Abstract
In Arabidopsis thaliana, lateral roots initiate in a process preceded by periodic gene expression known as the root clock. We identified the vesicle-trafficking regulator GNOM and its suppressor, ADENOSINE PHOSPHATE RIBOSYLATION FACTOR GTPase ACTIVATION PROTEIN DOMAIN3, as root clock regulators. GNOM is required for the proper distribution of pectin, a mediator of intercellular adhesion, whereas the pectin esterification state is essential for a functional root clock. In sites of lateral root primordia emergence, both esterified and de-esterified pectin variants are differentially distributed. Using a reverse-genetics approach, we show that genes controling pectin esterification regulate the root clock and lateral root initiation. These results indicate that the balance between esterified and de-esterified pectin states is essential for proper root clock function and the subsequent initiation of lateral root primordia.
Both plants and animals use biological clocks to estimate time and space during development. In vertebrates, the formation of presumptive vertebrae, known as somitogenesis, is one of the best characterized mechanisms involving periodic gene expression. In plants, before the initiation of lateral root primordia (LRP), the auxin response marker DR5 and other genes exhibit periodic expression in a region called the oscillation zone (1, 2). This rhythmic gene expression, known as the root clock, produces prebranch sites, DR5-expressing spots that develop into LRP (1). The root clock represents a flexible oscillator, producing prebranch sites approximately every 6 hours. Most genes implicated in the initiation of LRP are related to auxin signaling (3, 4) or metabolism (5). Recently, it has been shown that auxin-dependent changes in endodermis cells are required before the initiation of LRP in underlying pericycle cells (6). One candidate protein that putatively mediates signaling between cells and is also involved in LR initiation is GNOM (7).
GNOM is a trans-Golgi network-localized (8) ARF-GEF [ADP RIBOSYLATION FACTOR GUAANINE NUCLEOTIDE EXCHANGE FACTOR (9)] that regulates vesicle trafficking to the plasma membrane. It had been suggested that the reduced LR phenotype of the weak gnom allele, fwr, is mediated by one of the PIN-FORMED (PIN) proteins. However, the fwr mutant has properly localized PIN1 (7). In addition, there are no known PIN mutants with LR phenotypes. These data suggest that the lack of LRs in the gnom mutant might depend on other factors that are not directly involved in auxin signaling. Vesicle trafficking is necessary for the secretion of cell wall components including pectin, hemicelluloses, and cell wall-modifying enzymes (10). Here, we show that disrupting GNOM alters the distribution of pectin and blocks the functioning of the root clock. Furthermore, higher levels of esterified and de-esterified pectin are differentially localized around emerging LRP, whereas modulation of either form of pectin leads to a loss of prebranch site formation. On the basis of these data, we conclude that GNOM-dependent vesicle trafficking mediates the distribution of pectin in the cell wall, which in turn is essential for root clock function and subsequent LR formation.
LRP cell divisions initiate soon after oscillation
The root clock is defined as a 6-hour periodic DR5 expression in a region known as the oscillation zone near the border of the elongation and differentiation zones of the Arabidopsis thaliana root (1, 2). After peak DR5-driven luciferase expression, the region shrinks to form a confined spot defined as a prebranch site, where LRP emerge subsequently. All LRP emerge from prebranch sites and most prebranch sites develop into LRP (1). To better understand the relationship between LR initiation and prebranch site formation, we examined roots expressing both the DR5:: LUC reporter, which marks prebranch sites, and pCLE44::GFP, which marks early LRP (fig. S1, A and B; see below for details on the CLE44 marker). In most cases (21/26), the most recent DR5::LUC-marked prebranch site was associated with an early-stage LRP. We conclude that LRP initiation starts concurrent with or soon after the formation of a prebranch site.
RNA-sequencing screen identifies oscillating genes
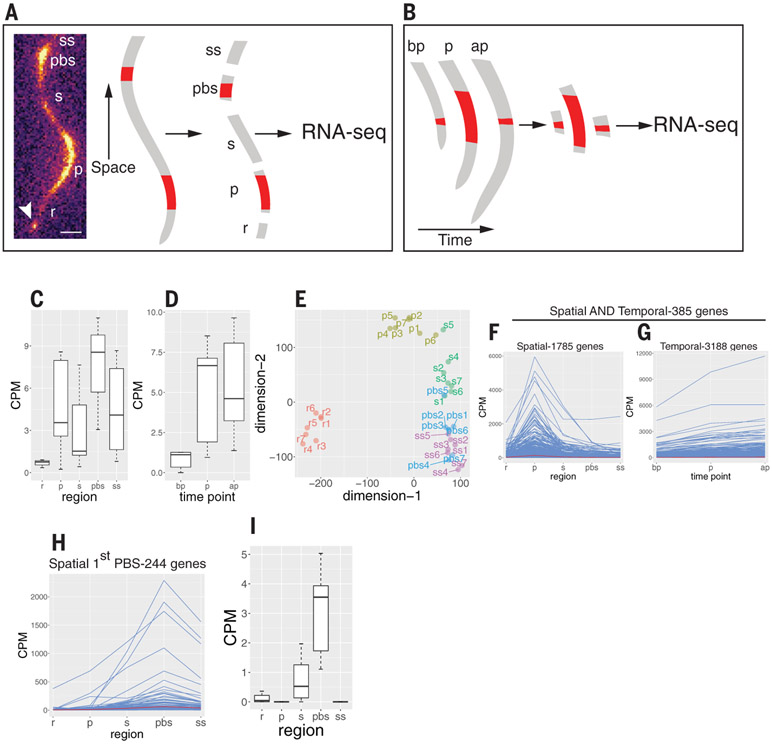
To identify genes with elevated expression in the oscillation zone and/or in the most recently formed prebranch site, we used RNA-sequencing (RNA-seq) analysis of sections from individual plants using the modified Smart-seq2 method (11). In a spatial experiment, we performed RNA-seq on transverse sections from the oscillation zone, the first prebranch site, and their flanking regions (Fig. 1A). In a complementary, temporal experiment, we performed RNA-seq on DR5::LUC-expressing sections before, during, and after peak expression (Fig. 1B and supplementary materials). To validate the quality of the data, we compared RNA-seq reads of the luciferase transgene (driven by the DR5 promoter) with luciferase imaging and found a high correlation (Fig. 1, C and D). Multidimensional scaling analysis and other statistical analyses provided further evidence for the fidelity of the data (Fig. 1E and fig. S2). To identify putative root clock regulators, we selected genes that were overrepresented in the oscillation zone-spatial, the oscillation zone-spatial together with the oscillation zone-temporal, or the prebranch site-spatial datasets (Fig. 1, F to I, and table S1, A to D) and used these three lists to perform gene ontology (GO) analyses (table S2). We identified CLE44 as one of the genes expressed in incipient prebranch sites with similar frequency as DR5 (fig. S1, C to E, and movie S1) and used it as a prebranch site marker together with DR5 in subsequent experiments. Some of the most common GO terms were related to cell wall biogenesis and vesicle trafficking (Fig. 2A). Of particular interest was a term for pectin-metabolic process, including genes that code for pectin lyases, pectate lyases, pectin methylesterases (PMEs), and their inhibitors (PMEIs) (P = 0.005; Fig. 2, B and C, and table S2). PMEs de-methyl-esterify galacturonic acid residues of homogalacturonan pectin. Pectin is synthesized in the Golgi in a highly methyl-esterified form and delivered to the apoplast, where it can be de-methyl-esterified. Blockwise de-esterified homogalacturonan chains can be crossed-linked by calcium, leading to stiffening of the cell wall and/or promoting adhesion between cells, whereas randomly de-esterified homogalacturonan can be degraded by pectinases, resulting in wall loosening and/or cell separation (12, 13). All nine oscillation zone-spatial PMEs belong to group 1 (14) within the type I class of PMEs, which contain a PRO domain at the N terminus that requires pre-processing in the Golgi before secretion to the apoplast (15). Another group of genes overrepresented in the oscillation zone-spatial dataset is involved in auxin transport and signal transduction. Mutations in five of these genes (LAX3, AUX1, ARF19, IAA14, and IAA28) directly affect LR formation [fig. S3A and (3, 16-18)]. A third group overrepresented in the oscillation zone-spatial dataset is involved in the transport, metabolism, and response to nitrogen-related compounds (fig. S3B), all processes that have been shown to affect LR density and outgrowth (19, 20). For example, the tonoplast aquaporin TONOPLAST INTEGRAL PROTEIN2; 1/DELTA-TIP1 (TIP2;1), which is involved in NH3 transport to the vacuole and response to NH3 accumulation, is redundantly required, together with two other TIP proteins, for LR initiation (21, 22). We also identified two additional genes that were not included in the dataset because they did not fulfill the log-fold change (logFC) > 0.5 criteria but still had high expression in the oscillation zone: AMT2;1, an ammonium transporter required for ammonium-dependent LR branching (20), and TGA4, a basic leucine zipper transcription factor controlling nitrate-dependent LR initiation (19). Another group of genes with high expression in the oscillation zone belong to the cysteine-rich receptor-like protein kinase (crRLK) subfamily (fig. S3C). FERONIA, one of the members of this family, was recently shown to bind pectin and to regulate pavement cell morphogenesis (23). Finally, our approach also identified 10 genes in a GO term for LR morphogenesis (fig. S3D), which involve auxin transport (LAX3 and AUX1), auxin signaling (ARF11/ARF19, IAA14 and IAA28), exocytosis (EXO70A1), and the promotion of protein degradation by ubiquitin ligase activity (XBAT32). Together, these results suggest that the oscillation zone functions as a transcriptional hub for multiple pathways required for emergence of LRP.
Fig. 1. Experimental design and analysis of RNA-seq experiments.
(A) Spatial experiment. Seven DR5::LUC roots were imaged using the Lumazone system (left). Once peak expression was detected in the oscillation zone, the root was dissected into five sections: r, a section proximal to the root meristem and distal to the oscillation zone; p, the oscillation zone; s, a section proximal to the oscillation zone and distal to the youngest prebranch site; pbs, the youngest prebranch site; and ss, a section proximal to the prebranch site. Each section was used to generate a transcriptome profile (35 total). Arrowhead indicates the root tip. Scale bar, 0.3 mm. (B) Same as in (A) but a root section was collected from the oscillation zone before peak expression (bp), during peak expression (p), or after peak expression (ap). (C and D) CPM reads for luciferase RNA in the spatial experiment (C) and the temporal experiment (D). (E) Multidimensional scaling plot of the spatial experiment. Note that the regions (r, p, s, pbs, and ss) mostly form distinctive clades. (F) Expression pattern of all 1785 genes in the spatial experiment with a log2FC > 0.5 (FC > 1.414) in the p region compared with each of the other zones. (G) Expression pattern of all 3188 genes in the temporal experiment with a log2FC > 0.5 (FC > 1.414) in the p region (peak luciferase expression) compared with the bp region (before peak luciferase expression). (H) Expression pattern of all 244 genes in the spatial experiment with a log2FC > 0.5 (FC > 1.414) in the pbs zone compared with each of the other zones. (I) Expression profile of CLE44, which was later used as a prebranch site marker. CPM, counts per million.
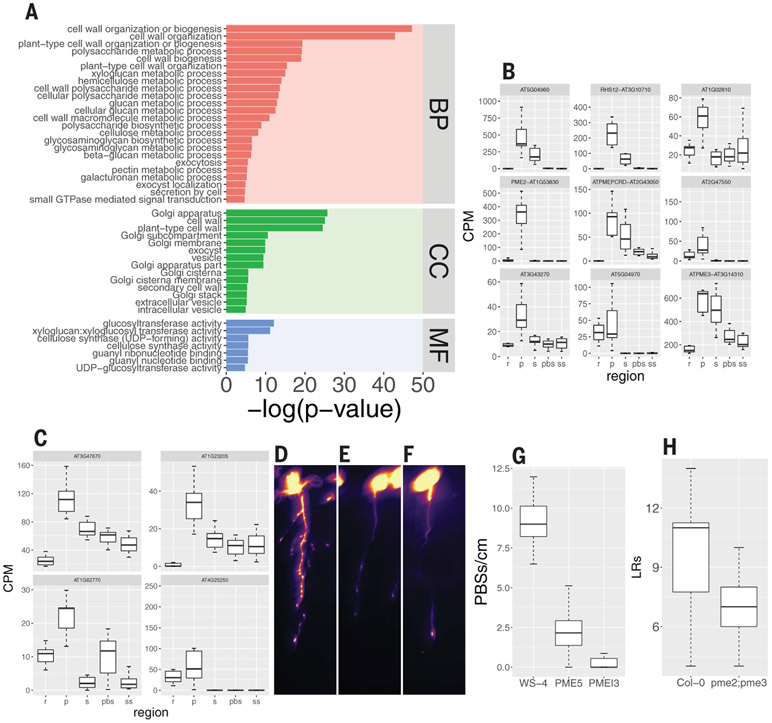
Fig. 2. Cell wall-, vesicle trafficking-, and membrane-related genes are overrepresented in the oscillation zone and prebranch site and required for its formation.
(A) GO term analysis of the 1785 genes described in Fig. 1F (BP, biological process; CC, cellular component; MF, molecular function). The plot depicts vesicle trafficking-, Golgi-, cell wall-, small GTPase–mediated signal transduction-, and carbohydrate metabolism-related GO terms with P ≤ 0.01 (hypergeometric test using the gProfiler R package; see table S2 for the complete list). (B and C) Expression pattern of nine PME (B) and four PMEI (C) genes in the spatial experiment. (D to F) pCLE44::LUC expression in WT [WS-4, (D)], PME5-overexpressing (E), and PMEI3-overexpressing (F) seedlings after EtOH induction. Scale bar, 0.2 cm. (G) Quantification of prebranch site number [(D) to (F)]. P < 10−14, Wilcoxon rank sum test. (H) LR number of pme2;pme3 compared with Col-0 WT. P = 0.007, Wilcoxon rank sum test.
PMEs and PMEIs are required for prebranch site and LR formation
Because PME5 and PMEI3 have been implicated in phyllotaxis (24), we decided to explore the role of in LR branching. To determine whether changing the pectin esterification state also affects rhyzotaxis, we inducibly overexpressed PME5 and PMEI3 and found a severe reduction in prebranch site number in both cases (Fig. 2, D to G, and fig. S4, A to D), as well as a decrease in LR number (fig. S4, E to G). On the basis of our RNA-seq results, we identified two PME genes, the mutants of which, pme2 and pme3, as well as the double mutant combination, showed reduced numbers of LRs (Fig. 2H and fig. S4, H to K). These results suggest that the balance between pectin esterification and de-esterification under-pins the functionality of the root clock.
Vesicle-trafficking genes mediate root clock and LR formation
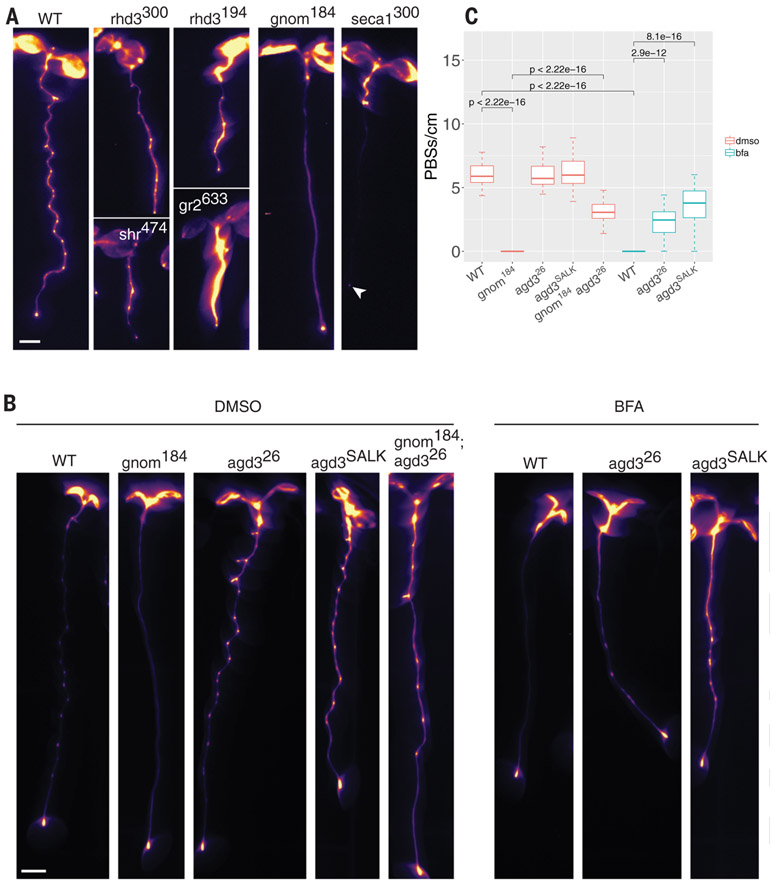
To complement our reverse-genetics approach, we mutagenized DR5::LUC seeds and screened for alterations in root clock function based on imaging. We identified six mutations with modified root clock function (table S3A) and mapped the affected genes [table S3 and (25)]. Two independent mutations were found in ROOT HAIR DEFECTIVE 3 (RHD3) and one each in GNOM, SHORTROOT, GLUTHATIONE REDUCTASE, and SECA1 (Fig. 3A and table S3). Because half of the mutations affect genes coding for vesicle-trafficking-related proteins (RHD3 and GNOM) and the gnom mutant allele (gnom184) did not appear to be highly pleiotropic, we focused our analysis on this mutant (Fig. 3 and fig. S5, A to I). This allele could be partially complemented by its native promoter and 3′ untranslated region or with the G1090-inducible overexpression promoter [fig. S5J and (26)]. To better understand the gnom184 phenotype, we grew wild-type (WT) seedlings on brefeldin A (BFA), a vesicle-trafficking inhibitor that targets a subset of plant ARF-GEFs, including GNOM (27). A single amino acid substitution in the catalytic Sec7 domain of GNOM can convert WT seedlings to BFA-resistant seedlings (27), which produce LRs despite exposure to BFA, suggesting that BFA specifically targets GNOM. Seedlings grown on low BFA concentrations (3 to 5 μM) have a similar phenotype to gnom184 with no LRs [fig. S5I and (27)]. We next tested the effect of BFA on the root clock using the DR5::LUC and the pCLE44::LUC lines. The expression pattern of these markers was strongly altered, phenocopying the gnom184 allele (Fig. 3, B and C, and fig. S5, G and H). To further identify root-clock-regulated genes that function in the GNOM pathway, we used the mutagenized DR5::LUC population and screened for seedlings that could develop LRs under BFA treatment. We identified two BFA-insensitive mutants (table S3) displaying both LRs and prebranch sites upon BFA treatment. The first affected gene, LEUCINE CARBOXYLMETHYLTRANSFERASE1/SUPPRESSOR OF BRI1 (LCMT/SBI1), is involved in terminating the brassinosteroid signaling cascade during endocytosis by promoting degradation of the hormone receptor BRI1 [fig. S5, K to M, and (28)]. The second gene, ARF-GTP ACTIVATING PROTEIN DOMAIN (AGD3), is required for scission during vesicle formation (29). GNOM is an ARF-GEF that facilitates the exchange of GDP to GTP, leading to membrane recruitment of the ARF protein. AGD3 acts in the opposite direction by activating ARF-dependent GTP hydrolysis, allowing vesicle budding and eventually membrane dissociation of ARF-GDP (30). When introduced into the gnom184 background, our mutant agd3 allele (agd326) partially rescued the loss of root clock function and the LR phenotype (Fig. 3, B and C, and fig. S5, G to I). These results indicate that the vesicle-trafficking GNOM-AGD3 circuit is required for LRP initiation and root clock function.
Fig. 3. Perturbation of vesicle trafficking affects the root clock.
(A) Root clock phenotype of six ethyl methanesulfonate mutants. Line name is shown as a superscript and the gene name is in lowercase letters. Arrowhead indicates DR5::LUC expression in the root tip. (B) Expression of pCLE44::LUC in different genotypes under mock treatment [dimethyl sulfoxide (DMSO), left five panels] or BFA treatment (right three panels). (C) Quantification of pCLE44::LUC prebranch site number in gnom184 and its suppressor, agd3 (SALK and the EMS mutant agd326), with or without 3 μM BFA (t test). Scale bar, 0.2 cm.
GNOM regulates homogalacturonan distribution
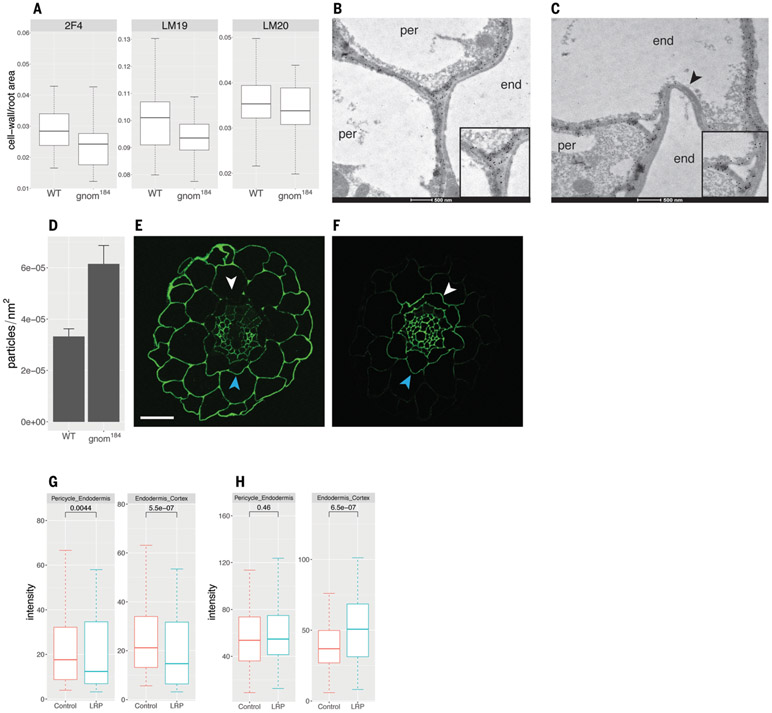
Because pectic homogalacturonan is synthesized in its esterified form in the Golgi (covalently linked to a methyl group through an ester bond) and then secreted to the cell wall, where it can be de-esterified by PMEs, we sought to determine whether its involvement in the root clock and LRP initiation requires GNOM-dependent vesicle trafficking. We used immunohistochemistry to determine whether gnom184 has altered distribution of three homogalacturonan forms (31): de-esterified and Ca2+ cross-linked [2F4 antibody; (32)], de-esterified (LM19 antibody), and esterified [LM20 antibody; (33)]. The mean intensity signal of homogalacturonan in all three experiments did not differ between WT and gnom184 (fig. S6). Using xyloglucanase to digest hemicellulose did not unmask additional pectin sites or change the intensity of the antipectin antibody staining (fig. S6), as previously shown for mannan polysaccharide (34). However, the cell wall area occupied by the low-esterified form was significantly lower in gnom184 than in WT (P = 0.0028; Fig. 4A). A severe gnom allele has been shown to alter the distribution of de-esterified homogalacturonan in the cell wall (35). Because the localization of de-esterified pectin is altered in gnom184 and the boundary between the pericycle and the endodermis is relevant for the emergence of LRP, we focused our comparison of de-esterified homogalacturonan distribution on pericycle-endodermis junctions. Using an antibody that preferentially binds low-esterified homogalacturonan (36), immunogold labeling (37, 38) followed by transmission electron microscopy identified two types of gold particle distribution in three-way pericycle-endodermis junctions (junctions of two pericycle cells and one endodermis cell or one pericycle cell and two endodermis cells). gnom184 junctions were more particle dense compared with WT junctions (Fig. 4, B to D, and fig. S7, B to E), which may prevent LR emergence. A more qualitative analysis showed a similar trend (fig. S7A). This indicates that GNOM mediates homogalacturonan distribution in pericycle-endodermis junctions.
Fig. 4. Low-esterified homogalacturonan distribution is affected in gnom184 and negatively correlates with sites of LR inception.
(A) Immunohistochemical quantification of cell wall area per root cross-section area with the antibody against de-esterified and Ca2+ cross-linked homogalacturonan 2F4 (P2F4 = 0.31, Wilcoxon rank sum test), de-esterified homogalacturonan LM19 (PLM19 = 0.0028, t test), and esterified homogalacturonan LM20 (PLM20 = 0.042, t test). False discovery rate method adjustment was applied. (B and C) Transmission electron microscopy (TEM) images of LM19 immunogold localization. De-esterified homogalacturonan is marked by round black gold particles. Cell types (end, endodermis; per, pericycle) are indicated in the cytoplasmic region of each cell. Insets show the three-way dense (B) and particle-free (C) junctions. Note the collapsed and antibody-free endodermis-endodermis cell wall marked by the arrowhead. (D) Quantification of gold particles in pericycle-endodermis junctions; genotypes show different particle densities (P = 0.001, Wilcoxon test). (E and F) Immunohistochemistry with LM19 (E) and LM20 (F) antibodies of root cross-sections in regions of emerging LRP. White arrowheads indicate the side facing LRP; blue arrowheads indicate the opposite (control) side. Scale bar, 20 μm. (G and H) Quantification and statistics of (E) and (F), respectively. Wilcoxon paired test statistics are given above boxes.
Homogalacturonan subtypes are differentially distributed at sites of LRP initiation
We next investigated whether we could identify a direct link between homogalacturonan distribution and LRP inception. Using immunohistochemistry on transverse sections at the site of LRP inception, we found that pericycle-endodermis and endodermis-cortex circumference boundaries adjacent to the LRP have reduced levels of de-esterified homogalacturonan compared with the opposite (control) side (Fig. 4, E and G). By contrast, we identified higher levels of high-esterified homogalacturonan in the endodermis-cortex boundary (but not in the pericycle-endodermis) adjacent to the LRP compared with the opposite (control) side (Fig. 4, F and H). These results show that de-esterified homogalacturonan, which is usually associated with increased cell wall stiffness, is reduced near emerging LRP, and that pectin subtypes are differentially distributed at sites of initiating LRP. Given that LRP emerge by pushing through overlying cell layers, loss of de-esterified pectin could reduce cell adhesion at prebranch sites, providing a competence to form LRP.
Discussion
A minimum requirement for an oscillating system is delayed negative feedback, which can produce a periodic pattern with as few as two components (39). However, the somitogenesis clock requires the function of multiple components arranged in three complex networks (40). In Arabidopsis, more than 30 genes have been implicated in LR development, most of which are related to auxin signaling, transport, and homeostasis (41). A recent study showed that endodermis cells overlying the pericycle undergo volume reduction, which is required for the first cell division during LRP initiation (6). Although the precise mechanism by which pectin modification modulates the root clock and LRP initiation is still not known, we show here that LRP initiation involves differential distribution of pectin species, with reduced de-esterified pectin levels in overlying primordia. This distribution could allow the physical emergence of LRP while preventing emergence on the opposite side of the root. Additional support for the hypothesis that a balanced esterification level is essential for the root clock and LRP initiation is demonstrated by overexpression of either PME5 or PMEI3, which confers similar phenotypes in the root (this study) and in pavement cells (42). The fact that two proteins (GNOM and AGD3) with opposing functionalities also have an opposite phenotype provides additional evidence for the importance of balanced pectin components. One of the remaining questions is how the status of extracellular pectin is conveyed to the cell to initiate LR formation. One possibility is that biomechanical changes in the cell wall are translated into signal transduction that leads to LRP initiation. Alternatively, pectin might bind and activate a membranous receptor such as the FERONIA crRLK to initiate a signaling cascade that eventually results in the formation of LRP.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Wilson and Y. Rui for technical support and advice; A. Peaucelle for seeds; M. Plue (Duke, SMif), R. Vancini and H. Mekeel (Duke Pathology), and K. Pryer for assistance with immunotissue processing and TEM microscopy; the Duke Center for Genomic and Computational Biology; and I. W. Taylor, R. Shahan, J. Vermeer, and J. K. Polko for critical review of the manuscript.
Funding: This work was supported by grants to P.N.B. from the National Institutes of Health (grant nos. R01-GM043778 and R35-GM131725), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation (grant no. GBMF3405). M.A.M.-R. was funded by Ministerio de Economía y Competitividad of Spain (MINECO) and ERDF (grant no. BFU2016-80315-P).
Footnotes
Competing interests: The authors declare no competing interests. P.N.B. is the cofounder and chair of the scientific advisory board of Hi Fidelity Genetics, Inc., a company that works on crop root growth.
Data and materials availability: All materials are available upon request. All data are available in the main paper or the supplementary materials. The RNA-seq and DNA-seq datasets are available in the Sequence Read Archive repository with the accession numbers PRJNA478564 and PRJNA490692, respectively.
REFERENCES AND NOTES
- 1.Moreno-Risueno MA et al. , Science 329, 1306–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Smet I et al. , Development 134, 681–690 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Fukaki H, Tameda S, Masuda H, Tasaka M, Plant J. 29, 153–168 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Wilmoth JC et al. , Plant J. 43, 118–130 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Xuan W et al. , Science 351, 384–387 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Vermeer JEM et al. , Science 343, 178–183 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Okumura K et al. , Plant Cell Physiol. 54, 406–417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naramoto S et al. , Plant Cell 26, 3062–3076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shevell DE et al. , Cell 77, 1051–1062 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Kim S-J, Brandizzi F, Glycobiology 26, 940–949 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Picelli S et al. , Nat. Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Micheli F, Trends Plant Sci. 6, 414–419 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Willats WG et al. , J. Biol. Chem 276, 19404–19413 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Louvet R et al. , Planta 224, 782–791 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Wolf S, Rausch T, Greiner S, Plant J. 58, 361–375 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Tatematsu K et al. , Plant Cell 16, 379–393 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rybel B et al. , Curr. Biol 20, 1697–1706 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Swarup K et al. , Nat. Cell Biol 10, 946–954 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Alvarez JM et al. , Plant J. 80, 1–13 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Lima JE, Kojima S, Takahashi H, von Wirén N, Plant Cell 22, 3621–3633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loqué D, Ludewig U, Yuan L, von Wirén N, Plant Physiol. 137, 671–680 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhardt H et al. , Plant Physiol. 170, 1640–1654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W, Tang W, Anderson CT, Yang Z, FERONIA’s sensing of cell wall pectin activates ROP GTPase signaling in Arabidopsis. bioRxiv 269647 [Preprint]. 22 February 2018. 10.1101/269647. [DOI] [Google Scholar]
- 24.Peaucelle A et al. , Curr. Biol 18, 1943–1948 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Wachsman G, Modliszewski JL, Valdes M, Benfey PN, Plant Physiol. 174, 1307–1313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siligato R et al. , Plant Physiol. 170, 627–641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geldner N et al. , Cell 112, 219–230 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Wu G et al. , Sci. Signal 4, ra29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koizumi K et al. , Development 132, 1699–1711 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Donaldson JG, Jackson CL, Nat. Rev. Mol. Cell Biol 12, 362–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rui Y et al. , Plant Cell 29, 2413–2432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liners F, Letesson J-J, Didembourg C, Van Cutsem P, Plant Physiol. 91, 1419–1424 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP, Carbohydr. Res 344, 1858–1862 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Verhertbruggen Y, Walker JL, Guillon F, Scheller HV, Front. Plant Sci 8, 1505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevell DE, Kunkel T, Chua NH, Plant Cell 12, 2047–2060 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcus SE et al. , Plant J. 64, 191–203 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Ruel K, Nishiyama Y, Joseleau J-P, Plant Sci. 193-194, 48–61 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Polko JK et al. , Curr. Biol 28, 3174–3182.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Gierer A, Meinhardt H, Kybernetik 12, 30–39 (1972). [DOI] [PubMed] [Google Scholar]
- 40.Goldbeter A, Pourquié O, J. Theor. Biol 252, 574–585 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Péret B et al. , Trends Plant Sci. 14, 399–408 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Haas KT, Wightman R, Meyerowitz EM, Peaucelle A, Science 367, 1003–1007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.