Abstract
Accumulating evidence have suggested that long non-coding RNAs (lncRNAs) act as a critical regulator in tumorgenesis. LncRNA KCNQ1OT1 (KCNQ1OT1) has been recently shown to be dysregulated in many cancers. This study was aimed to explore the biological role of KCNQ1OT1 in hepatocellular carcinoma (HCC). In our study, we first observed the expression level of KCNQ1OT1 was distinctly up-regulated in HCC tissues and cell lines compared with adjacent non-cancer tissues and normal liver cell line. And clinical results indicated that higher expression of KCNQ1OT1 was correlated with poor prognosis of patients with HCC. Next, functional studies revealed that knockdown of KCNQ1OT1 induced apoptosis and repressed proliferation, migration and invasion of HCC cells. In addition, knockdown of KCNQ1OT1 suppressed xenograft tumor growth in vivo. Mechanically, we found that KCNQ1OT1 can promote the expression of IGF1R by functioning as a competing endogenous RNA of miR-148a-3p. In conclusion, our results shown the oncogenic role of KCNQ1OT1 in HCC by regulating the miR-148a-3p/IGF1R axis and may provide a new insight and a potential therapeutic target for HCC.
Keywords: hepatocellular carcinoma, KCNQ1OT1, miR-148a-3p, IGF1R, tumor
Introduction
Hepatocellular carcinoma (HCC) is one of the most common human malignancies worldwide and as the second most common cause of cancer-related death.1 It is estimated that HCC accounts for 90% of liver cancer.2 The incidence of HCC is closely related to chronic liver inflammation, some main reasons that cause hepatitis including hepatitis B and C viruses, alcohol and aflatoxin, etc.3 In recent years, despite advances in prognosis and treatment to HCC, such as surgery, chemotherapy, radiotherapy. The prognosis of HCC patients is still poor, unfortunately, the recurrence rate of HCC patients is more than 80%.4 Therefore, it is urgent to comprehend the underlying pathogenesis of HCC and find novel targets for examination and treatment of HCC.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs with over 200 nucleotides in length and no protein coding ability.5,6 Studies disclosed that lncRNAs are involved in diverse pathophysiological processes, and the dysregulation of lncRNAs expression was implicated in the occurrence and development of cancers. For instance, long noncoding RNA GMAN promotes HCC progression by regulating the expression of eIF4B.7 Silencing of long noncoding RNA HOXA11-AS inhibits proliferation, invasion, and self-renewal of hepatocellular carcinoma stem cells by preventing the Wnt signaling pathway.8 LncRNA ID2-AS1 suppresses metastasis of hepatocellular carcinoma by activating the HDAC8/ID2 pathway.9 LncRNA MCM3AP-AS1 promotes cancer progression by regulating miR-194-5p/FOXA1 axis in hepatocellular carcinoma.10 In recent years, numerous studies shown KCNQ1OT1 presents a carcinogenic role in multiple cancers. For instance, KCNQ1OT1 promote osteosarcoma growth by ALDOA enhancing aerobic glycolysis and sponging miR-34c-5p.11 In tongue cancer, KCNQ1OT1 regulates proliferation and displaying resistance via modulating miR-211-5p/Ezrin/Fak/Src signaling.12 LncRNA KCNQ1OT1 inhibits gastric cancer cell progression by mediating miR-9/LMX1A axis.13 However, the biological role of KCNQ1OT1 in HCC remains to be further identified.
It is well known that miRNAs work as key regulators in human cancers through directly binding to 3′UTR of their targets. For example, MiR-129-5p inhibited gastric cancer progression through modulating HOXC10/Cyclin D1 to impel cell cycle arrest.14 In HCC, miR-17-5p and miR-20a-5p suppress postoperative metastasis by blocking HGF/ERBB3-NF-κB positive feedback loop.15 Although the role of miR-148a-3p in HCC has been reported, the relationship between miR-148a-3p and KCNQ1OT1 remains barely known in cancers. Here, we found that KCNQ1OT1 was aberrantly up-regulated in HCC tissues and cell lines. Further, we predicted that miR-148a-3p was a potential target of KCNQ1OT1 by starBase, and dual-luciferase reporter gene assays confirmed this prediction. In addition, we found that the effects of miR-148a-3p on apoptosis, proliferation, migration and invasion of HCC cells were abolished by KCNQ1OT1. And the negative correlation between KCNQ1OT1 and miR-148a-3p was determined by correlation analysis. Simultaneously, KCNQ1OT1 promoted IGF1R expression through suppressing miR-148a-3p. Therefore, our study revealed the carcinogenic role of KCNQ1OT1 in HCC by miR-148a-3p/IGF1R axis.
Materials and Methods
Clinical Tissues
Clinical HCC tissues and matched adjacent normal tissues were collected from 60 HCC patients without any treatments before surgery at Tianjin Medical University Second Hospital. Specimens were frozen in liquid nitrogen. This study was approved by the Ethics Committee on Human Research of the Tianjin Medical University Second Hospital. And written informed consents were obtained from all patients.
Cell Culture and Transfection
HCC cell lines (SMMC-7721, Huh-7, HepG2 and Hep3B), normal liver cell line LO2 and 293 T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were maintained in Roswell Park Memorial Institute RPMI-1640 medium (Gibco, CA) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a 5% CO2 incubator at 37°C.
To obtain overexpressed KCNQ1OT1 vector, full-length sequence of KCNQ1OT1 was amplified and cloned into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). For knockdown of KCNQ1OT1, 2 short hairpin RNA (shRNA) (sh-KCNQ1OT1#1 and KCNQ1OT1#2) targeting KCNQ1OT1 were purchased from GenePharma (Shanghai, China). The miR-148a-3p mimics and inhibitors, small interfering RNA-IGF1R (si-IGF1R, for knockdown IGF1R) and their respective negative control (NC) were purchased from Ribobio (Guangzhou, China). HepG2 and Hep3B cells were transfected with indicated sh-RNAs, plasmid vectors, mimics or inhibitors by Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
QRT-PCR Assay
Total RNA in HCC tissues and cells were extracted with TRIzol reagent (Invitrogen). RNA reverse transcription was carried out using Reverse Transcription System Kit (Takara, Dalian, China). Then qRT-PCR assay was performed using SYBR Premix EX Taq kit (Takara Biotech, Japan) on an ABI 7900 system (Applied Biosystems, Foster City, CA). GAPDH and U6 were used as endogenous control. The relative expression levels of genes were calculated by 2−ΔΔCt method.
CCK8 Assay
To assess the proliferation ability of cells, cell counting kit-8 (CCK-8) assay were performed. In brief, transfected HCC cells were placed in a 96-well plate (5000 cells/well). Then, cells were incubated for additional 24 hours. After incubated with CCK8 reagents (10 μL) for 4 hours. The absorbance at the 450 nm was measured using a microplate reader machine.
Apoptosis Assay
Cell apoptosis was evaluated by an Annexin V kit (Solarbio, Beijing, China). After treatments, cells were collected and washed with PBS. Then cells were resuspended with 1 × binding buffer. 5 µL of FITC-Annexin V and 5 µL PI (PI) were added into cells and incubated for 10 min in the dark. Subsequently, the cell apoptosis rates were assessed by a flow cytometry (BD Biosciences, USA).
Transwell Assay
The migration and invasion capacities of HCC cells were evaluated using transwell assay (8 µm pore size, BD Biosciences, San Jose, CA). In brief, in migration analysis, transfected HCC cells were seeded into the upper chamber with a serum-free medium. The lower chambers were added with medium containing 10% FBS. After incubation for 24 hours, the migrating and invading cells were fixed using 4% paraformaldehyde and stained with 0.1% crystal violet, then cells were counted by a microscope. For invasion analysis, the upper chamber was pre-coated with Matrigel. Other procedures were performed in accordance with the migration analysis.
Western Blot Analysis
After transfection, total proteins were extracted from HCC tissues and cells using RIPA lysis buffer (Beyotime, Beijing, China). Protein concentrations were calculated by BCA Protein Assay Kit (Beyotime, Beijing, China). Then, equal amount of protein samples were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membrane (Invitrogen). Next, membrane was blocking with 5% non-fat milk for 1 hour at room temperature and incubated with primary antibodies (anti-IGF1R antibody, 1:1000, Abcam; anti-GAPDH antibody, 1:10 000 Abcam) overnight at 4°C. In addition, the membrane was incubated with corresponding secondary antibodies at room temperature for 1 hour. The immunoreactive signal was detected by an ECL immunoblotting kit (Millipore, USA).
Luciferase Reporter Assay
The potential binding sites of miR-148a-3p on KCNQ1OT1 and IGF1R were predicted using starBase (http://starbase.sysu.edu.cn/). Luciferase reporter plasmids containing wild-type (Wt) or mutant (Mut) 3′UTR of KCNQ1OT1 or IGF1R were cotransfected with miR-148a-3p mimics or inhibitors and respective negative control (NC) into HCC or 293 T cells by Lipofectamine 2000 (Invitrogen). After 48 hours of transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s instructions.
Xenograft Tumor Assay in Nude Mice
Female 4-week-old BALB/c nude mice purchased from Central Laboratory of Animal Science, Xi’an jiaotong University and randomly divided into 2 groups. HepG2 cells (3 × 106) transfected with sh-NC or sh-KCNQ1OT1#1 were subcutaneously injected into nude mice. The tumor formation was observed every 5 days. Tumor volume was calculated according to the formula (0.5 × length × width2). Thirty days later, all mice were sacrificed and tumor tissue were used for further research. This study was carried out in accordance with institutional guidelines and approved by the Ethics Committee of the Tianjin Medical University Second Hospital.
Statistical Analysis
Statistical analyses were performed by GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA). Results were presented as mean ± SD. Statistical analyses were conducted using Student’s t test or one-way ANOVA analysis. p < 0.05 was considered statistically significant.
Results
KCNQ1OT1 Was Overexpressed in HCC Tissues and Cell Lines
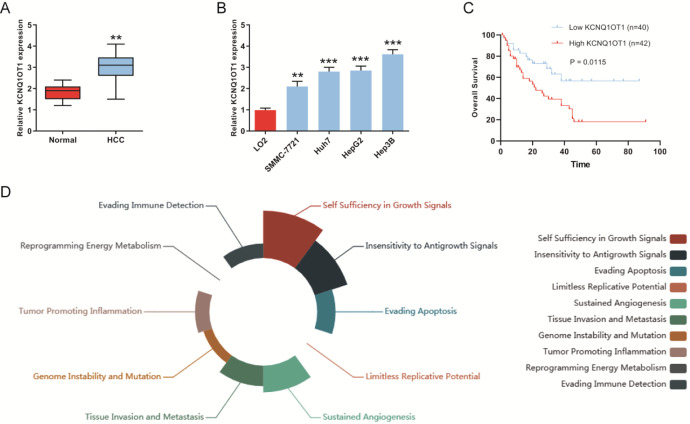
To determine KCNQ1OT1 was related to the progression of HCC, we firstly detected the expression of KCNQ1OT1 in 60 pairs of HCC cancer tissues and adjacent normal tissues. We found that KCNQ1OT1 was remarkably overexpressed in HCC cancer tissues (Figure 1A). As presented in Figure 1B, the expression levels of KCNQ1OT1 were also significantly increased in HCC cell lines (SMMC-7721, Huh-7, HepG2 and Hep3B) compared with normal liver LO2 cells. In addition, Kaplan-Meier analysis suggested that HCC patients with low KCNQ1OT1 expression have longer overall survival (OS) than those with high KCNQ1OT1 expression (Figure 1C). And the dysregulation of KCNQ1OT1 expression was involved in different cancer hallmark which was analyzed by LnCeVar (Figure 1D). These results showed that KCNQ1OT1 expression was increased in HCC tissues and cell lines.
Figure 1.
KCNQ1OT1 was overexpressed in HCC tissues and cell lines. A, The expression of KCNQ1OT1 was detected in HCC cancer tissues and adjacent normal tissues by qRT-PCR. B, KCNQ1OT1 expression was detected in HCC cell lines (SMMC-7721, Huh-7, HepG2 and Hep3B) and normal liver cell line LO2 by qRT-PCR. C, Overall survival rate was analyzed in HCC patients with differential KCNQ1OT1 expression. D, Dysregulation of KCNQ1OT1 was involved in different cancer hallmark which was analyzed by LnCeVar.
Knockdown of KCNQ1OT1 Repressed the Growth of HCC Cells In Vitro
Based on the results above, we next explored the biological role of KCNQ1OT1 in HCC cells. We knocked down or overexpressed KCNQ1OT1 in HepG2 and Hep3B cells (Figure 2A and B). sh-KCNQ1OT1#1 shown a better knockdown efficiency and was selected for other experiments. First, cell apoptosis assay shown that apoptosis rates were increased in HCC cells transfected with sh-KCNQ1OT1#1 (Figure 2C). Next, the effect of KCNQ1OT1 on cell proliferation was detected using CCK-8 assay. We found that knockdown of KCNQ1OT1 inhibited proliferation of HepG2 and Hep3B cells (Figure 2D). Furthermore, similar results were obtained from transwell assay. Knockdown of KCNQ1OT1 attenuated migration and invasion capacities of HepG2 and Hep3B cells (Figure 2E and F). Taken together, these results indicated that lower expression of KCNQ1OT1 inhibited proliferation, migration and invasion capacities of HCC cells.
Figure 2.
Knockdown of KCNQ1OT1 inhibited proliferation of HCC cells. A and B, Efficiency of knockdown and overexpression of KCNQ1OT1 was measured in HepG2 and Hep3B cells by qRT-PCR. C, Flow cytometry was used to analyze apoptosis of HepG2 and Hep3B cells transfected with sh-NC or sh-KCNQ1OT1#1. D, Cell proliferation capacity was assessed in HepG2 and Hep3B cells transfected with sh-NC or sh-KCNQ1OT1#1 by CCK-8 assay. E and F, Transwell assay was carried out to study the effects of KCNQ1OT1 on migration and invasion abilities of HCC cells, respectively.
KCNQ1OT1 Is a Molecular Sponge of miR-148a-3p
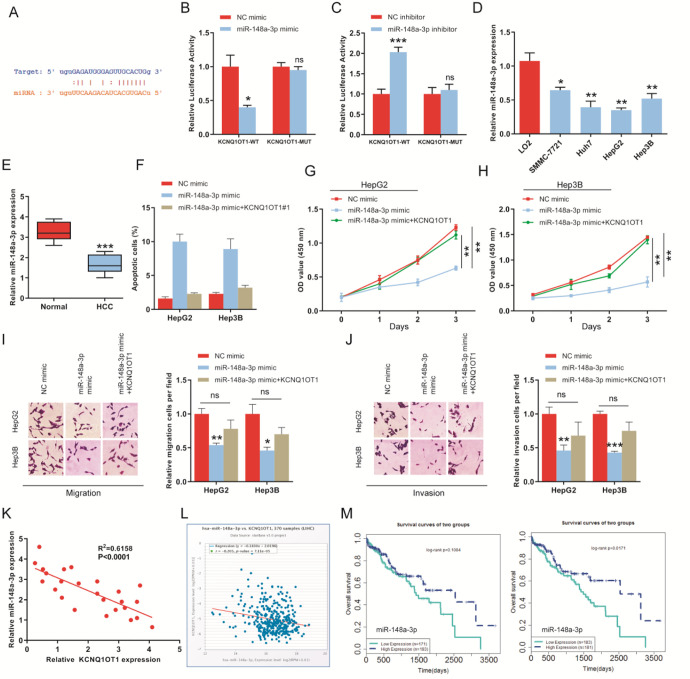
Mechanistically, we predicted that miR-148a-3p might be a potential target of KCNQ1OT1 by starBase (Figure 3A). Then, dual-luciferase assays were carried out, and results shown that miR-148a-3p mimics reduced the luciferase activity (Figure 3B). However, luciferase activity was greatly increased by miR-148a-3p inhibitors (Figure 3C). We also detected the expression of miR-148a-3p in HCC tissues and cell lines. We found that the expression of miR-148a-3p was significantly down-regulated in HCC cell lines and tissues as compared to normal liver cells and tissues (Figure 3D and E). Further, to determine whether KCNQ1OT1-miR-148a-3p axis regulated the HCC oncogenesis. Cell proliferation, apoptosis and transwell assays were performed. We observed that high expression of miR-148a-3p remarkably promoted apoptosis of HepG2 and Hep3B cells (Figure 3F). And proliferation, migration and invasion abilities were suppressed in HCC cells transfected miR-148a-3p mimics (Figure 3G-J). However, cotransfection of KCNQ1OT1 and miR-148a-3p mimics rescued these effects of miR-148a-3p mimics on cell apoptosis, proliferation, migration and invasion (Figure 3F-J). Besides, we found that the expression of KCNQ1OT1 was negatively related to miR-148a-3p expression in HCC tissues (Figure 3K). Similarly, the negative correlation between KCNQ1OT1 and miR-148a-3p was also proved by starBase database (Figure 3L). Finally, the clinical data from LnVeCar suggested that patients with high miR-148a-3p expression have longer survival time than those with low expression of miR-148a-3p (Figure 3M).
Figure 3.
miR-148a-3p is a target of KCNQ1OT1. A, The binding sites of miR-148a-3p on KCNQ1OT1 was predicted by starBase database. B and C, Dual-luciferase reporter gene assays were employed to determine the combination between KCNQ1OT1 and miR-148a-3p. D, The expression level of miR-148a-3p was measured in normal liver cell line and HCC cell lines using qRT-PCR. E, Expression of miR-148a-3p in HCC tissues and adjacent normal tissues. F, KCNQ1OT1 rescued the promotion of miR-148a-3p on apoptosis of HCC cells. G and H, Overexpression of miR-148a-3p inhibited the proliferation of HepG2 and Hep3B cells, but, KCNQ1OT1 abolished the inhibition. I and J, Transwell assay was employed to determine migration and invasion abilities of HepG2 and Hep3B cells transfected with miR-148a-3p mimics or miR-148a-3p mimics+sh-KCNQ1OT1. K, The negative correlation between KCNQ1OT1 and miR-148a-3p in HCC tissues. L, The correlation analysis between KCNQ1OT1 and miR-148a-3p was performed by starBase database. M, Kaplan-Meier analysis was performed to study the role of miR-148a-3p on overall survival of HCC patients by LnCeVar.
KCNQ1OT1 Upregulates the Expression of IGF1R via Competitively Interacting With miR-148a-3p in HCC Cells
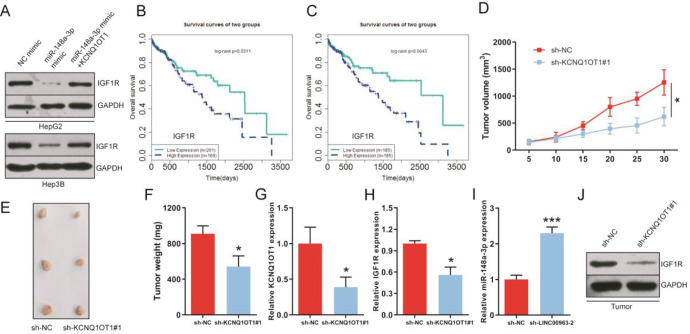
Next, we further explored the potential molecular mechanism of KCNQ1OT1 regulating the progression of HCC. We predicted that IGF1R as a candidate target of miR-148a-3p using starBase (Figure 4A). To confirm the predicted result, dual-luciferase reporter assay was performed and results proffered that miR-148a-3p mimics obviously reduced the luciferase activity of 293 T cells transfected with IGF1R-WT 3′-UTR (Figure 4B), while miR-148a-3p inhibitor exerted significant enhancement on luciferase activity (Figure 4C). Further, we observed that IGF1R expression level was extremely up-regulated in HCC cell lines and cancer tissues (Figure 4D and E). In addition, data from apoptosis assay shown that the inhibition of IGF1R enhanced apoptosis of HCC cells, but, the inhibition was abolished by miR-148a-3p inhibitors (Figure 4F). CCK8 and transwell assay indicated that the knockdown of IGF1R repressed cells proliferation, migration and invasion capacities, similarly, these results were greatly reversed by miR-148a-3p inhibitors (Figure 4G-J). In addition, correlation analysis in HCC tissues shown that the relative expression of IGF1R was inversely or positively correlated with miR-148a-3p or KCNQ1OT1 expression (Figure 4K and L). Bioinformatics analysis has also proved the existence of a negative regulatory relationship between IGF1R and miR-148a-3p via starBase (Figure 4M). In addition, western blot shown that knockdown of KCNQ1OT1 obviously decreased IGF1R expression in HCC cells (Figure 4N). And we found that the inhibition of miR-148a-3p mimics on protein expression of IGF1R was reversed by KCNQ1OT1 (Figure 5A). Survival analysis shown that the overall survival was extended in HCC patients with low IGF1R expression by LnCeVar (Figure 5B and C). Collectively, these results shown that KCNQ1OT1 promoted expression of IGF1R by competitively targeting miR-148a-3p in HCC cells.
Figure 4.
KCNQ1OT1 promotes HCC progression by up-regulating the expression of IGF1R via miR-148a-3p in vitro. A, Bioinformatics software predicted that IGF1R is a target of miR-148a-3p, and interaction region between miR-148a-3p and IGF1R was displayed. B and C, Interaction region between miR-148a-3p and IGF1R was determined by dual-luciferase reporter gene assays. D and E, IGF1R expression levels in normal liver cell line, HCC cell lines, normal tissues and HCC tissues, respectively. F, Regulation between miR-148a-3p and IGF1R on apoptosis of HepG2 and Hep3B cells. G and H, Cell proliferative capacity was suppressed in HCC cells with low IGF1R expression, correspondingly, it was reversed by miR-148a-3p inhibitors. I and J, Transwell assay confirmed the inhibition of si-IGF1R on migration and invasion of HCC cells. Similarly, miR-148a-3p inhibitors rescued the inhibition. K, IGF1R expression was reversely associated with miR-148a-3p expression in HCC patients. L, Kaplan-Meier analysis between IGF1R expression and KCNQ1OT1 expression in HCC patients. M, The correlation analysis of miR-148a-3p and IGF1R in HCC patients by starBase database. N, Knockdown of KCNQ1OT1 suppressed IGF1R expression in protein level in HepG2 and Hep3B cells.
Figure 5.
KCNQ1OT1 regulates the growth of HCC tumor by miR-148a-3p/IGF1R axis. A, Western blot assay shown that overexpression of KCNQ1OT1 abolished the inhibition of miR-148a-3p on IGF1R expression in HepG2 and Hep3B cells. B and C, Overall survival was analyzed of HCC patients with high or low IGF1R expression by LnCeVar. D-F, Tumor volume and weight were measured, respectively. G-I, The expression levels of KCNQ1OT1, IGF1R and miR-148a-3p were detected in HCC tumor tissues using qRT-PCR, respectively. (J) Expression of IGF1R in protein levels in tumor tissues.
Knockdown of KCNQ1OT1 Inhibited HCC Tumorigenesis
To evaluate the effects of KCNQ1OT1 on tumor growth of HCC, HepG2 cells transfected with sh-KCNQ1OT1#1 were subcutaneously injected into nude mice to construct tumor xenograft model. Results shown that knockdown of KCNQ1OT1 suppressed HCC tumor growth (Figure 5D and E) and tumor weight were reduced compared with sh-NC group (Figure 5F). Further, results from HCC tumor tissues indicated that the expression levels of KCNQ1OT1 and IGF1R were down-regulated while miR-148a-3p expression was greatly increased (Figure 5G-I). Besides, expression of IGF1R in protein level was suppressed in tumor tissues of sh-KCNQ1OT1#1 group (Figure 5J). In conclusion, these results suggested that the knockdown of KCNQ1OT1 inhibited tumor growth of HCC.
Discussion
HCC always remains a serious health problem and caused a high death rate despite the advanced in diagnosis and treatments.16 Thus, it is urgent to discover new biomarkers of diagnostic, prognostic indicators and more advanced treatment methods to HCC. In recent years, mounting studies have identified many functional lncRNAs that play key roles in various cancers including HCC.17-20 In this study, we reported a cancer-related lncRNA KCNQ1OT1 and its biological functions in HCC were largely studied. In previous reports, Wang et al observed that KCNQ1OT1 was up-regulated in non-small cell lung cancer and promoted cell proliferation, migration and invasion via regulating miR-129-5p/JAG1 axis.21 Sun et al indicated that LncRNA KCNQ1OT1 promoted progression and chemoresistance of acute myeloid leukemia by modulating the miR-193a-3p/Tspan3 axis.22 However, there are few studies for the biological role of KCNQ1OT1 in HCC. Here, we observed that KCNQ1OT1 expression was distinctly up-regulated in HCC tissues and cell lines. And high expression of KCNQ1OT1 was closely related the poor prognosis of HCC patients. Moreover, in this study, we found that knockdown of KCNQ1OT1 repressed proliferation, migration and invasion while enhanced apoptosis of HCC cells. Therefore, our study demonstrated that KCNQ1OT1 was highly up-regulated in HCC cancer tissues and cell lines, it served as a tumor promoter in the progression of HCC.
LncRNAs can act as competing endogenous (ceRNAs) to mediate gene expression through sponging miRNAs in the occurrence and development of cancers.23-26 For instance, in Glioblastoma, LncRNA RPSAP52 can increase cancer cell stemness and predict postoperative survival by up-regulating the expression of TGF-β1.27 The long non-coding RNA PIK3CD-AS2 inhibited p53 pathway to induce the progression of lung adenocarcinoma via mediating YBX1.28 In our study, bioinformatics methods validated that miR-148a-3p was a target of KCNQ1OT1. Besides, the results from dual-luciferase reporter assays demonstrated that miR-148a-3p can directly bind to KCNQ1OT1. Furthermore, the opposite relationship between KCNQ1OT1 and miR-148a-3p was confirmed in HCC tissues. In other studies, miR-148a-3p has been reported as a key tumor regulator in various cancers. For, instance, Wang et al identified that miR-148a-3p represses proliferation and EMT of bladder cancer by ERBB3/AKT2/c-myc and DNMT1 pathway.29 Wang et al indicated that the expression level of miR-148a-3p was decreased in ovarian cancer, and miR-148a-3p overexpression restrains progression of ovarian cancer by targeting c-Met.30 Correspondingly, in our study, we also found that miR-148a-3p as a tumor suppressor to suppress HCC progression. Therefore, our data indicated that KCNQ1OT1 contributed to HCC progression by modulating miR-148a-3p expression.
Normally, miRNAs exerted its biological functions in cancers through directly targeting mRNAs. Here, we first predicted and determined that IGF1R is a target of miR-148a-3p. Several evidence have confirmed that IGF1R functions as a key oncogene in various cancers including HCC. For instance, the inhibition of IGF-1R/PI3K/Akt signaling suppressed the progression of liver cancer by paracrine factors of human fetal MSCs.31 IGF1R have critical clinicopathological and prognostic roles in hepatocellular carcinoma.32 Besides, resistance to mTORC1/2 inhibition was mediated by focal adhesion- and IGF1R-dependent survival and migratory pathways in tumor.33 In our study, we found that the expression level of IGF1R was increased in HCC tissues and cell lines. In addition, we found that the silence of IGF1R repressed proliferation while promoted apoptosis of HCC cells. However, these roles of IGF1R on HCC cells were reversed by miR-148a-3p inhibitors.
In conclusion, our results indicated that KCNQ1OT1 plays an oncogene role in HCC by promoting IGF1R expression via sponging miR-148a-3p. Further, we uncovered a new KCNQ1OT1/miR-148a-3p/IGF1R axis for the first time. Our study may provide a novel candidate diagnostic, prognostic or therapeutic target for HCC.
Abbreviations
- CCK-8
Cell Counting Kit-8
- ceRNAs
competing endogenous
- FBS
fetal bovine serum
- HCC
hepatocellular carcinoma
- IGF1R
insulin like growth factor 1 receptor
- LncRNAs
Long non-coding RNAs
- miRNA
MicroRNA
- mRNA
messenger RNA
- MUT
mutant
- qRT-PCR
quantitative real-time polymerase chain reaction
- WT
wild type.
Footnotes
Authors’ Note: Guoping Xu, PhD, and Yungang Zhu, PhD, contributed equally to this work. Human study in this study was approved by the Ethics Committee on Human Research of the Tianjin Medical University Second Hospital (No.TJYEY20180143). Animal study in this study was approved by the Ethics Committee of the Tianjin Medical University Second Hospital (No. 2019N134).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Xuening Zhang  https://orcid.org/0000-0002-4621-5731
https://orcid.org/0000-0002-4621-5731
References
- 1. Li W, Xu W, Song JS, Wu T, Wang WX. LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma. 2019;66(3):397–404. doi:10.4149/neo_2018_180720N504 [DOI] [PubMed] [Google Scholar]
- 2. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–458. doi:10.1038/nrgastro.2010.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13(17):2436–2441. doi:10.3748/wjg.v13.i17.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):835–853. doi:10.1053/j.gastro.2015.12.041 [DOI] [PubMed] [Google Scholar]
- 5. Xu M, Xu X, Pan B, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18(1):135 doi:10.1186/s12943-019-1063-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu J, Lu Y, Liu Q, et al. Long noncoding RNA GMAN promotes hepatocellular carcinoma progression by interacting with eIF4B. Cancer Lett. 2020;473:1–12. doi:10.1016/j.canlet.2019.12.032 [DOI] [PubMed] [Google Scholar]
- 8. Guo JC, Yang YJ, Zheng JF, et al. Silencing of long noncoding RNA HOXA11-AS inhibits the Wnt signaling pathway via the upregulation of HOXA11 and thereby inhibits the proliferation, invasion, and self-renewal of hepatocellular carcinoma stem cells. Exp Mol Med. 2019;51(11):1–20. doi:10.1038/s12276-019-0328-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Y, Huan L, Wu Y, et al. LncRNA ID2-AS1 suppresses tumor metastasis by activating the HDAC8/ID2 pathway in hepatocellular carcinoma. Cancer Lett. 2020;469:399–409. doi:10.1016/j.canlet.2019.11.007 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Molecular Cancer. 2019;18(1):28 doi:10.1186/s12943-019-0957-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen Y, Xu J, Pan X, et al. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020;11(4):278 doi:10.1038/s41419-020-2485-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9(7):742 doi:10.1038/s41419-018-0793-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng L, Li H, Li F, Bei S, Zhang X. LncRNA KCNQ1OT1 regulates microRNA-9-LMX1A expression and inhibits gastric cancer cell progression. Aging. 2020;12(1):707–717. doi:10.18632/aging.102651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He J, Ge Q, Lin Z, et al. MiR-129-5p induces cell cycle arrest through modulating HOXC10/Cyclin D1 to inhibit gastric cancer progression. FASEB J. 2020;34(6):8544–8557. doi:10.1096/fj.201903217 R [DOI] [PubMed] [Google Scholar]
- 15. Liu DL, Lu LL, Dong LL, et al. miR-17-5p and miR-20a-5p suppress postoperative metastasis of hepatocellular carcinoma via blocking HGF/ERBB3-NF-kappaB positive feedback loop. Theranostics. 2020;10(8):3668–3683. doi:10.7150/thno.41365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anestopoulos I, Voulgaridou GP, Georgakilas AG, Franco R, Pappa A, Panayiotidis MI. Epigenetic therapy as a novel approach in hepatocellular carcinoma. Pharmacol Therap. 2015;145:103–119. doi:10.1016/j.pharmthera.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 17. Song W, Zou SB. Prognostic role of lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim Acta. 2016;463:169–173. doi:10.1016/j.cca.2016.10.035 [DOI] [PubMed] [Google Scholar]
- 18. Ma M, Xu H, Liu G, et al. Metabolism-induced tumor activator 1 (MITA1), an energy stress-inducible long noncoding rNA, promotes hepatocellular carcinoma metastasis. Hepatology. 2019;70(1):215–230. doi:10.1002/hep.30602 [DOI] [PubMed] [Google Scholar]
- 19. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi:10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 20. Qin G, Tu X, Li H, et al. Long noncoding RNA p53-stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology. 2020;71(1):112–129. doi:10.1002/hep.30793 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Zhang L, Yang J, Sun R. LncRNA KCNQ1OT1 promotes cell proliferation, migration and invasion via regulating miR-129-5p/JAG1 axis in non-small cell lung cancer. Cancer Cell Int. 2020;20:144 doi:10.1186/s12935-020-01225-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun H, Sun Y, Chen Q, Xu Z. LncRNA KCNQ1OT1 contributes to the progression and chemoresistance in acute myeloid leukemia by modulating Tspan3 through suppressing miR-193a-3p. Life Sci. 2020;241:117161 doi:10.1016/j.lfs.2019.117161 [DOI] [PubMed] [Google Scholar]
- 23. Quagliata L, Matter MS, Piscuoglio S, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi:10.1002/hep.26740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shih JW, Chiang WF, Wu ATH, et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat Commun. 2017;8:15874 doi:10.1038/ncomms15874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi Y, Ma Y, Peng Z, et al. Long noncoding RNA PENG upregulates PDZK1 expression by sponging miR-15b to suppress clear cell renal cell carcinoma cell proliferation. Oncogene. 2020;39(22):4404–4420. doi:10.1038/s41388-020-1297-1 [DOI] [PubMed] [Google Scholar]
- 26. Wang M, Mao C, Ouyang L, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ 2019;26(11):2329–2343. doi:10.1038/s41418-019-0304-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang S, Guo X, Lv W, et al. LncRNA RPSAP52 upregulates TGF-beta1 to increase cancer cell stemness and predict postoperative survival in glioblastoma. Cancer Manag Res. 2020;12:2541–2547. doi:10.2147/CMAR.S227496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng X, Zhang J, Fang T, et al. The long non-coding RNA PIK3CD-AS2 promotes lung adenocarcinoma progression via YBX1-mediated suppression of p53 pathway. Oncogenesis. 2020;9(3):34 doi:10.1038/s41389-020-0217-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X, Liang Z, Xu X, et al. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis. 2016;7(12):e2503 doi:10.1038/cddis.2016.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W, Dong J, Wang M, et al. miR-148a-3p suppresses epithelial ovarian cancer progression primarily by targeting c-Met. Oncol Lett. 2018;15(5):6131–6136. doi:10.3892/ol.2018.8110 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Yulyana Y, Ho IA, Sia KC, et al. Paracrine factors of human fetal MSCs inhibit liver cancer growth through reduced activation of IGF-1R/PI3K/Akt signaling. Molecular Therapy 2015;23(4):746–756. doi:10.1038/mt.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin SB, Zhou L, Liang ZY, Zhou WX, Jin Y. Expression of GRK2 and IGF1 R in hepatocellular carcinoma: clinicopathological and prognostic significance. J Clin Pathol. 2017;70(9):754–759. doi:10.1136/jclinpath-2016-203998 [DOI] [PubMed] [Google Scholar]
- 33. Yoon SO, Shin S, Karreth FA, et al. Focal adhesion- and IGF1R-dependent survival and migratory pathways mediate tumor resistance to mTORC1/2 inhibition. Mol Cell. 2017;67(3):512–527 e4. doi:10.1016/j.molcel.2017.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]