ABSTRACT
Temperature is one of the most impactful environmental factors to which plants adjust their growth and development. Although the regulation of temperature signaling has been extensively investigated for the aerial part of plants, much less is known and understood about how roots sense and modulate their growth in response to fluctuating temperatures. Here, we found that shoot and root growth responses to high ambient temperature are coordinated during early seedling development in Arabidopsis. A shoot signaling module that includes HY5, the phytochromes and the PIFs exerts a central function in coupling these growth responses and maintaining auxin levels in the root. In addition to the HY5/PIF-dependent shoot module, a regulatory axis composed of auxin biosynthesis and auxin perception factors controls root responses to high ambient temperature. Taken together, our findings show that shoot and root developmental responses to temperature are tightly coupled during thermomorphogenesis and suggest that roots integrate energy signals with local hormonal inputs.
KEY WORDS: Root development, Temperature, Thermomorphogenesis, Phytochromes, HY5, Arabidopsis
Summary: HY5 and phytochrome signaling in the shoot modulates coupling between shoot and root thermomorphogenesis. Auxin biosynthesis and signaling form a further regulatory axis for root responses to temperature.
INTRODUCTION
Over the course of their life, plants are subjected to constant environmental fluctuations. Consequently, plants have evolved tremendous developmental plasticity that allows them to precisely adjust their development to environmental conditions and therefore to thrive in dynamically and often unpredictably changing environments. In particular, the early stage of seedling development constitutes a crucial moment at which plants need to sense their environment and respond quickly to fine-tune their developmental programs and successfully establish themselves as autotrophic seedlings (reviewed by Ha et al., 2017). Not surprisingly, early life stages have been shown to strongly contribute to local acclimation (reviewed by Donohue et al., 2010).
Temperature is a pervasive environmental parameter influencing biological systems at all scales from the rate of biochemical reactions to the timing of developmental transitions (reviewed by Penfield, 2008). In addition, temperature shows important geographical, diurnal and seasonal variation. Importantly, plants are equipped with sophisticated molecular machineries to perceive temperature fluctuations, which allows them to sense and translate these signals into appropriate developmental responses. Accordingly, raised ambient temperature leads to increased elongation of the hypocotyl and root – a process called thermomorphogenesis (reviewed by Quint et al., 2016).
The molecular mechanisms underlying shoot thermo-responses have largely been investigated (Quint et al., 2016). In this context, the photoreceptor phytochrome B (phyB) enables perception of higher ambient temperature by switching from an active to an inactive form (Legris et al., 2016). This process of phytochrome thermal reversion subsequently prevents sequestration and degradation of transcription factors such as the PHYTOCHROME INTERACTING FACTORs (PIFs) that can accumulate and promote the expression of downstream regulatory genes (Jung et al., 2016; Kumar et al., 2012; Park et al., 2018).
Among the PIF clade, PIF4 acts as a central signaling hub during shoot thermomorphogenesis (Quint et al., 2016; Koini et al., 2009), and recent studies showed that PIF7 also has a role in this process (Chung et al., 2020; Fiorucci et al., 2020). Upon higher ambient temperature, PIF4 directly positively regulates the expression of a battery of genes, including the auxin biosynthetic genes YUCCA8 (YUC8) and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), thereby promoting an elevation of auxin levels and increased hypocotyl cell elongation (Franklin et al., 2011; Sun et al., 2012). This regulatory circuit also integrates inputs from the transcription factor LONG HYPOCOTYL5 (HY5) that can act antagonistically to PIF4 by repressing PIF4 expression or by directly regulating key PIF4 target genes including YUC8 (Delker et al., 2014; Gangappa and Kumar, 2017). Both HY5 and PIF4 expression levels and protein abundance are tightly regulated by a plethora of factors (reviewed by Lau and Deng, 2012; Quint et al., 2016). Among those, CONSTITUTIVE PHOTOMORPHOGENESIS PROTEIN1 (COP1) and DEETIOLATED1 (DET1) trigger HY5 degradation and promote both PIF4 expression and protein stabilization (Gangappa and Kumar, 2017; Osterlund et al., 2000; Saijo et al., 2003; Yanagawa et al., 2004). The collective genetic activity of PIF4, HY5, COP1 and DET1 defines an intertwined regulatory module that acts at the interface between light and temperature signaling (Delker et al., 2014; Gangappa and Kumar, 2017). Interestingly, HY5 protein has also been shown to translocate from the shoot to the root and to coordinate carbon fixation with nitrogen uptake (Chen et al., 2016).
Importantly, roots can autonomously sense and respond to temperature (Bellstaedt et al., 2019), which might allow them to reach deeper and cooler layers of the soil under warm surface conditions (Illston and Fiebrich, 2017). However, in contrast to the shoot, the molecular mechanisms underlying plant root thermo-responses have so far remained elusive. Similar to the shoot, maintenance of auxin homeostasis is crucial for the root response to temperature (Wang et al., 2016). In line with this idea, auxin signaling increases upon perception of higher ambient temperature (Hanzawa et al., 2013; Wang et al., 2016). In this context, the auxin efflux transporters PIN2 and PILS6 mediate auxin transport and local accumulation at the root, which in turn triggers developmental response to temperature in the root (Feraru et al., 2019; Hanzawa et al., 2013). Furthermore, the auxin receptors TIR1 and AFB2 are stabilized upon increased ambient temperature by forming a protein complex with HEAT SHOCK PROTEIN 90 (HSP90) and its co-chaperone SUPPRESSOR OF G2 ALLELE SKP1 (SGT1). The accumulation of TIR1 and AFB2 subsequently activates auxin signaling and mediates root thermo-sensory elongation (Wang et al., 2016).
Although root and shoot thermomorphogenesis occur simultaneously during early seedling development (Bellstaedt et al., 2019), it is still unclear whether these responses are coordinated at the whole plant level. In this study, we leveraged a genetic approach combined with comprehensive phenotypic analyses, transcriptional profiling and metabolic measurements to further characterize the molecular circuits mediating root thermomorphogenesis. We found that a shoot regulatory module including HY5, phytochromes and PIFs can also regulate the root growth response upon perception of higher ambient temperature, demonstrating that shoot and root growth responses are coupled during early seedling development. Furthermore, we show that an additional regulatory axis composed of auxin biosynthesis and perception genes is required during root thermomorphogenesis and propose that the relative abundance of auxin and its downstream signaling activity in both the shoot and root are crucial to coordinately control growth response to temperature in these organs.
RESULTS
HY5 controls the root thermo-response
The impact of increased temperature on plant development has been extensively investigated (reviewed by Quint et al., 2016); however, it is still unclear whether a core regulatory network governs temperature sensing and signaling in multiple developmental contexts and whether these responses are coordinated across multiple organs. To assess how ambient temperature modulates root development, we grew plants at 21°C and analyzed their growth until 3 days after transfer to either 21°C or 27°C. In line with previous reports (Feraru et al., 2019; Martins et al., 2017; Wang et al., 2016), wild-type plants grown at 27°C displayed an increased primary root growth compared with that of plants kept at 21°C (Fig. 1A,B). Having established this experimental setup to analyze the root response to temperature shifts, we went on to further characterize the genetic mechanisms underlying this process.
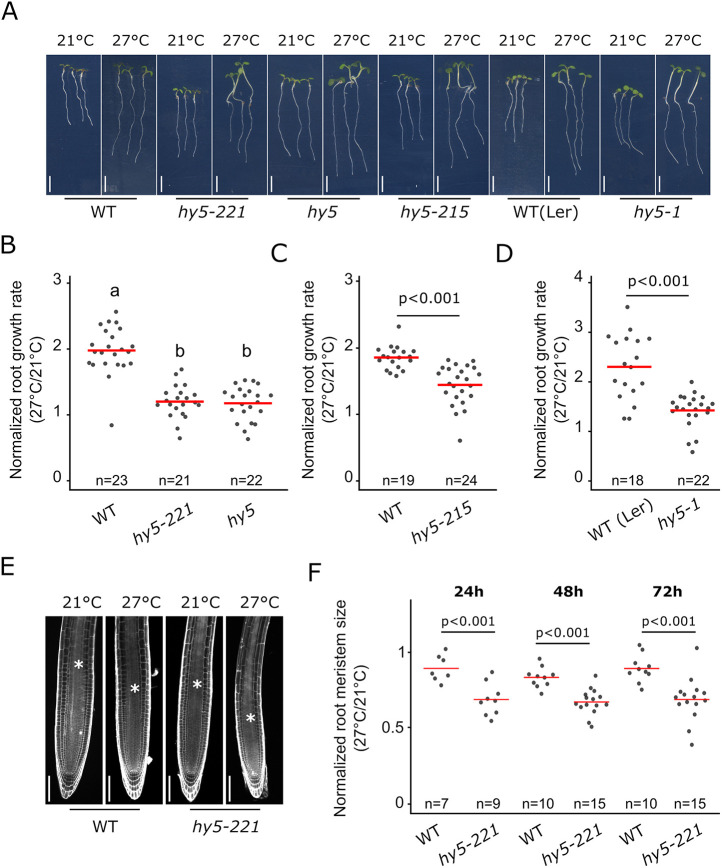
Fig. 1.
HY5 mediates the root response to higher ambient temperature. (A) Wild-type (WT) and hy5 allelic mutant seedlings at 6 days after germination (DAG) and 3 days after transfer to 21°C or 27°C. WT(Ler) and hy5-1 are in the Ler ecotype background. (B-D) Normalized root growth (27°C/21°C) in WT, hy5 and hy5-221 (B), hy5-1 (D) and hy5-215 (C) seedlings. (E) Root meristem in WT and hy5-221 mutant at 5 DAG and 2 days after transfer to 21°C or 27°C. Asterisks mark the root transition zone. (F) Normalized root meristem size (27°C/21°C) in WT and hy5-22 seedlings at 24, 48 and 72 h after temperature shift. n indicates the number of individual seedlings measured. Measured seedlings were obtained in one (F) or two (B,C,D) independent replications of the experiment. In B, groups denoted by different letters are significantly different from one another (P<0.05; one-way ANOVA and Tukey HSD post hoc test). Other P-values were calculated using a two-tailed, unpaired Student's t-test (C,D,F). Red bar represents the mean (B-D,F). Scale bars: 5 mm (A); 100 µm (E).
The transcription factor HY5 is a key regulator of shoot thermomorphogenesis, while at the same time regulates root development and hormonal signaling pathways (reviewed by Gangappa and Botto, 2016). Thus, we hypothesized that HY5 could regulate the root response to increased ambient temperature. We analyzed the relative root growth of hy5 mutant and wild-type plants grown at 21°C and 27°C (Fig. 1A-D), and, in line with our hypothesis, four different allelic versions of hy5 mutants displayed reduced root growth response to temperature compared with that of wild-type plants. Whereas wild-type plants increased root growth by 80-120%, hy5 mutants displayed an increase of only 20-40% (Fig. 1A-D). This reduced response was also observed under a different growth condition with reduced light intensity (see Materials and Methods; Table S1) as well as when roots were grown in the dark or on medium not supplemented with sucrose (Fig. S1A-C), indicating that the reduced response observed in hy5 mutants was not dependent on light or nutrient conditions. To test whether this reduced response was also associated with changes in root apical meristem activity, we measured the dynamics of the root meristem size after temperature shift. Interestingly, hy5 mutants displayed a lower relative root meristem size at all time points analyzed – from 24 h to 72 h after temperature shift – and showed an earlier onset of cell elongation. This indicates that their meristem is hypersensitive to increased ambient temperature compared with those of wild-type plants (Fig. 1E,F; Fig. S1D). Taken together, these data demonstrate that HY5 is required to mediate root responses to temperature.
While analyzing the root phenotypes of hy5 mutants, we observed that plants with a lower root growth frequently displayed longer hypocotyls than plants with a higher root growth, suggesting that shoot and root responses to temperature could be functionally connected. To test this observation, we simultaneously measured hypocotyl and root growth of individual plants and calculated the relative hypocotyl or root growth. Raising ambient temperature strongly promoted hypocotyl growth while decreasing root growth response in the hy5 mutant (Fig. S1E), supporting the idea that these two processes could be coordinated during early seedling development.
Phytochromes and PIF activity regulate the root response to higher ambient temperature
The phenotypic relation between hypocotyl and root growth responses in the hy5 mutant suggested that additional regulators of shoot thermomorphogenesis might also modulate the root growth response. Previous studies have demonstrated a crucial role of PHYB in sensing temperature in the shoot and mediating hypocotyl growth (Jung et al., 2016; Legris et al., 2016), leading us to hypothesize that the phytochromes might also regulate root thermo-responses. Accordingly, both phyA and phyB single mutant plants displayed a reduction in the root response to temperature compared with that of wild-type plants. This difference was further enhanced in phyAB double mutants, showing that PHYA and PHYB co-regulate this process (Fig. 2A,B; Fig. S1F). The reduced root growth in the phyAB plants was also associated with a decreased relative root meristem size, demonstrating that root meristematic activity was hypersensitive to increased ambient temperature, similar to what we observed in hy5 mutant plants (Fig. 2C,D; Fig. S1D). Collectively, these data demonstrate that, in addition to their function in the shoot, the phytochromes are also required for root thermomorphogenesis.
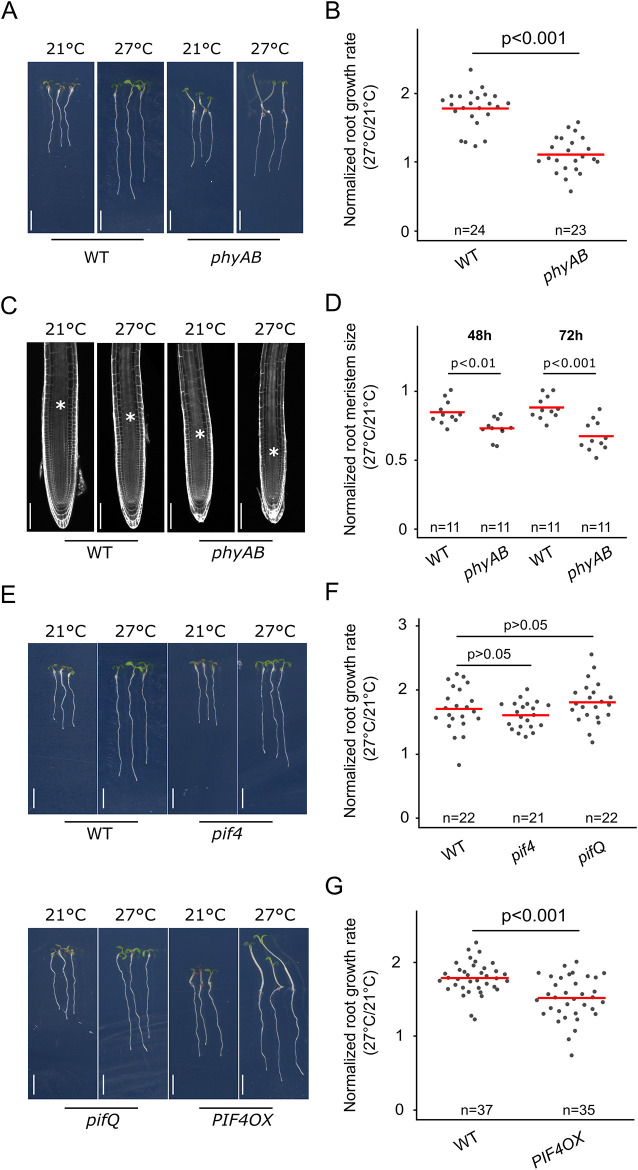
Fig. 2.
Phytochrome signaling regulates the root response to higher ambient temperature. (A) Wild-type (WT) and phyAB mutant seedlings at 6 DAG and 3 days after transfer to 21°C or 27°C. (B) Normalized root growth (27°C/21°C) in WT and phyAB seedlings. (C) Root meristems in WT and phyAB seedlings, 5 DAG and 2 days after transfer to 21°C or 27°C. Asterisks mark the root transition zone. (D) Normalized root meristem size (27°C/21°C) in WT and phyAB seedlings, 48 and 72 h after temperature shift. (E) WT, pif4, pifQ and PIF4 OX mutant seedlings at 6 DAG and 3 days after transfer to 21°C or 27°C. (F,G) Normalized root growth (27°C/21°C) in WT, pif4, pifQ (F) and PIF4 OX (G) seedlings. n indicates the number of individual seedlings measured. Measured seedlings were obtained in one (F) or two (B,D,G) independent replications of the experiment. P-values in F were calculated using one-way ANOVA and Tukey HSD post hoc test. Other P-values were calculated using a two-tailed, unpaired Student’s t-test (B,D,G). Red bar represents the mean (B,D,F,G). Scale bars: 5 mm (A,E); 100 µm (C).
Phytochromes mediate the phosphorylation of downstream factors including the PIFs, which are then targeted for degradation (Lorrain et al., 2008). Because PIF4 functionally interacts with HY5 during shoot thermomorphogenesis (Delker et al., 2014; Gangappa and Kumar, 2017), we reasoned that PIF4 might also modulate root responses to temperature downstream of the phytochromes. Thus, we tested whether PIF4 and other PIF family members could control the root growth response to temperature. Similarly to previous studies (Martins et al., 2017), pif4 mutants did not show an impaired root response (Fig. 2E,F). Moreover, simultaneously interfering with the function of multiple PIFs such as PIF1, PIF3, PIF4, PIF5 and PIF7 in pifq (a pif1 pif3 pif4 pif5 quadruple mutant) or pif7 pifq mutants had no effect on the root response compared with that of wild-type plants, indicating that the PIFs were not required to regulate this process (Fig. 2E,F; Fig. S1G). Although the loss-of-function mutants did not display impaired root response to higher temperature, we reasoned that because phytochromes are negative regulators of PIFs, PIF activity might be increased in phytochrome mutants, and that, in turn, might contribute to the reduction of the root thermo-response in phyAB mutants. Thus, we next tested whether promoting PIF function could be sufficient to modulate root growth response. In line with this idea, the gain-of-function pPIF4:PIF4-FLAG mutant line (PIF4OX; Gangappa and Kumar, 2017) showed a significant reduction in the root response to higher temperature (Fig. 2G), demonstrating that although PIF4 function is not required, it is indeed sufficient to modulate this developmental response. PIF activity is promoted in phytochrome mutants (Park et al., 2018, 2004), and our results further suggest that increased PIF4 activity in the phyAB mutant could lead to a reduction of the root thermo-response.
HY5 and PIF activity co-regulates root thermomorphogenesis
Having shown that HY5 or phytochrome/PIF activity can modulate shoot and root responses to temperature, we next hypothesized that HY5 and PIFs could co-regulate this process. To test this idea, we first impaired HY5 function together with DET1 and COP1, which are regulators of PIF4 expression and the hypocotyl response to temperature (Fig. S2A; Gangappa and Kumar, 2017). In accordance with a previous report (Gangappa and Kumar, 2017), both hy5 det1 and hy5 cop1 double mutations suppressed the enhanced hypocotyl response observed for hy5 mutants (Fig. 3A,B). Interestingly, these lines also displayed a significant increase in root growth temperature response compared with that of the hy5 mutant (Fig. 3C). The genetic interaction of HY5 and COP1 was highly significantly dependent on temperature (two-way ANOVA, P=1.85×10−6), whereas the genetic interaction of HY5 and DET1 was only marginally significant (two-way ANOVA, P=0.0553). Overall, these results demonstrated that impairing DET1 and COP1 function can partially rescue root growth in response to higher ambient temperature. Importantly, neither det1 nor cop1 single mutants displayed an increased root growth response to temperature, suggesting that the genetic interaction between HY5 and DET1 or COP1 is crucial for modulation of root thermomorphogenesis (Fig. 3C). To directly test whether HY5 and PIFs could co-regulate this process, we next simultaneously interfered with HY5 and PIF function using the hy5 pifQ quintuple mutant and analyzed growth responses to elevated temperature (Fig. 3D-F). Consistent with this idea, both hypocotyl and root growth responses were significantly rescued compared with the response of hy5 mutants, demonstrating that HY5 and PIF pathways functionally interact to regulate shoot and root responses to temperature (Fig. 3D-F). These results demonstrate that the activity of a shoot signaling module including HY5 and PIF genes mediates root response to temperature.
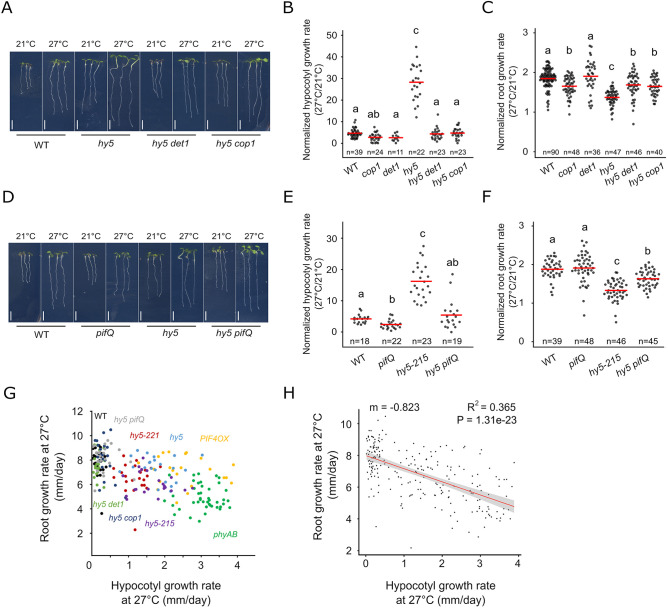
Fig. 3.
The HY5-PIF module regulates the root response to temperature. (A) Wild-type (WT), hy5, hy5 det1 and hy5 cop1 mutant seedlings at 6 DAG and 3 days after transfer to 21°C or 27°C. (B,C) Normalized hypocotyl (B) and root growth (C) (27°C/21°C) in WT, cop1, det1, hy5, hy5 det1 and hy5 cop1 seedlings. (D) WT, pifQ, hy5 and hy5 pifQ mutant seedlings at 6 DAG and 3 days after transfer to 21°C or 27°C. (E,F) Normalized hypocotyl (E) and root growth (F) (27°C/21°C) in WT, pifQ, hy5-215 and hy5 pifQ seedlings. (G,H) Relation between root and hypocotyl growth rate at 27°C, as shown with measurements on individual WT (n=23), hy5-221 (n=24), phyAB (n=43), PIF4OX (n=22), hy5 (n=22), hy5 det1 (n=20), hy5 cop1 (n=22), hy5-215 (n=23) and hy5 pifQ (n=22) plants (G) and after non-parametric regression analysis (H). Shaded region in H indicates a point-wise 95% confidence interval on the fitted values (red line). n indicates the number of individual seedlings measured. Measured seedlings were obtained in one (G,H) or three (B,C,E,F) independent replications of the experiment. In B,C,E,F, groups denoted by different letters are significantly different from one another (P<0.05; one-way ANOVA and Tukey HSD post hoc test in C,F; one-way ANOVA after log10 transformation in B,E). Linear regression method, Pearson correlation (H). Red bar represents the mean (B,C,E,F). Scale bars: 5 mm.
Taken together, our phenotypic analyses showed that enhanced shoot growth response was associated with a decreased root response to temperature. We observed a similar trend when wild-type plants were grown in the dark and shifted to higher ambient temperature (Fig. S2B,C). These results suggested that shoot and root thermomorphogenesis could be quantitatively negatively correlated. To test this idea, we combined measurements of hypocotyl and root growth of individual plants for nine different genotypes (wild type, hy5-221, hy5, hy5-215, hy5 pifQ, hy5 cop1, hy5 det1, phyAB and PIF4OX) as well as for wild-type and hy5-221 mutant plants under a different light environment. We then analyzed the relation between hypocotyl and root growth rate at 21°C and 27°C, as well as the relation between their normalized growth (Fig. 3G; Fig. S2D-I). Remarkably, we observed that at 27°C individual genotypes formed distinct groups, with root growth rate decreasing as the hypocotyl growth increased, supporting the idea that these traits could be negatively correlated (Fig. 3G; Fig. S2D). We next applied a linear regression model and observed a negative correlation between the root and hypocotyl growth rates at 27°C (R2=0.365; Fig. 3H). We also observed this negative correlation under the second light environment (R2=0.623; Fig. S2E) indicating that root growth rate negatively correlates with hypocotyl growth rate at 27°C. Interestingly, we did not observe this relation at 21°C (R2=0.064) or when analyzing temperature responses (R2=0.035) (Fig. S2F-I), indicating that this hypocotyl-root growth correlation is specific to higher ambient temperature conditions. Taken together, these results show that upon increased ambient temperature, the HY5-PIF module is required to balance hypocotyl with root growth responses, and further suggest that a developmental trade-off governs hypocotyl and root growth responses at higher ambient temperature.
A shoot-to-root developmental trade-off in response to higher ambient temperature
The observation that shoot and root thermomorphogenesis were negatively correlated was intriguing and prompted us to test whether modulating shoot thermo-response was sufficient to impact root growth. To investigate this idea, we used a genetic chimera approach by taking advantage of an HA-YFP-HA-HY5 fusion protein (DoF-HY5) that shows restricted cell-to-cell movement, with the aim of driving its expression specifically in the shoot of hy5 mutants using CAB3 or CER6 promoters (Burko et al., 2020b; Procko et al., 2016). In line with previous studies (Chen et al., 2016; Procko et al., 2016), we detected strong accumulation of DoF-HY5 in leaves, petioles and at a weaker level in hypocotyls for both constructs, confirming that our CAB3 and CER6 promoters were driving expression in the shoot (Fig. S3A-F; Burko et al., 2020b). Although we detected DoF-HY5 accumulation in the root of the pCER6:DOF-HY5 line, we did not detect fluorescence signal in the root of the pCAB3:DOF-HY5 lines, indicating that expression driven from the CAB3 promoter was specific to the shoot and that our tagged version of HY5 was not able to move from the shoot to the root (Fig. 4A-C; Fig. S3A-F). To further confirm these observations, we assessed the accumulation of DoF-HY5 fusion protein either in the root or in the shoot using immunoblotting with a HY5 or HA antibody (Fig. 4D,E). Consistent with our microscopy observations, we observed that DoF-HY5 protein accumulated in the shoot of pCAB3:DOF-HY5 lines, whereas the detected protein levels were similar to those of the hy5 mutant in the root or were accumulating ubiquitously in the pCER6:DOF-HY5 line (Fig. 4D,E). This provided us with valuable genetic material to further test whether HY5 local activity in the shoot could regulate the root response to temperature.
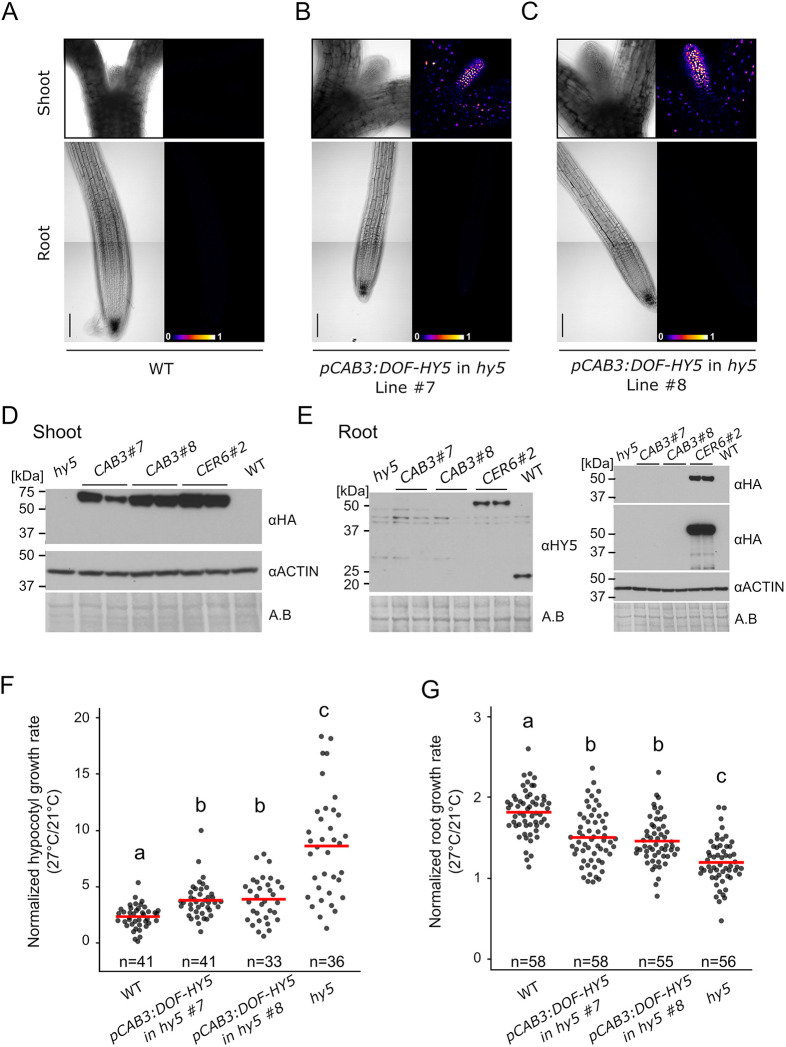
Fig. 4.
Shoot response to temperature is sufficient to modulate root growth response. (A-C) Brightfield and false color view of DoF-Hy5 fluorescence at 6 DAG of wild-type (WT) seedlings (A) and two independent lines of hy5 carrying pCAB3:DOF-HY5 (B,C). (D,E) Immunoblotting of shoot (D) or root tissues (E) in WT, hy5, two independent lines of hy5 carrying pCAB3:DOF-HY5 (CAB3#7 and CAB3#8) and hy5 carrying pCER6:DOF-HY5 and pCAB3:DOF-HY5 (CER6#2) at 27°C. DoF-HY5 protein was detected using anti-HA or anti-HY5 antibodies. For anti-HA, the same blot was displayed at short (upper panel) and longer exposure time (lower panel). Amido Black (A.B.) staining and anti-actin antibody were used as controls. DoF-HY5 samples were run in duplicate. (F) Normalized hypocotyl growth (27°C/21°C) in WT, hy5 and pCAB3:DOF-HY5 rescue lines. (G) Normalized root growth (27°C/21°C) in WT, hy5 and pCAB3:DOF-HY5 rescue lines. n indicates the number of individual seedlings measured. Measured seedlings were obtained in two (D-G) or three (A-C) independent replications of the experiment. In F and G, groups denoted by different letters are significantly different from one another (P<0.05; one-way ANOVA and Tukey HSD post hoc test). Red bar represents the mean (F,G). Scale bars: 100 µm.
We went on to analyze the functionality of the DoF-HY5 fusion protein by measuring hypocotyl and root growth upon response to increased ambient temperature in the pCER6:DOF-HY5 line. Although the hy5 mutant displayed increased relative hypocotyl growth and a reduced root growth response, these responses were rescued to levels similar to those of wild-type plants in the pCER6:DOF-HY5 line, demonstrating that the DoF-HY5 fusion protein was functional (Fig. S3G-I). These results prompted us to next investigate the local function of HY5 in the shoot during temperature response by analyzing the pCAB3:DOF-HY5 chimera rescue lines (Fig. 4F,G). In line with DoF-HY5 accumulation in the shoot, both pCAB3:DOF-HY5 lines displayed a partial rescue of the relative hypocotyl growth observed in hy5 mutants (Fig. 4F). Strikingly, these two independent lines also showed a significant rescue of the root growth response compared with the response of hy5 plants, demonstrating that HY5 function in the shoot was sufficient to modulate the root growth response to temperature (Fig. 4G). However, the partial rescue of these lines also suggested that HY5 function could be required locally for root thermomorphogenesis. To test this idea, we removed the shoots of seedlings and assessed the root growth response upon temperature shift on the isolated roots. Although root elongation in isolated roots of wild-type plants was lower than in intact plants, their relative responses to the temperature shift were similar (Fig. S3J-L, Fig. S6, Table S1). The root growth response in whole seedlings and isolated roots of hy5 mutants was lower than that of wild type, suggesting that HY5 function is indeed also required locally in the root (Fig. S3K). In contrast to hy5 mutants, the relative response was fully rescued in isolated roots of phyAB mutants, suggesting that the phytochromes act mainly in the shoot during thermomorphogenesis (Fig. S3L). Together, these results reveal that modulating shoot thermomorphogenesis by local HY5 rescue is sufficient to regulate root growth, but that local root action of HY5 is required for the full root growth temperature response.
Transcriptional change of metabolic genes in response to temperature
Having shown that a developmental trade-off quantitatively couples shoot and root thermomorphogenesis, we wanted to further delineate the regulatory mechanisms underlying this process. To this end, we used a genome-wide approach and profiled the transcriptomes of isolated shoots and roots after a short (4 h) or a more prolonged (18 h) temperature treatment using RNA-seq.
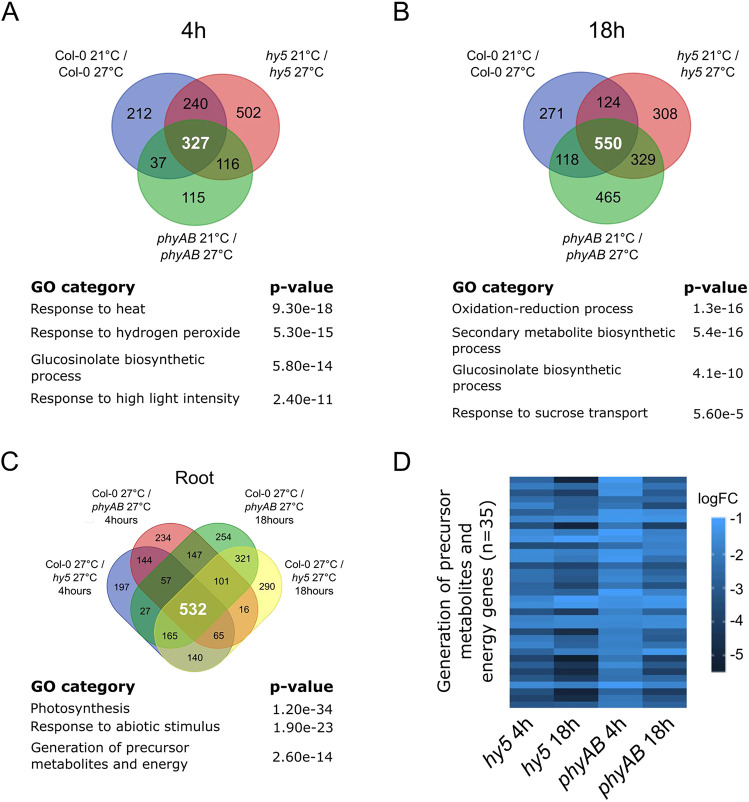
We first asked whether a core regulatory network could mediate responses to temperature in the shoot and in the root. To strengthen our approach and to alleviate the influence of the genotypes on the response, we compared the transcriptional changes in wild-type, hy5 and phyAB plants. Using this method, we identified 327 genes in the shoot and in the root that were temperature regulated in all genotypes for the early time point, whereas we found that 550 and 904 genes were commonly regulated at the late time point in the root and shoot, respectively (Fig. 5A,B; Fig. S4A,B). Consistent with the temperature treatment imposed on the plants, the shared regulatory signatures were associated with heat response [‘response to heat’, ‘response to hydrogen peroxide’ and ‘response to high light intensity’ gene ontology (GO) categories] (Fig. 5A; Fig. S4A,B). We also observed contrasting regulatory responses in the shoot and root, which were mainly related to metabolism. In particular, we detected a significant enrichment for members of the glucosinolate biosynthetic process among genes specifically responding in the root, whereas we observed that flavonoid biosynthesis genes were enriched among those responding in the shoot (Fig. 5A,B; Fig. S4A,B). Moreover, we observed a greater proportion of genes involved in sucrose transport in the root and sucrose response in the shoot, suggesting that increased ambient temperature modulates energy metabolism (Fig. 5B; Fig. S4A,B). Taken together, these results show that, in addition to common core regulatory signatures, shoots and roots display specific responses to elevated temperature, with roots differentially re-adjusting their metabolism in response to higher temperature.
Fig. 5.
Genome-wide analysis of root response to temperature. (A,B) Genes regulated at 4 h (A) or 18 h (B) after temperature shift in wild-type (Col-0), hy5 and phyAB roots. Gene ontologies (GO) characterize the biological processes enriched among the temperature-regulated genes that are shared between wild-type, hy5 and phyAB samples. (C) Overlapping misregulated genes in hy5 and phyAB roots at 27°C. Gene ontologies characterize the biological processes shared among HY5 and phytochrome co-regulated genes in the root at higher ambient temperature. (D) Differentially regulated genes belonging to the GO category ‘generation of precursor metabolites and energy genes’ in hy5 and phyAB roots at 27°C. Biological triplicates were analyzed; P-values were calculated using AgrigoV2 (A-C).
To further characterize the regulatory function of HY5 and phytochromes during root thermomorphogenesis, we next identified genes misregulated in hy5 and phyAB mutants compared with the wild type at 27°C (Fig. 5C). Strikingly, we observed significant overlap (hypergeometric test; P<0.001) in the sets of genes that were upregulated or downregulated in hy5 and phyAB mutant roots or shoots at both time points (Fig. S4C,D). This overlap supports the findings of our previous genetic analyses and demonstrates that HY5 and phytochromes regulate a set of common genes in the root (Fig. 5C; Fig. S4C). Among the co-regulated genes, we identified known HY5 target genes – such as HY5 HOMOLOG (HYH), the SUPPRESSOR OF PHYA (SPA) gene family and FHY1-LIKE (FHL) – as well as known light-signaling genes, which confirmed the quality of our dataset (Fig. S4E; Burko et al., 2020a; Ciolfi et al., 2013; Lee et al., 2007; Li et al., 2010). Importantly, we also detected an enrichment for misregulated genes involved in the generation of precursor metabolites, suggesting that the metabolic status was altered in hy5 and in phyAB mutant roots (P=2.6e-14; Fig. 5C). Accordingly, all genes belonging to the GO category ‘generation of precursor metabolites and energy precursor’ were significantly downregulated either in hy5 or phyAB mutants at both time points, indicating that HY5 and phytochrome activities are required for the expression of energy metabolism genes in the root (n=35/35; Fig. 5D). These results also show that the reduced root growth response observed in hy5 and phyAB mutants correlates with a substantial downregulation of genes involved in the chemical reactions and pathways resulting in the formation of substances from which energy is derived or genes involved in releasing energy from these metabolites. Taken together, the analysis of transcriptional responses suggests that HY5 and phytochrome activity regulates root growth at higher temperature by modulating energy metabolism.
Auxin perception, signaling and biosynthesis are involved in root thermomorphogenesis
Having shown that HY5 and phytochromes are required for the expression of energy precursor genes in the root, we next wanted to investigate whether other signals could regulate root thermomorphogenesis downstream of the HY5-PIF module. Some reports have demonstrated that auxin transport and signaling are required for the root response to higher ambient temperature (Feraru et al., 2019; Hanzawa et al., 2013; Wang et al., 2016); however, these regulatory interactions have also been challenged and debated (Martins et al., 2017). This prompted us to first confirm the function of auxin homeostasis in our root growth assays.
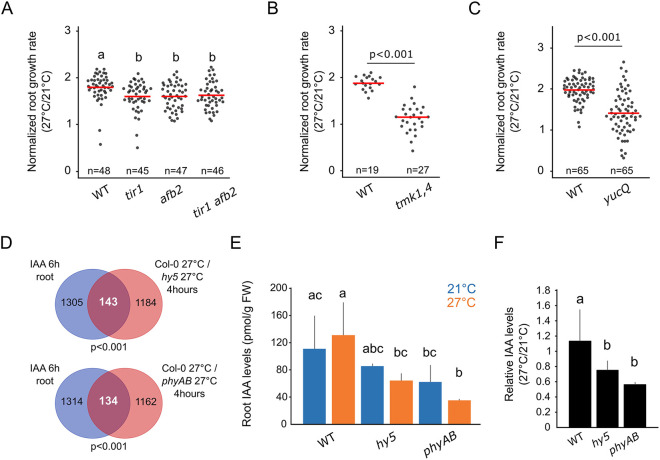
Shifting plants from 21°C to 27°C led to increased auxin signaling, as shown by the increased signal of the pDR5v2:3×YFP-NLS transcriptional reporter at the root tip and increased IAA29 gene expression (Fig. S5A-C; Wang et al., 2016). Genetically interfering with the auxin receptors TIR1 and AFB2 in tir1, afb2 and tir1 afb2 mutants also led to a slight but significant reduction when considering the root growth response to temperature (Fig. 6A; Fig. S5D). However, we note that when considering root growth rates without normalizing, the single knockouts did not significantly respond to temperature, and the genetic interaction of TIR1 and AFB2 interacted with temperature in only a marginally significant manner (two-way ANOVA for AFB1:TIR1:temperature interaction; P=0.0967). To complement these data, we impaired another branch of auxin signaling by interfering with the function of TMK proteins, which are membrane-localized receptor-like kinases involved in the perception of auxin independently of the TIR/AFB system (Cao et al., 2019; Xu et al., 2014). As suggested by the TIR/AFB-related mutants, we found reduced root elongation in the tmk1,4 double mutant compared with that of wild-type plants during the root temperature response (Fig. 6B; Fig. S5D). Taken together, these results confirmed that auxin perception and signaling are required for root thermomorphogenesis.
Fig. 6.
Auxin homeostasis regulates root thermomorphogenesis. (A-C) Normalized root growth (27°C/21°C) in wild-type (WT), tir1, afb2, tir1 afb2 (A), tmk1,4 (B) and yucQ (C) seedlings. (D) Differentially regulated genes in hy5 and phyAB roots at 27°C that are auxin (IAA) responsive according to Omelyanchuk et al. (2017), 4 h after temperature shift. (E) IAA concentration [pmol/g of fresh weight (FW)] in roots of seedlings at 6 DAG, 12 h after transfer to 21°C or 27°C (n>3). (F) Relative IAA content in roots compared with those in shoot tissues of seedlings 6 DAG, 12 h after transfer to 21°C or 27°C (n>3). n indicates the number of individual seedlings measured (A-C) or the number of biological replicates (E,F). Measured seedlings were obtained in three (A-C) independent replications of the experiment. In A,E,F, groups denoted by different letters are significantly different from one another [P<0.05; one-way ANOVA and Tukey HSD post hoc test (A,E) or Student–Newmann–Keuls post hoc test (F)]. Other P-values were calculated using a two-tailed, unpaired Student's t-test (B,C) or hypergeometric test (D). Red bar represents the mean (A-C).
We next hypothesized that auxin biosynthesis and the control of the hormone level at the root might also modulate root thermomorphogenesis. Thus, we examined the function of auxin biosynthesis by genetically interfering with YUC gene activity in the yuc3,5,7,8,9 (yucQ) quintuple mutant. Accordingly, the yucQ mutant displayed a reduced root growth response compared with that of wild-type plants, demonstrating that auxin biosynthesis, through the activity of the YUCs, is also required for root elongation upon higher ambient temperature (Fig. 6C; Fig. S5D). Taken together, these data demonstrate that auxin biosynthesis is required for root thermomorphogenesis, and further suggest that auxin perception and signaling play a role in this process.
HY5 and phytochromes regulate auxin homeostasis at the root
Having confirmed the function of auxin signaling during root thermomorphogenesis, we next asked whether HY5 and phytochromes could regulate this hormonal pathway. In our root transcriptome, we analyzed the overlap between genes misregulated in hy5 or phyAB mutants at 27°C and auxin-responsive genes in the root, as obtained from RNA-seq after 6 h of indole-3-acetic acid (IAA) treatment (Omelyanchuk et al., 2017). Interestingly, we found a significant overlap of genes that transcriptionally responded to IAA treatment and were misregulated in hy5 and phyAB mutants at both early and late time points (Fig. 6D; Fig. S5E). We also found that a significant proportion of genes whose transcriptional response to temperature was differentially regulated in hy5 or phyAB mutants also responded to auxin in the root (Fig. S5F). Taken together, these results demonstrate that HY5 and phytochrome activities converge with the auxin regulatory network and further suggested that these factors might control auxin homeostasis during root thermomorphogenesis.
To further examine this idea, we assessed the state of the auxin metabolic pathway by measuring the concentration of IAA and its precursors in roots of wild-type plants and of hy5 and phyAB mutants 12 h after a temperature shift. Surprisingly, we did not observe a change in total auxin level after temperature shift in wild-type roots, suggesting that an increase in total auxin level is not required for root thermomorphogenesis, unlike what has been reported for the shoot (Fig. 5E; Gray et al., 1998). Although we observed a slight decrease in levels of IAA and some of the auxin precursors in hy5 and phyAB roots compared with levels in wild-type roots, demonstrating that HY5 and phytochromes are required to maintain IAA levels in the root independently of temperature (Fig. 6E; Fig. S5G,H), the interaction of the mutant genotypes with temperature was not statistically significant (two-way ANOVA; Table S1). When comparing the ratio of root IAA levels at the two temperatures, we observed a decrease in the relative IAA level in hy5 and phyAB mutants compared with that in wild-type roots upon increased ambient temperature, indicating that the dynamics of auxin accumulation in the root might be impaired upon loss of HY5 and phytochrome activity (Fig. 6F). Taken together, these results show that HY5 and phytochrome are required to maintain auxin levels, but that further data will be required to thoroughly test the role of HY5- and phytochrome-dependent auxin levels in root thermomorphogenesis.
DISCUSSION
In this study, we investigated the regulatory mechanisms controlling root thermomorphogenesis. Using a genetic approach combined with phenotypic analyses, we have found that a regulatory module including HY5 and phytochromes modulates the shoot-to-root growth coordination of responses to higher temperature. In addition, we have gained insight on the function of the auxin signaling pathway and its connection with HY5 and phytochromes during root thermomorphogenesis (Fig. 7). Taken together, our findings highlight that a developmental trade-off governs shoot and root growth responses and further suggests that roots integrate energy signals with hormonal inputs during thermomorphogenesis.
Fig. 7.
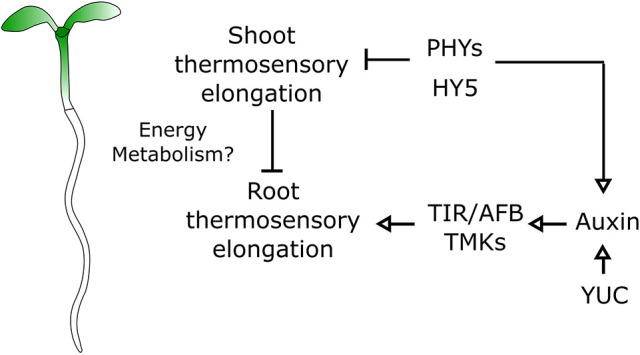
A genetic model for organ growth coordination during plant thermomorphogenesis. Model of root thermosensory response. Roots integrate regulatory signals coming from the shoot through the activity of phytochromes and HY5 with auxin signals mediated by biosynthetic genes (YUC) and signaling (TIR, AFB, TMK).
We showed that HY5 and the phytochromes are required for the root response to temperature. In line with published reports, interfering with PIF activity did not lead to impaired root growth responses to temperature. Previously, such observations led to the conclusion that PIFs do not regulate root responses to temperature (Martins et al., 2017). However, we observed that a PIF4 gain-of-function line phenocopies the hy5 and phyAB mutant phenotypes, showing that PIF4 is sufficient to regulate root thermomorphogenesis. Furthermore, HY5 acts antagonistically to PIF4 at the promoter of multiple target genes, and interfering with HY5 function could enhance PIF4-mediated gene regulation (Gangappa and Kumar, 2017). Accordingly, shoot and root phenotypes in hy5 mutants are suppressed by dampening PIF expression, demonstrating that HY5 genetically interacts with PIFs during shoot and root thermomorphogenesis. Thus, our results support a model where PIF4 acts downstream of the phytochromes and functionally converges with HY5 to regulate root thermomorphogenesis. Future experiments interfering with the function of phytochromes and PIFs in higher-order mutants will be important to further dissect the function of this regulatory circuit during thermomorphogenesis.
In this context, HY5 also genetically interacts with COP1 and DET1, as shown by the suppression of hy5 phenotypes in hy5 det1 and hy5 cop1 mutants. Interestingly, whereas the det1-1 mutant responds similarly to control plants, cop1-4 shows decreased root growth in response to temperature. Moreover, although the genetic interaction of COP1 and HY5 is statistically highly significant, the DET1 HY5 genetic interaction with temperature is only marginally significant. Taken together, these results are intriguing, because DET1 and COP1 act together in order to promote HY5 degradation (reviewed by Lau and Deng, 2012). Thus, our results also suggest that COP1 can signal independently of HY5 during root thermomorphogenesis.
Our finding that a shoot regulatory module can control hypocotyl growth response and can concomitantly modulate root growth raises interesting questions as to how these two processes are coordinated. Our current data suggest two putative mechanisms that could act in parallel to coordinate shoot and root thermomorphogenesis.
First, we observed that a reduced root growth response is associated with a strong promotion of hypocotyl growth. Such an increase in growth is promoted by the temperature shift and could impact the availability of compounds, including water, as well as having an effect on root metabolism. Accordingly, our data suggest that temperature responses are tightly connected with energy metabolism. The observed negative correlation between hypocotyl and root growth responses and the associated downregulation of metabolic precursor genes that play a role in chemical reactions and pathways from which energy is released indicate that these two processes could be coordinated by a limitation of metabolic resources that are required during enhanced hypocotyl growth. This hypothesis is consistent with classical studies on biomass allocation between shoots and roots (Shipley and Meziane, 2002; Thornley, 1972). In this context, one possible relevant energy signal could be sucrose, which is produced in the shoot through photosynthesis and has been shown to act as a long-distance signal to promote root growth (Kircher and Schopfer, 2012). Interestingly, in our genome-wide expression analysis of root responses to temperature, we found that a significant proportion of genes involved in sucrose transport was enriched, suggesting that changes in sugar availability could regulate shoot-to-root growth coordination upon increased ambient temperature. In addition, HY5 and PIF4 have been shown to directly regulate the expression of photosynthetic genes and, consequently, the production of chlorophyll in young seedlings. Accordingly, hy5 mutants display lower chlorophyll content than wild-type plants at 27°C (Toledo-Ortiz et al., 2014), which could have a direct impact on the production of photosynthesis-derived sucrose and consequently on root growth. Given that hy5 mutants still displayed a reduced root response to increased ambient temperature in medium that was not supplemented with sucrose (Fig. S1C), we believe that external sucrose would have limited impact on this process. Taken together, these data suggest that shoot growth could influence the availability of energy signals and in turn modulate root growth response upon increased ambient temperature. To uncouple growth mechanism from energy balance, it would be interesting to analyze root growth response to temperature upon overexpression of expansins in the shoot (Cosgrove, 2000).
In parallel to this pathway, another important signal could be the phytohormone auxin. Previous studies have shown that auxin transport and signaling regulate root growth upon a shift to higher temperature (Feraru et al., 2019; Hanzawa et al., 2013; Wang et al., 2016), whereas the brassinosteroid pathway regulates this process upon long-term exposure (Martins et al., 2017). Furthermore, in the studies from Feraru et al. (2019) and Wang et al. (2016), plants were shifted to 29°C, which could also be perceived as a stress by the plants, thereby possibly confounding root responses to higher ambient temperature with the temperature stress pathway response (Bielach et al., 2017). These studies highlight the important function of auxin in controlling plant response to various environmental fluctuations (Kazan, 2013; Zhao, 2018). By conducting our temperature shifts at 27°C, we have now confirmed that auxin perception and signaling are required for root responses to higher ambient temperature. We also obtained genetic evidence for the requirement of auxin biosynthesis for the root growth response to elevated temperature, suggesting that the control of auxin levels is crucial for regulation of root thermomorphogenesis. Our measurements of auxin levels showed that hy5 and phytochrome mutants display lower auxin levels at 21°C and at 27°C than wild-type plants. However, although the levels seemed to mildly decrease upon increased ambient temperature in these mutants, these changes were not statistically significantly different to those of wild-type plants with our limited experimental replication at a single time point. Given our genetic evidence that suggests that auxin signaling is required for root thermormorphogenesis and has a permissive role rather that an inductive role, it is possible that auxin levels might be controlled in a more complex and dynamic manner. Moreover, auxin signaling output is tightly connected to its transport within and across tissues (reviewed by Benjamins and Scheres, 2008). For instance, during shoot responses to temperature, auxin is produced in the cotyledons and transported to the hypocotyl to promote cell elongation (Bellstaedt et al., 2019). Furthermore, the modulation of auxin long-distance transport from the shoot to the root can regulate root developmental responses to environmental light conditions (Salisbury et al., 2007; Sassi et al., 2012). Because the molecular mechanisms controlling the shoot-to-root auxin transport during thermomorphogenesis remains elusive, it will be crucial to further investigate the dynamics of auxin production, signaling and transport as well as to study how it is coordinated between shoot and root.
Together with our findings, these hypotheses open new avenues to further characterize the communication between shoot and root, which could have important implications for plant growth and biomass allocation upon environmental challenges. Studies have commonly used micro-grafting experiments to investigate long distance signaling between the shoot and the root (Chen et al., 2006, 2016). Given that we analyzed growth response to temperature at early seedling stage, this strategy remains technically challenging, because the impact of sectioning on the growth response might override the effect of the genetic backgrounds used as scions. Instead, we have used a domain-specific rescue approach (Hacham et al., 2011; Kang et al., 2017) by driving a tagged version of HY5 under a shoot-specific promoter. In line with the specificity of the shoot expression, we did not detect fluorescence signal or observe HY5 protein accumulation in the root by immunoblotting. Although these experimental methods cannot fully exclude that traces of HY5 protein are still present, the levels would be considerably lower than wild-type levels and unlikely to have strong impact on the observed phenotype. The use of large tags fused to HY5, such as 3×YFP, could further immobilize the protein and could be used in combination with organ-specific promoters to probe the HY5 domain-specific function. To complement the chimera approach, we used a mechanical approach by removing shoots, and observed that HY5 function might be locally required in the root for thermomorphogenesis, whereas phytochrome might act mainly from the shoot. This observation could be further tested using shoot-specific or root-specific genetics, with tools such as the CAB3 or INORGANIC PHOSPHATE TRANSPORTER 1-1 (PHT1-1) promoters (Procko et al., 2016; Vijaybhaskar et al., 2008) being valuable to further elucidate how the shoot and the root communicate during thermomorphogenesis.
Based on our results, we propose a model wherein roots integrate systemic signals modulated by a shoot module including HY5 and phytochromes with more locally acting auxin signaling during thermomorphogenesis (Fig. 7). The integration of signals that are relayed from the shoot as well as more local ones in the root could constitute a flexible system to adapt growth in response to changes in air temperature perceived in the shoot, while at the same time tuning growth locally by modulating hormonal homeostasis. Thus, it will be important in the future to further understand to what extent these two signaling pathways interact and how they are coupled at the temporal level.
MATERIALS AND METHODS
Plant material and growth conditions
In this study we used the following published lines: Col-0 and Ler ecotypes as wild type, phyAB (Zheng et al., 2013), hy5 (Jia et al., 2014), hy5-221, hy5-215, hy5-1 (Oyama et al., 1997), phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993), pif4-101 (Lorrain et al., 2008), pif1,3,4,5 (pifQ) (Leivar et al., 2008), PIF4-OX (pPIF4:PIF4-FLAG) (Gangappa and Kumar, 2017), hy5 pifQ (Jia et al., 2014), det1-1 (Pepper et al., 1994), cop1-4 (McNellis et al., 1994), hy5 det1 (Gangappa and Kumar, 2017), hy5 cop1 (Rolauffs et al., 2012), tir1-1, afb2-3, tir afb2 (Parry et al., 2009), tmk1 tmk4 (Dai et al., 2013), yucca3,5,7,8,9 (yucQ) (Chen et al., 2014), DR5v2 (pDR5v2:3×YFP-NLS ) (Liao et al., 2015). CAB3 and CER6 promoters have been previously described (Procko et al., 2016), and the DoF (HA-YFP-HA) tag was described in Burger et al. (2017). HY5 rescue lines were generated by inserting pCAB3:HA-YFP-HA-HY5 and pCER6:HA-YFP-HA-HY5 in the hy5 background (Lian et al., 2011), as described in (Burko et al., 2020b).
Unless specified otherwise, plants were grown in long-day conditions (16 h light/8 h dark) in walk-in growth chambers (Conviron) at 21°C or 27°C, 60% humidity, at 146 photosynthetic active radiation (PAR) (see Table S1 for light spectra). During nighttime, temperature was decreased to 15°C and 21°C for the low and high temperature conditions, respectively. In our growth condition 2, plants were grown in reach-in growth chambers at 60% humidity, 122 PAR (see Table S1 for light spectra), temperature was kept constant at either 21°C or 27°C. Environmental conditions were established and monitored using commercial software (Valoya).
Plants were cultivated on plates containing 0.5× Murashige and Skoog (Caisson), 1% MES (Acros Organic, Hampton, NH, USA), 1% sucrose (Fisher Bioreagents) and 0.8% agar powder (Caisson). For temperature shift experiments, plants were germinated and grown until 3 days after germination at 21°C to synchronize their development. On the third day, plants were shifted at zeitgeber 1-3 (ZT1-3) at 27°C and grown for an additional 3 days at 21°C or 27°C.
For dark-grown seedlings, the plates were left in the light for 24 h at 21°C, then isolated from light using aluminium foil and grown at 21°C for an additional 2 days. After 3 days, half of the plates were moved to 27°C. After 24 h, half of the plates from 21°C or 27°C were scanned, then after 72 h, the rest of the plates were scanned.
Roots grown on plates in the dark were isolated from light using metal combs that contained holes, and plates were wrapped with aluminium foil.
Shoot sectioning was performed by cutting at the apex of the hypocotyl to prevent damage of the root. Sections were cut 3 days after germination and were then transferred to 21°C or 27°C.
Root measurements and analysis
Root images were acquired using a multiplex scanning system, as described in Slovak et al. (2014). Images were processed using the Fiji software (https://fiji.sc/). Root and hypocotyl lengths were measured at 3 days after germination (DAG; before temperature shift) and at 6 DAG. Growth rates were obtained by subtracting the length at 6 DAG from the length at 3 DAG. Normalized growth was calculated by dividing root growth rate at 27°C by the average growth rate at 21°C. Root growth at individual temperatures can be found in Fig. S6. Raw values for individual temperatures can be found in Table S1.
Statistical analysis was performed using Excel (Microsoft) or R software (https://www.r-project.org/). Linear regression was performed using the lm function in R and graphs were displayed in R using ggplot2.
Confocal images were acquired on a Zeiss 710 inverted microscope (Zeiss) or on a Zeiss CSU spinning disk confocal microscope (Salk Biophotonics Core). Pictures were processed using Fiji software. Roots were stained with propidium iodide (Sigma-Aldrich) at 0.1 mg/ml. Root meristem size was measured from the quiescent center to the first cortical cell that was twice as long as it was wide, as previously described (Feraru et al., 2019). Dot plots were generated using the plots of data online tool (Postma and Goedhart, 2019).
For the timecourse analysis of normalized root growth, plates were scanned at 0, 24, 48 and 72 h after temperature shift. Images were stacked using ImageJ, and root length was measured at individual time points. Cumulative root meristem cell size was conducted in Fiji using the Cell-O-Tape plugin (French et al., 2012).
Immunoblotting
Western blots were performed as described in Li et al. (2012), with minor modifications. A total of 25 roots and 20 shoots were harvested at 6 DAG and extracted in 2× loading buffer [18 μl β-mercaptoethanol to 1 ml 2× NuPAGETM LDS sample buffer (NP008, Thermo Fisher Scientific)]. Loading buffer was added to roots (70 µl) and shoots (140 µl), and the samples were then boiled for 5 min. Bis-tris gels (4-12%; Invitrogen) and semi-dry transfer (Pierce G2 Fast Blotter, Thermo Fisher Scientific) were used. Primary antibodies used were anti-HA-HRP (1:2000; 12013819001, Roche), anti-HY5(N) (1:5000; R1245-1b, ABicode) and anti-actin (1:30,000; A0408, Sigma-Aldrich). Secondary antibodies used were goat anti-mouse (1:70,000; 1706516, Bio-Rad) for αActin and goat anti-rabbit (1:5000; 1706515, Bio-Rad) for αHY5(N).
Gene expression analysis
Biological triplicates were analyzed. Total RNA was extracted from roots or shoots of plants 6 DAG using an RNeasy kit (Qiagen). RNA was treated with DNAse using the Turbo DNA-free kit (Invitrogen) and further purified on columns from the RNeasy kit.
Next generation sequencing (NGS) libraries were generated using TruSeq Stranded mRNA library prep kits (Illumina). Libraries were sequenced on a HiSeq2500 (Illumina) as single-end 50 bp reads.
NGS analysis was performed using Tophat2 for mapping reads on the Arabidopsis genome (TAIR10) (Kim et al., 2013), HT-seq for counting reads (Anders et al., 2015) and EdgeR for quantifying differential expression (Robinson et al., 2009). We set a threshold for differentially expressed genes [fold change (FC)>2 or FC<−2, false discovery rate (FDR)<0.01]. Genotype×environment interaction analysis was performed using a linear model and type II ANOVA in R. Gene ontology analysis was performed using AgriGOv2 online tool (Tian et al., 2017). Venn diagrams were generated with the VIB online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Auxin measurements
For auxin measurement, plants were shifted at ZT1-3 at 27°C, grown at 21°C or 27°C and harvested at ZT 13-15.
The extraction, purification and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of endogenous IAA, and its precursors and metabolites, were carried out according to Novák et al. (2012). Briefly, ∼10 mg of frozen material per sample was homogenized using a bead mill (27 Hz, 10 min, 4°C; MixerMill, Retsch GmbH) and extracted in 1 ml of 50 mM sodium phosphate buffer containing 1% sodium diethyldithiocarbamate and a mixture of 13C6- or deuterium-labeled internal standards. After centrifugation (18,000 g, 15 min, 4°C), the supernatant was divided in two aliquots, the first aliquot was derivatized using cysteamine (0.25 M, pH 8; 1 h; room temperature; Sigma-Aldrich), the second aliquot was immediately further processed as follows. The pH of the sample was adjusted to 2.5 by addition of 1 M HCl, and the sample was then applied to a preconditioned solid-phase extraction column Oasis HLB (30 mg, 1 cc; Waters Inc., Milford, MA, USA). After sample application, the column was rinsed with 2 ml 5% methanol. Compounds of interest were then eluted with 2 ml 80% methanol. The derivatized fraction was purified in the same way. Mass spectrometry analysis and quantification were performed using an LC-MS/MS system comprising of a 1290 Infinity Binary LC System coupled to a 6490 Triple Quad LC/MS System with Jet Stream and Dual Ion Funnel technologies (Agilent Technologies). Raw measurements for individual temperatures can be found in Table S1.
Supplementary Material
Acknowledgements
We would like to thank Yvon Jaillais (ENS, Lyon, France), Mark Estelle (UCSD, La Jolla, USA), Yunde Zhao (UCSD, La Jolla, USA) and Adam Seluzicki (Salk Institute, La Jolla, USA) for kindly sharing published mutant plant lines with us. We would also like to thank members of the Busch laboratory for critically reading the manuscript. K.L and J.S. thank the Swedish Metabolomics Centre (http://www.swedishmetabolomicscentre.se/) for access to instrumentation.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.G., W.B.; Methodology: C.G.; Formal analysis: L.Z.; Investigation: C.G., Y.B., M.P.P., J.S., K.L.; Resources: B.W., S.V.K., W.B.; Data curation: L.Z.; Writing - original draft: C.G., W.B.; Writing - review & editing: C.G., W.B.; Visualization: C.G., Y.B.; Supervision: K.L., J.C., W.B.; Project administration: W.B.; Funding acquisition: K.L., J.C., W.B.
Funding
This study was funded by the National Institute of General Medical Sciences of the National Institutes of Health (grant number R01GM127759 to W.B.) and start-up funds from the Salk Institute for Biological Studies. J.C. is an investigator of the Howard Hughes Medical Institute. This study was supported by the HHS NIH National Institute of General Medical Sciences (grant 5R35GM122604-02_05 to J.C.), the Howard Hughes Medical Institute (to J.C.), the European Molecular Biology Organization (grant ALTF 785-2013 to Y.B.), the United States - Israel Binational Agricultural Research and Development Fund (grant FI-488-13 to Y.B.) and the Human Frontier Science Program (LT000222/2013-L to B.W). K.L. and J.S. acknowledge the Swedish research councils VINNOVA, Vetenskapsrådet (VR) and the Knut och Alice Wallenbergs Stiftelse (KAW). Deposited in PMC for release after 12 months.
Data availability
Raw RNA-seq reads generated as part of this study are deposited at GEO under accession number GSE138133.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.192625.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.192625.reviewer-comments.pdf
References
- Anders S., Pyl P. T. and Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellstaedt J., Trenner J., Lippmann R., Poeschl Y., Zhang X., Friml J., Quint M. and Delker C. (2019). A Mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 180, 757-766. 10.1104/pp.18.01377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R. and Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443-465. 10.1146/annurev.arplant.58.032806.103805 [DOI] [PubMed] [Google Scholar]
- Bielach A., Hrtyan M. and Tognetti V. B. (2017). Plants under stress: involvement of auxin and Cytokinin. Int. J. Mol. Sci. 18, 1427 10.3390/ijms18071427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M., Willige B.C. and Chory J. (2017). A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nat. Commun. 8, 2201 10.1038/s41467-017-02347-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y., Seluzicki A., Zander M., Pedmale U. V., Ecker J. R. and Chory J. (2020a). Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32, 967-983. 10.1105/tpc.19.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y., Gaillochet C., Seluzicki A., Chory J. and Busch W. (2020b). Local HY5 activity mediates hypocotyl growth and shoot-to-root communication. Plant Commun. 1, 100078 10.1016/j.xplc.2020.100078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Chen R., Li P., Yu Y., Zheng R., Ge D., Zheng W., Wang X., Gu Y., Gelová Z. et al. (2019). TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568, 240-243. 10.1038/s41586-019-1069-7 [DOI] [PubMed] [Google Scholar]
- Chen A., Komives E. A. and Schroeder J. I. (2006). An improved grafting technique for mature arabidopsis plants demonstrates long-distance shoot-to-root transport of phytochelatins in arabidopsis. Plant Physiol. 141, 108-120. 10.1104/pp.105.072637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y. and Zhao Y. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in arabidopsis roots. Plant Cell Physiol. 55, 1072-1079. 10.1093/pcp/pcu039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yao Q., Gao X., Jiang C., Harberd N. P. and Fu X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640-646. 10.1016/j.cub.2015.12.066 [DOI] [PubMed] [Google Scholar]
- Chung B. Y. W., Balcerowicz M., Di Antonio M., Jaeger K. E., Geng F., Franaszek K., Marriott P., Brierley I., Firth A. E, Wigge P. A. et al. (2020). An RNA thermoswitch regulates daytime growth in arabidopsis. Nat. Plants 6, 522-532. 10.1038/s41477-020-0633-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolfi A., Sessa G., Sassi M., Possenti M., Salvucci S., Carabelli M., Morelli G. and Ruberti I. (2013). Dynamics of the shade-avoidance response in arabidopsis. Plant Physiol. 163, 331-353. 10.1104/pp.113.221549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321-326. 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- Dai N., Wang W., Patterson S. E. and Bleecker A. B. (2013). The TMK subfamily of receptor-like kinases in arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS ONE 8, e60990 10.1371/journal.pone.0060990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., Sonntag L., James G. V., Janitza P., Ibañez C., Ziermann H., Peterson T., Denk K., Mull S., Ziegler J. et al. (2014). The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9, 1983-1989. 10.1016/j.celrep.2014.11.043 [DOI] [PubMed] [Google Scholar]
- Donohue K., Rubio de Casas R., Burghardt L., Kovach K. and Willis C. G. (2010). Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 41, 293-319. 10.1146/annurev-ecolsys-102209-144715 [DOI] [Google Scholar]
- Feraru E., Feraru M. I., Barbez E., Waidmann S., Sun L., Gaidora A. and Kleine-Vehn J. (2019). PILS6 is a temperature-sensitive regulator of nuclear auxin input and organ growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 116, 3893-3898. 10.1073/pnas.1814015116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci A.-S., Galvão V. C., Ince Y. Ç., Boccaccini A., Goyal A., Allenbach Petrolati L., Trevisan M. and Fankhauser C. (2020). PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in arabidopsis seedlings. New Phytol. 226, 50-58. 10.1111/nph.16316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., Lee S. H., Patel D., Kumar S. V., Spartz A. K., Gu C., Ye S., Yu P., Breen G., Cohen J. D. et al. (2011). PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108, 20231-20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A. P., Wilson M. H., Kenobi K., Dietrich D., Voß U., Ubeda-Tomás S., Pridmore T. P. and Wells D. M. (2012). Identifying biological landmarks using a novel cell measuring image analysis tool: cell-o-tape. Plant Methods 8, 7 10.1186/1746-4811-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S. N. and Botto J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353-1365. 10.1016/j.molp.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Gangappa S. N. and Kumar S. V. (2017). DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 18, 344-351. 10.1016/j.celrep.2016.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. M., Östin A., Sandberg G., Romano C. P. and Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7197-7202. 10.1073/pnas.95.12.7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J.-H., Han S.-H., Lee H.-J. and Park C.-M. (2017). Environmental adaptation of the heterotrophic-to-autotrophic transition: the developmental plasticity of seedling establishment. Crit. Rev. Plant Sci. 36, 128-137. 10.1080/07352689.2017.1355661 [DOI] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M. J., Chory J. and Savaldi-Goldstein S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138, 839-848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa T., Shibasaki K., Numata T., Kawamura Y., Gaude T. and Rahman A. (2013). Cellular auxin homeostasis under high temperature is regulated through a sorting NEXIN1-dependent endosomal trafficking pathway. Plant Cell 25, 3424-3433. 10.1105/tpc.113.115881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illston B. G. and Fiebrich C. A. (2017). Horizontal and vertical variability of observed soil temperatures. Geosci. Data J. 4, 40-46. 10.1002/gdj3.47 [DOI] [Google Scholar]
- Jia K.-P., Luo Q., He S.-B., Lu X.-D. and Yang H.-Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in arabidopsis. Mol. Plant 7, 528-540. 10.1093/mp/sst093 [DOI] [PubMed] [Google Scholar]
- Jung J.-H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A. K., Box M. S., Charoensawan V., Cortijo S. et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886-889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Kang Y. H., Breda A. and Hardtke C. S. (2017). Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development 144, 272-280. 10.1242/dev.145623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. (2013). Auxin and the integration of environmental signals into plant root development. Ann.Bot. 112, 1655-1665. 10.1093/aob/mct229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. and Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S. and Schopfer P. (2012). Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 11217-11221. 10.1073/pnas.1203746109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M. A., Alvey L., Allen T., Tilley C. A., Harberd N. P., Whitelam G. C. and Franklin K. A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408-413. 10.1016/j.cub.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Kumar S. V., Lucyshyn D., Jaeger K. E., Alós E., Alvey E., Harberd N. P. and Wigge P. A. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242-245. 10.1038/nature10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S. and Deng X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584-593. 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X. W. et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731-749. 10.1105/tpc.106.047688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M., Klose C., Burgie E. S., Rojas C. C. R., Neme M., Hiltbrunner A., Wigge P. A., Schäfer E., Vierstra R. D., Casal J. J. et al. (2016). Phytochrome B integrates light and temperature signals in arabidopsis. Science 354, 897-900. 10.1126/science.aaf5656 [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E. and Quail P. H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815-1823. 10.1016/j.cub.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.-H., Zhang H., Shen Y. et al. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 Plays a role in the feedback regulation of phytochrome a signaling. Plant Cell 22, 3634-3649. 10.1105/tpc.110.075788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ljung K., Breton G., Schmitz R. J., Pruneda-Paz J., Cowing-Zitron C., Cole B. J., Ivans L. J., Pedmale U. V., Jung H.-S. et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785-790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.-L., He S.-B., Zhang Y.-C., Zhu D.-M., Zhang J.-Y., Jia K.-P., Sun S.-X., Li L. and Yang H.-Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023-1028. 10.1101/gad.2025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.-Y., Smet W., Brunoud G., Yoshida S., Vernoux T. and Weijers D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207-210. 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P. D., Whitelam G. C. and Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312-323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Martins S., Montiel-Jorda A., Cayrel A., Huguet S., Roux C. P.-L., Ljung K. and Vert G. (2017). Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nat. Commun. 8, 309 10.1038/s41467-017-00355-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T. W., von Arnim A. G., Araki T., Komeda Y., Miséra S. and Deng X. W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487-500. 10.1105/tpc.6.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O., Hényková E., Sairanen I., Kowalczyk M., Pospíšil T. and Ljung K. (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72, 523-536. 10.1111/j.1365-313X.2012.05085.x [DOI] [PubMed] [Google Scholar]
- Omelyanchuk N. A., Wiebe D. S., Novikova D. D., Levitsky V. G., Klimova N., Gorelova V., Weinholdt C., Vasiliev G. V., Zemlyanskaya E. V., Kolchanov N. A. et al. (2017). Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci. Rep. 7, 2489 10.1038/s41598-017-02476-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M. T., Hardtke C. S., Wei N. and Deng X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462-466. 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y. and Okada K. (1997). The arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983-2995. 10.1101/gad.11.22.2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.-I., Liu J. R. and Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968-975. 10.1093/pcp/pch125 [DOI] [PubMed] [Google Scholar]
- Park E., Kim Y. and Choi G. (2018). Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30, 1277-1292. 10.1105/tpc.17.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Calderon-Villalobos L. I., Prigge M., Peret B., Dharmasiri S., Itoh H., Lechner E., Gray W. M., Bennett M., Estelle M. et al. (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 106, 22540-22545. 10.1073/pnas.0911967106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008). Temperature perception and signal transduction in plants. New Phytol. 179, 615-628. 10.1111/j.1469-8137.2008.02478.x [DOI] [PubMed] [Google Scholar]
- Pepper A., Delaney T., Washburnt T., Poole D. and Chory J. (1994). DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell 78, 109-116. 10.1016/0092-8674(94)90577-0 [DOI] [PubMed] [Google Scholar]
- Postma M. and Goedhart J. (2019). PlotsOfData—A web app for visualizing data together with their summaries. PLoS Biol. 17, e3000202 10.1371/journal.pbio.3000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C., Burko Y., Jaillais Y., Ljung K., Long J. A. and Chory J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30, 1529-1541. 10.1101/gad.283234.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Delker C., Franklin K. A., Wigge P. A., Halliday K. J. and van Zanten M. (2016). Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2, 15190 10.1038/nplants.2015.190 [DOI] [PubMed] [Google Scholar]
- Reed J. W., Nagpal P., Poole D. S., Furuya M. and Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147-157. 10.1105/tpc.5.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J. W., Nagatani A., Elich T. D., Fagan M. and Chory J. (1994). Phytochrome A and Phytochrome B have overlapping but distinct functions in arabidopsis development. Plant Physiol. 104, 1139-1149. 10.1104/pp.104.4.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. and Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolauffs S., Fackendahl P., Sahm J., Fiene G. and Hoecker U. (2012). Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol. 160, 2015-2027. 10.1104/pp.112.207233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J. A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U. and Deng X. W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642-2647. 10.1101/gad.1122903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury F. J., Hall A., Grierson C. S. and Halliday K. J. (2007). Phytochrome coordinates arabidopsis shoot and root development. Plant J. 50, 429-438. 10.1111/j.1365-313X.2007.03059.x [DOI] [PubMed] [Google Scholar]
- Sassi M., Lu Y., Zhang Y., Wang J., Dhonukshe P., Blilou I., Dai M., Li J., Gong X., Jaillais Y. et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in arabidopsis. Development 139, 3402-3412. 10.1242/dev.078212 [DOI] [PubMed] [Google Scholar]
- Shipley B. and Meziane D. (2002). The balanced-growth hypothesis and the allometry of leaf and root biomass allocation. Funct. Ecol. 16, 326-331. 10.1046/j.1365-2435.2002.00626.x [DOI] [Google Scholar]
- Slovak R., Göschl C., Su X., Shimotani K., Shiina T. and Busch W. (2014). A scalable open-source pipeline for large-scale root phenotyping of arabidopsis. Plant Cell 26, 2390-2403. 10.1105/tpc.114.124032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J. and Li C. (2012). PIF4–mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594 10.1371/journal.pgen.1002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornley J. H. M. (1972). A balanced quantitative model for root: shoot ratios in vegetative plants. Ann. Bot. 36, 431-441. 10.1093/oxfordjournals.aob.a084602 [DOI] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W. and Su Z. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122-W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K. P., Bou-Torrent J., Stewart K., Steel G., Rodríguez-Concepción M. and Halliday K. J. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10, e1004416 10.1371/journal.pgen.1004416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaybhaskar V., Subbiah V., Kaur J., Vijayakumari P. and Siddiqi I. (2008). Identification of a root-specific glycosyltransferase from arabidopsis and characterization of its promoter. J. Biosci. 33, 185-193. 10.1007/s12038-008-0036-5 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang Y., Kieffer M., Yu H., Kepinski S. and Estelle M. (2016). HSP90 regulates temperature-dependent seedling growth in arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 7, 10269 10.1038/ncomms10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Dai N., Chen J., Nagawa S., Cao M., Li H., Zhou Z., Chen X., De Rycke R., Rakusová H. et al. (2014). Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343, 1025-1028. 10.1126/science.1245125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y., Sullivan J. A., Komatsu S., Gusmaroli G., Suzuki G., Yin J., Ishibashi T., Saijo Y., Rubio V., Kimura S. et al. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172-2181. 10.1101/gad.1229504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2018). Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417-435. 10.1146/annurev-arplant-042817-040226 [DOI] [PubMed] [Google Scholar]
- Zheng X., Wu S., Zhai H., Zhou P., Song M., Su L., Xi Y., Li Z., Cai Y., Meng F. et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25, 115-133. 10.1105/tpc.112.107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.