.
Funding. This research was funded by National Key R&D Program of China, grant numbers: 2017YFD0101903 and 2016YFE0205500, National Natural Science Foundation of China, grant number: 31401868, Natural Science Foundation of Hubei Province, grant number: 2019CFB577, and China Agriculture Research System, grant number: CARS-23-G28.
References
- Agrios G. N., Walker M. E., Ferro D. N. (1985). Effect of cucumber mosaic virus inoculation at successive weekly intervals on growth and yield of pepper (Capsicum annuum) plants. Plant Dis. 69 52–55. 10.1094/PD-69-52 [DOI] [Google Scholar]
- Altpeter F., Springer N. M., Bartley L. E., Blechl A. E., Brutnell T. P., Citovsky V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28 1510–1520. 10.1105/tpc.16.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilla C., Collar J. L., Duque M., Perez P., Fereres A. (1997). Impact of floating rowcovers on bell pepper yield and virus incidence. Hortscience 32 882–883. 10.21273/HORTSCI.32.5.882 [DOI] [Google Scholar]
- Bansal R. O., Aulakh R. K., Hundal J. S. (1992). “Reaction of different genotypes of pepper (Capsicum annuum) to cucumber mosaic virus,” in Proceedings of the EUCARPIA VIIIth Meeting on Genetics and Breeding on Capsicum and Eggplant, Rome, 132–137. [Google Scholar]
- Barrios E. P., Mosokar H. I., Black L. L. (1971). Inheritance of resistance to tobacco etch and cucumber mosaic viruses in Capsicum frutescens. PHYTAJ 61:1318 10.1094/Phyto-61-1318 [DOI] [Google Scholar]
- Blevins T., Rajeswaran R., Shivaprasad P. V., Beknazariants D., Si-Ammour A., Park H. S., et al. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34 6233–6246. 10.1093/nar/gkl886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G., Voinnet O., Li W., Ji L., Ding S., Baulcombe D. C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17 6739–6746. 10.1093/emboj/17.22.6739 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cai W., Fang R., Shang H., Wang X., Zhang F., Li Y., et al. (2003). Development of CMV-and TMV-resistant chili pepper: field perfermance and biosafety assessment. Mol. Breeding 11 25–35. 10.1023/A:1022655204552 [DOI] [Google Scholar]
- Caranta C., Palloix A., Lefebvre V., Daubeze A. M. (1997). QTLs for a component of partial resistance to cucumber mosaic virus in pepper: restriction of virus installation in host-cells. Theor. Appl. Genet. 94 431–438. 10.1007/s001220050433 [DOI] [Google Scholar]
- Caranta C., Pflieger S., Lefebvre V., Daubèze A. M., Thabuis A., Palloix A. (2002). QTLs involved in the restriction of cucumber mosaic virus (CMV) long-distance movement in pepper. Theor. Appl. Genet. 104 586–591. 10.1007/s001220100753 [DOI] [PubMed] [Google Scholar]
- Chaim A. B., Grube R. C., Lapidot M., Jahn M., Paran I. (2001a). Identification of quantitative trait loci associated with resistance to cucumber mosaic virus in Capsicum annuum. Theor. Appl. Genet. 102 1213–1220. 10.1007/s001220100581 [DOI] [Google Scholar]
- Chaim A. B., Paran I., Grube R. C., Jahn M., Wijk R. V., Peleman J. (2001b). QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 102 1016–1028. 10.1007/s001220000461 [DOI] [Google Scholar]
- Chen J., Chai L., Li Q., Feng M. (2000). Relationship between the rampancy of cucumber mosaic virus and climate warming and counter-measures. Ecoagriculture Res. 8 23–26. 10.1016/j.ajem.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Choi S., Lee J., Kang W., Kim J., Huy H. N., Park S., et al. (2018). Identification of cucumber mosaic resistance 2 (cmr2) that confers resistance to a new cucumber mosaic virus isolate P1 (CMV-P1) in pepper (Capsicum spp.). Front. Plant Sci. 9:1106. 10.3389/fpls.2018.01106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M., Caciagli P., Casetta A. (1979). Infection sources and aphid vectors in relation to the spread of cucumber mosaic virus in pepper crops. Phytopath. Medit. 18 123–128. 10.2307/42684442 [DOI] [Google Scholar]
- Ding S. W., Anderson B. J., Haase H. R., Symons R. H. (1994). New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198 593–601. 10.1006/viro.1994.1071 [DOI] [PubMed] [Google Scholar]
- Dogimont C., Daubeze A. M., Palloix A. (1994). Expression of resistance to CMV migration in pepper seedlings. J. Phytopathol. 141 209–216. 10.1111/j.1439-0434.1994.tb01463.x [DOI] [Google Scholar]
- Doolittle S. P. (1916). A new infectious mosaic disease of cucumber. PHYTAJ 6 145–147. [Google Scholar]
- Dufour O., Palloix A., Selassie K. G., Pochard E., Marchoux G. (1989). The distribution of cucumber mosaic virus in resistant and susceptible. Can. J. Bot. 67 655–660. 10.1139/b89-088 [DOI] [Google Scholar]
- Edwardson J. R., Christie R. G. (1986). “Viruses infecting Forage Legumes,” in Viruses Infecting Forage Legumes, eds Edwardson J. R., Christie R. G. (Gainesville: Florida Agric. Expt. Sta; ). [Google Scholar]
- Feng L., Cai S., Tian R. (1986). Method of inoculation of virus on tomato and pepper. Plant Protection 12 24–25. [Google Scholar]
- Florini D. A., Zitter T. A. (1987). Cucumber mosaic virus (CMV) in pepper (Capsicum annuum L.) in New York and associated yield losses. PHYTAJ 77:652. [Google Scholar]
- Fulton J. P. (1950). Studies on strains of cucumber virus 1 from spinach. Phytopathology 40 729–736. [Google Scholar]
- Fusaro A. F., Matthew L., Smith N. A., Curtin S. J., Dedic-Hagan J., Ellacott G. A., et al. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7 1168–1175. 10.1038/sj.embor.7400837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Wang Y., Zhang C., Zhang A., Zhu X. (2016). Investigation and pathogen preliminary identification of pepper virus disease in Tianjin. Shandong Agricult. Sci. 48 91–94. 10.14083/j.issn.1001-4942.2016.03.023 [DOI] [Google Scholar]
- Garcia-Ruiz H., Murphy J. F. (2011). Age-related resistance in bell pepper to cucumber mosaic virus. Ann. Appl. Biol. 139 307–317. 10.1111/j.1744-7348.2001.tb00144.x [DOI] [Google Scholar]
- Goldbach R. (1987). Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 4 197–202. [PubMed] [Google Scholar]
- Green S. K., Kim J. S. (1991). ”Characteristics and Control of Viruses Infecting Peppers: a Literature Review”, Technical Bulletin No.18. Taipei: Asian Vegetable Research and Development Center. [Google Scholar]
- Green S. K., Kim J. S. (1994). Sources of Resistance to Viruses of Pepper (Capsicum spp.): a Catalog”, Technical Bulletin No.20. Taipei: Asian Vegetable Research and Development Center. [Google Scholar]
- Grube R. C., Zhang Y. P., Murphy J. F., Loaiza-Figueroa F., Lackney V. K., Provvidenti R., et al. (2000). New source of resistance to cucumber mosaic virus in Capsicum frutescens. Plant Dis. 84 885–891. 10.1094/PDIS.2000.84.8.885 [DOI] [PubMed] [Google Scholar]
- Guo G., Wang S., Liu J., Pan B., Diao W., Ge W., et al. (2017). Rapid identification of QTLs underlying resistance to cucumber mosaic virus in pepper (Capsicum frutescens). Theor. Appl. Genet. 130 41–52. 10.1007/s00122-016-2790-3 [DOI] [PubMed] [Google Scholar]
- Guo J. (1994). A new disease-resistant sweet pepper variety recently bred in China. Technol. Plant Protection Popularization 1 36–36. [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. 10.1126/science.286.5441.950 [DOI] [PubMed] [Google Scholar]
- Herison C., Rustikawati, Sudarsono (2014). Genetics of resistance against cucumber mosaic virus (CMV) in hot pepper (Capsicum annuum L.). Capsicum Eggplant Newslett. 23 113–116. [Google Scholar]
- Hollings M., Stone O. M., Brunt A. A. (1967). Cucumber mosaic virus. Glasshouse Crops Res. Inst. Ann. Rept. 95–98. [Google Scholar]
- Ito Y., Nishizawa-Yokoi A., Endo M., Mikami M., Toki S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467 76–82. 10.1016/j.bbrc.2015.09.117 [DOI] [PubMed] [Google Scholar]
- Jacquemond M. (2012). Cucumber mosaic virus. Adv. Virus Res. 84 439–504. 10.1016/B978-0-12-394314-9.00013-0 [DOI] [PubMed] [Google Scholar]
- Jaganathan D., Ramasamy K., Sellamuthu G., Jayabalan S., Venkataraman G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9:985. 10.3389/fpls.2018.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger I. C. (1916). Experiments with the cucumber mosaic disease. PHYTAJ 6 148–151. [Google Scholar]
- Joshi R. D., Dubey L. N. (1973). Assessment of losses due to CMV on chili. Sci. Cult. 39 521–522. [Google Scholar]
- Juan O., Jaime E. A., Ricardo R., Paulina S. R. (2006). Datura genus weeds as an epidemiological factor of alfalfa mosaic virus (AMV), cucumber mosaic virus (CMV), and potato virus y (PVY) on Solanaceus crops. Chilean J. Agricult. Res. 66 333–341. [Google Scholar]
- Kang W. H., Hoang N. H., Yang H. B., Jinkyung K., Sunghwan J., Jangkyun S., et al. (2010). Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor. Appl. Genet. 120 1587–1596. 10.1007/s00122-010-1278-9 [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Waterworth H. E. (1981). Handbook of Plant Virus Infections and Comparative Diagnosis, ed. Kurstak E. (North-Holland, NY: Elsevieri; ), 257–332. [Google Scholar]
- Kazinczi G., Horváth J., Takács A. P., Gáborjányi R., Béres I. (2004). Experimental and natural weed host-virus relations. Commun. Agric. Appl. Biol. Sci. 69 53–60. [PubMed] [Google Scholar]
- Kenyon L., Kumar S., Tsai W. S., Hughes J. D. A. (2014). Virus diseases of peppers (Capsicum spp.) and their control. Adv. Virus Res. 90 297–354. 10.1016/B978-0-12-801246-8.00006-8 [DOI] [PubMed] [Google Scholar]
- Kim H., Yoon J. B., Lee J. (2017). Development of Fluidigm SNP type genotyping assays for marker-assisted breeding of chili pepper (Capsicum annuum L.). Hortic. Sci. Technol. 35 465–479. 10.12972/kjhst.20170050 [DOI] [Google Scholar]
- Kim S., Lee S., Kim B., Paek K. (1997). Satellite-RNA-mediated resistance to cucumber mosaic virus in transgenic plants of hot pepper (Capsicum annuum cv. Golden Tower). Plant Cell Rep. 16 825–830. 10.1007/s002990050328 [DOI] [PubMed] [Google Scholar]
- Lapidot M., Paran I., Benjoseph R., Benharush S., Pilowsky M., Cohen S., et al. (1997). Tolerance to cucumber mosaic virus in pepper: development of advanced breeding lines and evaluation of virus level. Plant Dis. 81 185–188. 10.1094/PDIS.1997.81.2.185 [DOI] [PubMed] [Google Scholar]
- Lecoq H., Pochard E., Pitrat M. L., Marchoux G. (1982). Identification et exploitation de résistances aux virus chez les plantes maraîchères. Cryptog. Mycol. 3 333–345. [Google Scholar]
- Lee Y., Jung M., Shin S., Lee J., Choi S., Her N., et al. (2009). Transgenic peppers that are highly tolerant to a new CMV pathotype. Plant Cell Rep. 28 223–232. 10.1007/s00299-008-0637-3 [DOI] [PubMed] [Google Scholar]
- Li G., Qian W., Zhang S., Zhang S., Sun R. (2016). Development of gene-based markers for the Turnip mosaic virus resistance gene retr02 in Brassica rapa. Plant Breeding 135 466–470. 10.1111/pbr.12372 [DOI] [Google Scholar]
- Li N., Yin Y., Wang F., Yao M. (2018). Construction of a high-density genetic map and identification of QTLs for cucumber mosaic virus resistance in pepper (Capsicum annuum L.) using specific length amplified fragment sequencing (SLAF-seq). Breed. Sci. 68 233–241. 10.1270/jsbbs.17063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Fu D., Zhu B., Luo Y., Zhu H. (2018). CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 94 513–524. 10.1111/tpj.13872 [DOI] [PubMed] [Google Scholar]
- Li S. (1995). Advances in Main Vegetable Crops Breeding for Diseases Resistance in China. Beijing: Science Press. [Google Scholar]
- Li X., He Q. (2000). Identification and evaluation of virus-resistant materials of hot (sweet) pepper. Hunan Agricult. Sci. 3 49–50. 10.3969/j.issn.1006-060X.2000.03.028 [DOI] [Google Scholar]
- Lockhart B. E. L., Fischer H. U. (1976). Cucumber mosaic virus infections of pepper in Morocco. Plant Dis. Rep. 60 262–264. [Google Scholar]
- Lot H., Marrou J., Quiot J. B., Esvan C. (1972). Contribution a I’etude du virus de la mosaique du comcombre (CMV). II. Method de purification rapide du virus. Ann. Phytopathol. 4 25–38. [Google Scholar]
- Maher M. F., Nasti R. A., Vollbrecht M., Starker C. G., Clark M. D., Voytas D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38 84–89. 10.1038/s41587-019-0337-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A., Geng S., Yan X., Chen B. (2004). Identification of TMV, CMV and blight resistant materials of hot (sweet) Pepper. Yangtze River Vegetables 48–50. 10.3865/j.issn.1001-3547.2004.02.033 [DOI] [Google Scholar]
- Mazourek M., Moriarty G., Glos M., Fink M., Kreitinger M., Henderson E., et al. (2009). ’Peacework’: a cucumber mosaic virus-resistant early red bell pepper for organic systems. Hortscience 44 1464–1467. 10.21273/HORTSCI.44.5.1464 [DOI] [Google Scholar]
- Mihailova B., Stoimenova E., Petrov P., Stoynova-Bakalova E. (2013). Movement of cucumber mosaic virus in resistant pepper lines. Comp. Rendus l’Acadeìmie Bulgare Sci. 66 911–918. 10.7546/CR-2013-66-6-13101331-19 [DOI] [Google Scholar]
- Min H. E., Han J. H., Yoon J. B., Lee J. (2016). QTL mapping of resistance to the cucumber mosaic virus P1 strain in pepper using a genotyping-by-sequencing analysis. Hortic. Environ. Biotechnol. 57 589–597. 10.1007/s13580-016-0128-3 [DOI] [Google Scholar]
- Min W. K., Jaehwang R., Suhyeon A. (2014). Developmental changes of recessive genes-mediated cucumber mosaic virus (CMV) resistance in peppers (Capsicum annuum L.). Korean J. Hortic. Sci. Technol. 32 235–240. 10.7235/hort.2014.13092 [DOI] [Google Scholar]
- Mochizuki T., Ohki S. T. (2005). Amino acid 129 in the coat protein of cucumber mosaic virus primarily determines invasion of the shoot apical meristem of tobacco plants. J. Gen. Plant Pathol. 71 326–332. 10.1007/s10327-005-0207-2 [DOI] [Google Scholar]
- Monma S., Sakata Y. (1996). Screening of Capsicum accessions for resistance to cucumber mosaic virus. J. Japan Soc. Hortic. Sci. 65 769–776. 10.2503/JJSHS.65.769 [DOI] [Google Scholar]
- Moury B., Verdin E. (2012). Viruses of pepper crops in the Mediterranean Basin: A remarkable stasis. Adv Virus Res 84 127–162. 10.1016/b978-0-12-394314-9.00004-x [DOI] [PubMed] [Google Scholar]
- Naresh P., Reddy M. K., Reddy P. H. C., Reddy K. M. (2016). Screening chilli (Capsicum spp.) germplasm against cucumber mosaic virus and chilli veinal mottle virus and inheritance of resistance. Eur. J. Plant Pathol. 146 1–14. 10.1007/s10658-016-0930-x [DOI] [Google Scholar]
- Nono-Womdim R., Marchoux G., Pochard E., Palloix A., Gebre-Selassie K. (1991). Resistance of pepper lines to the movement of cucumber mosaic virus. J. Phytopathol. 132 21–32. 10.1111/j.1439-0434.1991.tb00090.x [DOI] [Google Scholar]
- Nono-Womdim R., Palloix A., Gebre-Selassie K., Marchoux G. (1993). Partial resistance of bell pepper to cucumber mosaic virus movement within plants: field evaluation of its efficiency in Southern France. J. Phytopathol. 137 125–132. 10.1111/j.1439-0434.1993.tb01332.x [DOI] [Google Scholar]
- Palukaitis P., Garcíaarenal F. (2003). Cucumoviruses. Adv. Virus Res. 62 241–323. 10.1016/S0065-3527(03)62005-1 [DOI] [PubMed] [Google Scholar]
- Palukaitis P., Roossinck M. J., Dietzgen R. G., Francki R. I. B. (1992). Cucumber mosaic virus. Adv. Virus Res. 41 281–348. 10.1016/S0065-3527(03)62005-1 [DOI] [PubMed] [Google Scholar]
- Pares R. D., Gunn L. V. (1989). The role of non-vectored soil transmission as a primary source of infection by pepper mild mottle and cucumber mosaic viruses in glasshouse-grown Capsicum in Australia. J. Phytopathol. 126 353–360. 10.1111/j.1439-0434.1989.tb04498.x [DOI] [Google Scholar]
- Park S. W., Hoang N. H., Kang W. H., Schrieck M., Lee H. R., Choi D., et al. (2011). “Development of molecular markers for a single recessive resistance gene against CMV in pepper (Capsicum annuum L.),” in Proceedings of the International Symposium Commemorating the 20th Anniversary of Korea Capsicum Research Cooperative: Pepper Disease and Resistance Breeding, (Cheongju: Chungbuk National University; ). [Google Scholar]
- Pinaki A. (1999). Screening of chilli germplasm against strains of cucumber mosaic virus. Environ. Ecol. 17 484–487. [Google Scholar]
- Pochard E. (1977). “Methods for the study of partial resistance to cucumber mosaic virus in pepper,” in Capsicum 77, Proceedings of 3rd EUCARPIA Meeting, Capbreton, 99–104. 10.5423/rpd.2015.21.2.099 [DOI] [Google Scholar]
- Pochard E. (1982). A major gene with quantitative effect on two different viruses: CMV and TMV. Capsicum Eggplant Newslett. 1 54–56. [Google Scholar]
- Pochard E., Daubèze A. (1989). “Progressive construction of a polygenic resistance to cucumber mosaic virus in the pepper,” in Proceedings of Eucarpia meeting on genetics and breeding of Capsicum and Eggplant, 7th), Kragujevac, 187–192. [Google Scholar]
- Pochard E., Dumas D. V. R., Florent A. (1983). Linkage between partial resistance to CMV and susceptibility to TMV in the line Perennial: analysis on androgenetic homozygous lines. Capsicum Eggplant Newslett. 2 34–35. [Google Scholar]
- Pooggin M. M. (2018). Small RNA-omics for plant virus identification, virome reconstruction, and antiviral defense characterization. Front. Microbiol. 9:2779. 10.3389/fmicb.2018.02779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyott D. E., Sheehan E., Molnar A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17 1276–1288. 10.1111/mpp.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. S., Akanda A. M., Mian I. H., Bhuiyan M. K. A., Hossain M. M. (2016). New sources of resistance to Cucumber mosaic virus in Capsicum annuum. J. Crop Sci. Biotechnol. 19 249–258. 10.1007/s12892-016-0044-1 [DOI] [Google Scholar]
- Rast A. T. B., Stijger C. C. M. M. (1987). Disinfection of pepper seed infected with different strains of capsicum mosaic virus by trisodium phosphate and dry heat treatment. Plant Pathol. 36 583–588. 10.1111/j.1365-3059.1987.tb02277.x [DOI] [Google Scholar]
- Reddy M. K., Sadhashiva A. T., Reddy K. M., Chalam C., Deshpande A. A., Chandro A. (2001). “Integrated disease and pest management: leaf curl and other viruses of tomato and peppers,” in Proceedings of the South Asia Vegetable Research Network, Tainan, 3–8. [Google Scholar]
- Ridzuan R., Rafii M. Y., Ismail S. I., Yusoff M. M., Miah G., Usman M. G. (2018). Breeding for anthracnose disease resistance in chili: progress and prospects. Int. J. Mol. Sci. 19:3122. 10.3390/ijms19103122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohini N., Lakshmanan V. (2017). Evaluation studies of hot pepper hybrids (Capsicum annuum L.) for yield and quality characters. Electronic J. Plant Breeding 8 643–651. 10.5958/0975-928X.2017.00098.9 [DOI] [Google Scholar]
- Roossinck M. J., Zhang L., Hellwald K. H. (1999). Rearrangements in the 5’ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 73 6752–6758. 10.1128/jvi.73.8.6752-6758.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusco J., Csillery G. (1980). “Selection for CMV resistance in pepper by the method developed by Pochard,” in Proceedings of the IVth Eucarpia Capsicum Meeting, Wageningen, 37–39. [Google Scholar]
- Scott H. (1963). Purification of cucumber mosaic virus. Virology 20 103–106. 10.1016/0042-6822(63)90144-2 [DOI] [PubMed] [Google Scholar]
- Shifriss C., Cohen S. (1987). A progress in breeding for resistance to CMV in pepper. Capsicum Eggplant Newslett. 6:60. [Google Scholar]
- Shin J., Xu S., Kim J., Woo J., Kim H., Park Y., et al. (2013). CMV-P1 resistance evaluation using enzyme-linked immunosorbent assay of pepper genetic sources (Capsicum spp.). Korean J. Hortic. Sci. Technol. 31 764–771. 10.7235/hort.2013.13021 [DOI] [Google Scholar]
- Singh J., Thakur M. R. (1977). “Genetics of resistance to tobacco mosaic virus, cucumber mosaic virus and leaf curl virus in hot pepper (Capsicum annuum L.),” in Proceedings of the Capsicum 77. 3rd Eucarpia Meeting, Montfavet, 119–123. [Google Scholar]
- Sivakumaran K., Bao Y., Roossinck M. J., Kao C. C. (2000). Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of brome mosaic virus and cucumber mosaic virus. J. Virol. 74 10323–10331. 10.1128/JVI.74.22.10323-10331.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran K., Chen M., Roossinck M. J., Kao C. C. (2002). Core promoter for initiation of cucumber mosaic virus subgenomic RNA4A. Mol. Plant Pathol. 3 43–52. 10.1046/j.1464-6722.2001.00089.x [DOI] [PubMed] [Google Scholar]
- Sugiura M., Bandaranayake C. M., Hemachandra G. H. (1975). Chilli virus disease in Sri Lanka. Japan: Tropical Agriculture Research Center, Ministry of Agriculture and Forestry. [Google Scholar]
- Sun X., Lei J., Zhou S., Chen G., Cao B., Liu A. (2008). Identification of pepper CMV Guangzhou isolate and screening of pepper material resistance. China Vegetables 3 11–14. [Google Scholar]
- Suzuki K., Kuroda T., Miura Y., Murai J. (2003). Screening and field trials of virus resistant sources in Capsicum spp. Plant Dis. 87 779–783. 10.1094/PDIS.2003.87.7.779 [DOI] [PubMed] [Google Scholar]
- Thomson M. J. (2014). High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotech. 2 195–212. 10.9787/PBB.2014.2.3.195 [DOI] [Google Scholar]
- Tian R., Fang L., Cai S., Guo J., Li P. (1989). Identification of resistance of sweet (hot) pepper varieties (lines) to cucumber mosaic virus and tobacco mosaic virus at seedling stage. Crop Variety Resour. 4 32–33. [Google Scholar]
- Tien P. O., Zhang X., Qiu B., Qin B., Gusui W. U. (1987). Satellite RNA for the control of plant diseases caused by cucumber mosaic virus. Ann. Appl. Biol. 111 143–152. 10.1111/j.1744-7348.1987.tb01441.x [DOI] [Google Scholar]
- Troutman J. L., Fulton R. W. (1958). Resistance in tobacco to cucumber mosaic virus. Virology 6 303–316. 10.1016/0042-6822(58)90084-9 [DOI] [PubMed] [Google Scholar]
- Van Eck J. (2018). Genome editing and plant transformation of solanaceous food crops. Curr. Opin. Biotech. 49 35–41. 10.1016/j.copbio.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Wang K. H., Uchida J. (2014). Plant Disease Prevention and Management in Sustainable Agricultural Systems. Sustainable Horticultural Systems. Berlin: Springer International Publishing. [Google Scholar]
- Wang L., Ma Y., Zhang B. (2019). Market demand and breeding trend of pepper varieties in China. China Vegetables 8 1–4. 10.1079/9781786393814.0001 [DOI] [Google Scholar]
- Wang S., Wu X., Liu J., Pan B. (2009). The ISSR markers linked to CMV resistant gene in hot pepper. Mol. Plant Breed. 7 569–572. [Google Scholar]
- Wang S., Yuan X., Zou X., Ma Y., Li H., Yin D. (2001). Evaluation of excellent pepper germplasm resources in China. Jiangsu J. Agricult. 17 244–247. [Google Scholar]
- Wang X. (2016). QTL Analysis of Cucumber Mosaic Virus Resistance in Pepper. Master thesis, Chinese Academy of Agricultural Sciences, Beijing. [Google Scholar]
- Waterhouse P. M., Helliwell C. A. (2003). Exploring plant genomes by RNA-induced gene silencing. Nat. Rev. Genet. 4 29–38. 10.1038/nrg982 [DOI] [PubMed] [Google Scholar]
- Watterson J. (1993). Development and Breeding of Resistance to Pepper and Tomato Viruses. Oregon: Timber Press, Portland. [Google Scholar]
- Wu X., Wang S., Cao P. (2006). Progresses on resistance of cucumber mosaic virus in pepper. J. China Capsicum 4–9. [Google Scholar]
- Xu J., Hua K., Lang Z. (2019). Genome editing for horticultural crop improvement. Hortic Res. 6:113. 10.1038/s41438-019-0196-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Ju L. R., Xu X. R., Cui Y. H. (1996). Genetic analysis on resistance of sweet (hot) pepper to CMV. Acta Horticult. Sin. 23 45–48. [Google Scholar]
- Yang H., Shen H., Zhu X., Cheng J., Han Q., Xin X. (2006). A study on the relationship between the resistance to CMV of hot pepper and related defendant enzyme activity, the content of lignins, and total phenols. Chinese Agricult. Sci. Bull. 22 369–373. [Google Scholar]
- Yang Y., Yan S., Wang H., Chen Z. (1992). Screening and application of differential host of CMV strain on pepper. Virol. Sin. 7 317–327. [Google Scholar]
- Yang Z., Huang Y., Yang J., Yao S., Zhao K., Wang D. (2020). Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28 89–103. 10.1016/j.chom.2020.05.001 [DOI] [PubMed] [Google Scholar]
- Yao M. (2013). Inheritance and QTL Analysis for Resistance to Cucumber Mosaic Virus in Pepper. Doctoral thesis, Huazhong Agricultural University, Wuhan. [Google Scholar]
- Yao M., Li N., Wang F., Ye Z. (2013). Genetic analysis and identification of QTLs for resistance to cucumber mosaic virus in chili pepper (Capsicum annuum L.). Euphytica 193 135–145. 10.1007/s10681-013-0953-8 [DOI] [Google Scholar]
- Yasuo K. (1963). Studies on cucumber mosaic virus. J. Phytopathol. 28 131–138. 10.3186/jjphytopath.23.235 [DOI] [Google Scholar]
- Young N. D., Tanksley S. D. (1989). RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor. Appl. Genet. 77 353–359. 10.1007/bf00305828 [DOI] [PubMed] [Google Scholar]
- Yu H., Zhang Z., Cao Y., Zhang B., Wang L. (2019). Progress of resistance to cucumber mosaic virus in pepper. Acta Hortic. Sin. 46 1813–1824. [Google Scholar]
- Yu X., Kong Q. G., Wang Y. L. (2000). Inheritance of resistance to cucumber mosaic virus pepper isolate (CMV-P) in Capsicum annum L. J. Shenyang Agricult. Univ. 31 169–171. [Google Scholar]
- Zhang G., Guo G., Pan B., Diao W., Liu J., Ge W., et al. (2020). Functional identification of candidate genes of resistance to cucumber mosaic virus in pepper. Mol. Plant Breed. 18 5537–5543. [Google Scholar]
- Zhang X., Geng S., Chen B., Sun H. (2016). Molecular Marker genSSR3000 Closely Linked to Pepper CMV Resistance Gene and its Acquisition and Application. China patent application 11. [Google Scholar]
- Zhang Z. (2009). Study on Pepper Virus Disease. Doctoral thesis, Hunan Agricultural University, Changsha. [Google Scholar]
- Zhao J., Wang L., Yun X., Zhang B. (2009). Review and prospect of genetic breeding on CMV resistance in pepper. China Vegetables 1 1–7. [Google Scholar]
- Zhao J., Wang L. H., Mao S. L., Zhang Z. H., Yun X. F., Zhang B. X. (2011). QTL analysis of cucumber mosaic virus resistance in pepper (Capsicum annuum L.). China Vegetables 1 30–35. [Google Scholar]
- Zhao Z., Shi L., Tan G., Yan X. (2004). Study on pepper (Capsicum annuum L.) virus disease and its distribution in the Guanzhong area of Shaanxi province. Sci. Agricult. Sin. 37 1738–1742. [Google Scholar]
- Zhou X., Pu Z., Fang Z. (1994). Non-vectored soil transmission of cucumber mosaic virus. J. Nanjing Agricult. Univ. 17 39–42. [Google Scholar]
- Zhu C., Li X., Zheng J. (2018). Transcriptome profiling using Illumina- and SMRT-based RNA-seq of hot pepper for in-depth understanding of genes involved in CMV infection. Gene 666 123–133. 10.1016/j.gene.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Zitter T. A., Murphy J. F. (2009). Cucumber mosaic. Plant Health Instructor 10.1094/PHI-I-2009-0518-01 [DOI] [Google Scholar]
- Zou X. (2005). Studies on Inheritance of Main Quantitative Characters and Relative Mechanism of Male Sterility in Capsicum. Doctoral thesis, Nanjing Agricultural University, Nanjing. [Google Scholar]
- Zou X., Hou X., Chen W., Liu R., Zhang Z., Ma Y., et al. (2004). Diallel crossing analyses of resistances to main diseases in pepper (Capsicum annuum L.). Sci. Agricult. Sin. 37 1636–1640. [Google Scholar]