Abstract
Pseudomonas aeruginosa is one of the most common reasons for nosocomial infections. Given the high morbidity and mortality, as well as the cost of management, particularly in developing countries, burn injuries are considered important health concerns. Owing to the increased rate of resistance against antibiotics, this study aimed to isolate Pseudomonas aeruginosa strains from burn patient's wounds by analyzing antibiotic susceptibility and genetic profiling. In this regard, we explored the relationship between the nucleotide sequence and antibiotic susceptibility. In this cross-sectional study, 107 isolates of P. aeruginosa were collected from a major burn center in Tehran, Iran. The isolates were characterized with standard biochemical tests and examined by applying the Disk Diffusion method to find the patterns of sensitivity, and their genetic relationship was revealed by RAPD-PCR method. According to the antibiogram results, most of the isolates were resistant to 3 or more antibiotics tested and the most sensitivity was related to the Colistin antibiotic. RAPD-PCR method revealed a high polymorphism among P. aeruginosa isolates in Tehran. There was no significant association between the genotype groups and antibiotic susceptibility profiles. We evaluated the pattern of resistance to pathogenic organisms and identified multi-drug resistant organisms. Currently, Colistin antibiotic is the most suitable treatment option for burned patients. RAPD-PCR is a genotyping method with high efficiency for typing and categorizing different isolates of MDR-P. aeruginosa.
Keywords: Biotechnology, Genetics, Microbiology, Molecular biology, Infectious disease, Pseudomonas aeruginosa, RAPD-PCR, Nosocomial infections, Burn patients
Biotechnology; Genetics; Microbiology; Molecular Biology; Infectious disease; Pseudomonas aeruginosa; RAPD-PCR; Nosocomial infections; Burn patients.
1. Introduction
Pseudomonas aeruginosa is a life-threatening opportunistic bacterium found in hospitalized patients (Sorkh et al., 2017). In fact, P. aeruginosa causes nosocomial infections in immunosuppressed patients because of some diseases such as cystic fibrosis, neutropenia, especially burns (Diken Gur and Aksoz, 2016). This pathogen is the main reason for infections in burn patients owing to the destruction of skin as the first line of the innate immunity. In addition, it constitutes approximately 77% of mortality in burn patients during the past 25 years (Mahmmudi et al., 2016). Tissue injury due to burn that is caused by thermal energy is one of the most painful events suffered by people worldwide. Numerous subjects experience burn injuries and thousands of which ultimately demise annually (Mehta et al., 2017). Although therapeutic approaches for burns have greatly developed, infections remain one of the major causes of death in these patients, particularly critically-ill burn patients. Compared with other hospitalized individuals, burn patients are characterized by skin shortage, long hospital stays, and multiple invasive operations; therefore, they are more susceptible to infection (Dou et al., 2017). Multidrug-resistant (MDR) P. aeruginosa causes 4–60% of nosocomial infections, generating high mortality and morbidity in burn patients. Owing to its intrinsic resistance to a wide range of antibiotics, practically its capability of resisting to all effective antibiotics, P. aeruginosa is considered one of the major concerns for nosocomial infections in hospitals in recent years (Sorkh et al., 2017). The reasons for the emergence of MDR strains may be the use of antibiotics having high anti-pseudomonal activity and long-term antibiotic administration (Diken Gur and Aksoz, 2016).
Molecular methods, which are less influenced by environmental factors in comparison to phenotypic methods, play pivotal roles in probing the ways of pathogen transmission (Mahmmudi et al., 2016). The diversity of P. aeruginosa strains has been broadly investigated by molecular typing methods, including ribotyping, repetitive-element-based Polymerase Chain Reaction (rep-PCR), Arbitrarily Primed-PCR (AP- PCR), Amplified Fragment Length Polymorphism (AFLP), Restriction Fragment Length Polymorphic (RFLP), Random Amplified Polymorphic DNA (RAPD) assay, and Pulsed-Field Gel Electrophoresis (PFGE) (Wolska et al., 2012). Nowadays, PCR-based fingerprinting methods have gained attraction in epidemiological studies because of their rapid nature, power of discrimination, and successful typing (Diken Gur and Aksoz, 2016). The major benefit of RAPD-PCR is that it does not require DNA pre-sequencing. The wide range of useful primers makes the technique as a great diagnostic approach. Reproducible RAPD bands are detectable by a precise selection of primers and optimization of PCR circumstances for target species and replication to confirm that only reproducible bands are scored (Kumari and Thakur, 2014). Among these methods, RAPD-PCR method is useful in long-term studies as it can show the differences in the whole genome, rather than a specific region in the bacterial genome (Sorkh et al., 2017). Indeed, RAPD method has the advantage that it can be performed cost-effectively and will totally amplify a range of fragments of most DNA and show polymorphisms (Kumari and Thakur, 2014).
2. Materials and methods
2.1. Isolation of P. aeruginosa
In this cross-sectional study, 107 P. aeruginosa isolates were collected from burn patients hospitalized in Shahid Motahhari Hospital in Tehran, Iran from March 2017 to August 2018. The sampling procedure from wounds consisted of swabs taken from clinically deep areas of the burn wounds when clinical signs of wound infection were evident. The ethics committee of Iran University of Medical Sciences approved the study before collecting the samples. The samples were immediately transferred to the laboratory. The isolates were identified as P. aeruginosa by the application of culture and standard biochemical tests for identification of P. aeruginosa as follows: Gram-negative, non-lactose fermenting, oxidase-positive colonies, which oxidized glucose and maltose, grew on cetrimide agar at 42 °C, and pigment production in Mueller Hinton Agar (HiMedia, India). Then P. aeruginosa isolates were preserved at -80 °C in Trypticase Soy Broth medium (Merck, 1.05459, Germany) containing 18% glycerol until further processing.
2.2. Antibiotic susceptibility
Drug susceptibility was determined by the Kirby-Bauer disk diffusion method (Padtan Teb, Iran) on Mueller Hinton agar (HiMedia, India), in accordance with Clinical and Laboratory Standards Institute (CLSI, 2017) guidelines. A total of six antibiotics were selected for assessing the drug resistance of P. aeruginosa including, Amikacin (AN, 30 mg), Ceftazidime (CAZ, 30 mg), Ciprofloxacin (CP, 5 mg), Colistin (CO, 10 mg), Gentamicin (GM, 10 mg), Imipenem (IPM, 10 mg). Pseudomonas aeruginosa ATCC 27853 was used as the laboratory standard for these tests.
2.3. Isolation of P. aeruginosa genomic DNA
Genomic DNA was extracted from overnight cultures of P. aeruginosa by boiling method. Template DNA was prepared and a cell pellet obtained from 1.5 ml of overnight culture was resuspended in 500 μl of TE (10 mM Tris, 1 mM EDTA, pH 8.0). After centrifugation and boiling for 10 min the supernatant was used for PCR (Galli et al., 1992; Sabath, 1980). The DNA quality was evaluated by calculating the absorbance of the specimen at 260 nm using a spectrophotometer.
2.4. RAPD analysis
DNA amplification was performed on a thermocycler (Bio-Rad, USA) in a final volume of 15 ml containing Taq DNA Polymerase Master Mix (7.5 ml), double distilled water (4.5 ml), Template DNA (2ml), primer 272 (3′-AGCGGGCCAA-5′) and primer 208 (3′-ACGGCCGACC-5′) (1 ml) [2,6], according to the following protocol: initial denaturation (94 °C for 5 min) followed by 35 cycles of denaturation (94 °C for 1min), annealing (39 °C for 1 min), extension (72 °C for 2 min), and a final cycle of extension at 72 °C for 10 min. The RAPD-PCR products were loaded on a 1.5% (w/v) agarose gel with 0.5 mg/ml of Green Viewer and were analyzed by gel electrophoresis and banding patterns were observed in Gel-Documentation system (Uvitec, UK). We used a one-kilobase DNA ladder (Fermentas, Canada) as a molecular size standard.
2.5. Statistical analysis
Statistical analyses were carried out with SPSS version 23.0 (SPSS Inc., Chicago, Illinois, USA). The normality of the data was evaluated by Kolmogorov–Smirnov test. In descriptive analysis, central indicators such as mean and percentage of frequency and coefficient of dispersion such as Standard Deviation (SD) were used. The one-sample Chi-square and Fisher's exact tests were used to assess significant intergroup variations. A value of P less than 0.05 was considered statistically significant. The RAPD fingerprints were analyzed both by an observer and also by computer with GelCompar II software.
3. Results
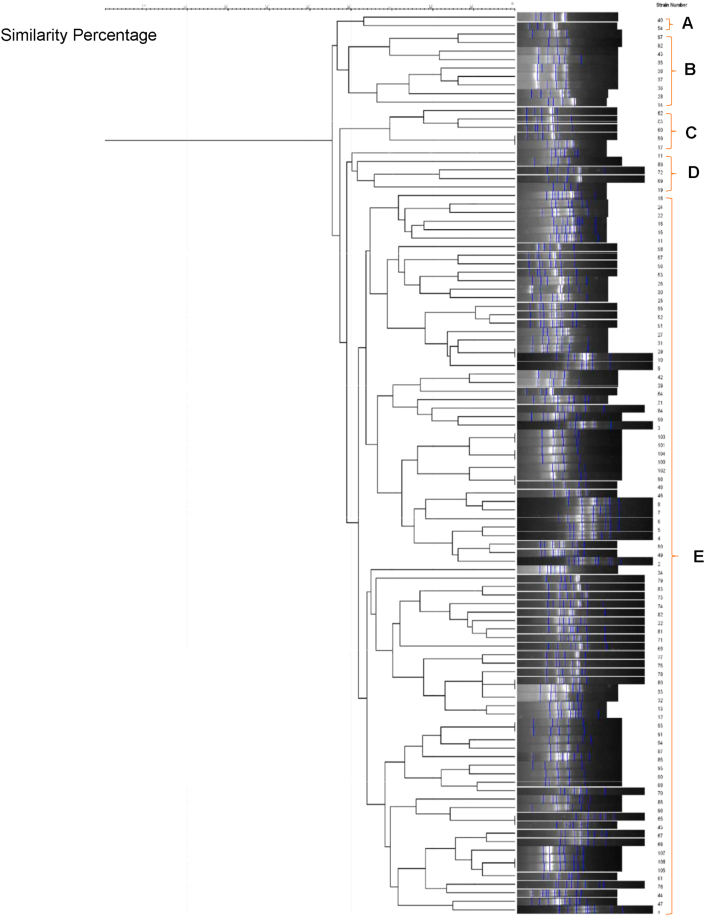
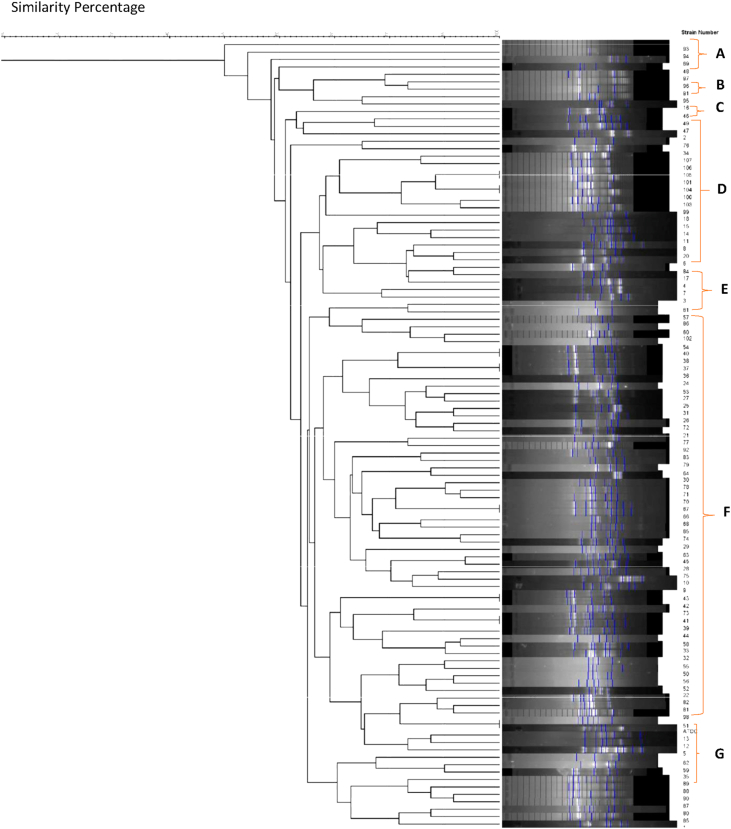
In the current study, 107 P. aeruginosa isolates were collected from the clinical wounds of the hospitalized burn patients, with different burn degrees from different wards of Motahhari Hospital. A total of 87.1% (n = 14) and 12.9% (n = 61) of P. aeruginosa strains were isolated from male and female patients with different burn percentages, respectively. The patients' ages ranged from 1 to 79 years, the majority (42.3%) of whom were between the ages of 20–40 years. The demographic and clinical characteristics of the burn patients (Table 1). In the following, 107 P. aeruginosa strains were typed phenotypically using antibiotic susceptibility test, and genotypically using the RAPD-PCR method. In this study, the strains were most frequently isolated from men's ward. According to the dendrogram obtained from GelCompar II software, the similarity percentage for primer 208 was considered to be 65% and accordingly, they were divided into 7 genotype groups (A-G) and for primer 272 (Figure 1), 60% was considered and accordingly, they were divided into 5 genotype groups (A-E) (Figure 2). No significant relationship was found between the genetic groups and the degree of burn and antibiotic resistance.
Table 1.
The demographic and clinical characteristics of the burn patients.
| Gender | Age | Cause of Burn | Burn Percentage | Burn Degree | |||||
|---|---|---|---|---|---|---|---|---|---|
| Male | 87.1% | Up to 10 years | 11.53% | Gas | 28.20% | <10% | 21.79% | Degree 1 | - |
| 10–20 years | 6.41% | Gasoline | 23.07% | 10–30% | 19.23% | ||||
| Female | 12.9% | 20–40 years | 42.30% | Electricity | 16.66% | 30–50% | 34.61% | Degree 2 | 34.54% |
| 40–60 years | 28.20% | Boiling water | 5.12% | 50–70% | 16.66% | ||||
| Over 60 years old | 11.53% | Alcohol | 3.84% | >70% | 7.69% | Degree 3 | 65.45% | ||
| Other | 23.07% | ||||||||
Figure 1.
Dendrogram of P. aeruginosa isolates based on RAPD-PCR Analysis (Primer 272).
Figure 2.
Dendrogram of P. aeruginosa isolates based on RAPD-PCR Analysis (Primer 208).
3.1. Antibiotic resistance
The findings of this study revealed that P. aeruginosa strains had the highest resistance to Gentamicin and Imipenem antibiotics (97.1%) followed by Ciprofloxacin (96.2%), Amikacin (95.3%), and Ceftazidime (66.3%), with the lowest resistance to Colistin (0.0 %) (Table 2). In other words, all isolates were sensitive to Colistin. There was no significant association between the genotype groups and antibiotic susceptibility profiles.
Table 2.
The Comparison of frequency of resistance and sensitivity to antibiotics studied in total isolates.
| Antibiotic | Sensitive |
Intermediate |
Resistant |
Total |
||||
|---|---|---|---|---|---|---|---|---|
| Total | % | Total | % | Total | % | Total | % | |
| Amikacin (30) (AN) |
5 | 4.67 | 0 | 0 | 102 | 95.3 | 107 | 100 |
| Ceftazidime (30) (CAZ) |
34 | 31.7 | 2 | 1.86 | 71 | 66.3 | 107 | 100 |
| Ciprofloxacin (5) (CP) |
3 | 2.80 | 1 | 0.93 | 103 | 96.2 | 107 | 100 |
| Gentamycin (10) (GM) |
3 | 2.80 | 0 | 0 | 104 | 97.1 | 107 | 100 |
| Imipenem (10) (IMP) |
3 | 2.80 | 0 | 0 | 104 | 97.1 | 107 | 100 |
| Colistin (10) (Cl) |
107 | 100% | 0 | 0 | 0 | 0 | 0 | 0 |
3.2. RAPD-PCR
The profiles were compared and clustered by the GelCompar II software. Analysis of genetic linkage showed 55%–100% similarity for primer 272 (Figure 1) and 50%–100% similarity for primer 208 (Figure 2) among P. aeruginosa isolates. The length sizes of RAPD fragments ranged from 100 bp to 3000 bp. Totally, 2 to 12 bands (primer 208) and 6 to 13 bands (primer 272) were detected on the gel electrophoresis of RAPD-PCR products. The RAPD-PCR results revealed a high polymorphism among P. aeruginosa isolates.
4. Discussion
In the current study, 107 isolates of P. aeruginosa were investigated using disk diffusion and RAPD-PCR methods for a better understanding of antibiotic resistance rates and P. aeruginosa clone diversity. RAPD is a method that does not essentially evaluate the whole genome. Nowadays, there are controversial findings whether such typing methods can confirm the broad diversity of bacterial populations in the environment (Lanotte et al., 2004). Our findings showed the most resistance to Ciprofloxacin, Imipenem, and Amikacin antibiotics. Considering that the Quinolones, Carbapenems, and Aminoglycosides are still the drug of choice for MDR P. aeruginosa, this high rate of resistance is very alarming.
The mortality rate as a result of infections caused by this bacterium is approximately 51%. Intrinsic and acquired resistance of this bacterium to antimicrobial factors increases the mortality rate of patients affected. Although among available antibiotics, Carbapenems (Meropenem, Ertapenem, Doripenem, Imipenem) are commonly used to treat P. aeruginosa infections, their widespread application has increased the resistance of this organism to these antibiotics. Nevertheless, some resistant strains to Carbapenem have been detected, which are defective in the expression of OprD (Li et al., 2012; Rossolini and Mantengoli, 2005).
By removing and disrupting the skin barrier as the first-line defense of innate immunity, burn injuries generate a suitable environment for different infections; thus microorganisms inevitably colonize even if topical antibacterial agents are administered (Vindenes and Bjerknes, 1995). The results of our research were similar to some reports from Iran and other countries (Fazeli and Momtaz, 2014; Vaez et al., 2015). For example, according to a study conducted in Tehran by Adabi et al., P. aeruginosa has been reported as the most prevalent bacterium found in the wounds of burn patients (Lari et al., 2000). In another study carried out by Zolfaghari et al. in Qom city on burn samples, P. aeruginosa was determined as the most common cause of nosocomial infection. The emergence of increased resistance in this organism to antimicrobial agents has raised concerns such as decreased therapeutic strategies and increased mortality (Zolfaghari, 2011).
In a study on the resistance pattern of 94 strains of P. aeruginosa isolated from wounds of burn patients showed that these strains had a high resistance to the antibiotics prescribed (Adabi et al., 2015). Numerous studies have been also conducted in this regard, e.g. Niitsuma et al. In Japan in 2001, reported P. aeruginosa strains isolated from sputum were resistant to Ceftazidime (4.6%), Imipenem (15.7%), and Meropenem (8.8%) (Niitsuma et al., 2001). There are many studies in this area in Iran. In the study of Bavasheh and Karmostaji in the South of Iran, the highest resistance rate which was observed to sulfamethoxazole and ceftazidime, were (94.44%) and (61.11%), respectively (35). In another similar study by Ranjbar et al. at Baqiyatallah Hospital in Tehran, the antibiotic resistance were as follows: ceftazidime (57.5%), Amikacin (90%), Ciprofloxacin (65%), Gentamycin (67.5%), and Imipenem (97.5%) (Ranjbar et al., 2011). In addition, in a study by Kianpour et al. in Isfahan, it was shown that resistance to antibiotics Ceftriaxone, Amikacin, Ceftazidime, Ciprofloxacin, Tazocin, and imipenem were 82.14%, 57.14%, 53.57%, 42.85%, 39.28%, and 14.28%, respectively (Kianpour et al., 2010). Contrary to our findings, SinemDiken Gur and Nilufer Aksoz in 2016 acclaimed Amikacin and Ceftazidime were the most effective antibiotics (Diken Gur and Aksoz, 2016). Sharma showed that all (100%) of the P. aeruginosa isolates were sensitive to Imipenem and Meropenem (Khosravi et al., 2016).
Reviewing previous studies indicates that the resistance to different antibiotics is relatively high in P. aeruginosa species, which is dependent on the time and location of the species. On the other hand, these resistance patterns are constantly changing, which should be taken into account. The results of this study showed that P. aeruginosa is highly resistant to beta-lactam antibiotics, including anti-pseudomonas penicillins, and this broad group of antibiotics is relatively ineffective on this organism. According to this study, the only antibiotic that has 100% effect on this bacterium is Colistin (polymyxin E), which is an antibiotic with partial neurotoxic and nephrotoxic properties and it is considered to be the last line of treatment in infections caused by multidrug-resistant organisms, including P. aeruginosa, Acinetobacter baumannii and carbapenamase-producing Enterobacters (Montero et al., 2009).
Another finding of the present study is the high percentage of MDR strains. The occurrence of MDR in Pseudomonas species is important and its upward trend has always been reported. For example, in a study by Mir Salehiian et al. on burn patients, multiple resistance was observed in 87% of cases (Mirsalehian et al., 2010). In another study conducted by Fazeli et al. in Isfahan, all isolates from a burn hospital were resistant to more than three antibiotics (Fazeli et al., 2012). A similar study was done by Mardaneh et al. that the highest antibiotic resistance of Imipenem was reported at 85% (Mardaneh et al., 2013), which the highest resistance to Imipenem and Gentamicin was reported at 97.0% in the present study.
Long-term use of antibiotics alone leads to the association of developing resistance and the appearance of cross-resistance among antibiotics, which results in the emergence of MDR P. aeruginosa strains, which can be transmitted from patient to patient and lead to nosocomial infections (Diken Gur and Aksoz, 2016). Active monitoring is mandatory to detect resistant strains of nosocomial infections, especially in burn patients. Therefore, quick and proper solutions should be considered to prevent the spread of pseudomonas species infection. Plasmid-mediated resistance is possible in many ways. This transmission will result in resistance to 3 or 5 antibiotics or even more. Multiple antibiotic resistance to effective antibiotic classes such as beta-lactams, aminoglycosides, and quinolones is generally prevalent and is widely seen in Gram-negative bacteria (Jafari et al., 2013; Mardaneh et al., 2013; Morita et al., 2012).
Genetic techniques along with phenotypic tests can now identify strains isolated from different sites in different parts of the hospital, patient samples and finally, identify the source of the infection. In order to find this link between clinical and environmental strains, some techniques such as endonuclease cleavage analysis, phenotypic testing, pulse-field gel electrophoresis or as in the present study, RAPD -PCR can be used.
The results showed that the isolates obtained in the present study had a different banding pattern in the two types of primers used 208 and 272. This variation in the number of bands can be related to the primer sequence used, access to multiple primer binding sites in the genome, or the genetic pattern quality. It seems that some random primers may work better than other primers and may produce repeatable results (Tyler et al., 1997). It seems that primers with relatively high C + G content perform best for RAPD-PCR (Oladunmoye et al., 2009). The two types of primers used in this study were previously used in other studies to compare the results with PFGE methods. The presence of polymorphic banding pattern using RAPD-PCR technique indicates that there is a high rate of polymorphism in the P. aeruginosa genome. This predicts true compatibility in this bacterium with a genome of 5,570 reads (Stover et al., 2000).
In the present study, which was carried out on genotyping of 107 isolates of P. aeruginosa, in banding analysis created by RAPD-PCR method based on percentage and similarity bands, repeatability different patterns of P. aeruginosa from different patients with Gel Compare software were observed. Although molecular genotyping of P. aeruginosa has been performed in many studies, the molecular genotyping of P. aeruginosa species by RAPD-PCR method has been carried out by a few researchers. Nanvazadeh et al. performed molecular genotyping of P. aeruginosa species isolated from 71 burn wounds by RAPD-PCR method and identified the dominant genotype and showed that this method is suitable for assay of polymorphism among genes of P. aeruginosa (Nanvazadeh et al., 2013). In the study of Nanvazadeh et al., in 2013, 50 clinical samples were taken from Taleghani Hospital in Ahvaz and 50 samples were taken from Pseudomonas aeruginosa isolates in which RAPD_PCR was performed with only primer 272, even though in our study, in addition to primer 272, primer 208 was also used. In their study, genotypes related to type 1, 2, and 6 were determined in patients with severe burns that were sampled from their wounds and blood samples, but in our study there was no relationship between the genotype and burn degree or burn percentage, and so on. Also, their study did not examine the antibiotic resistance pattern, while we also tested the diffusion disc. The Hachem et al. study was performed on cancer patients who were infected with resistant Pseudomonas aeruginosa (MDR) and treated with Colistin and other drugs other than Colistin for three years, while our study was conducted on burn patients infected with resistant Pseudomonas aeruginosa (MDR). The Hachem et al. study used the MIC (E-Test Method) method, but we used antibiogram test. Mahmmudi et al., (2016) showed that the polymorphism of 200 hospital specimens in different areas of the hospital using RAPD-PCR varied based on their antibiotic resistance pattern (Mahmmudi et al., 2016). Salimi et al. in Tehran, identified 8 different genotypes with 6% polymorphism in P. aeruginosa species isolated from burn patients by RAPD-PCR. Previous studies have reported high levels of polymorphism in the genome of this organism (Salimi et al., 2010). Khosravi et al. also reported the high level of genotypic heterogeneity in P. aeruginosa strains isolated from Taleghani burn center in Ahvaz by ERIC-PCR analysis method (Khosravi et al., 2016). Several studies reported genetic diversity and heterogeneity among P. aeruginosa clinical isolates using ERICPCR, rep-PCR, PFGE, and RAPD-PCR methods in hospitals of Iran as well as other countries (Nanvazadeh et al., 2013; Selim et al., 2015).
The results of the RAPD-PCR method used in this study indicates that the isolated P. aeruginosa isolates have a high polymorphism, presumably due to their high rate of genetic variation. This high diversity may be one of the selective benefits for a strain to adapt to the environment and may be useful for propagating and domesticating wild strains. Similar to different studies, the results of this study support the hypothesis that genetic variation can be due to cross-establishment or contamination with the same source or acquisition of genes independent of environmental strains. Patient-to-patient transmission rate is somehow low, but not zero. Cross-contamination transmission confirmation is not accurate and possible since different factors are involved, so each banding pattern in P. aeruginosa can represent the dominant environmental strain in one place.
5. Conclusion
Antibiotic susceptibility/resistance is caused by a change in genotype or gene expression. Given the rapid growth of antibiotic resistance due to the overuse of these drugs, the evaluation of the resistance pattern of pathogenic organisms and in particular, strains associated with hospital-acquired infections in wounded patients burn and identify multidrug-resistant organisms seem necessary. The analysis of antibiotic resistance profiles of the strains show that Colistin is the most effective antibiotic, and the highest resistance rates were detected for Gentamicin and Imipenem. There was no association between the genotype groups that were determined using RAPD-PCR analysis, and the antibiotype patterns that were determined for the strains. According to the results of this study, RAPD-PCR is an effective method to determine the clonal relationship between MDR P. aeruginosa strains. Compared with different methods of infectious agent genotyping, this method is important. On the other hand, the greater the number of bands created in a technique, the greater the possibility of differentiation between strains. Therefore, RAPD-PCR is a suitable method for typing and classifying different isolates of P. aeruginosa, however, more patients, more isolates, and further investigation on its presumptive clinical and environmental sources are needed.
Declarations
Author contribution statement
Parastoo Parsa: Performed the experiments; Wrote the paper.
Nour Amirmozafari: Conceived and designed the experiments.
Bahareh Nowruzi: Analyzed and interpreted the data.
Mohammad Ali Bahar: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We appreciate Iran University of Medical Sceinces for supporting this study.
References
- Adabi M., Talebi T.M., Arbabi L., Afshar M., Fathizadeh S., Minaeian S., Moghadam M.N., Majidpour A. 2015. Determination of Antibiotic Resistance Pattern of Pseudomonas aeruginosa Strains Isolated from Patients with Burn Wounds. [Google Scholar]
- Diken Gur S., Aksoz N. Molecular typing of clinical Pseudomonas aeruginosa strains by using RAPD-PCR. Minerva Biotecnol. 2016;28:104–113. [Google Scholar]
- Dou Y., Huan J., Guo F., Zhou Z., Shi Y. Pseudomonas aeruginosa prevalence, antibiotic resistance and antimicrobial use in Chinese burn wards from 2007 to 2014. J. Int. Med. Res. 2017;45:1124–1137. doi: 10.1177/0300060517703573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli H., Fatahi Bafghi M., Faghri M., Akbari R. Molecular study of PER and VEB genes is multidrug resistant Pseudomonas aeroginosa isolated from clinical specimens in Isfahan/Iran and their antibiotic resistance patterns. J. Kerman Univ. Med. Sci. 2012;19 [Google Scholar]
- Fazeli N., Momtaz H. Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran. Red Crescent Med. J. 2014;16 doi: 10.5812/ircmj.15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli E., Silver S., Witholt B. 1992. Pseudomonas: Molecular Biology and Biotechnology. Bacterium-Plant Interactions – Molecular Mechanisms of Animal Pathogenicity – Taxonomy and Identification – Cell Envelope and Transport – Metabolism and Regulation – Biotechnology: Biodegradation and Industrial Products – Biotechnology: Manipulation, Cloning, and Vectors – Rhodobacter: a Photosynthetic Pseudomonad – Environmental Release. [Google Scholar]
- Jafari S., Najafipour S., Kargar M., Abdollahi A., Mardaneh J., Fasihy Ramandy M., Abdollahi Kheirabadi S., Moravej A. Phenotypical evaluation of multi-drug resistant Acinetobacter baumannii. J. Fasa Univ. Med. Sci. 2013;2:254–258. [Google Scholar]
- Khosravi A.D., Hoveizavi H., Mohammadian A., Farahani A., Jenabi A. Genotyping of multidrug-resistant strains of Pseudomonas aeruginosa isolated from burn and wound infections by ERIC-PCR. Acta Cir. Bras. 2016;31:206–211. doi: 10.1590/S0102-865020160030000009. [DOI] [PubMed] [Google Scholar]
- Kianpour F., Havaei S.A., Hosseini M.M. Evaluation of Pseudomonas aeroginosa isolated from cutaneous infections and determination of drug resistance pattern in patients of Alzahra Hospital in Esfahan. J. Isfahan Med. School. 2010;28 [Google Scholar]
- Kumari N., Thakur S.K. Randomly amplified polymorphic DNA-A brief review. Am. J. Anim. Vet. Sci. 2014;9:6–13. [Google Scholar]
- Lanotte P., Watt S., Mereghetti L., Dartiguelongue N., Rastegar-Lari A., Goudeau A., Quentin R. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J. Med. Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- Lari A.R., Alaghehbandan R., Nikui R. Epidemiological study of 3341 burns patients during three years in Tehran, Iran. Burns. 2000;26:49–53. doi: 10.1016/s0305-4179(99)00102-3. [DOI] [PubMed] [Google Scholar]
- Li H., Luo Y.-F., Williams B.J., Blackwell T.S., Xie C.-M. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int. J. Med. Microbiol. 2012;302:63–68. doi: 10.1016/j.ijmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmmudi Z., Emami A., Gorzin A. 2016. Genotyping of Pseudomonas aeruginosa Strains Isolated from Burn Patients by RAPD-PCR Molecular Technique with Primers 287.https://www.researchgate.net/profile/Amir_Emami9/publication/317011776_Genotyping_of_Pseudomonas_aeruginosa_strains_Isolated_from_burn_patients_by_RAPD-PCR_molecular_technique_with_primer_287/links/595fab25458515a357be486b/Genotyping-of-Pseudomonas-aeruginosa-strains-Isolated-from-burn-patients-by-RAPD-PCR-molecular-technique-with-primer-287.pdf [Google Scholar]
- Mardaneh J., Abbas Poor S., Afrugh P. Prevalence of Shigella species and antimicrobial resistance patterns ofIsolated strains from infected pediatrics in Tehran. Int. J. Enteric. Pathog. 2013;1:28–31. [Google Scholar]
- Mehta S., Singh K., Sawhney N., Singh V.A., Goyal S. Time related changes in pathogenic bacterial patterns in burn wound infections and their antibiotic sensitivity traits. Bangladesh J. Med. Sci. 2017;16:295–301. [Google Scholar]
- Mirsalehian A., Feizabadi M., Nakhjavani F.A., Jabalameli F., Goli H., Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum β-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2010;36:70–74. doi: 10.1016/j.burns.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Montero M., Horcajada J., Sorli L., Alvarez-Lerma F., Grau S., Riu M., Sala M., Knobel H. Effectiveness and safety of colistin for the treatment of multidrug-resistant Pseudomonas aeruginosa infections. Infection. 2009;37:461–465. doi: 10.1007/s15010-009-8342-x. [DOI] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology. 2012;158:1071–1083. doi: 10.1099/mic.0.054320-0. [DOI] [PubMed] [Google Scholar]
- Nanvazadeh F., Khosravi A.D., Zolfaghari M.R., Parhizgari N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39:1409–1413. doi: 10.1016/j.burns.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Niitsuma K., Saitoh M., Kojimabara M., Kashiwabara N., Aoki T., Tomizawa M., Maeda J., Kosenda T. Antimicrobial susceptibility of Pseudomonas aeruginosa isolated in Fukushima Prefecture. Jpn. J. Antibiot. 2001;54:79–87. [PubMed] [Google Scholar]
- Oladunmoye M., Adetuyi F., Akinyosoye F. Effect of Cassia hirsuta (L) extract on DNA profile of some microorganisms. Afr. J. Biotechnol. 2009;8 [Google Scholar]
- Ranjbar R., Owlia P., Saderi H., Mansouri S., Jonaidi-Jafari N., Izadi M., Farshad S., Arjomandzadegan M. Characterization of Pseudomonas aeruginosa strains isolated from burned patients hospitalized in a major burn center in Tehran, Iran. Acta Med. Iran. 2011:675–679. [PubMed] [Google Scholar]
- Rossolini G., Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2005;11:17–32. doi: 10.1111/j.1469-0691.2005.01161.x. [DOI] [PubMed] [Google Scholar]
- Sabath L.D. Huber; 1980. Pseudomonas aeruginosa: the Organism, Diseases it Causes, and Their Treatment. [Google Scholar]
- Salimi H., Owlia P., Yakhchali B., Rastegar Lari A. Characterization of pseudomonas aeruginosa in burn patients using PCR-restriction frag-ment length polymorphism and random amplified polymorphic DNA analysis. Iran. J. Med. Sci. 2010;35:236–241. [Google Scholar]
- Selim S., El Kholy I., Hagagy N., El Alfay S., Aziz M.A. Rapid identification of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. Biotechnol. Biotechnol. Equip. 2015;29:152–156. doi: 10.1080/13102818.2014.981065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkh M.A.G., Shokoohizadeh L., Rashidi N., Tajbakhsh E. Molecular analysis of Pseudomonas aeruginosa strains isolated from burn patients by repetitive extragenic palindromic-PCR (rep-PCR) Iran. Red Crescent Med. J. 2017;19 [Google Scholar]
- Stover C., Pham X., Erwin A., Mizoguchi S., Warrener P., Hickey M., Brinkman F., Hufnagle W., Kowalik D., Lagrou M. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Tyler K., Wang G., Tyler S., Johnson W. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol. 1997;35:339. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaez H., Faghri J., Esfahani B.N., Moghim S., Fazeli H., Sedighi M., Safaei H.G. Antibiotic resistance patterns and genetic diversity in clinical isolates of Pseudomonas aeruginosa isolated from patients of a referral hospital, Isfahan, Iran. Jundishapur J. Microbiol. 2015;8 doi: 10.5812/jjm.20130v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindenes H., Bjerknes R. Microbial colonization of large wounds. Burns. 1995;21:575–579. doi: 10.1016/0305-4179(95)00047-f. [DOI] [PubMed] [Google Scholar]
- Wolska K., Kot B., Jakubczak A. Phenotypic and genotypic diversity of Pseudomonas aeruginosa strains isolated from hospitals in Siedlce (Poland) Braz. J. Microbiol. 2012;43:274–282. doi: 10.1590/S1517-838220120001000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari M. Bacterial elements affecting infections after burn in nequiee-hedaiati Burn hospital. Ghom. 2011 [Google Scholar]