Abstract
There remains an urgent need for assays to quantify humoral protective immunity to SARS-CoV-2 to understand the immune responses of COVID-19 patients, evaluate efficacy of vaccine candidates in clinical trials, and conduct large-scale epidemiological studies. The plaque-reduction neutralization test (PRNT) is the reference-standard for quantifying antibodies capable of neutralizing SARS-CoV-2. However, the PRNT is logistically demanding, time-consuming, and requires containment level-3 facilities to safely work with live virus. In contrast, a surrogate virus neutralization test (sVNT) manufactured by Genscript is a quick and simple assay that detects antibodies that inhibit the RBD-ACE2 interaction, crucial for virus entry into host cells. In this study, we evaluate the sensitivity, specificity, and cross-reactivity of the sVNT compared with the PRNT using both 50% and 90% SARS-CoV-2 neutralization as a reference-standard. We found that the sVNT provides a high-throughput screening tool prior to confirmatory PRNT testing for the evaluation of SARS-CoV-2 neutralizing antibodies.
Keywords: COVID-19, Neutralization test, Immunity
1. Introduction
COronaVIrus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was deemed a public health emergency of international concern in January 2020 (WHO Statement, 2020; Wu et al., 2020; Zhu et al., 2020). As of October 27, 2020, over 42 million cases and well over 1.1 million fatalities of SARS-CoV-2 have occurred globally since its emergence in December 2019 (WHO 2020; WHO Statement, 2020). Transmitted primarily by inhalation of secretions generated by infected individuals and direct contact with fomites (Van Doremalen et al., 2020; Wei et al., 2020), SARS-CoV-2 infection causes a wide range of clinical manifestations. Mild symptoms of SARS-CoV-2 infection include cough, fever, sore throat, malaise, and muscle weakness, which typically occur 4 to 5 days after infection (Lauer et al., 2020). Gastrointestinal symptoms, such as nausea, vomiting, abdominal pain, and diarrhea can occur in 3% to 4% of those infected (Buscarini et al., 2020). An estimated 15% develop severe pneumonia and approximately 5% progress to acute respiratory distress syndrome (ARDS), kidney injury, cardiac injury, liver dysfunction, and sometimes death (Huang et al., 2020; Xu et al., 2020). The reported case fatality rates of SARS-CoV-2 range from 1% to 7% (Onder et al., 2020). Despite ongoing clinical testing of vaccines and therapeutics (Tu et al., 2020), there remains no medical countermeasure approved for the prevention or treatment of SARS-CoV-2 infection (Sanders et al., 2020).
A crucial hurdle in the race to implement an effective vaccine is the need for a high-throughput means to quantify humoral protective immunity to SARS-CoV-2. Such assays are also crucial for assessing the immune response of recovered COVID-19 patients and conducting large-scale epidemiological studies. Several in-house and commercially-available enzyme-linked immunosorbent assays have been developed to detect anti-SARS-CoV-2 antibodies raised upon infection (Freeman et al., 2020; Lassaunière et al., 2020). While these platforms provide a high-throughput means of detecting antibodies against SARS-CoV-2, they are unable to measure the immunological function of SARS-CoV-2-specific antibodies. In contrast, the plaque-reduction neutralization test (PRNT) quantifies levels of antibodies capable of neutralizing SARS-CoV-2. However, due to the time-consuming and laborious nature of the PRNT, as well as the need for containment level 3 facilities to work with the Risk Group-3 pathogen, it is limited in its capacity to be implemented in large-scale immunity testing.
Virus neutralizing antibodies confer protection by blocking the interaction that mediates virus entry into susceptible host cells. For SARS-CoV-2, this interaction involves binding of the receptor binding domain (RBD) of the SARS-CoV-2 spike glycoprotein with the angiotensin-converting enzyme 2 (ACE2; Hoffmann et al., 2020). The Genscript SARS-CoV-2 surrogate virus neutralization test (sVNT) is a commercially-available assay that detects antibodies that specifically inhibit the RBD-ACE2 interaction without the use of live SARS-CoV-2 (Tan et al., 2020). While the assay is high-throughput and can be safely conducted in containment level 2 facilities, it remains unknown whether the assay accurately captures functional antibody-mediated neutralization to the same extent as the reference standard PRNT. Ideally, a surrogate neutralization test should demonstrate high sensitivity and a low false negative rate for samples eliciting neutralization by PRNT. Conversely, a surrogate neutralization test should demonstrate a low non-neutralizing antibody detection rate, which we define as the rate at which specimens that test positive by the assay are unable to neutralize SARS-CoV-2 by PRNT test positive by the assay.
In this study, we aimed to evaluate the commercially-available sVNT for SARS-CoV-2 compared with an in-house PRNT as a reference standard. We conducted a comprehensive comparison of the sVNT with the PRNT using a panel of serological samples from COVID-19 patients, healthy individuals, as well as non-COVID-19 patients. While the sVNT demonstrated similar or greater detection of COVID-19 specimens, it displayed a high non-neutralizing antibody detection rate for PRNT-negative COVID-19 samples, which may lead to over-estimation of a functional neutralizing antibody response. Furthermore, the sVNT demonstrated cross-reactivity for specimens collected from SARS-CoV-1 and syphilis patients compared with PRNT. However, due to its low false negative rate for specimens eliciting 90% SARS-CoV-2 neutralization, the sVNT may offer a high-throughput screening tool to prioritize samples for neutralizing antibody testing.
2. Materials and methods
2.1. Ethics statement and sample subset
This study was approved by the Public Health Agency of Canada's Research Ethics Board (Protocol #2020-004P). A total of 301 samples were selected for the evaluation of the commercially-available sVNT compared with the reference-standard PRNT. Samples from COVID-19 patients (n = 194) were provided to the Public Health Agency of Canada (Winnipeg, Canada) from Sunnybrook Hospital (Toronto, Canada) and Cadham Provincial Laboratory (Winnipeg, Canada). The COVID-19 subset comprised plasma and serum from patients who were admitted to the hospital and tested positive for SARS-CoV-2 by molecular testing. These specimens were collected from patients at 3 to 91 days postsymptom onset (DPO) from March to June 2020. This included some specimens drawn from the same individual during the course of disease. Serum samples collected from healthy, asymptomatic individuals prior to the COVID-19 outbreak (n = 47) were used to establish the specificity of the sVNT compared with the traditional PRNT. Samples collected from patients previously confirmed positive for SARS-CoV-1 by molecular testing (n = 20), human coronavirus OC43 by molecular testing (n = 1), human coronavirus 229E by molecular testing (n = 1), HIV by serological testing (n = 10), and syphilis by serological testing (n = 20) were used to evaluate the cross-reactivity of the assays.
2.2. PRNT
The SARS-CoV-2 PRNT was adapted from a previously described method for SARS-CoV-1 (Wang et al., 2005). Briefly, serological specimens were diluted 2-fold from 1:20 to 1:640 in DMEM supplemented with 2% FBS and challenged with 50 plaque forming units (PFU) of SARS-CoV-2 (hCoV-19/Canada/ON_ON-VIDO-01-2/2020, EPI_ISL_425177), which were titrated by plaque assay (Mendoza et al., 2020). After 1 hour of incubation at 37 °C and 5% CO2, the sera-virus mixtures were added to 12-well plates containing Vero E6 cells at 90% to 100% confluence and incubated at 37 °C and 5% CO2 for 1 hour. After adsorption, a liquid overlay comprising 1.5% carboxymethylcellulose diluted in MEM supplemented with 4% FBS, L-glutamine, nonessential amino acids, and sodium bicarbonate was added to each well and plates were incubated at 37 °C and 5% CO2 for 72 hours. The liquid overlay was removed and cells were fixed with 10% neutral-buffered formalin for 1 hour at room temperature. The monolayers were stained with 0.5% crystal violet for 10 minutes and washed with 20% ethanol. Plaques were enumerated and compared to controls. The highest serum dilution resulting in 50% and 90% reduction in plaques compared with controls were defined as the PRNT-50 and PRNT-90 endpoint titers, respectively. PRNT-50 titres and PRNT-90 titers ≥1:20 were considered positive for SARS-CoV-2 neutralizing antibodies.
2.3. sVNT
The SARS-CoV-2 sVNT Kit (L00847; GenScript, Piscataway, USA) was performed according to manufacturer instructions, with each sample tested in duplicate. Briefly, samples and controls were incubated with horseradish peroxidase-conjugated RBD (HRP-RBD) at 37 °C for 30 minutes. The mixtures were added to the hACE2-coated capture plate and incubated at 37 °C for 15 minutes. Plates were then washed, removing HRP-RBD:neutralizing antibody complexes and allowing unbound HRP-RBD and HRP-RBD:non-neutalizing antibody complexes to remain bound to hACE2. TMB solution was added and allowed to incubate at room temperature for 15 minutes and the reaction was stopped by Stop Solution. The OD of each well was measured by spectrophotometry at 450 nm. The percent inhibition of a sample was calculated as (1-Average OD of sample/Average OD of negative control) × 100%. A sample with a percent inhibition <20% was considered “negative” and ≥20% was considered “positive” for SARS-CoV-2 neutralizing antibodies.
2.4. Statistical analysis
Test results generated from the kit evaluation were categorized as either “positive” or “negative” for SARS-CoV-2-neutralizing antibodies as described above. Detection was defined as the proportion of samples collected from COVID-19 patients which tested positive in each assay. Sensitivity of the sVNT compared with PRNT was defined as the proportion PRNT positive samples from COVID-19 patients which tested positive by sVNT. Specificity was defined as the percentage of PRNT negative samples from healthy patients which tested negative in the assay. Cross-reactivity of the sVNT compared with PRNT was defined as the percentage of samples positive for viruses other than SARS-CoV-2 which tested positive. A 2-tailed McNemar's test (continuity corrected) or Pearson's Chi-square test was conducted using GraphPad Prism Software (GraphPad, La Jolla, CA, USA) where appropriate. To compute the Pearson correlation coefficient between sVNT % inhibition and PRNT titre levels, PRNT titres were first pre-processed and then log10-transformed. Specifically, samples with negative PRNT titres were assigned a titre of 1 to avoid taking the log of 0, and samples with ≥640, for example, were taken as 640. Log10 transformation was conducted on PRNT titres before computing the Pearson's product moment correlation coefficient to sVNT % inhibition. Coefficients of 0.00 to 0.30, 0.30 to 0.50, 0.50 to 0.70, 0.70 to 0.90, and 0.90 to 1.00 corresponded with negligible, low, moderate, high, and very high correlation (Mukaka, 2012). P values less than 0.05 were considered statistically significant.
3. Results
3.1. The sVNT demonstrates similar and increased detection of COVID-19 serological specimens compared with PRNT-50 and PRNT-90, respectively
Serological specimens collected from molecular-confirmed COVID-19 patients were tested to evaluate the detection by the sVNT compared with PRNT-50 and PRNT-90. The sVNT detected 78.9% of all serological specimens collected from molecular-confirmed COVID-19 patients (Table 1 ), which was similar compared with PRNT-50 (80.9%, P= 0.5708) and increased compared with PRNT-90 (50.5%, P< 0.0001). Detection by sVNT of serological specimens collected from COVID-19 patients at 1 to 7, 8 to 14, 15 to 21, 22 to 28, 29 to 35, 36 to 42, 43 to 49, and ≥50 days postsymptom onset (DPO) was 56.4%, 73.3%, 95.0%, 88.9%, 83.3%, 84.6%, 100%, and 90.5%, respectively. These were similar to detection by PRNT-50 (66.7%, 75.0%, 95.0%, 100%, 75.0%, 84.6%, 100%, and 85.7%, respectively). The sVNT detected samples similar to PRNT-90 for samples collected at 22 to 28, 29 to 35, and 36 to 42 DPO (66.7%, 66.7%, and 46.2%, respectively). However, detection by the sVNT was increased over the PRNT-90 for samples collected at 1 to 7, 8 to 14, 15 to 21, 43 to 49, and >50 DPO, which were 30.8%, 50.0%, 65.0%, 50.0%, and 61.9%, respectively. The corresponding P values were 0.0094, 0.0005, 0.0412, 0.0044, and 0.0412, respectively.
Table 1.
Detection of COVID-19 samples collected at different time points postsymptom onset by SARS-CoV-2 surrogate and conventional neutralization assays.
| Days postsymptom onset | sVNT | PRNT-50 | PRNT-90 |
|---|---|---|---|
| 1–7 (n = 39) |
56.4% (22; 39.6%–72.2%) |
66.7% (26; 49.8%–80.9%, P= 0.4795) |
30.8%a (12; 17.0%–47.6%, P= 0.0094) |
| 8–14 (n = 60) |
73.3% (44; 60.3%–83.9%) |
75.0% (45; 62.1%–85.3%, P = 1.000) |
50.0%b (30; 36.8%–63.2%, P = 0.0005) |
| 15–21 (n = 20) |
95.0% (19; 75.1%–99.9%) |
95.0% (19; 75.1%–99.9%, P = 1.000) |
65.0%a (13; 40.8%–84.6%, P = 0.0412) |
| 22–28 (n = 9) |
88.9% (8; 51.8%–99.7%) |
100% (9; 66.4%–100%, P = 0.4795) |
66.7% (6; 29.9%–92.5%, P = 0.4795) |
| 29–35 (n = 12) |
83.3% (10; 51.6%–97.9%) |
75.0% (9; 42.8%–94.5%, P = 1.000) |
66.7% (8; 34.9%–90.1%, P = 0.4795) |
| 36–42 (n = 13) |
84.6% (11; 54.6%–98.1%) |
84.6% (11; 54.6%–98.1%, P = 0.3428) |
46.2% (6; 19.2%–74.9%, P = 0.0736) |
| 43–49 (n = 20) |
100% (20; 83.2%–100%) |
100% (20; 83.2%–100%, P = 1.000) |
50.0%a (10; 25.2%–72.8%, P = 0.0044) |
| 50+ (n = 21) |
90.5% (19; 69.6%–98.8%) |
85.7% (18; 63.7%–97.0%, P = 1.000) |
61.9%a (13; 38.4%–81.9%, P = 0.0412) |
| Total (n = 194) |
78.9% (153; 72.4%–84.4%) |
80.9% (157; 74.7%–86.2%, P = 0.5708) |
50.5%c (98; 43.3%–57.8%, P< 0.0001) |
Number of specimens and 95% confidence intervals are indicated in parentheses. Two-tailed McNemar tests were conducted to analyze the increase in detection of COVID-19+ samples conferred by sVNT compared with PRNT-50 or PRNT-90. sVNT vs. PRNT-50 *P < 0.05, **P < 0.001, ***P < 0.0001. sVNT vs PRNT-90.
P < 0.05,
P < 0.001,
P < 0.0001.
3.2. The sVNT demonstrates a high non-neutralizing antibody detection rate but a low false negative rate for COVID-19 specimens when using PRNT-90 as a reference standard
The sensitivity of the sVNT for SARS-CoV-2 neutralizing antibodies was evaluated using samples that tested positive by SARS-CoV-2 PRNT-50 and PRNT-90. Sensitivity of the sVNT was 89.8% (P< 0.0001) and 99.0% (P< 0.0001) for PRNT-50+ and PRNT-90+ specimens, respectively (Table 2 ). Samples from healthy individuals were tested to establish the specificity of the assays – all assays demonstrated 100% specificity. The inter-rater agreement between sVNT and PRNT-50 was moderate (κ-value = 0.551); the interrater agreement between sVNT and PRNT-90 was also moderate (κ-value = 0.409). This was likely due to high non-neutralizing antibody detection rate of the sVNT of 32.4% and 58.3% for COVID-19 specimens which tested negative by PRNT-50 and PRNT-90, respectively. The false negative rates of the sVNT were 10.2% and 1.02% for COVID-19 specimens that tested positive by PRNT-50 and PRNT-90, respectively.
Table 2.
Assay characteristics of the surrogate virus neutralization test (sVNT) for SARS-CoV-2 PRNT-50 positives and PRNT-90 positives.
| sVNT vs PRNT-50 | sVNT vs PRNT-90 | |
|---|---|---|
| Sensitivity | 89.8% (141/157; 84.0%–94.1%) |
99.0%⁎⁎ (97/98; 94.5%–100%, P< 0.0001) |
| False negative rate | 10.2% (16/157; 5.94%–16.0%) |
1.02%⁎⁎ (1/98; 0.02%–5.56%; P< 0.0001) |
| Non-neutralizing antibody detection ratea | 32.4% (12/37; 18.0%–49.8%) |
58.3%* (56/96; 47.8%–68.3%, P= 0.0014) |
| Specificity | 100% (47/47; 92.5%–100%) |
100% (47/47; 92.5%–100%) |
Sensitivity: percentage of SARS-CoV-2 PRNT positive samples from COVID-19 patients that also tested positive by sVNT. False negative rate: percentage of SARS-CoV-2 PRNT positive samples from COVID-19 patients, which tested negative by sVNT.
Non-neutralizing antibody detection rate: percentage of SARS-CoV-2 PRNT negative samples from COVID-19 patients, which tested positive by sVNT. Specificity: percentage of SARS-CoV-2 PRNT negative samples from healthy individuals that also tested negative by sVNT. Pearson's Chi-square tests were conducted to analyze the difference in sensitivity of the sVNT for PRNT-50+ and PRNT-90+ COVID-19 specimens. Number of specimens of total subset and 95% confidence intervals are indicated in parentheses.
P < 0.05.
P < 0.0001.
3.3. The % inhibition generated by the sVNT demonstrated a high positive correlation with PRNT-50 titres and a moderately positive correlation with PRNT-90 titres
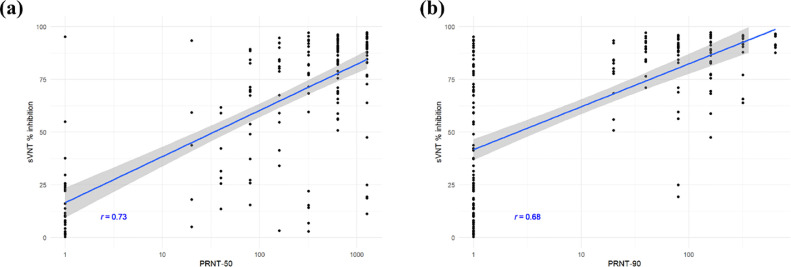
Serological specimens collected from molecular-confirmed COVID-19 patients were tested to evaluate the correlation between the % inhibition values generated by the sVNT and titres generated by PRNT-50 and PRNT-90 (Fig. 1 ). The Pearson's product-moment correlation between sVNT % inhibition and log-transformed PRNT-50 titres was 0.73, corresponding with a high degree of positive correlation. The Pearson's product-moment correlation between sVNT % inhibition and log-transformed PRNT-90 titres was 0.65, corresponding with a moderate degree of positive correlation.
Fig. 1.
Comparison between sVNT % inhibition and titres from (a) PRNT-50 and (b) PRNT-90. The x-axes are PRNT titres illustrated in log10-scale, whereas the y-axes are the sVNT % inhibition. Pearson correlation coefficients between sVNT % inhibition and log10-transformed PRNT titre values are reported in the figure. The blue lines and the grey bands show the fitted linear models and the 95% confidence level intervals, respectively.
3.4. The sVNT demonstrates higher cross-reactivity for specimens from SARS-CoV-1 and syphilis patients compared with PRNT
The cross-reactivity of the sVNT for SARS-CoV-1 specimens was 70%, which was significantly higher than the PRNT-50 (10%; P = 0.0015) and PRNT-90 (0%; P = 0.0005; Table 3 ). All assays demonstrated 0% cross-reactivity with hCoV-229E and hCoV-OC43 specimens. Compared with the sVNT and PRNT-90, which demonstrated 0% cross-reactivity for HIV specimens, the PRNT-50 demonstrated an increased cross-reactivity of 20%. However, this increase was not considered significant (P= 0.4795). The cross-reactivity of the sVNT for syphilis specimens was 50%, which was significantly higher (P= 0.0044) than PRNT-50 and PRNT-90, both of which demonstrated 0% cross-reactivity. The total cross-reactivity of the sVNT of 46.2% was significantly greater than the PRNT-50 (7.69%; P < 0.0001) and PRNT-90 (0%; P< 0.0001).
Table 3.
Cross-reactivity of SARS-CoV-2 neutralization assays for antibodies against other human coronaviruses.
| sVNT | PRNT-50 | PRNT-90 | |
|---|---|---|---|
| SARS-CoV-1 (n = 20) | 70.0% | 10.0%* | 0%b |
| hCoV-OC43 (n = 1) | 0% | 0% | 0% |
| hCoV-229E (n = 1) | 0% | 0% | 0% |
| HIV (n = 10) | 0% | 20% | 0% |
| Syphilis (n = 20) | 50% | 0%* | 0%a |
| Total n = 52 | 46.2% | 7.69%⁎⁎ | 0%c |
Cross-reactivity: percentage of SARS-CoV-1, seasonal coronavirus, and syphilis specimens which gave a positive result by neutralization assay. 95% confidence intervals are indicated in parentheses. Two-tailed McNemar tests were conducted to analyze the cross-reactivity sVNT compared with PRNT-50 or PRNT-90. Cross-reactivity of sVNT compared with PRNT-50.
P < 0.05.
P < 0.0001. Cross-reactivity of sVNT compared with PRNT-90.
P < 0.05.
P < 0.001.
P < 0.0001.
4. Discussion
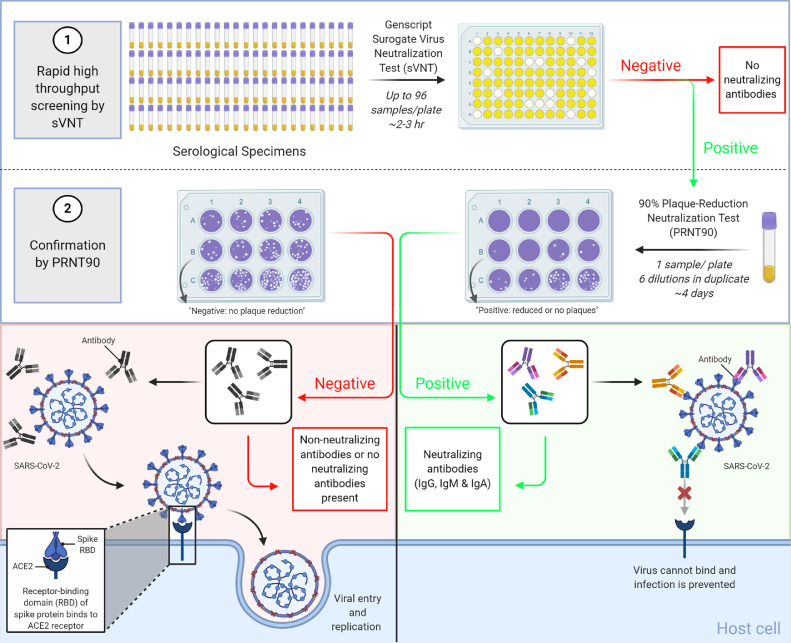
The COVID-19 pandemic has warranted an urgent need to understand both vaccine-mediated and infection-induced immunity to SARS-CoV-2. In this study, we validated a commercially-available sVNT and established a high-throughput algorithm for COVID-19 neutralizing antibody testing as depicted in Fig. 2 . The sVNT demonstrated similar detection of specimens collected from COVID-19 patients as the PRNT-50 and higher detection of specimens collected from COVID-19 patients than the PRNT-90. Furthermore, the sVNT generated % inhibition values that positively correlated with PRNT-50 and PRNT-90 titres and demonstrated excellent specificity when testing sera from healthy individuals collected prior to the COVID-19 pandemic. While we anticipated the sVNT would demonstrate some cross-reactivity with SARS-CoV-1 sharing ∼75% homology with SARS-CoV-2 Spike (Jaimes et al., 2020), it did not demonstrate cross-reactivity with sera from hCoV-OC43 (β-coronavirus) and hCoV-229E (α-coronavirus) patients. However, since these samples were only confirmed by molecular testing, it is possible the sera was collected prior to the development of antibodies against hCoV-OC43 and hCoV-229E. It is also important to note only one sample was tested for each of these subsets and future investigation will be required to understand the generalizability of this observation. Interestingly, the sVNT demonstrated cross-reactivity with specimens from syphilis patients. Cross-reactivity with sera from syphilis patients has also been demonstrated in Borrelia burgdorferi (Wong et al., 2004) and West Nile virus (Wong et al., 2004) assays. However, due to the number of samples used in the cross-reactivity study, further testing of larger subsets will be needed to understand the relevance of the cross-reactivity of the sVNT.
Fig. 2.
Schematic of a high-throughput algorithm for SARS-CoV-2 neutralizing antibody testing. The sVNT can process a large number of specimens in CL-2 within 2 to 3 hours. Samples testing positive by sVNT are subsequently confirmed by PRNT, which can take up to 7 days in CL-3. Created with BioRender.com.
Since sVNTs are intended to be used as surrogate assays to measure functional humoral immunity, we investigated the ability of the Genscript sVNT kit to differentiate between non-neutralizing and neutralizing COVID-19 specimens. To do so, we evaluated the non-neutralizing antibody detection rate of the sVNT for COVID-19 specimens that were unable to functionally neutralize 50% and 90% of SARS-CoV-2 by PRNT. We found that the sVNT demonstrated a high non-neutralizing antibody detection rate, generating positive results for 32.4% and 58.3% of samples confirmed negative by PRNT-50 and PRNT-90, respectively. The inability of some sVNT positive specimens to neutralize SARS-CoV-2 by PRNT could be attributed to the detection of poorly neutralizing anti-RBD antibodies, which have been documented in several studies (Rogers et al., 2020; Alsoussi et al., 2020). Due to this, caution should be exercised if the sVNT is used as an alternative to the PRNT because specimens that test positive by sVNT may not be able to neutralize SARS-CoV-2 in vitro and may translate to inability to neutralize SARS-CoV-2 in vivo. However, further studies may evaluate more stringent cut-off values to enhance specificity and reduce the non-neutralizing antibody detection rate of the sVNT.
We wished to investigate whether the sVNT could instead be used as high-throughput method of screening samples for subsequent testing by the laborious and time-consuming PRNT. We evaluated the sensitivity of the sVNT for COVID-19 specimens that were confirmed capable of neutralizing 50% and 90% of SARS-CoV-2 by PRNT. We found that the sVNT demonstrated high sensitivity for both PRNT-50+ and PRNT-90+ specimens. However, the sVNT demonstrated a significantly higher false negative rate for PRNT-50+ specimens compared with PRNT-90. This could be attributed to the inability of the sVNT to detect neutralizing antibodies that target non-RBD epitopes of SARS-CoV-2 (Rogers et al., 2020), including the N-terminal domain of the spike protein (Chi et al., 2020). It could also fail to detect antibodies targeting either or both of two different cleavage sites of the Spike protein that may neutralize SARS-CoV-2 by blocking priming by the serine protease, TMPRSS2, crucial for entry into the cell via ACE2 (Hussain et al., 2020). Future research assessing the limit of detection and analytical sensitivity will be important in understanding the false negative rate of the sVNT for samples that test positive by PRNT-50. Given the significantly higher sensitivity and lower false negative rate of the sVNT for PRNT-90+ specimens over PRNT-50+ specimens, we concluded that a negative result produced by the sVNT could accurately predict a negative result by PRNT-90 and be used to screen out samples for further testing. Meanwhile, specimens giving a positive result by the sVNT could be subsequently tested by PRNT-90 to confirm the presence of neutralizing antibodies (Fig. 2). Such an algorithm could significantly optimize neutralizing antibody testing compared with solely testing by PRNT, which can take up to seven days from cell preparation to process 32 samples at a time by highly trained containment level 3 staff. In contrast, the sVNT can process a large number of specimens in containment level 2 within 2 to 3 hours, significantly reducing the number of samples requiring confirmation by PRNT.
There remain limitations to consider for the implementation of the described algorithm for analyzing humoral protective immunity against SARS-CoV-2. First, the algorithm uses 90% neutralization as a reference-standard, which may prevent the detection of specimens only capable of neutralizing 50% of SARS-CoV-2. While much remains unknown about the characteristics of the neutralizing antibody responses required for protection, conclusions about neutralizing antibodies vary in studies using 50% neutralization as a reference-standard. On one hand, high neutralizing titres capable of 50% neutralization correlated with protection in immunized nonhuman primates (Yu et al., 2020) and COVID-19 patients receiving convalescent plasma transfusions (Duan et al., 2020). In contrast, other studies did not observe a correlation between SARS-CoV-2 neutralizing antibodies and clinical course of disease when using 50% neutralization as a reference-standard (Wölfel et al., 2020). Given that heterogeneity in B cell responses gives rise to antibodies of varying affinity and avidity within a single host (Brouwer et al., 2020), it could be possible that an antibody response capable of 50% neutralization lacks the collective potency to effectively block entry and replication of SARS-CoV-2 compared with an antibody response capable of eliciting 90% neutralization. A present, but suboptimal antibody response may be detrimental for disease course of COVID-19 (Iwasaki and Yang, 2020). For example, a study in nonhuman primates suggested that antibodies specific for other S protein epitopes could induce antibody-dependent enhancement (ADE; Wang et al., 2016). Further investigations are needed to determine whether using 90% neutralization as a reference-standard could provide additional insight into the relationship between neutralizing antibody responses and protection. Some evidence suggests that neutralizing antibodies against SARS-CoV-2 wane weeks after infection (Seow et al., 2020). Therefore, the timing at which a patient's sample is assessed for neutralizing antibodies will be an important factor to consider when investigating whether or not a person has protective immunity against the virus. Furthermore, future research could compare the sVNT with other previously described surrogate platforms for use in such an algorithm for the detection of SARS-CoV-2 neutralizing antibodies (Abe et al., 2020; Tan et al., 2020).
5. Conclusions
The Genscript sVNT provides a high-throughput screening tool prior to confirmatory PRNT testing for the evaluation of SARS-CoV-2 neutralizing antibodies. Such an algorithm used to understand the immune responses of COVID-19 patients, evaluate efficacy of vaccine candidates in clinical trials, and conduct large-scale epidemiological studies.
Author Contributions
Conceptualization, E.J.V. and H.W.; methodology, E.J.V; validation, E.J.V.; formal analysis, E.J.V. and J.C.; investigation, E.J.V., K.M., A.R., K.D., C.P. L.L., E.M.; resources, H.W., Z.S., J.C.; data curation, E.J.V.; writing - original draft preparation, E.J.V; writing - review and editing, H.W, M.D.; visualization, E.J.V., Z.S., J.C.; supervision, H.W.; project administration, E.J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Declaration of Competing Interests
The authors declare no conflict of interest.
Acknowledgments
This project was supported by the Public Health Agency of Canada (PHAC). The authors would like to thank Dr. Darwyn Kobasa, Dr. Amrit Boese, and Anders Leung for providing the SARS-CoV-2 stock (PHAC). We wish to thank Dr. Allison McGreer (Mount Sinai Hospital), Dr. Samira Mubareka (Sunnybrook Health Science Centre), and Dr. Derek Stein (Cadham Provincial Laboratory) for providing serological specimens from COVID-19 patients, as well as Dr. Robbin Lindsay and Antonia Dibernardo (PHAC) for organizing the sample subset. We would also like to thank Dr. Todd Hatchette (Nova Scotia Health Authority), Dr. John Kim (PHAC), Dr. Tim Booth (PHAC), and Dr. Vanessa Tran (Public Health Ontario) for providing serological specimens for cross-reactivity studies. Fig. 2 was created with BioRender.com.
References
- Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5(19) doi: 10.1172/jci.insight.142362. Oct 2PMID: 32870820; PMCID: PMC7566699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi WB., Turner JS, Case JB, Zhao H, Schmitz AJ, Zhou JQ. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J Immunol. 2020 doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer PJM., Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarini E, Manfredi G, Brambilla G, Menozzi F, Londoni C, Alicante S. GI symptoms as early signs of COVID-19 in hospitalised Italian patients. Gut. 2020;69(8):1547–1548. doi: 10.1136/gutjnl-2020-321434. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Liu B, Li C, Zhang H, Yu T, Qu J. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B, Lester S, Mills L, Rasheed MAU, Moye S, Abiona O. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. bioRxiv Prepr Serv Biol. 2020 [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Jabeen N, Amanullah A, Baig AA, Aziz B, Shabbir S, Raza F, Uddin N. Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions[J] AIMS Microbiology. 2020;6(3):350–360. doi: 10.3934/microbiol.2020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20:1–3. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaunière R, Frische A, Harboe ZB, Nielsen AC, Fomsgaard A, Krogfelt KA. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020 2020.04.09.20056325. Pre-print. [Google Scholar]
- Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;10:M20–0504. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza EJ, Manguiat K, Wood H, Drebot M. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr Protoc Microbiol. 2020;57:ecpmc105. doi: 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaka MM. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323(18):1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W-T. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19) JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020 doi: 10.1038/s41564-020-00813-8. Oct 26Epub ahead of print. PMID: 33106674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CW, Chia WN, Chen MI-C, Hu Z, Young BE, Tan Y-J. 2020. A SARS-CoV-2 surrogate virus neutralization test (sVNT) based on antibody-mediated blockage of ACE2-spike (RBD) protein-protein interaction. [DOI] [PubMed] [Google Scholar]
- Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. Sep. [DOI] [PubMed] [Google Scholar]
- Tu Y-F, Chien C-S, Yarmishyn AA, Lin Y-Y, Luo Y-H, Lin Y-T. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doremalen N, Bushmaker T, Morris DH., Holbrook MG, Gamble A, Williamson BN. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1 (Supplementary Appendix) N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang L, Kuwahara K, Li L, Liu Z, Li T. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sakhatskyy P, Chou T-HW, Lu S. Assays for the assessment of neutralizing antibody activities against severe acute respiratory syndrome (SARS) associated coronavirus (SCV) J Immunol Methods. 2005;301:21–30. doi: 10.1016/j.jim.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei WE, Li Z, Chiew CJ, Yong SE, Toh MP, Lee VJ. Presymptomatic transmission of SARS-CoV-2 — Singapore, January 23–March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Statement on the second meeting of the International Health Regulations . 2020. Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV)https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) (accessed Mar 29, 2020) [Google Scholar]
- WHO . 2020. COVID-19 Weekly Epidemiological Update.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed October 30, 2020) [Google Scholar]
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wong SJ, Demarest VL, Boyle RH, Wang T, Ledizet M, Kar K. Detection of human anti-flavivirus antibodies with a west nile virus recombinant antigen microsphere immunoassay. J Clin Microbiol. 2004;42:65–72. doi: 10.1128/JCM.42.1.65-72.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020 doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. China novel coronavirus investigating and research team a novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]