Abstract
Introduction:
The research objective was to systematically review evidence on neurotrophic tyrosine receptor kinase (NTRK) gene fusion frequency in solid tumors.
Methods:
Using Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, a systematic literature review (SLR) was conducted of studies published from January 1987 to 2 January 2020. Selected studies were appraised for use in meta-analysis, with frequency reported as a point estimate with confidence intervals, to estimate NTRK gene fusion tumor incidence and prevalence.
Results:
The SLR identified 222 studies from North America (n = 122), Europe (n = 33), Asia (n = 41), Brazil (n = 5), Australia (n = 2), and multi-continental (n = 19) reporting NTRK gene fusion frequencies across 101 histologies. Studies were prospective (n = 43) and retrospective (n = 179). Testing methods involved DNA (n = 93), RNA (n = 72), combined DNA/RNA (n = 48), protein [immunohistochemistry (IHC), n = 5], and unreported (n = 5). Sample sizes ranged from 1 to 66,871. Of the 222 studies, 107 were suitable for meta-analysis. Highest NTRK gene fusion frequencies were reported in rare cancers: infantile/congenital fibrosarcoma (90.56%, 95% CI 67.42–100.00), secretory breast cancer (92.87%, 95% CI 72.62–100.00), and congenital mesoblastic nephroma (21.52%, 95% CI 13.06–32.20). Lower frequencies were reported in non-small cell lung cancer (0.17%, 95% CI 0.09–0.25), colorectal adenocarcinoma (0.26%, 95% CI 0.15–0.36), cutaneous melanoma (0.31%, 95% CI 0.07–0.55), and non-secretory breast carcinoma (0.60%, 95% CI 0.00–1.50). Reported frequency was ~0% for some cancers: mesothelioma, renal cell carcinoma, prostate cancer, and bone sarcoma. Estimated global overall NTRK gene fusion tumour incidence and 5-year prevalence in 2018 was 0.52 and 1.52 per 100,000 persons, respectively.
Conclusion:
This research confirms the rarity and varying frequency of NTRK gene fusion across tumor types. Limitations included relatively low historic NTRK gene fusion testing and reporting, limited study samples for some cancers, and suboptimal molecular testing methods. In this rapidly developing area, gold-standard testing methods and companion diagnostics are needed to capture all NTRK gene fusions.
Keywords: gene fusion, meta-analysis, NTRK, SLR, solid tumor
Introduction
In recent years, cancer treatment has evolved from nonspecific cytotoxic chemotherapy to a greater role for therapies that target molecular features of cancer cells. Precision medicine aims to identify and address the genetic features of each patient’s cancer to maximize effectiveness while minimizing toxicity to healthy cells. Targeting specific genetic alterations allows for more individualized cancer treatment, but this has previously required identifying the specific oncogenic drivers in each histology.1 There are now, however, several tumor-agnostic treatments that are specific for genetic features seen across tumor types rather than being tailored to molecular features of a particular histology.2
Cancer cells harbor a variety of genetic abnormalities, including point mutations, chromosomal rearrangements, and gene fusions. Some gene fusions that have been identified in cancer involve ALK, ROS1, NRG1, EGFR, MET, and NTRK.3 Gene fusions involving NTRK are found in a broad range of solid tumors. Fusions involving the three NTRK genes (NTRK1, NTRK2, and NTRK3) result in the overexpression or constitutive activation of tropomyosin receptor kinases (TRKs) that can promote oncogenesis.4 Tumors derived from an NTRK gene fusion are commonly referred to as “TRK fusion cancers”.5–9 Based on their putative role in cancer cell proliferation, TRK fusion proteins are an active area of investigation and are the molecular target of several approved drugs, including larotrectinib and entrectinib.4
NTRK gene fusions are rare in more common solid tumors (e.g. colorectal, bladder, breast, and non-small cell lung cancers) but have been detected at high frequencies in some rare cancer types (e.g. secretory breast carcinoma, secretory salivary gland cancer, also known as mammary analogue secretory carcinoma of the salivary gland, and congenital mesoblastic nephroma) and in some pediatric cancers.4,10 Primary brain tumors have been noted as an area of interest for studying NTRK gene fusions, given the lack of effective treatment options and the relative frequency of the fusion in some brain tumor subtypes.11 This variation across multiple histologies means that it is difficult to quantify the patient population for drugs such as larotrectinib, which is approved in the United States (US) and Europe as a tumor-agnostic therapy for advanced cancers with an NTRK gene fusion.1 While immunohistochemistry (IHC) testing is sometimes used, a 2019 European Society for Molecular Oncology (ESMO) panel recommended that confirmatory testing for NTRK gene fusions should be performed at the molecular level [next-generation DNA or RNA sequencing, fluorescence in situ hybridization and reverse transcription polymerase chain reaction (RT-PCR)].12 These molecular testing methods may, however, be time consuming, costly, and material dependent.13 Lack of a defined gold-standard testing method means that NTRK fusions are not always detected, or even tested for in solid tumor studies, leading to added difficulty in evaluating the true prevalence of these mutations.
It is important for healthcare decision-makers to have a clear sense of the size – and the range of tumor types – of the population that might be eligible for treatments targeted for tumors harboring NTRK gene fusions. Without applying rigorous methodologies to determining the incidence of NTRK gene fusions across histologies, it is possible to make erroneous assumptions about incidence based on a subset of the published literature. The objective of this research was therefore to conduct a systematic literature review (SLR) of studies and a meta-analysis to gain a better understanding of the frequency of NTRK gene fusions in various tumor histologies and across the population. Based on the current literature knowledge of NTRK gene fusion frequency, the TRK fusion cancer incidence worldwide was estimated.
Methods
This systematic review and meta-analysis were performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. The methods outlined for the SLR followed those published by the Cochrane Collaboration and the United Kingdom (UK)’s National Institute for Health and Care Excellence (NICE), following principles outlined in Cochrane Handbook for Systematic Reviews of Interventions, CRD’s Guidance for Undertaking Reviews in Health Care, and Methods for the Development of NICE Public Health Guidance.14–16 The methods outlined for the meta-analysis followed those published by the Cochrane Collaboration guidelines, the Agency for Healthcare Research and Quality (AHRQ), and the UK’s National Institute for Health and Care Excellence (NICE).14,17 Studies included in the SLR underwent appraisal to determine suitability for meta-analysis as outlined below.
SLR study selection
Two groups of authors independently reviewed the titles and abstracts of the identified articles to determine their eligibility, based on previously established inclusion/exclusion criteria that were built using the PICOS statement (Table 1). All methods of identifying NTRK gene fusion were included.
Table 1.
SLR study eligibility criteria (PICOS statement).
| Category | Inclusion criteria | Exclusion criteria |
|---|---|---|
| Patient population | • Any patient who was diagnosed with any solid tumors at any age, after any length of time | • Non-human • Not fulfilling inclusion criteria |
| Intervention | • No criteria on interventions | • No criteria on interventions |
| Outcomes measures | • Frequency/percentage, incidence, and prevalence data on NTRK gene fusions among solid tumors • NTRK gene fusion types • Information on test methods and fusion partners was also collected |
• Studies not including at least one of the outcomes listed in the inclusion criteria |
| Study design | • Prospective and retrospective studies, such as observational studies, registries, retrospective studies, database analyses, and non-interventional studies • Pathology-based analyses of stored tumor biopsies were considered • Case-series were considered |
• Case reports were excluded |
| Restrictions | • English language • Year limitation: 1 January 1987–6 December 2018 (first search); first update on 25 November 2019; second update on 2 January 2020 |
• Non-English language studies • Published prior to 1987 |
PICOS, population, intervention, comparison, outcomes and study design; SLR, systematic literature review.
Systematic literature search
Ovid, a search platform that provides standardized access to a wide range of clinical literature databases and is an accepted tool by health technology assessment (HTA) agencies for use in SLR, was used to conduct searches for all literature databases. The literature databases searched were key biomedical literature databases [Medical Literature Analysis and Retrieval System Online (MEDLINE®) and Excerpta Medica Database (Embase®)] and Cochrane from 1 January 1987 to 2 January 2020, using a combination of keywords (Supplemental Table S1). PubMed was also searched using a combination of keywords (Table S2). Studies referenced in other SLRs and meta-analyses in NTRK gene fusion frequencies among solid tumors were reviewed to ensure that all relevant studies were captured. Furthermore, bibliographies from selected studies were reviewed to identify studies relevant for the SLR. This process ensured that papers and articles not picked up in the original search were included in the review.
Three separate searches were conducted. The original search was conducted on 6 December 2018. The first updated search was conducted on 25 November 2019. Finally, the second updated search was conducted on 2 January 2020.
Data extraction
The following information was extracted from each selected article: author, publication year, country of the study, study design, sample size (patient numbers for testing), institution type, database name, registry name, population, inclusion criteria, patient demographics [i.e. age, gender, prior therapies, Eastern Cooperative Oncology Group (ECOG) performance status (PS), and tumor type], testing methods, details of the testing methods, patient number with NTRK gene fusion by NTRK gene fusion type (NTRK1, NTRK2, NTRK3) and by tumor types, gene fusion partners, and tumor histology. Tumor type and histology were extracted using the exact nomenclature reported in the publication.
Meta-analysis study selection
Studies selected in the SLR underwent appraisal for suitability to include in the meta-analysis (MA). To reduce sources of bias across trials and generate more reliable results, the MA was performed on studies published in a peer-reviewed journal or scientific congress where a full poster was available, published after 2010, with irredundant samples and a population size of at least 20 patients evaluated for NTRK fusion. Studies had to represent patients, not samples, that is, studies were excluded if the sample size reflected the number of samples and not necessarily the patient number. An exception was made to the patient population threshold for secretory salivary gland cancer (also known as mammary analogue secretory carcinoma) and secretory breast cancer, as these tumors are extremely rare. Congress abstracts without posters were excluded. Regarding testing methodology, the meta-analysis included studies in which the NTRK testing methodology was specific and included NTRK1, NTRK2, and/or NTRK3. If the NTRK testing methodology was not specified, the publication was excluded. Publications reporting NTRK fusion only in pre-selected samples [e.g. epidermal growth factor receptor (EGFR)- and anaplastic lymphoma kinase (ALK)-negative NSCLC; MSI-H colorectal cancer] were excluded. When publications with overlapping populations were available, the publication with the most complete and relevant set of data was chosen for data extraction.
Meta-analysis methodology
The meta-analysis was conducted in Excel (Version 2006). The weights were calculated using the sample size or the variance of each study.18 Full details of the methods and formulae used to conduct the MA are detailed in a previously published research paper.19
Results for a histology were often reported in more than one study. Likewise, some studies presented data on NTRK gene fusion frequencies in several different tumor groups and histologies. Therefore, the NTRK gene fusion frequencies of patients with similar tumor histology were pooled, and the heterogeneity, the frequency, the confidence intervals (CIs), and standard error were measured (Table S3). Within each tumor histology, NTRK gene fusion frequencies were pooled using the inverse variance method per Cochrane guidelines, which assumes a normally distributed variance of treatment effect. This approach weights the importance of each study result within the meta-analysis according to variance from each study’s NTRK frequency.
There are two widely used statistical effect models for meta-analysis: the fixed effect and random effect models. The fixed effect model assumes that all the studies in the meta-analysis have one true effect size, and the observed variation among studies is caused by sampling errors or chance.20 The random effect model assumes that different studies exhibit substantial diversity, and the true effect size may vary from study to study.21 Consequently, the fixed effect model assesses only intra-study sampling errors (intra-study variation), while the random effect model assesses both intra-study sampling errors and inter-study variance (between-study variation).22 Statistical heterogeneity estimates (I2, Q) tested for rate differences across studies. Q values >0 informed that studies did observe different rates and I2>30% represented at least moderate heterogeneity.
For randomized studies, direct results were included from single arms; per Cochrane guidelines, methodological and clinical heterogeneity were considered. This led to prioritizing results from random effects meta-analyses (where feasible) over fixed effects.
To avoid computational error, while estimating the standard error and therefore the weights, a standard Cochrane correction (fixed value of 0.5) was applied to studies that observed zero counts of NTRK.15 For studies with the Cochrane correction applied, the normal approximation method was used to obtain CIs around point estimates. For studies without the Cochrane correction, the binomial distribution (Clopper-Pearson exact approach) was used to obtain two-sided 95% CIs around individual study point estimates. These methods are from evidence leaders in SLR and meta-analysis techniques: Cochrane Collaboration Guidelines, the AHRQ, and NICE.14,17 Per these evidence guidelines, appropriate study comparison and robustness of final meta-analysis results were evaluated according to the potential for clinical and methodological heterogeneity across studies.14–16
Overall NTRK gene fusion incidence and prevalence methodology
The objective of estimating the overall NTRK gene fusion incidence rate was to provide a single estimate that encompasses the diverse NTRK gene fusion frequencies across various tumor entities. To calculate the NTRK gene fusion frequency for the individual tumor entities reported in the SLR, the broad tumor type incidence rate was multiplied by the histology proportion of the individual tumor entity and then multiplied by the reported SLR meta-analysis NTRK gene fusion frequency. The summation of the individual NTRK gene fusion tumor entities then resulted in the overall NTRK gene fusion incidence across all tumor entities (Table S4).
The broad tumor type crude incidence rate in 2018 was obtained from the Global Cancer Observatory (GCO) Cancer Today.23 Where applicable, incidence rates were adjusted to reflect both genders (e.g. breast cancer) and all ages (e.g. pediatric cancers). Estimates of rare cancers that were unavailable from GCO were instead obtained from population-based studies in the literature, primarily reflecting US and European data.24,25 The histology proportions (e.g. proportion of lung cancers that are non-small cell lung cancer) were obtained from the most recent ESMO clinical guidelines, representing putative histology distribution at cancer diagnosis.26 There was no filtering for disease stage, that is, all samples were included regardless of the stage of the patient’s cancer. Lastly, the NTRK gene fusion frequency for each tumor entity was obtained from the SLR meta-analysis. To prevent overlap in the summation of incidence rates, only the NTRK gene fusion frequency of the broadest tumor entity was included in the overall NTRK gene fusion incidence rate.
The objective and methods used in estimating the overall NTRK gene fusion 5-year limited duration point prevalence rate were similar to those for the incidence rate. To calculate the 5-year prevalence, broad tumor type prevalence rates in 2018 were selected from the GCO, and the prevalence of the rare cancers was calculated by applying tumor-specific 5-year overall survival curves obtained from US-based Surveillance, Epidemiology, and End Results (SEER).27
Results
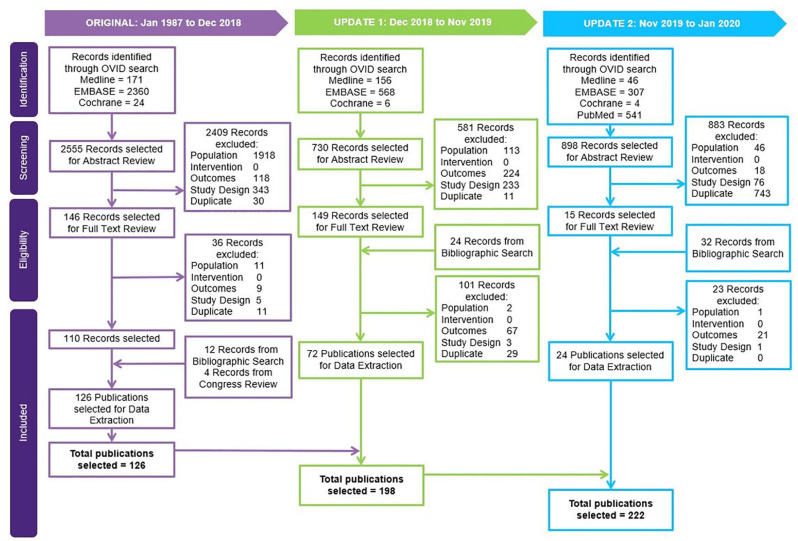
SLR study selection and characteristics: A total of 4183 records were identified in Ovid and PubMed searches (Figure 1). Title and abstract screening led to the exclusion of 3873 records based on PICOS discrepancy or duplication. The full text versions of the remaining 366 records, including 56 records from bibliographic search, were reviewed, with 160 records excluded at the full-text review stage due to PICOS discrepancies or duplication. This led to selection of 222 articles from the Ovid and PubMed searches for data extraction, including an additional four articles that were selected via congress review and twelve from bibliographic searches.
Figure 1.
Study selection (PRISMA) diagram.
Of the 222 selected publications in the SLR, 179 were retrospective and 43 were prospective. Of the 179 retrospective studies, 41 were database analyses, 79 were single-center studies, 56 were multicenter studies, two were registry studies, and one was a retrospective study of tumor samples that did not report the number of centers involved. Of the 43 prospective studies, 21 were single-center studies, and 22 were multicenter studies. Most included studies (122) were conducted in North America; 33 were conducted in Europe, 41 in Asia, five in Brazil, two in Australia, and 19 at sites across multiple continents. Testing methods involved DNA (n = 93), RNA (n = 72), combined DNA/RNA (n = 48), protein [immunohistochemistry (IHC), n = 5], and not reported (n = 5). Note that some studies used more than one method.
Meta-analysis and overall frequency: Of the 222 studies included in the SLR, 107 studies that met the meta-analysis inclusion and exclusion criteria outlined in the methods section were considered. Five studies were excluded due to not reporting NTRK testing methodology. Based on data extracted from these 107 studies, the NTRK gene fusion frequency of 98 different histologies from 29 different tumor groups was calculated (Table S3). There was significant variability in the frequencies of NTRK gene fusion reported across tumor histology in the included studies, ranging from 0% to 92.87%, in keeping with what has been reported in the literature about the variability of NTRK gene fusion frequency across tumor types.4,10 A total of 30 tumor histologies had an NTRK frequency of zero taken from single studies. These single-study frequencies are reported without a CI and standard error.
The fixed effect model for meta-analysis was chosen to measure the NTRK frequency of 37 tumor histologies from 24 tumor groups. In these groups, the frequency of NTRK gene fusion ranged from 0% to 92.87%. The random effect model was used to measure the NTRK frequency of thirteen tumor histologies from eight tumor groups. In these groups, the frequency of NTRK gene fusion ranged from 0.10% to 50.47%. The NTRK gene fusion frequencies of 18 histologies came from single studies that did not report a frequency of zero. The CIs for these studies were calculated using the binomial distribution exact approach.
Of the 98 tumor histologies, only 12 had ⩾10% frequency (Table 2). The highest NTRK gene fusion frequencies per tumor histology were calculated in rare cancers such as infantile/congenital fibrosarcoma, secretory salivary gland cancer (also known as mammary analogue secretory carcinoma), secretory breast cancer, and congenital mesoblastic nephroma (all subtypes). Lower frequencies were reported in some more common cancers such as cervical carcinoma, uterine soft tissue sarcoma, cutaneous melanoma, pancreatic adenocarcinoma, colorectal adenocarcinoma, neuroendocrine tumors, non-small cell lung cancer, non-secretory breast carcinoma, and some primary brain tumors (including glioma, low grade glioma, high grade glioma, ependymal, and dysembryoplastic neuroepithelial tumors). Reported frequency was close to 0% for some cancers, including adenoid cystic carcinoma of the salivary gland, retinoblastoma, uveal melanoma, anaplastic astrocytoma, diffuse astrocytoma, pilocytic astrocytoma, anaplastic oligodendroglioma, oligodendroglioma, pediatric ependymoma, pediatric choroid plexus carcinoma, pediatric neuroblastoma, pediatric medulloblastoma, follicular thyroid carcinoma, medullary thyroid carcinoma, focal anaplastic thyroid carcinoma/poorly differentiated thyroid carcinoma, thymoma, lung adenosquamous carcinoma, squamous cell carcinoma of the lung, large cell carcinoma of the lung, mesothelioma, gastric and esophageal adenocarcinoma, gastric/stomach adenocarcinoma, rectal carcinoma, bladder carcinoma, rectum adenocarcinoma, adult adrenocortical carcinoma, pediatric adrenocortical carcinoma, pheochromocytoma and paraganglioma of the adrenal gland, renal clear cell carcinoma, renal papillary cell carcinoma, renal chromophobe rumor, Wilm’s tumor of the kidney, invasive lobular carcinoma of the breast, uterine/endometrial carcinoma, ovarian carcinoma, ovarian serous cystadenocarcinoma, prostate cancer, prostate adenocarcinoma, testicular germ cell tumors, osteosarcoma, Ewing sarcoma, rhabdomyosarcoma, and pheochromocytoma and paraganglioma of the neuroendocrine. Primary brain tumors, comprising multiple histologies, showed a wide range of NTRK gene fusion frequency, from close to 0% to over 20%. The complete list of NTRK gene fusion frequencies of the 98 different histologies evaluated is presented in the supplementary materials (Table S3).
Table 2.
Frequency of NTRK gene fusions in selected tumor histologies.
| Histology | Frequency NTRK gene fusions | 95% CI |
|---|---|---|
| Secretory breast carcinoma | 92.87% | 72.62–100.00 |
| Fibrosarcoma, infantile (congenital) | 90.56% | 67.42–100.00 |
| Secretory salivary gland cancer (also known as mammary analogue secretory carcinoma) | 79.68% | 62.84–96.51 |
| Pigmented spindle cell nevus of Reed | 56.52% | 34.49–76.81 |
| Pleomorphic adenoma | 50.47% | 0.00–100.00 |
| Papillary thyroid carcinoma, pediatric | 25.93% | 11.11–46.28 |
| Differentiated thyroid cancer, pediatric | 22.22% | 6.41–47.64 |
| Congenital mesoblastic nephroma (all subsets) | 21.52% | 13.06–32.20 |
| High grade glioma | 21.21% | 8.98–38.91 |
| Low grade mucoepidermoid carcinoma | 20.00% | 5.73–43.66 |
| Acinic cell carcinoma of salivary gland | 11.11% | 4.19–22.63 |
| Diffuse leptomeningeal glioneuronal tumor | 10.00% | 2.11–26.53 |
| Frequency of NTRK gene fusions in common tumor types | ||
| Cervical carcinoma | 0.36% | 0.00–0.81 |
| Uterine soft tissue sarcoma | 0.34% | 0.00–0.78 |
| Cutaneous melanoma | 0.31% | 0.07–0.55 |
| Pancreatic adenocarcinoma | 0.31% | 0.09–0.53 |
| Colorectal adenocarcinoma | 0.26% | 0.15–0.36 |
| Neuroendocrine tumors | 0.26% | 0.07–0.44 |
| Non-small cell lung cancer | 0.17% | 0.09–0.25 |
| Invasive breast carcinoma | 0.10% | 0.03–0.18 |
| Examples of primary brain tumors | ||
| High grade glioma | 21.21% | 8.98–38.91 |
| Diffuse leptomeningeal glioneuronal tumor | 10.00% | 2.11–26.53 |
| High grade glioma, pediatric | 6.19% | 3.11–9.28 |
| Glial, glioneuronal, and ependymal | 3.28% | 0.40–11.35 |
| Dysembryoplastic neuroepithelial tumors, pediatric | 3.03% | 0.08–15.76 |
| Low grade glioma, pediatric | 1.61% | 0.00–3.33 |
| Glioma | 0.99% | 0.00–2.79 |
| Low grade glioma | 0.88% | 0.22–1.54 |
| Glioma/neuroepithelial tumor | 0.55% | 0.24–1.07 |
CI, confidence interval.
Globally, the calculated overall NTRK gene fusion incidence and 5-year prevalence were 0.52 and 1.52 per 100,000 persons, respectively.
Discussion
Based on our meta-analysis of literature available at the time of this research, the overall incidence of NTRK gene fusion in solid tumors is estimated to be 0.52 per 100,000 persons globally in 2018. The overall 5-year prevalence of NTRK gene fusion in solid tumors is estimated to be 1.52 per 100,000 persons globally in 2018. While there is no formal consensus on the definition of an ultra-rare disease, a generally accepted European Union (EU) definition is a disease with a prevalence of less than 1 in 50,000 persons.28 Thus, cancers with NTRK gene fusions fall below the threshold for an ultra-rare disease.
The evidence that we captured in our SLR is based on published literature, which has limitations. Genomic profiling of tumor samples is a relatively new practice, and thus gene fusion frequencies are not historically a standard component of reporting in oncology studies. Moreover, the literature provides limited information regarding tumor histologies. In general, studies do not consistently report on biomarker-directed subtypes (e.g. MSI-high versus MSS, RAS mutant versus RAS wildtype in colorectal cancer, or methylation status in brain tumors). Studies may not clearly distinguish between number of samples and number of patients, and they may not provide sufficient detail on testing methods and rationale, or on pan-TRK testing practices. Moreover, genomic profiling data came from a variety of study types, with varying numbers of patients per study; in some cases, results were reported per patient, and in other cases per sample. This inconsistent reporting poses challenges in evaluating the quality of publications and adds some limitations to our interpretation of the data.
We noted substantial variability in NTRK gene fusion frequencies, even for studies reporting on the same tumor types. There are several factors that may influence the inter- and intra-variability in the frequencies of NTRK gene fusion reported in the included studies. One explanation is that results are limited by the sample size that was tested. Due to the rarity of NTRK gene fusions, particularly in the more prevalent cancer types, a small sample size tested may not accurately reflect the true NTRK gene fusion frequency. Some of the higher NTRK gene fusion frequencies that we report are based on single studies, that is, we were unable to conduct meta-analyses on these histologies. Moreover, some studies were excluded based on small sample size or limited testing information provided. The second explanation relates to the heterogeneity in testing methods employed to determine the NTRK fusion frequency and the lack of a universally used, gold-standard testing method. Per 2019 ESMO recommendations, fluorescence in situ hybridization (FISH), reverse transcription polymerase chain reaction (RT-PCR), and RNA-based sequencing panels should be used to confirm the presence of NTRK fusions.12 While RNA-based tests are generally favorable due to the ability to detect novel and known NTRK fusions, NTRK fusion detection capability of DNA-based tests is highly dependent on the assay design and intron coverage of the NTRK genes (and fusion gene partners).29 The majority of studies selected in this SLR used the currently recommended molecular genetic techniques; of the 222 selected studies, 5 did not specify the testing method and thus were excluded from the meta-analysis, while 5 that were included used IHC, potentially introducing some uncertainty.
With the comprehensive literature survey, this study also provides critical information for pan-tumor companion diagnostic development. The NTRK fusion prevalence estimation is a pivotal step in calculating population-based diagnostic performance measures. The NTRK fusion prevalence calculated in this manuscript depends on the distribution assumption and statistical modeling used in the meta-analysis. Different meta-analysis procedures may lead to different estimated values. A comprehensive evaluation of varying meta-analysis methods will be conducted in our future work, which will also support NTRK fusion companion diagnostic development.
Additionally, the study is further limited by the lack of confidence intervals for the overall NTRK gene fusion tumour incidence and prevalence estimates. To provide CIs for the overall NTRK gene fusion tumour incidence and prevalence estimate, the confidence levels of the incidence and prevalence estimate, as well as the NTRK gene fusion frequency need to be considered. Though the NTRK gene fusion frequency CI is available through the meta-analysis (please see Table S3), the CIs for the tumour incidence and prevalence estimates are not reported by cancer registries. Hence, a CI for the overall NTRK gene fusion tumour incidence and prevalence based solely on the NTRK gene fusion frequency confidence was deemed potentially misleading and therefore not constructed. Future studies may explore the ability to improve this limitation.
Despite these limitations, to our knowledge, this study is the first to systematically and rigorously review the literature on NTRK gene fusion frequencies across tumor histologies. These findings support the characterization of cancers with NTRK gene fusions as a rare disease, which is relevant to healthcare decision-makers evaluating the potential impact of precision medicines targeting this abnormality. Our findings are among the first to assess the incidence of a cancer genetic feature independent of tumor histology. This is an important step in the evolution of tumor-agnostic and precision medicine treatment strategies.
Conclusion
In the first comprehensive SLR and meta-analysis on the frequency of NTRK fusions in solid tumors, we demonstrated the variability of the fusion across tumor types and even across studies of a single histology. Although the NTRK gene fusion is rare in most of the more prevalent tumor types, it has higher frequency in several rare tumor histologies, leading to a worldwide incidence of 0.52 per 100,000 persons and a 5-year prevalence below 1 per 50,000 persons, which is below the current threshold definition for an ultra-rare disease. In the rapidly evolving field of precision medicine, changes in testing practices and techniques, as well as more rigorous reporting of genetic profiling results in studies, may lead to changes in NTRK gene fusion frequency estimates.
Supplemental Material
Supplemental material, sj-pdf-1-tam-10.1177_1758835920975613 for A systematic review and meta-analysis of neurotrophic tyrosine receptor kinase gene fusion frequencies in solid tumors by Anna Forsythe, Wei Zhang, Uwe Phillip Strauss, Marc Fellous, Maesumeh Korei and Karen Keating in Therapeutic Advances in Medical Oncology
Footnotes
Conflict of interest statement: Anna Forsythe and Maesumeh Korei are employees of Purple Squirrel Economics; hired as consultants to Bayer Pharmaceuticals, Inc. Wei Zhang, Marc Fellous and Karen Keating are employees of Bayer Pharmaceuticals, Inc. Uwe Phillip Strauss is an employee of Bayer Vital GmbH, a fully consolidated subsidiary of Bayer Pharmaceuticals, Inc.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was fully funded by Bayer Pharmaceuticals, Inc.
ORCID iD: Anna Forsythe  https://orcid.org/0000-0001-6017-1926
https://orcid.org/0000-0001-6017-1926
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Anna Forsythe, Purple Squirrel Economics, 4 Lexington Ave, Suite 15K, New York, NY 10010, USA.
Wei Zhang, Bayer Pharmaceuticals, Inc., Whippany, NJ, USA.
Uwe Phillip Strauss, Strauss Bayer Vital GmbH, Leverkusen, Nordrhein-Westfalen, Germany.
Marc Fellous, Bayer Pharmaceuticals, Inc., Whippany, NJ, USA.
Maesumeh Korei, Purple Squirrel Economics, New York, NY, USA.
Karen Keating, Bayer Pharmaceuticals, Inc., Whippany, NJ, USA.
References
- 1. Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016; 1: e000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lavacchi D, Roviello G, D’Angelo A. Tumor-agnostic treatment for cancer: when how is better than where. Clin Drug Investig. Epub ahead of print 19 April 2020. DOI: 10.1007/s40261-020-00915-5. [DOI] [PubMed] [Google Scholar]
- 3. Pan Y, Zhang Y, Ye T, et al. Detection of novel NRG1, EGFR, and MET fusions in lung adenocarcinomas in the Chinese population. J Thorac Oncol 2019; 14: 2003–2008. [DOI] [PubMed] [Google Scholar]
- 4. Gatalica Z, Xiu J, Swensen J, et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 2019; 32: 147–153. [DOI] [PubMed] [Google Scholar]
- 5. Albert CM, Davis JL, Federman N, et al. TRK fusion cancers in children: a clinical review and recommendations for screening. J Clin Oncol 2018; 37: 513–524. [DOI] [PubMed] [Google Scholar]
- 6. Federman N, McDermott R. Larotrectinib, a highly selective tropomyosin receptor kinase (TRK) inhibitor for the treatment of TRK fusion cancer. Expert Rev Clin Pharmacol 2019; 12: 931–939. [DOI] [PubMed] [Google Scholar]
- 7. Lassen U, Albert CM, Kummar S, et al. Larotrectinib efficacy and safety in TRK fusion cancer: an expanded clinical dataset showing consistency in an age and tumor agnostic approach. Ann Oncol 2018; 29(Suppl. 9): ix23–ix27. [Google Scholar]
- 8. Penault-Llorca F, Rudzinski ER, Sepulveda AR. Testing algorithm for identification of patients with TRK fusion cancer. J Clin Pathol 2019; 72: 460–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 2020; 33: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ling Q, Li B, Wu X, et al. The landscape of NTRK fusions in Chinese patients with solid tumor. Ann Oncol 2018; 29: viii22–viii23. [Google Scholar]
- 11. Gambella A, Senetta R, Collemi G, et al. NTRK fusions in central nervous system tumors: a rare, but worthy target. Int J Mol Sci. Epub ahead of print 23 January 2020. DOI: 10.3390/ijms21030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marchiò C, Scaltriti M, Ladanyi M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol 2019; 30: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 13. Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 2017; 41: 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Methods for the development of NICE public health guidance (third edition), http://nice.org.uk/process/pmg4 (2012, accessed 1 September 2020). [PubMed]
- 15. Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions (version 5.1.0), http://handbook-5-1.cochrane.org (2011, Accessed 1 September 2020).
- 16. CRD’s guidance for undertaking reviews in health care (third edition). https://www.york.ac.uk/crd/SysRev/!SSL!/WebHelp/SysRev3.htm. (accessed 1 September 2020) [Google Scholar]
- 17. Morton SC, Murad MH, O’Connor E, et al. Quantitative synthesis–an update. Methods guide for comparative effectiveness reviews. Rockville, MD: Agency for Healthcare Research and Quality; (AHRQ), 2018. [Google Scholar]
- 18. Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ 1997; 315: 1533–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neyeloff JL, Fuchs SC, Moreira LB. Meta-analyses and forest plots using a microsoft excel spreadsheet: step-by-step guide focusing on descriptive data analysis. BMC Res Notes 2012; 5: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davey Smith G, Egger M. Meta-analyses of randomised controlled trials. Lancet 1997; 350: 1182. [DOI] [PubMed] [Google Scholar]
- 21. DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 2007; 28: 105–114. [DOI] [PubMed] [Google Scholar]
- 22. Hedges LV, Vevea JL. Fixed- and random-effects models in meta-analysis. Psychol Methods 1998; 3: 486–504. [Google Scholar]
- 23. Ferlay J, Ervik M, Lam F, et al. International Agency for Research on Cancer. Global cancer observatory: cancer today, http://gco.iarc.fr/today/home (2018, accessed 28 April 2020).
- 24. von Holstein SL, Therkildsen MH, Prause JU, et al. Lacrimal gland lesions in Denmark between 1974 and 2007. Acta Ophthalmol 2013; 91: 349–354. [DOI] [PubMed] [Google Scholar]
- 25. Marmor S, Portschy PR, Tuttle TM, et al. The rise in appendiceal cancer incidence: 2000-2009. J Gastrointest Surg 2015; 19: 743–750. [DOI] [PubMed] [Google Scholar]
- 26. Grégoire V, Lefebvre J-L, Licitra L, et al. Squamous cell carcinoma of the head and neck: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010; 21(Suppl. 5): v184–v186. [DOI] [PubMed] [Google Scholar]
- 27. National Cancer Institute. Cancer statistics. Surveillance, Epidemiology, and End Results (SEER) program, https://seer.cancer.gov/statistics/index.html (accessed 15 June 2020).
- 28. Harari S. Why we should care about ultra-rare disease. Eur Respir Rev 2016; 25: 101–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res 2019; 25: 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tam-10.1177_1758835920975613 for A systematic review and meta-analysis of neurotrophic tyrosine receptor kinase gene fusion frequencies in solid tumors by Anna Forsythe, Wei Zhang, Uwe Phillip Strauss, Marc Fellous, Maesumeh Korei and Karen Keating in Therapeutic Advances in Medical Oncology