SUMMARY
Post-translational modifications are efficient means to rapidly regulate protein function in response to a stimulus. Although ubiquitination events and the E3 ubiquitin ligases involved are increasingly characterized in many signaling pathways, their regulation by deubiquitinating enzymes remains less understood. The C-type lectin receptor (CLR) signaling adaptor CARD9 was previously reported to be activated via TRIM62-mediated ubiquitination. Here, we identify the deubiquitinase USP15 as a novel regulator of CARD9, demonstrating that USP15 constitutively associates with CARD9 and removes TRIM62-deposited ubiquitin marks. Furthermore, USP15 knockdown and knockout specifically enhance CARD9-dependent CLR signaling in both mouse and human immune cells. Altogether, our study identifies a novel regulator of innate immune signaling and provides a blueprint for the identification of additional deubiquitinases that are likely to control these processes.
Keywords: CARD9, USP15, deubiquitination
INTRODUCTION
Human genetic studies have uncovered that CARD9 is important for immune-microbial interactions at mucosal surfaces, as CARD9 polymorphisms are associated with disorders such as immunodeficiency, inflammatory bowel diseases (IBD), and increased susceptibility to fungal infection (1-3). To date, more than two dozen CARD9 coding mutations have been reported in clinical studies to predispose individuals to diseases associated with fungal or microalgal infection (4-9). CARD9 has also been reported to be critical during bacterial and viral infections in mouse models (10-14), although it remains unclear whether CARD9 is important in defense against microbes other than fungi and microalgae in humans.
In IBD, CARD9 variants have been associated with both risk and protection. A rare splicing mutation IVS11+1G>C, resulting in a C-terminally truncated CARD9, plays a protective role against IBD and ankylosing spondylitis (15, 16). A common coding variant, CARD9 S12N (SNP rs4077515), results in higher CARD9 mRNA expression (17) and is associated with increased risk of IBD (15, 17), ankylosing spondylitis (18), immunoglobulin A nephropathy (19) and primary sclerosing cholangitis (20). This variant, however, also appears to have protective effects in some contexts, such as primary immune thrombocytopenia (21). Mechanistically, when expressed at equivalent levels, the S12N allele enhances CARD9-mediated cytokine production in murine bone marrow-derived dendritic cells (BMDCs) stimulated with fungal products relative to the wild-type allele (22), indicating that S12N is a gain-of-function mutant. CARD9 S12N reportedly enhances IL-5 production in macrophages; accordingly, peripheral blood mononuclear cells (PBMCs) from three S12N homozygous patients with allergic bronchopulmonary aspergillosis (ABPA) produced higher levels of IL-5 than those from an ABPA patient carrying wild-type CARD9 (23).
CARD9 is an essential component of innate immune signaling through multiple C-type lectin receptors (CLRs), such as Dectin-1, Dectin-2, Dectin-3 and Mincle (2). These receptors are generally expressed on myeloid cells and recognize a broad range of fungal cell wall components (24). Mincle is additionally activated by nuclear protein SAP130 released from damaged cells (25) and trehalose dimycolate (TDM, also known as cord factor) present in the cell membranes of mycobacteria (26), suggesting a broad role for CLRs in innate immune recognition of fungi, damaged cells and certain bacteria. Upon engagement, CLRs activate the tyrosine kinase Syk to facilitate the formation of the CARD9-BCL10-MALT1 (CBM) complex (27), which is essential for downstream NF-κB signaling and related immune responses. Notably, CBM architecture in signaling cascades is not limited to CARD9 in myeloid cells, but also exists with CARD10 in non-hematopoietic cells, CARD11 in lymphoid cells and CARD14 in keratinocytes. Although these CBM complexes are highly specific to cell types and stimuli, their activity likely involves similar regulatory events.
Post-translational modifications (PTMs) are known to directly regulate the assembly of CBM complexes. In the case of CARD9, phosphorylation directly regulates the formation of the CBM complex: protein kinase C delta phosphorylates CARD9 on T231, facilitating the interaction between CARD9 and BCL10 (28). In contrast, protein phosphatase 1 (PP1) interacts with CARD9 via DOK3 to dephosphorylate CARD9 threonine phosphorylation, which negatively regulates CARD9-dependent signaling in neutrophils (29). Furthermore, casein kinase 2 negatively regulates CARD9 via phosphorylation of CARD9 C-terminus at T531 and T533 (30). We previously uncovered that the E3 ubiquitin ligase TRIM62 is a binding partner of CARD9 that might associate with the CBM complex (22). TRIM62-mediated, K27-linked CARD9 ubiquitination at K125 is critical for CARD9-mediated NF-κB signaling and cytokine production in myeloid cells as well as antifungal immunity in vivo. These findings revealed a previously unknown ubiquitin-dependent regulatory mechanism for CARD9 function. However, the regulation of this process and the involvement of potential deubiquitinases for CARD9 remained unclear. In this study, we identified ubiquitin specific peptidase 15 (USP15) as a novel CARD9 interactor by proteomics. We show that USP15 removes TRIM62-mediated CARD9 ubiquitination. Additionally, we directly show the impact of USP15 on signaling downstream of CARD9, suggesting that it is an important regulator of this innate immune signaling pathway.
EXPERIMENTAL PROCEDURES
Reagents and plasmids.
Whole glucan particles (WGP) were purchased from Invivogen (Cat. No: tlrl-wgp). Trehalose 6,6’-dimycolate (TDM) was obtained from Enzo Life Sciences (Cat. No: AL-581-210-M001). TDM was dissolved in chloroform:methanol:water (90:10:1, 1 mg/ml) and further diluted with isopropanol. Lipopolysaccharide (LPS) was purchased from Invivogen (Cat. No: tlrl-peklps).
Lentivirus-based shRNA constructs were generated in a pLKO.1 vector by the Genetic Perturbation Platform at the Broad Institute of MIT and Harvard. A lentiviral vector pCDH-CMV from Addgene (#72265) was used to generate lenti-based USP15 constructs with a puromycin-T2A-3xHA tag in the N-terminus. All other constructs were either cloned into a pCMV vector (Clontech) or pcDNA4/TO-FLAG-StrepII as described previously (22). USP15 variants were originally amplified from a Clontech immune cDNA panel and subcloned into indicated tagged vectors by PCR.
Immunoprecipitation and western blotting.
The experimental methods were previously described in detail (22). Briefly, cells were lysed in standard lysis buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% Nonidet P-40, Halt phosphatase inhibitor single-use cocktail (Pierce), and protease inhibitor tablets (Roche)]. For immunoprecipitation of Flag-StrepII-tagged protein, cell lysates were incubated with StrepII Sepharose beads (IBA, Cat. No: 2-1201-010) for 1 h at 4°C. For immunoprecipitation of endogenous CARD9 in BMDCs, cell lysates were incubated with anti-CARD9 antibody or control IgG overnight followed by pulldown with protein A/G magnetic beads (Millipore, Cat. No: LSKMAGAG02) incubation for 30 minutes. For ubiquitination blots, cells were lysed in standard lysis buffer with 10mM N-ethylmaleimide (Sigma, Cat. No: E3876-5G).
The following antibodies were used: anti-FLAG M2 antibody (Sigma, Cat No: F1804); anti-β-actin antibody (Abcam, Cat No: 8226); anti-hemagglutinin (HA) antibody (BioLegend, Cat. No: 901502); anti-Myc antibody (BioLegend, Cat. No: 626802); anti-V5 antibody (BioLegend, Cat.: No: 903801); anti-CARD9 (A-8) antibody (Santa Cruz, Cat. No: sc0374569); anti-CARD9 antibody (mouse preferred, Cell Signaling, Cat. No: 12283); anti-USP15 (D1K6S) antibody (Cell Signaling, Cat. No: 66310); anti-USP15 (2D5) antibody (Santa Cruz, Cat. No: sc-100629); and normal mouse IgG2b isotype control (Santa Cruz, Cat. No: sc-3879). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies (Dako) were used as secondary antibodies.
Cell culture and lentiviral production.
HEK293T cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal calf serum and 15 μg/ml gentamycin sulfate. To prepare lentivirus for infection, protocols from the Broad Institute’s RNAi Consortium shRNA Library were used as described previously (22). USP15 shRNA constructs in pLKO.1-puro vector were obtained from the Broad Institute. Control shRNA construct in pLKO.1-puro vector was from Sigma (Cat. No: SHC002).
Mouse USP15 shRNA 1 target sequence: TGTCTATGGAGATGAAGTTAT
Mouse USP15 shRNA 2 target sequence: TGAGAGGTGAAATAGCTAAAT
Human USP15 shRNA A target sequence: CCTTGGAAGTTTACTTAGTTA
Human USP15 shRNA B target sequence: GCTGACACAATAGATACAATT
BMDC culture and functional assay.
Preparation and infection of BMDCs were described previously (22). Functional assays were performed on day 9 in 96-well plates. BMDCs were stimulated with WGP, LPS, pre-coated TDM or corresponding solvent. Twenty-four hours after stimulation, cytokine production in the supernatant was detected using appropriate ELISA kits (BD Sciences) according to the manufacturer’s protocol.
CRISPR-generated USP15−/− THP-1 cell lines and NF-κB luciferase assay
The USP15−/− THP-1 cell lines were generated by a CRISPR-based approach using USP15 sgRNA A1 (CCAAGTTACTTAGGCCACAG) and sgRNA B2 (AAGGTGTTCCTTAAGTGACT). Briefly, THP-1 cells were infected with lentivirus carrying CARD9 sgRNA. tracrRNAs for USP15 sgRNAs A and B were purchased from IDT. IDT Alt-R CRISP-Cas9 system was applied for preparation of RNP complex containing tracrRNA, crRNA (IDT) and Cas9 protein (PNA Bio). RNP complex and 0.5ug pmaxGFP were transfected with 1M THP-1 cells by nucleofection according to the manufacturer’s protocol (Lonza). Two days post nucleofection, GFP-positive THP-1 cells were sorted by flow cytometry in the flow cytometry facility at the Broad Institute. Seven days post sorting, cells were diluted and plated into wells at a ratio of 1 cell per well in 96-well plates. USP15−/− cell lines were validated by western blot.
For the NF-κB luciferase assay, three independent pools of the same THP-1 cells were transduced with Dectin-1 (NM_022570) and a lentivirus-based NF-κB luciferase reporter (31). Three independent pools of the same USP15−/− cells overexpressing Dectin-1 and the NF-κB luciferase reporter were reconstituted with indicated USP15 constructs in parallel. Cells with re-expressed USP15 were expanded for at least four passages prior to luciferase assay. Cells were treated with WGP for 24 hours prior to luciferase assay. Reporter luciferase activity was examined by according to the manufacturer’s protocol (Steadylite Plus, PerkinElmer). The luciferase activity of each sample was normalized by calculating the fold change to the luciferase activity of the corresponding untreated samples.
In vitro deubiquitination assay.
In vitro ubiquitination reactions were set up on a 50 μL scale as previously described (22) and contained 2.5 nM TRIM62, 5 μM HA-Ubiquitin (Boston Biochem), 25 nM CARD9, 12.5 nM UBE1 (LifeSensors), and 200 nM UBE2D2 (LifeSensors). After incubation at room temperature for 30 min the reactions were quenched by the addition of 1 μL 0.5 M EDTA, followed by the addition of 1 μL of 2 μM His6-USP15 (BostonBiochem, E-594, lot 30271815A). Incubation was continued for 1 h, after which the reaction was quenched by the addition of 2X SDS loading buffer.
CARD9 proteomics.
JAWSII dendritic cells overexpressed with Flag-StrepII-tagged human CARD9 were harvested and washed twice with PBS. Cellular extracts were prepared by cell disruption through 15 strokes in a tight-fitting Dounce homogenizer following by two freeze-thaw cycles in standard lysis buffer. The cell extract was incubated with 25 μl MagStrep “type3” XT beads (IBA) for 1 h at 4°C. Beads and bound proteins were washed three times with lysis buffer and eluted with SDS-PAGE loading buffer. Proteins of interest were separated by SDS-PAGE and stained using Colloidal Coomassie GelCode Blue (Pierce). Gel sections were cut from the gel and sent for mass spectrometry analysis at the Taplin Biological Mass Spectrometry Facility of Harvard Medical School.
Statistical analysis.
Unpaired two-tailed Student’s t-tests were used for comparisons. All statistical comparisons were made between multiple independent experiments performed in parallel.
RESULTS
USP15 is a CARD9 binding partner
Previously, we identified TRIM62 as an E3 ubiquitin ligase responsible for optimal CARD9 activation through K27-linked ubiquitination at the CARD9 K125 residue (22). We speculated that there must be a deubiquitinase to reverse TRIM62-mediated ubiquitination and activation of CARD9. To identify potential deubiquitinases, we overexpressed Flag-StrepII-tagged CARD9 in the murine bone marrow-derived dendritic cell (BMDC) line JAWSII and employed a proteomics approach (Experimental Procedures, Supplemental Figure 1). In addition to the known binding partner TRIM62, immunoprecipitation of overexpressed CARD9 and subsequent mass spectrometry analysis identified the deubiquitinase USP15 as a top hit based on high peptide counts and percentage of protein coverage (Supplemental Figure 1).
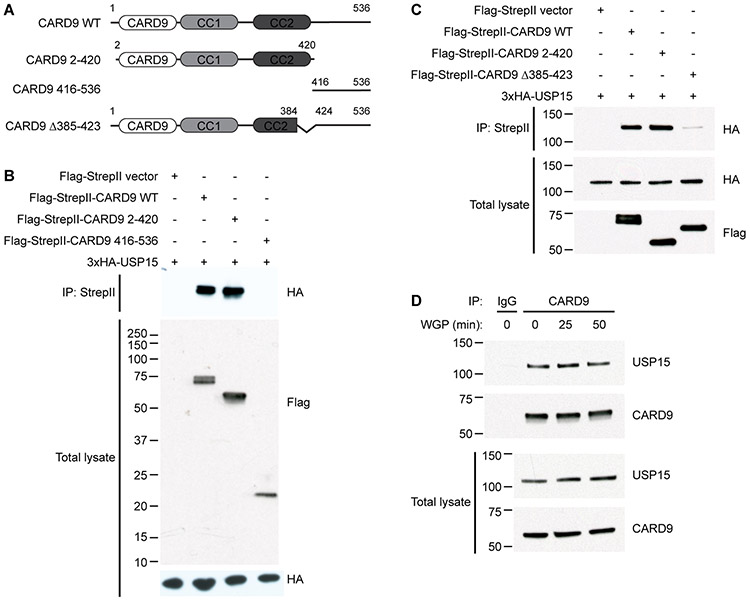
To validate the interaction between USP15 and CARD9, we overexpressed Flag-StrepII-tagged full length CARD9 as well as N-terminal and C-terminal fragments together with USP15 in HEK293T cells (Fig. 1A). Full length CARD9 and the N-terminal fragment, but not the C-terminal fragment, co-immunoprecipitated with USP15 (Fig. 1B). This result supports that CARD9 interacts with USP15 and suggests that the N-terminus of CARD9 harbors the USP15 interaction domain. Furthermore, we found that CARD9 Δ385-423(22)(22) (Fig. 1A) largely lost binding affinity with USP15 (Fig. 1C), indicating that this very short end region of the CARD9 coiled-coil domain is critical for interaction with USP15. We next tested whether USP15 binds to CARD9 endogenously in primary murine BMDCs. We immunoprecipitated endogenous USP15 with endogenous CARD9, demonstrating that the USP15-CARD9 interaction occurs under physiological conditions. In BMDCs, this interaction appeared constitutive and largely unaffected by stimulation with the Dectin-1 agonist WGP (whole glucan particle) (Fig. 1D).
Figure 1. USP15 binds the N-terminus of CARD9.
(A) Schematics of CARD9 variants used. WT, wild-type. CC, coiled-coil domain. Δ385-423, deletion of amino acids 385-423.
(B-C) HEK293T cells were transfected with HA-tagged USP15 and Flag-StrepII-tagged CARD9 WT, CARD9 2-420 (N-terminus), or either CARD9 416-536 (C-terminus) (B) or CARD9 Δ385-423 (C). Samples were immunoprecipitated with anti-StrepII and probed for HA (USP15). Total lysates were probed for HA and FLAG (CARD9).
(D) Wild-type, primary murine bone marrow-derived dendritic cells (BMDCs) were unstimulated or stimulated with 100ug/ml of the Dectin-1 agonist WGP (whole glucan particle) for 25 and 50 minutes. The unstimulated sample was divided and immunoprecipitated with normal IgG2b isotype and anti-CARD9 (A-8). Stimulated samples were immunoprecipitated with anti-CARD9 (A-8). All samples were probed for endogenous USP15 and CARD9.
USP15 removes ubiquitin marks deposited by TRIM62
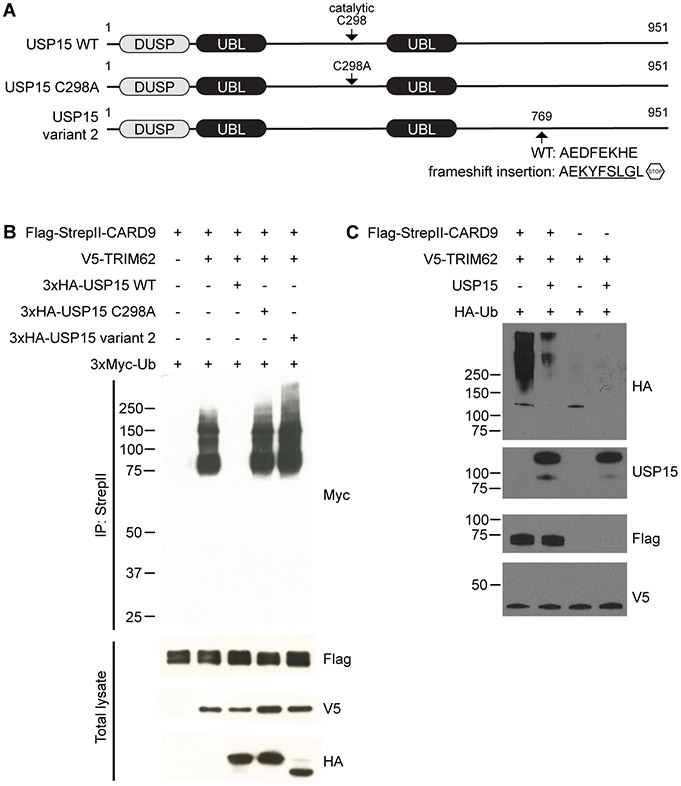
Given the well-characterized role of USP15 as a deubiquitinase (32), we examined whether USP15 regulates TRIM62-mediated CARD9 ubiquitination. To this end, we co-expressed CARD9, TRIM62 and ubiquitin with either the wild-type USP15, catalytically inactive USP15 C298A mutant, or USP15 variant 2 in HEK293T cells (Fig. 2A). USP15 variant 2 is a C-terminal truncation that lacks a zinc-binding motif Cys-x-x-Cys (aa809-812) required for the activity of ubiquitin-specific proteases (USPs) (32). We confirmed ubiquitination of CARD9 by TRIM62 at K125 and further showed that wild-type USP15 drastically reduces CARD9 ubiquitination (Fig. 2B, Supplemental Figure 2). Co-expression with neither the USP15 C298A mutant nor USP15 variant 2 affected CARD9 ubiquitination, indicating that the removal of ubiquitin is linked to the catalytic activity of USP15.
Figure 2. USP15 regulates TRIM62-mediated CARD9 ubiquitination.
(A) Schematics of USP15 variants used. The catalytic C298 residue and catalytically inactivating C298A mutation are indicated. USP15 variant 2 was identified from a Clontech immune cDNA library harboring the indicated frameshift insertion. WT, wild-type. DUSP, domain present in ubiquitin-specific protease. UBL, ubiquitin-like.
(B) HEK293T cells were transfected with Flag-StrepII-tagged CARD9, V5-tagged TRIM62, Myc-tagged ubiquitin (Ub), and HA-tagged USP15 WT, USP15 C298A, or USP15 variant 2. Samples were immunoprecipitated with anti-StrepII and probed for Myc. Total lysates were probed for Flag (CARD9), V5 (TRIM62), and HA (USP15).
(C) Ubiquitination of CARD9 was initiated by incubating purified Flag-StrepII-tagged CARD9 with V5-tagged TRIM62 and required ubiquitination machinery UBE1, UBE2D2 and ubiquitin. Ubiquitinated CARD9 was then incubated with and without USP15, and USP15-mediated CARD9 deubiquitination was assessed by western blot.
We next investigated the deubiquitinase activity of USP15 on CARD9 in a reconstituted in vitro system. CARD9 was incubated with TRIM62 and ubiquitination machinery UBE1, UBE2D2 and ubiquitin. Ubiquitinated CARD9 was then incubated with and without USP15. We observed that incubation with USP15 led to marked cleavage of the polyubiquitin chains present on CARD9 (Fig. 2C), confirming that USP15 is sufficient for the removal of ubiquitin from CARD9.
USP15 suppresses CARD9-mediated signaling
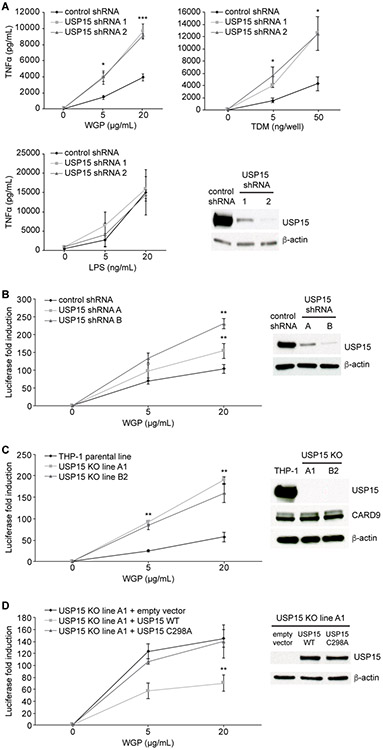
To determine whether USP15 can directly modulate CARD9-mediated signaling, we used shRNA to knockdown endogenous USP15 in wild-type BMDCs (Fig. 3A). USP15 knockdown enhanced TNFα production upon stimulation with WGP and TDM but not lipopolysaccharide (LPS), demonstrating that USP15 has a specific inhibitory effect on the CARD9-dependent WGP- and TDM-induced pathways but is not involved in the regulation of MyD88- and TRIF-dependent LPS recognition in primary murine cells.
Figure 3. USP15 suppresses CARD9-mediated signaling.
(A) Wild-type, primary murine bone marrow-derived dendritic cells (BMDCs) were transduced with lentivirus expressing the indicated control or USP15 shRNAs. BMDCs were stimulated with the Dectin-1 agonist WGP (whole glucan particle), pre-coated Mincle agonist TDM (trehalose dimycolate), or TLR4 agonist LPS (lipopolysaccharide) for 24 hours before TNFα levels were assessed by ELISA. Knockdown of protein expression was confirmed by western blot. Data were obtained from three independent experiments (N=3 mice). Error bars represent mean ± standard deviation. *P<0.05, ***P<0.005. Comparisons are relative to stimulated cells transduced with control shRNA.
(B-C) Endogenous USP15 in human THP-1 cells was knocked down by shRNA (B) or knocked out (KO) with a CRISPR-based approach (C). Indicated THP-1 cells were stimulated with WGP for 24 hours, and NF-κB reporter activity was assessed by a luciferase assay. Western blots show protein levels of USP15 (B, C) and CARD9 (C) expressed in the cells used. Three independent pools of the same cells (THP-1 in B and USP15−/− in C) were transduced with Dectin-1 and NF-κB luciferase reporter and utilized in parallel for the subsequent treatments and assays (N=3). Error bars represent mean ± standard deviation. *P<0.05, **P<0.01.
(D) USP15 KO THP-1 cells were reconstituted with either empty vector, USP15 WT or USP15 C298A. Cells were stimulated with WGP for 24 hours, and NF-κB reporter activity was assessed by a luciferase assay. The western blot shows USP15 protein levels expressed in the reconstituted cells. Three independent cell pools were transduced with Dectin-1 and NF-κB luciferase reporter and utilized in parallel for the subsequent treatments and assays (N=3). Error bars represent mean ± standard deviation. *P<0.05, **P<0.01.
We next tested whether USP15 is also able to inhibit CARD9-dependent signaling in human cells. We first knocked down USP15 by shRNA in THP-1 cells overexpressing Dectin-1 and an NF-κB luciferase reporter. As in murine BMDCs, we observed that knockdown of USP15 increased WGP-induced signaling measured through NF-κB luciferase activity (Fig. 3B). We further explored the effect of USP15 on CARD9 function in USP15 knockout (KO) THP-1 cells generated by a CRISPR-based approach (Experimental Procedures). USP15 KO also resulted in a higher fold induction of NF-κB luciferase upon WGP treatment (Fig. 3C). To further confirm that the observation in USP15 KO THP-1 cells is specific, we reconstituted the cells with either empty vector, wild-type USP15 or USP15 C298A. As expected, reconstitution with wild-type USP15 dramatically reduced WGP-induced NF-κB luciferase activity, while the catalytically inactive USP15 C298A mutant had no effect (Fig. 3D). These results suggest that USP15 catalytic activity counters that of TRIM62, inhibiting CARD9 via the removal of TRIM62-mediated ubiquitination.
DISCUSSION
Post-translational modifications play a critical role in virtually all signaling pathways, and ubiquitination in particular is crucial for the regulation of innate immune signaling (33). Previously, we identified the E3 ligase TRIM62 as a key activator of CARD9 through the deposition of K27-linked ubiquitin. This finding raised the question of the existence and identity of the deubiquitinase involved in the regulation of CARD9 activation. Here, through a mass spectrometry approach using CARD9 as bait, we demonstrate that USP15 is the deubiquitinase responsible for this regulation. Our in vitro and cell-based assays confirm that USP15 removes TRIM62-deposited ubiquitin and limits the extent of CARD9-dependent NF-κB activation and cytokine production. While we only assessed the effects of USP15 on CLR-mediated signaling, it is important to note that the architecture of the CBM complex is conserved across multiple cell types and signaling pathways, suggesting that these or other E3 ligase/deubiquitinase pairs may regulate their signaling.
Several deubiquitinases have been directly associated with immune processes through the identification of mutations in human patients, for example A20/TNFAIP3, USP18 and OTULIN (34), but overall our knowledge of the deubiquitinases involved in specific immune processes remains more limited. This is likely in part because the human genome harbors a comparatively low number of deubiquitinases (~100) relative to E3 ligases (~500-1,000), suggesting a requirement for reduced substrate specificity in deubiquitinases. Accordingly, USP15 is a ubiquitously expressed member of the ubiquitin specific peptidase family and has a broad range of functions (32). USP15 is a host factor for hepatitis C virus propagation by regulating viral RNA translation and lipid metabolism (35). It acts as a counterpart to the ER-located E3 ligase RNF26: RNF26 mediated-ubiquitination of SQSTM1 retains vesicles in the perinuclear cloud, while deubiquitination of SQSTM1 by USP15 facilitates the release of vesicles for fast transport (36). USP15 also deubiquitinates and stabilizes the E3 ligase MDM2, which results in the inhibition of antitumor T cell responses (37). Within innate immune pathways, USP15 facilitates RIG-I-mediated anti-viral signaling and type I interferon responses by deubiquitinating TRIM25 (38, 39). USP15 directly interacts with SMAD7 and other SMAD family members to deubiquitinate and stabilize type 1 TGF-β receptors, promoting TGF-β signaling (40, 41).
Importantly, we observed that USP15 binds to CARD9 in BMDCs in the absence of stimulation and that this interaction is largely unaffected by Dectin-1 ligand binding, indicating that USP15 constitutively interacts with CARD9 and may act to prevent aberrant CARD9 signaling at steady state. The negative regulation demonstrated in this study mediated by the USP15-CARD9 association could occur at multiple levels – deactivation of CARD9 after ubiquitination and modulation of CARD9-mediated signaling intensity, duration, or both – with the dynamic balance between TRIM62 and USP15 enzymatic activities likely critical in controlling the overall output of this pathway. Interestingly, mutations in NEMO that affect the binding of A20 can lead to immune phenotypes (42), raising the interesting possibility that some CARD9 mutations alter USP15 binding. More broadly, compounds that modulate the strength of CARD9-USP15 interactions may also represent an avenue for the specific regulation of this signaling pathway in fungal infections and inflammatory or autoimmune syndromes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nonghua Lu (the First Affiliated Hospital of Nanchang University, China) who supervised W.X. throughout the project, Jacques Deguine for guidance with manuscript preparation, and Theresa Reimels for manuscript preparation.
This work was supported by funding from the Helmsley Charitable Trust and the National Institutes of Health (grant AI137325 to R.J.X.). W.X. was supported by the China Scholarship Council.
REFERENCES
- 1.Drummond RA, Franco LM, and Lionakis MS. 2018. Human CARD9: A Critical Molecule of Fungal Immune Surveillance. Front Immunol 9: 1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drummond RA, and Lionakis MS. 2016. Mechanistic Insights into the Role of C-Type Lectin Receptor/CARD9 Signaling in Human Antifungal Immunity. Front Cell Infect Microbiol 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartjes L, and Ruland J. 2019. CARD9 Signaling in Intestinal Immune Homeostasis and Oncogenesis. Front Immunol 10: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quan C, Li X, Shi RF, Zhao XQ, Xu H, Wang B, Wang XP, Hu WG, Cao H, and Zheng J. 2019. Recurrent fungal infections in a Chinese patient with CARD9 deficiency and a review of 48 cases. Br J Dermatol 180: 1221–1225. [DOI] [PubMed] [Google Scholar]
- 5.Sari S, Dalgic B, Muehlenbachs A, DeLeon-Carnes M, Goldsmith CS, Ekinci O, Jain D, Keating MK, and Vilarinho S. 2018. Prototheca zopfii Colitis in Inherited CARD9 Deficiency. J Infect Dis 218: 485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaezi A, Fakhim H, Abtahian Z, Khodavaisy S, Geramishoar M, Alizadeh A, Meis JF, and Badali H. 2018. Frequency and Geographic Distribution of CARD9 Mutations in Patients With Severe Fungal Infections. Front Microbiol 9: 2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Xing H, Jiang X, Zeng J, Liu Z, Chen J, and Wu Y. 2019. Cerebral Phaeohyphomycosis Caused by Exophiala dermatitidis in a Chinese CARD9-Deficient Patient: A Case Report and Literature Review. Front Neurol 10: 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Zhang Y, Song Y, Wan Z, Wang X, and Li R. 2019. Phaeohyphomycosis caused by Phialophora americana with CARD9 mutation and 20-year literature review in China. Mycoses 62: 908–919. [DOI] [PubMed] [Google Scholar]
- 9.Arango-Franco CA, Moncada-Velez M, Beltran CP, Berrio I, Mogollon C, Restrepo A, Trujillo M, Osorio SD, Castro L, Gomez LV, Munoz AM, Molina V, Del Rio Cobaleda DY, Ruiz AC, Garces C, Alzate JF, Cabarcas F, Orrego JC, Casanova JL, Bustamante J, Puel A, Arias AA, and Franco JL. 2018. Early-Onset Invasive Infection Due to Corynespora cassiicola Associated with Compound Heterozygous CARD9 Mutations in a Colombian Patient. J Clin Immunol 38: 794–803. [DOI] [PubMed] [Google Scholar]
- 10.Dorhoi A, Desel C, Yeremeev V, Pradl L, Brinkmann V, Mollenkopf HJ, Hanke K, Gross O, Ruland J, and Kaufmann SH. 2010. The adaptor molecule CARD9 is essential for tuberculosis control. J Exp Med 207: 777–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamas B, Michel ML, Waldschmitt N, Pham HP, Zacharioudaki V, Dupraz L, Delacre M, Natividad JM, Costa GD, Planchais J, Sovran B, Bridonneau C, Six A, Langella P, Richard ML, Chamaillard M, and Sokol H. 2017. Card9 mediates susceptibility to intestinal pathogens through microbiota modulation and control of bacterial virulence. Gut. [DOI] [PubMed] [Google Scholar]
- 12.Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, and Ruland J. 2010. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol 11: 63–69. [DOI] [PubMed] [Google Scholar]
- 13.Roth S, Rottach A, Lotz-Havla AS, Laux V, Muschaweckh A, Gersting SW, Muntau AC, Hopfner KP, Jin L, Vanness K, Petrini JH, Drexler I, Leonhardt H, and Ruland J. 2014. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1beta production. Nat Immunol 15: 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagener M, Hoving JC, Ndlovu H, and Marakalala MJ. 2018. Dectin-1-Syk-CARD9 Signaling Pathway in TB Immunity. Front Immunol 9: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, Latiano A, Goyette P, Green T, Halfvarson J, Haritunians T, Korn JM, Kuruvilla F, Lagace C, Neale B, Lo KS, Schumm P, Torkvist L, D. National Institute of, C. Digestive Kidney Diseases Inflammatory Bowel Disease Genetics, C. United Kingdom Inflammatory Bowel Disease Genetics, C. International Inflammatory Bowel Disease Genetics, Dubinsky MC, Brant SR, Silverberg MS, Duerr RH, Altshuler D, Gabriel S, Lettre G, Franke A, D'Amato M, McGovern DP, Cho JH, Rioux JD, Xavier RJ, and Daly MJ. 2011. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43: 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, Park YR, Raychaudhuri S, Pouget JG, Hubenthal M, Folseraas T, Wang Y, Esko T, Metspalu A, Westra HJ, Franke L, Pers TH, Weersma RK, Collij V, D'Amato M, Halfvarson J, Jensen AB, Lieb W, Degenhardt F, Forstner AJ, Hofmann A, I. B. D. G. C. International, C. International Genetics of Ankylosing Spondylitis, P. S. C. S. G. International, C. Genetic Analysis of Psoriasis, E. Psoriasis Association Genetics, Schreiber S, Mrowietz U, Juran BD, Lazaridis KN, Brunak S, Dale AM, Trembath RC, Weidinger S, Weichenthal M, Ellinghaus E, Elder JT, Barker JN, Andreassen OA, McGovern DP, Karlsen TH, Barrett JC, Parkes M, Brown MA, and Franke A. 2016. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 48: 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, I. B. D. G. C. International, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, and Cho JH. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pointon JJ, Harvey D, Karaderi T, Appleton LH, Farrar C, Stone MA, Sturrock RD, Brown MA, and Wordsworth BP. 2010. Elucidating the chromosome 9 association with AS; CARD9 is a candidate gene. Genes Immun 11: 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerova D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Paczek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, and Gharavi AG. 2014. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janse M, Lamberts LE, Franke L, Raychaudhuri S, Ellinghaus E, Muri Boberg K, Melum E, Folseraas T, Schrumpf E, Bergquist A, Bjornsson E, Fu J, Jan Westra H, Groen HJ, Fehrmann RS, Smolonska J, van den Berg LH, Ophoff RA, Porte RJ, Weismuller TJ, Wedemeyer J, Schramm C, Sterneck M, Gunther R, Braun F, Vermeire S, Henckaerts L, Wijmenga C, Ponsioen CY, Schreiber S, Karlsen TH, Franke A, and Weersma RK. 2011. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL, and CARD9. Hepatology 53: 1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng Z, Li J, Wang Y, Li S, Hou M, Peng J, and Feng Q. 2019. A CARD9 single-nucleotide polymorphism rs4077515 is associated with reduced susceptibility to and severity of primary immune thrombocytopenia. Ann Hematol 98: 2497–2506. [DOI] [PubMed] [Google Scholar]
- 22.Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortiz ZG, Nedelsky NB, Huang H, Ng A, Gardet A, Cheng SC, Shamji AF, Rioux JD, Wijmenga C, Netea MG, Means TK, Daly MJ, and Xavier RJ. 2015. Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-fungal Immunity and Intestinal Inflammation. Immunity 43: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Xu JF, Zheng G, Lu HW, Duan JL, Rui W, Guan JH, Cheng LQ, Yang DD, Wang MC, Lv QZ, Li JX, Zhao X, Chen CX, Shi P, Jia XM, and Lin X. 2018. CARD9(S12N) facilitates the production of IL-5 by alveolar macrophages for the induction of type 2 immune responses. Nat Immunol 19: 547–560. [DOI] [PubMed] [Google Scholar]
- 24.Shiokawa M, Yamasaki S, and Saijo S. 2017. C-type lectin receptors in anti-fungal immunity. Current opinion in microbiology 40: 123–130. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, and Saito T. 2008. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol 9: 1179–1188. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, and Yamasaki S. 2009. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med 206: 2879–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruland J, and Hartjes L. 2019. CARD-BCL-10-MALT1 signalling in protective and pathological immunity. Nat Rev Immunol 19: 118–134. [DOI] [PubMed] [Google Scholar]
- 28.Strasser D, Neumann K, Bergmann H, Marakalala MJ, Guler R, Rojowska A, Hopfner KP, Brombacher F, Urlaub H, Baier G, Brown GD, Leitges M, and Ruland J. 2012. Syk kinase-coupled C-type lectin receptors engage protein kinase C-sigma to elicit Card9 adaptor-mediated innate immunity. Immunity 36: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh JT, Xu S, Huo JX, Kim SS, Wang Y, and Lam KP. 2019. Dok3-protein phosphatase 1 interaction attenuates Card9 signaling and neutrophil-dependent antifungal immunity. J Clin Invest 129: 2717–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, Zhang L, Kim WY, Olumi AF, and Kaelin WG Jr. 2007. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell 28: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connell DJ, Kolde R, Sooknah M, Graham DB, Sundberg TB, Latorre I, Mikkelsen TS, and Xavier RJ. 2016. Simultaneous Pathway Activity Inference and Gene Expression Analysis Using RNA Sequencing. Cell Syst 2: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou CK, Chang YT, Korinek M, Chen YT, Yang YT, Leu S, Lin IL, Tang CJ, and Chiu CC. 2017. The Regulations of Deubiquitinase USP15 and Its Pathophysiological Mechanisms in Diseases. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu H, and Sun SC. 2016. Ubiquitin signaling in immune responses. Cell research 26: 457–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Etzioni A, Ciechanover A, and Pikarsky E. 2017. Immune defects caused by mutations in the ubiquitin system. The Journal of allergy and clinical immunology 139: 743–753. [DOI] [PubMed] [Google Scholar]
- 35.Kusakabe S, Suzuki T, Sugiyama Y, Haga S, Horike K, Tokunaga M, Hirano J, Zhang H, Chen DV, Ishiga H, Komoda Y, Ono C, Fukuhara T, Yamamoto M, Ikawa M, Satoh T, Akira S, Tanaka T, Moriishi K, Fukai M, Taketomi A, Yoshio S, Kanto T, Suzuki T, Okamoto T, and Matsuura Y. 2019. USP15 Participates in Hepatitis C Virus Propagation through Regulation of Viral RNA Translation and Lipid Droplet Formation. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jongsma ML, Berlin I, Wijdeven RH, Janssen L, Janssen GM, Garstka MA, Janssen H, Mensink M, van Veelen PA, Spaapen RM, and Neefjes J. 2016. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell 166: 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, Nakaya M, Zhou X, Cheng X, Yang P, Lozano G, Zhu C, Watowich SS, Ullrich SE, and Sun SC. 2014. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol 15: 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, and Gack MU. 2014. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal 7: ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torre S, Polyak MJ, Langlais D, Fodil N, Kennedy JM, Radovanovic I, Berghout J, Leiva-Torres GA, Krawczyk CM, Ilangumaran S, Mossman K, Liang C, Knobeloch KP, Healy LM, Antel J, Arbour N, Prat A, Majewski J, Lathrop M, Vidal SM, and Gros P. 2017. USP15 regulates type I interferon response and is required for pathogenesis of neuroinflammation. Nat Immunol 18: 54–63. [DOI] [PubMed] [Google Scholar]
- 40.Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, Cuartas I, Jimenez J, Garcia-Dorado D, Sahuquillo J, Bernards R, Baselga J, and Seoane J. 2012. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med 18: 429–435. [DOI] [PubMed] [Google Scholar]
- 41.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, Cordenonsi M, and Piccolo S. 2011. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol 13: 1368–1375. [DOI] [PubMed] [Google Scholar]
- 42.Zilberman-Rudenko J, Shawver LM, Wessel AW, Luo Y, Pelletier M, Tsai WL, Lee Y, Vonortas S, Cheng L, Ashwell JD, Orange JS, Siegel RM, and Hanson EP. 2016. Recruitment of A20 by the C-terminal domain of NEMO suppresses NF-kappaB activation and autoinflammatory disease. Proceedings of the National Academy of Sciences of the United States of America 113: 1612–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.