Abstract
The transmembrane glycoprotein Trop2 plays important roles in various types of human cancers, especially lung cancer. Although it has been found to form clusters on cancer cell membranes, the factors that affect its clustering are not yet fully understood. Here, using direct stochastic optical reconstruction microscopy (dSTORM), we found that Trop2 generated more, larger, and denser clusters on apical cell membranes than on basal membranes and that the differences might be related to the different membrane structures. Moreover, dual-color dSTORM imaging revealed significant colocalization of Trop2 and lipid rafts, and methyl-β-cyclodextrin disruption dramatically impaired the formation of Trop2 clusters, indicating a key role of lipid rafts in Trop2 clustering. Additionally, depolymerization of the actin cytoskeleton decreased Trop2 cluster numbers and areas, revealing that actin can stabilize the clusters. More importantly, stimulation of Trop2 in cancer cells hardly changed the cluster morphology, suggesting that Trop2 is activated and forms clusters in cancer cells. Altogether, our work links the spatial organization of Trop2 to different membrane structures and Trop activation and uncovers the essential roles of lipid rafts and actin in Trop2 cluster maintenance.
1. Introduction
Human trophoblast cell–surface antigen 2 (Trop2) is a transmembrane glycoprotein originally identified in human placental trophoblasts.1,2 It is highly expressed in stem cells and in many organs during development, such as during fetal lung growth.3,4 More importantly, Trop2 is weakly expressed or not expressed in normal tissues of the human body but is overexpressed in various cancer tissues, including lung, colorectal, and ovarian cancer tissues.5,6 Its high expression is closely associated with tumor proliferation, migration, and aggressiveness.7,8
In addition to Trop2, many other membrane proteins have been reported to be abnormally expressed in cancers, such as EGFR in lung cancer and EpCAM in breast cancer.9,10 Of note, the spatial distributions of EGFR and EpCAM have the same clustering characteristics. Similarly, our previous study revealed that Trop2 forms clusters on membranes of lung cancer cells.11 It is thus possible that the clustered distribution of Trop2 is related to the role of this protein in tumorigenesis. Therefore, uncovering the cellular organizations that participate in Trop2 clustering is crucial.
Among the important microdomains on the membrane are lipid rafts, in which lipids, cholesterol, and specific proteins are tightly packed.12,13 The structures of these microdomains can maintain protein aggregation14 and provide platforms for interactions between lipids and specific proteins,15,16 and they are beneficial for signaling responses and transduction.17,18 Thus, research on the connection between lipid rafts and Trop2 clusters is important. In addition, actin participates in tumor morphogenesis, development, invasion, and migration.19 Some transmembrane proteins bind to actin directly or indirectly via actin-binding proteins so that actin can regulate protein recruitment and organization.20 However, it remains unclear whether actin filaments influence Trop2 distribution and clustering. In addition, particular proteins with aberrant activation have been implicated in many cancers,21 such as EGFR in lung cancer22 and vascular endothelial growth factor in hepatocellular carcinoma.23 Therefore, we sought to investigate whether activation of such proteins causes cluster formation.
Addressing the uncertainties regarding this issue requires information on the fine morphology and accurate stoichiometry of Trop2 clusters. However, traditional biochemical methods such as Western blot analysis, real-time quantitative polymerase chain reaction, and immunohistochemistry typically reveal changes only in the overall levels of proteins,24,25 and conventional fluorescence microscopy is limited by the diffraction of light.26 Fortunately, super-resolution fluorescence microscopy methods, such as stochastic optical reconstruction microscopy (STORM),27,28 direct STORM (dSTORM),29,30 and photoactivated localization microscopy,31 have broken the diffraction barrier and are suitable for research on the detailed spatial organization of clusters on the nanometer scale.32−34
Hence, we applied dSTORM to observe the spatial distribution of Trop2 on A549 cell membranes with different treatments. We found that different membrane properties affected the cluster size and number and that lipid rafts and the actin cytoskeleton played critical roles in cluster formation and stability. Moreover, activation did not change the cluster distribution in lung cancer cells but promoted clustering in normal cells, indicating that Trop2 is always in an activated state in cancer cells.
2. Results and Discussion
2.1. Heterogeneous Distribution of Trop2 on Apical and Basal Cell Membranes
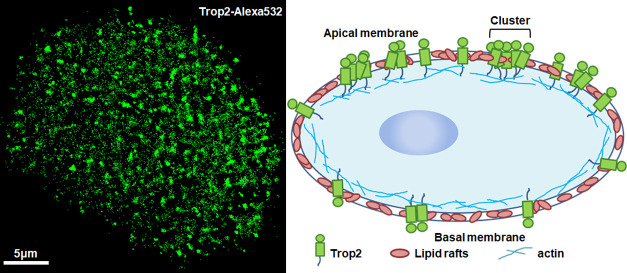
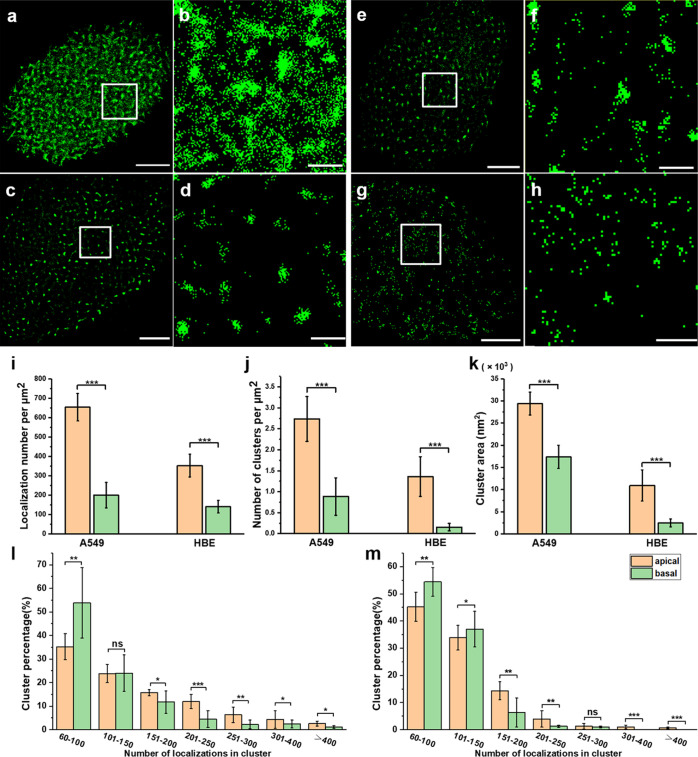
The plasma membrane is the first physical barrier by which cells distinguish themselves from the external environment.35,36 The membrane structure can change the distribution of membrane proteins.9 When cells are cultured with the adherent method, the medium-exposed (apical) and adherent (basal) membranes of the cells have different structural organizations; thus, we wondered whether the different membranes could influence Trop2 clustering. To answer this question, we used dSTORM to investigate Trop2 on the apical and basal membranes of A549 and HBE cells (Figure 1). The imaging method is shown in the Supporting Information (Figure S1). The reconstructed dSTORM images of A549 and HBE cells showed that there were more Trop2 proteins on the apical membrane than on the basal membrane (Figure 1a,c,e,g), and the corresponding magnified images showed that there were more and larger clusters on the apical membrane than on the basal membrane (Figure 1b,d,f,h).
Figure 1.
dSTORM imaging of Trop2 on apical and basal membranes of A549 and HBE cells. (a–d) Reconstructed dSTORM images of Trop2 on apical (a) and basal (c) membranes of A549 cells. The magnified images are shown in (b,d). (e–h) Reconstructed dSTORM images of Trop2 on apical (e) and basal (g) membranes of HBE cells. The magnified images are shown in (f,h). Scale bars: 5 μm in original images and 1 μm in magnified images. (i) Number of Trop2 localizations per μm2 on apical and basal membranes of both kinds of cells. (j) Number of clusters per μm2. (k) Cluster area. (l,m) Percentage of clusters containing different number of localizations on (l) A549 cells and (m) HBE cells. All results were obtained from 10 cell samples in 5 independent experiments. Statistical significance was processed by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ns means not significant.
To further quantitatively analyze the cluster characteristics, we used a graphic processing method named SR-Tesseler,37 which can precisely segment and quantify localization-based super-resolution imaging data. As Trop2 clusters have unequal size and irregular shape, it is suitable to use SR-Tesseler to analyze the clusters. Through this analysis method, we identified features of the clusters including the cluster size, number of clusters, and number of localizations in each cluster (Supporting Information, Figure S2). First, we analyzed the localization density of Trop2 on the apical and basal membranes of both cell lines (Figure 1i). The localization density on the apical membrane of A549 cells was three times greater than that on the basal membrane (634 ± 86 vs 200 ± 66 per μm2). A significant difference existed in HBE cells as well (352 ± 59 vs 140 ± 32 per μm2). Next, we analyzed Trop2 clustering with SR-Tesseler. The A549 cell data showed that there were significantly more Trop2 clusters on the apical membrane (2.5 ± 0.6 per μm2) than on the basal membrane (0.9 ± 0.4 per μm2). The number of Trop2 clusters on the apical membrane (1.4 ± 0.5 per μm2) of HBE cells was more than 9 times that on the basal membrane (0.15 ± 0.1 per μm2) (Figure 1j). Moreover, the cluster area on the apical membrane was also much larger than that on the basal membrane (Figure 1k). To further explore the molecular organization within the clusters, we quantified the percentages of clusters containing different numbers of localizations. The statistical results showed that the numbers of localizations in clusters ranged from 60 to more than 400. For A549 cells (Figure 1l), small clusters with fewer than 100 localizations on the basal membrane accounted for approximately 54% of clusters, much higher than those on the apical membrane. In contrast, large clusters with more than 150 localizations accounted for a higher percentage on the apical membrane than on the basal membrane. Additionally, only one group of clusters (100 < localizations < 150) did not significantly differ between the two membranes. The difference between the apical and basal membranes in HBE cells was roughly consistent with that in A549 cells, and there were no clusters containing more than 300 localizations on the basal membranes (Figure 1m).
Overall, we found that Trop2 had higher density and tended to form more and larger clusters on the apical membranes than on the basal membranes in both A549 cells and HBE cells, and we found that the apical clusters contained more molecules than the basal clusters. The differences in Trop2 clustering may have been due to the differences in membrane structures; therefore, the apical membrane which was exposed to the medium was easier to contact with various factors, such as ligands and hormones. Previous studies have also reported that more EGFR clusters are located on apical surfaces than on basal surfaces so that they can capture more signals and dimerize more quickly.9 Thus, we conclude that Trop2 distribution and clustering are potentially influenced by the different structures of the apical and basal membranes.
2.2. Colocalization of Trop2 and Lipid Rafts
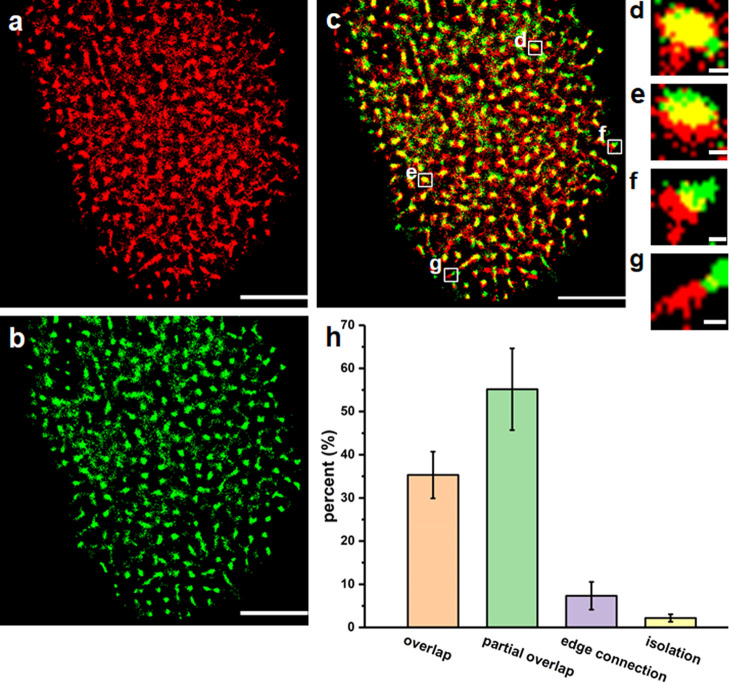
As mentioned above, lipid rafts, as important microdomains on the membrane, may be associated with Trop2 distribution. To assess this possibility, we employed dual-color dSTORM imaging to study the relationship between lipid rafts and Trop2 clusters. We labeled the lipid rafts with Alexa 647-conjugated cholera toxin B (CT-B), which binds glycosphingolipids in lipid rafts,38 and fluorescently labeled Trop2 with primary antibodies and Alexa 532-conjugated secondary antibodies. Since the contact between the apical membranes and the staining solution was greater than that between the basal membranes and the staining solution, we observed the apical membranes in all subsequent experiments. The dual-color image in Figure 2 shows the degree of colocalization between Trop2 and lipid rafts. We applied Mander’s coefficients, M1 and M2, to sort the relationships into four types: overlap (M1/M2 > 0.66, Figure 2d), partial overlap (0.33 < M1/M2 < 0.66, Figure 2e), edge connection (0 < both M1 and M2 < 0.33, Figure 2f), and isolation (M1/M2 = 0, Figure 2g). We randomly collected 500 cluster pairs of lipid rafts and Trop2 in 10 cells to analyze the colocalization (Figure 2h). The overlap relationship type had the highest percentage (55.2%), followed by the partial overlap (35.3%) and edge connection types (7.3%); the isolation type had the lowest proportion (2.2%). Overlap and partial overlap were collectively defined as colocalization, which characterized 90.5% of the relationships. The results indicated that there was a high degree of colocalization between Trop2 clusters and lipid rafts, confirming that lipid rafts serve as vital factors in the formation of Trop2 clusters.
Figure 2.
Colocalization of Trop2 and lipid rafts on the apical membranes of A549 cells. (a,b) dSTORM images of lipid rafts (a) and Trop2 (b). (c) Merging images of lipid rafts (red) and Trop2 (green). (d–g) Magnified view of four representative location relationships of lipid rafts and Trop2 boxed in (c): overlap (d), partial overlap (e), edge connection (f), and isolation (g). Scale bars: 5 μm in (a–c) and 200 nm in (d–g). (h) Percentage of the four types of location states. Data were from 500 random cluster pairs in 10 cells in 5 independent experiments.
2.3. Depletion of Lipid Rafts Disrupts the Formation of Trop2 Clusters
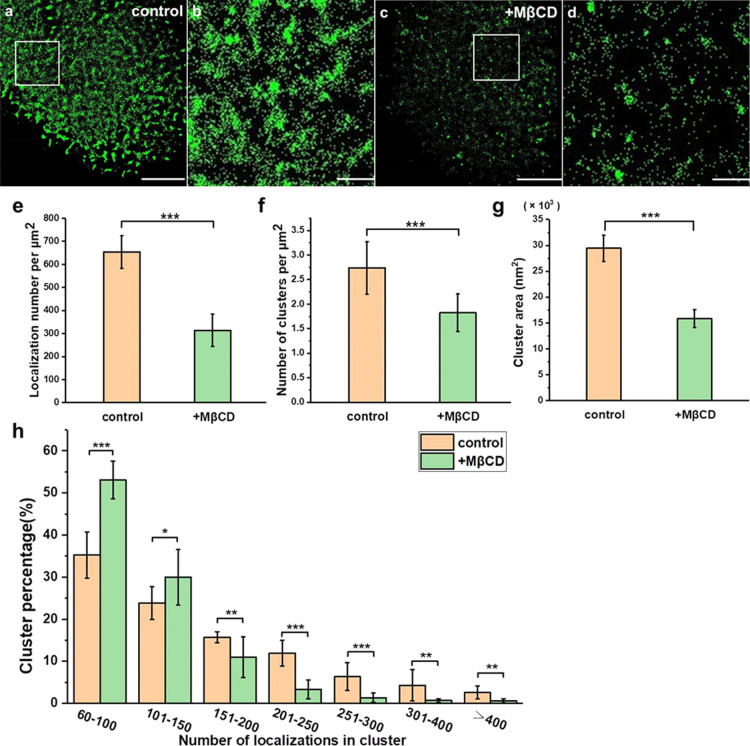
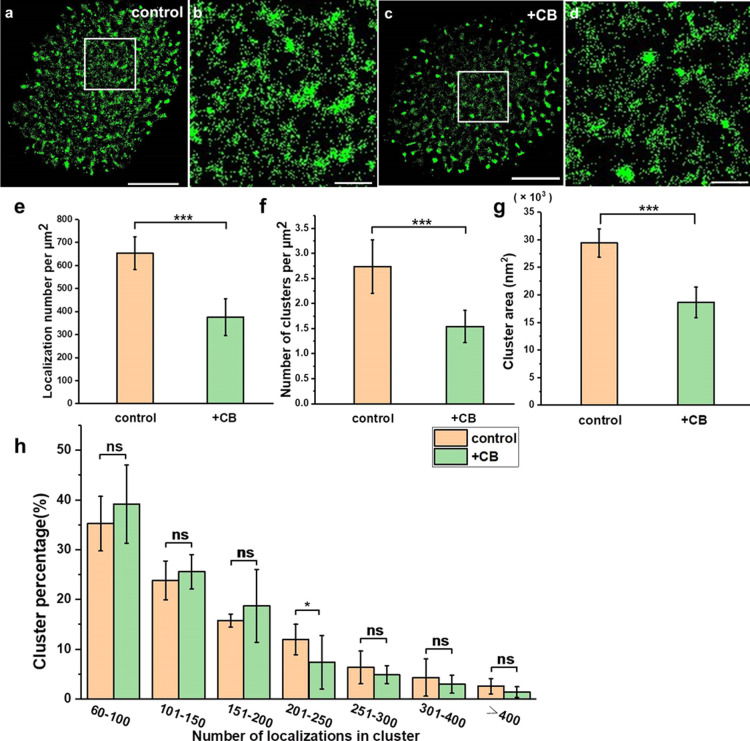
Cholesterol is the most abundant lipid in the plasma membrane, and it is also an important ingredient in lipid rafts.39,40 To study its influence on Trop2 cluster formation, we treated A549 cells with methyl-β-cyclodextrin (MβCD) for 30 min to remove cholesterol and disrupt lipid rafts.41 As shown in Supporting Information Figure S3, the expression of the lipid rafts (labelled with CT-B) was significantly decreased after MβCD treatment, which verified that MβCD disrupted lipid rafts. The reconstructed dSTORM images showed that Trop2 expression on the cell surface was dramatically decreased and that the clusters became smaller or even disappeared after MβCD treatment (Figure 3a–d). The localization density of Trop2 dropped from 653 ± 70 to 314 ± 71 per μm2 (an approximately 108% decrease) (Figure 3e). Furthermore, statistical analysis showed that the number of clusters decreased from 2.7 ± 0.5 to 1.8 ± 0.4 per μm2 (Figure 3f), and the cluster area was reduced by nearly half (Figure 3g). The number of small clusters with fewer than 150 localizations increased sharply after addition of MβCD, but the number of large clusters with more than 150 localizations decreased (Figure 3h).
Figure 3.
MβCD treatment disrupted Trop2 clustering on apical membranes. (a–d) Reconstructed dSTORM images of Trop2 on control (a) and MβCD-treated (c) A549 cell membranes and the corresponding magnified images (b,d). Scale bars: 5 μm in (a,c) and 1 μm in (b,d). (e) Number of Trop2 localizations per μm2 on control and MβCD-treated A549 cell membranes. (f) Number of clusters per μm2. (g) Cluster area. (h) Percentage of clusters containing different number of localizations. All results were obtained from 10 cell samples in 5 independent experiments. Statistical significance was processed by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ns means not significant.
Our data showed that depletion of lipid rafts decreased the total amount of surface Trop2 and greatly reduced the degree of clustering; for example, it reduced the cluster number and area. These results, together with the results regarding colocalization of lipid rafts and Trop2, demonstrated that lipid rafts participated in the spatial aggregation of Trop2. Of note, cholesterol depletion also disrupts vesicular transport to the plasma membrane,42 and it might affect Trop2 distribution on cell membranes.
2.4. Depolymerization of the Actin Cytoskeleton Impairs Trop2 Clustering
Actin is thought to influence the spatial organization of membrane components. In order to test whether actin can help organize Trop2 on lung cancer cells, we utilized dSTORM to observe the changes of Trop2 clusters after depolymerization of actin. We treated cells with 20 μg/mL cytochalasin B (CB) for 30 min to destroy the actin cytoskeleton. After treatment, the expression level of Trop2 seemed to decrease, and the clusters seemed to be smaller and less (Figure 4a–d). Further statistical analysis indicated that the localization density decreased from 653 ± 70 to 376 ± 80 per μm2 (Figure 4e). Both the cluster number and area tended to decrease after addition of CB (Figure 4f,g). Moreover, although the percentage of small clusters (localizations < 200) increased and that of large clusters (localizations > 200) decreased after actin depolymerization, the changes were not significant (Figure 4h). Only clusters with localizations between 200 and 250 showed significant differences between the control and CB-treated groups (Figure 4h). This outcome was different from what happened within clusters after lipid rafts were disrupted.
Figure 4.
CB treatment weakened Trop2 clustering on apical membranes. (a–d) Reconstructed dSTORM images of Trop2 on control (a) and CB-treated (c) A549 cell membranes and the corresponding magnified images (b,d). Scale bars: 5 μm in (a,c) and 1 μm in (b,d). (e) Number of Trop2 localizations per μm2 on control and CB-treated A549 cell membrane. (f) Number of clusters per μm2. (g) Cluster area. (h) Percentage of clusters containing different number of localizations. All results were obtained from 10 cell samples in 5 independent experiments. Statistical significance was processed by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ns means not significant.
These findings demonstrated that Trop2 clustering was limited when actin was disrupted. Notably, depolymerization of actin did not impair Trop2 expression level and clustering as strongly as depletion of lipid rafts, implying that actin has a smaller impact on Trop2 clustering than lipid rafts.
2.5. IGF-1 Stimulates Trop2 in Cancer Cells and Normal Cells
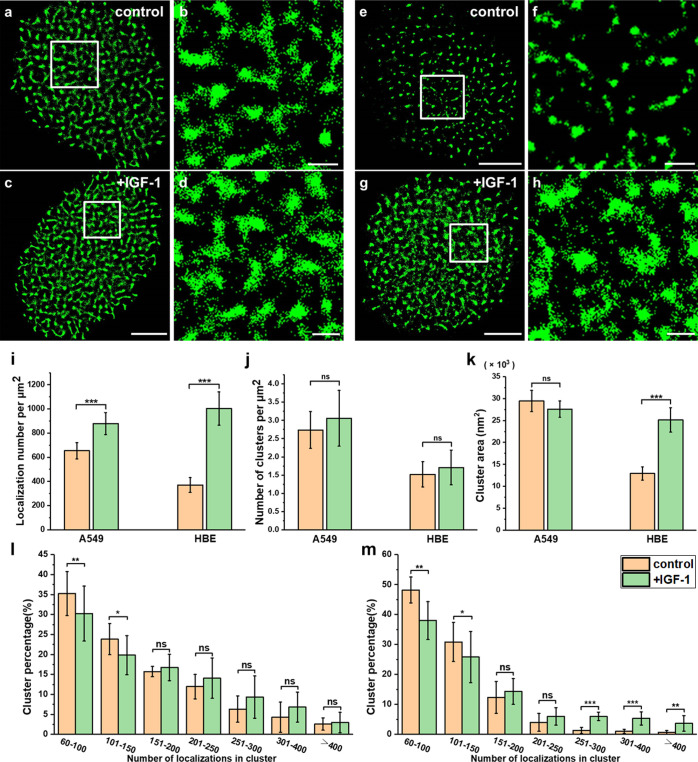
Abnormal activation of proteins is a common phenomenon in cancers. However, whether Trop2 activation influences Trop2 clustering is not clear. Several ligands, such as claudin-1, claudin-7, cyclin D1, and insulin-like growth factor 1 (IGF-1), have been reported to bind to and activate Trop2 and subsequently trigger downstream signaling pathways.1,43 One of the important pathways that promotes the proliferation and migration of lung cancer cells involves activation of Trop2 by IGF-1 and subsequent triggering of PIP2 and Ca2+ signaling. Therefore, we chose IGF-1 for activation and explored the link between Trop2 activation and clustering.1,2 To ensure that the binding of IGF-1 was sufficient, we used 1 ng/mL IGF-1 in our experiments, which is within the bioactive concentration range of 0.5–1.5 ng/mL,44 and used an optimized activation time of 120 min (Supporting Information, Figure S4). Stimulation with IGF-1 at 37 °C for 120 min did not seem to change the cluster morphology (Figure 5a–d). The statistical results indicated that the localization density increased from 648 ± 62 to 879 ± 91 per μm2 after IGF-1 stimulation (Figure 5i), but there were no significant differences in the cluster area and number (Figure 5j,k). These findings suggest that Trop2 in cancer cells is in an active state and exists as clusters and that IGF-1 stimulation can barely promote the formation of Trop2 clusters. Next, to determine whether the situation is the same in normal cells, we performed an experiment in HBE cells. In these cells, IGF-1 stimulation significantly promoted the formation of clusters (Figure 5e–h). Although the number of clusters did not increase, the localization density and cluster size increased dramatically after IGF-1 treatment (Figure 5i–k). Moreover, the cluster composition of A549 cells decreased significantly only when there were fewer than 150 localizations (Figure 5l). However, in HBE cells, the percentage of small clusters (localizations < 150) decreased and that of large clusters (250 < localizations) increased markedly (Figure 5m).
Figure 5.
IGF-1 stimulation caused different changes of Trop2 clusters in cancer cells and normal cells. (a–d) Reconstructed dSTORM images of Trop2 on control (a) and IGF-1-treated (c) A549 cell membranes and the corresponding magnified images (b,d). (e–h) Reconstructed dSTORM images of Trop2 on control (e) and IGF-1 treated (g) HBE cell membranes and the corresponding magnified images (f,h). Scale bars: 5 μm in original images and 1 μm in magnified images. (i) Number of Trop2 localizations per μm2 on A549 and HBE cells with or without IGF-1 stimulation. (j) Cluster area. (k) Number of clusters per μm2. (l,m) Percentage of clusters containing different number of localizations on (l) A549 cells and (m) HBE cells. All results were obtained from 10 cell samples in 5 independent experiments. Statistical significance was processed by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ns means not significant.
Taken together, the results indicated that IGF-1 stimulation promoted the expression level of Trop2, induced Trop2 to form larger clusters, and increased the number of molecules in clusters in normal cells. However, IGF-1 stimulation did not change the clustering level in cancer cells that had formed larger clusters. We reason that the clusters may facilitate the interactions of molecules within the clusters and may promote fast signal transduction. Therefore, we conclude that Trop2 is in an active state in cancer cells and exists in the form of clusters.
3. Conclusions
In summary, we found that Trop2 had different distribution patterns between apical and basal membranes. The majority of Trop2 aggregated in larger and denser clusters on the apical surfaces of cells than on the basal surfaces, suggesting that differences in membrane organization could affect Trop2 clustering. In addition, we illustrated the association between Trop2 and lipid rafts by dual-color imaging and removal experiments and revealed that actin was another key factor that maintained Trop2 cluster distribution. IGF-1 stimulation increased the number of localizations and the sizes of Trop2 clusters in normal cells; however, there were nearly no changes in cancer cells. The results suggest that Trop2 is persistently activated in cancer cells and that clusters may facilitate signal transduction. Overall, our findings identify several important factors that regulate Trop2 clustering on cell membranes, which will pave the way for further study of the relationship between the spatial structure and function of Trop2 in lung cancer.
4. Experimental Section
4.1. Cell Culture
The A549 and HBE cell lines were purchased from the Shanghai Institute of Biological Sciences (Shanghai, China). The cell lines were cultured individually in suitable medium (A549: Ham’s F-12K; HBE: RPMI 1640; Biological Industries, BI) with 10% fetal bovine serum (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin (BI). Both cell lines were incubated in a humidified environment with 5% CO2 at 37 °C. Before dSTORM imaging, they were passaged into a dish containing a precleaned cover slip (22 mm × 22 mm, Fisher). Cells that reached a confluence of 50–70% were selected for use.
4.2. Sample Preparation
The culture medium was discarded, and cell samples were washed with phosphate-buffered saline (PBS) three to four times. Then, they were fixed with 4% paraformaldehyde (PFA) at room temperature for 10 min to effectively immobilize the plasma membrane molecules. After the samples were washed with PBS three times, human Trop2 antibody (R&D Systems, U.S.) was diluted in 4% bovine serum albumin (BSA) to a final concentration of 1 μg/mL, and the samples were incubated with the antibody at 4 °C overnight. The primary antibody was then removed, and the cells were washed with PBS three times. Next, the cells were stained with Alexa Fluor 532-conjugated goat anti-mouse IgG (1 μg/mL in 4% BSA; Invitrogen, 1857666) for 1 h in the dark and washed with PBS three times. Then, 50 μL of imaging buffer containing Tris (50 mM, pH 8.0), NaCl (10 mM), glucose (10% w/v), glucose oxidase (500 μg/mL; Sigma), catalase (40 μg/mL; Sigma), and β-mercaptoethanol (β-ME; 1% v/v; Sigma) was dropped onto a large slide (24 mm × 50 mm). The imaging buffer was adjusted to pH 7.7 for Alexa Fluor 647 and to pH 7.9 for Alexa Fluor 532.45 Finally, the small slide where the cells were seeded was covered on the large one and sealed with nail polish.
For dual-color imaging of Trop2 and lipid rafts, the cells were fixed and incubated with Trop2 primary antibody overnight. After washing three to four times, the cells were labeled with two staining solutions at the same time for 1 h in the dark: Alexa Fluor 532-conjugated goat anti-mouse IgG (1 μg/mL in 4% BSA; Invitrogen) and Alexa Fluor 647-conjugated CT-B (20 μg/mL in 4% BSA; Invitrogen). To deplete membrane cholesterol, the cells were treated with 10 mM MβCD (Sigma) in PBS for 30 min before fixation with 4% PFA. To depolymerize the actin cytoskeleton, the cells were treated with 30 μg/mL CB (Sigma) for 30 min before fixation. To stimulate Trop2, both HBE and A549 cells were treated with IGF-1 (1 ng/mL in 4% BSA; R&D Systems) for 120 min at 37 °C. This stimulation time was optimized in HBE cells treated with IGF-1 for different durations. After that, the samples were washed with PBS three times and fixed. The subsequent steps were the same as those for Trop2 fluorescent staining.
4.3. dSTORM Imaging
The dSTORM imaging system included a Nikon Ti-E inverted fluorescence microscope (Nikon, Japan) equipped with a 100× TIRF lens (numerical aperture 1.49). The beam path consisted of an excitation filter (ZT405/488/532/647×, Chroma), its matched dichromic mirror, and an emission filter. Cells were selected under bright-field illumination. For single-color imaging of Trop2, a 532 nm laser was utilized to excite the fluorophores Alexa Fluor 532 (typically 70–80 mW). For dual-color imaging, a 640 nm laser was first used to image the lipid rafts, and then a 532 nm laser was used to image Trop2. A low-powered 405 nm laser (0.5 mW) was used to increase the number of on-state fluorophores in all samples. An electron-multiplying charge-coupled device camera (Andor Ixon Ultra 888) was used for imaging, and Micro-Manager software was used to record images. By calculating the localization density of reconstructed images with increasing frame number, 5000 frames were selected for recording of each cell, as this frame number enabled complete and good visualization of the morphology (Supporting Information, Figure S5). The exposure time was 25 ms, and the total time for capturing one cell was usually less than 5 min. During this time, a Perfect Focus System provided by Nikon microscopy was used to correct the z drift. 100 nm TetraSpeck microspheres (Invitrogen) were added as fiducials to correct the x–y drift.
4.4. Data Analysis
The raw dSTORM data were reconstructed using a free plugin called QuickPALM in ImageJ.46 We also obtained the resolution of the dSTORM image by block-wise Fourier ring correlation (FRC) resolution mapping in NanoJ-SQUIRREL.47Supporting Information, Figure S6 shows that the FRC value varied from small to large; the corresponding best resolution in our image was approximately 29 nm. SR-Tesseler was applied for cluster analysis. Raw dSTORM images were imported into the software, and a reconstructed image was obtained. After the cell membrane was delineated as a region of interest, the cell was divided into quantities of polygonal regions. The edges of the polygons were perpendicular bisectors from the two nearest localizations. When a polygon satisfied the condition that its localization density δi was higher than the average localization density of the cell δ, it was defined as an object. Finally, objects with higher localization density than the average localization density of all objects were extracted as clusters. Thus, the information of clusters such as the cluster number, the cluster area, and the localization number in clusters could be obtained. Further detailed information can be accessed at http://www.iins.u-bordeaux.fr/team-sibarita.
4.5. Dual-Color Colocalization Analysis
Fifty 0.5 μm2 regions were randomly selected in each cell to quantify the spatial association of Trop2 and lipid raft clusters. Mander’s colocalization coefficients M1 and M2 were calculated by Image-Pro Plus. Cluster pairs were classified into four location relationships according to the values of M1 and M2: (1) isolation (M1 or M2 is equal to zero), (2) edge connection (both M1 and M2 are less than 0.33 but larger than zero), (3) overlap (M1 or M2 is greater than 0.66), and (4) partial overlap (M1 and M2 were the other values).
Acknowledgments
We acknowledge the financial support from the Department of Finance of Jilin Province of China (no. 201817260815 to T.T., no. 3D5204883429 to T.T., no. 2020SCZT084 and no. 2019SCZT048 to P.H.), the Department of Science and Technology of Jilin Province (no. 20190201238JC to T.T.; 20190103113JH to J.G.), the National Key R&D Program of China (2017YFA0505300 to H.W.), the National Natural Science Foundation of China (21727816, 21525314, and 21721003 to H.W.; 21703231 to J.G.), and Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao) (MS2018NO08 to H.W.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c04597.
Schematic diagram of imaging the apical and basal membranes with TIRF illumination; flow diagram of analyzing clusters by SR-Tesseler method; effects of cholesterol depletion on the distribution of lipid rafts; optimizing the stimulation time of IGF-1; reconstructed dSTORM images of Trop2 with increasing frame number; and measurement of the imaging resolution by NanoJ-SQUIRREL (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Shvartsur A.; Bonavida B. Trop2 and its overexpression in cancers: regulation and clinical/therapeutic implications. Genes Cancer 2015, 6, 84–105. 10.18632/genesandcancer.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall A. R. A.; Tolcos M.; Hooper S. B.; Cole T. J.; Wallace M. J. Trop2: from development to disease. Dev. Dyn. 2015, 244, 99–109. 10.1002/dvdy.24242. [DOI] [PubMed] [Google Scholar]
- Tang G.; Tang Q.; Jia L.; Xia S.; Li J.; Chen Y.; Li H.; Ding X.; Wang F.; Hou D.; Kuai X.; Feng Z.; Fan Y. High expression of TROP2 is correlated with poor prognosis of oral squamous cell carcinoma. Pathol., Res. Pract. 2018, 214, 1606–1612. 10.1016/j.prp.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Garraway I. P.; Sun W.; Tran C. P.; Perner S.; Zhang B.; Goldstein A. S.; Hahm S. A.; Haider M.; Head C. S.; Reiter R. E.; Rubin M. A.; Witte O. N. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate 2010, 70, 491–501. 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heist R. S.; Guarino M. J.; Masters G.; Purcell W. T.; Starodub A. N.; Horn L.; Scheff R. J.; Bardia A.; Messersmith W. A.; Berlin J.; Ocean A. J.; Govindan S. V.; Maliakal P.; Mudenda B.; Wegener W. A.; Sharkey R. M.; Goldenberg D. M.; Camidge D. R. Therapy of Advanced Non-Small-Cell Lung Cancer With an SN-38-Anti-Trop-2 Drug Conjugate, Sacituzumab Govitecan. J. Clin. Oncol. 2017, 35, 2790–2797. 10.1200/jco.2016.72.1894. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang K.; Grabowska D.; Li A.; Dong Y.; Day R.; Humphrey P.; Lewis J.; Kladney R. D.; Arbeit J. M.; Weber J. D.; Chung C. H.; Michel L. S. Loss of Trop2 promotes carcinogenesis and features of epithelial to mesenchymal transition in squamous cell carcinoma. Mol. Cancer Res. 2011, 9, 1686–1695. 10.1158/1541-7786.mcr-11-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai X.; Jia L.; Yang T.; Huang X.; Zhao W.; Zhang M.; Chen Y.; Zhu J.; Feng Z.; Tang Q. Trop2 Promotes Multidrug Resistance by Regulating Notch1 Signaling Pathway in Gastric Cancer Cells. Med. Sci. Monit. 2020, 26, e919566 10.12659/msm.919566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G.; Tang Q.; Jia L.; Chen Y.; Lin L.; Kuai X.; Gong A.; Feng Z. TROP2 increases growth and metastasis of human oral squamous cell carcinoma through activation of the PI3K/Akt signaling pathway. Int. J. Mol. Med. 2019, 44, 2161–2170. 10.3892/ijmm.2019.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.; Wang Y.; Cai M.; Pan Y.; Xu H.; Jiang J.; Ji H.; Wang H. Mechanistic insights into EGFR membrane clustering revealed by super-resolution imaging. Nanoscale 2015, 7, 2511–2519. 10.1039/c4nr04962d. [DOI] [PubMed] [Google Scholar]
- Jing Y.; Zhou L.; Chen J.; Xu H.; Sun J.; Cai M.; Jiang J.; Gao J.; Wang H. Quantitatively Mapping the Assembly Pattern of EpCAM on Cell Membranes with Peptide Probes. Anal. Chem. 2020, 92, 1865–1873. 10.1021/acs.analchem.9b03901. [DOI] [PubMed] [Google Scholar]
- Fu Y.; Jing Y.; Gao J.; Li Z.; Wang H.; Cai M.; Tong T. Variation of Trop2 on non-small-cell lung cancer and normal cell membranes revealed by super-resolution fluorescence imaging. Talanta 2020, 207, 120312. 10.1016/j.talanta.2019.120312. [DOI] [PubMed] [Google Scholar]
- Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2015, 71, 3–18. 10.1107/s2053230x14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Herrera F.; Taoro-González L.; Valdés-Baizabal C.; Diaz M.; Marín R. Lipid and Lipid Raft Alteration in Aging and Neurodegenerative Diseases: A Window for the Development of New Biomarkers. Int. J. Mol. Sci. 2019, 20, 3810. 10.3390/ijms20153810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q.; Lu Y.; Zhou L.; Chen J.; Xu H.; Cai M.; Shi Y.; Jiang J.; Xiong W.; Gao J.; Wang H. Mechanistic insights into GLUT1 activation and clustering revealed by super-resolution imaging. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 7033–7038. 10.1073/pnas.1803859115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. B.; Shelby S. A.; Veatch S. L. Super-Resolution Microscopy: Shedding Light on the Cellular Plasma Membrane. Chem. Rev. 2017, 117, 7457–7477. 10.1021/acs.chemrev.6b00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. M.; Magenau A.; Williamson D.; Gaus K. The lipid raft hypothesis revisited--new insights on raft composition and function from super-resolution fluorescence microscopy. BioEssays 2012, 34, 739–747. 10.1002/bies.201200044. [DOI] [PubMed] [Google Scholar]
- Köberlin M. S.; Snijder B.; Heinz L. X.; Baumann C. L.; Fauster A.; Vladimer G. I.; Gavin A.-C.; Superti-Furga G. A Conserved Circular Network of Coregulated Lipids Modulates Innate Immune Responses. Cell 2015, 162, 170–183. 10.1016/j.cell.2015.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J.; Zhao H.; Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 2010, 90, 259–289. 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- Mattila P. K.; Feest C.; Depoil D.; Treanor B.; Montaner B.; Otipoby K. L.; Carter R.; Justement L. B.; Bruckbauer A.; Batista F. D. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 2013, 38, 461–474. 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- De Luca A.; Carotenuto A.; Rachiglio A.; Gallo M.; Maiello M. R.; Aldinucci D.; Pinto A.; Normanno N. The role of the EGFR signaling in tumor microenvironment. J. Cell. Physiol. 2008, 214, 559–567. 10.1002/jcp.21260. [DOI] [PubMed] [Google Scholar]
- Aran V.; Omerovic J. Current Approaches in NSCLC Targeting K-RAS and EGFR. Int. J. Mol. Sci. 2019, 20, 5701. 10.3390/ijms20225701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto A.; De Re V.; Steffan A.; Ravaioli M.; Miolo G.; Leone P.; Racanelli V.; Uzzau A.; Baccarani U.; Cescon M. Carcinogenesis and Metastasis in Liver: Cell Physiological Basis. Cancers 2019, 11, 1731. 10.3390/cancers11111731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K.; Yokouchi Y.; Kobayashi M.; Ninomiya H.; Sakakibara R.; Subat S.; Nagano H.; Nomura K.; Okumura S.; Shibutani T.; Ishikawa Y. Association of tumor TROP2 expression with prognosis varies among lung cancer subtypes. Oncotarget 2017, 8, 28725–28735. 10.18632/oncotarget.15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Jiang X.; Zhang W. TROP2 overexpression promotes proliferation and invasion of lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016, 470, 197–204. 10.1016/j.bbrc.2016.01.032. [DOI] [PubMed] [Google Scholar]
- Son S.; Shin S.; Rao N. V.; Um W.; Jeon J.; Ko H.; Deepagan V. G.; Kwon S.; Lee J. Y.; Park J. H. Anti-Trop2 antibody-conjugated bioreducible nanoparticles for targeted triple negative breast cancer therapy. Int. J. Biol. Macromol. 2018, 110, 406–415. 10.1016/j.ijbiomac.2017.10.113. [DOI] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–796. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R.; Mhlanga M. M. PALM and STORM: what hides beyond the Rayleigh limit?. Biotechnol. J. 2009, 4, 846–857. 10.1002/biot.200900024. [DOI] [PubMed] [Google Scholar]
- Bates M.; Jones S. A.; Zhuang X. Stochastic optical reconstruction microscopy (STORM): a method for superresolution fluorescence imaging. Cold Spring Harbor Protoc. 2013, 2013, 498–520. 10.1101/pdb.top075143. [DOI] [PubMed] [Google Scholar]
- van de Linde S.; Löschberger A.; Klein T.; Heidbreder M.; Wolter S.; Heilemann M.; Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011, 6, 991–1009. 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- Henriques R.; Griffiths C.; Hesper Rego E.; Mhlanga M. M. PALM and STORM: unlocking live-cell super-resolution. Biopolymers 2011, 95, 322–331. 10.1002/bip.21586. [DOI] [PubMed] [Google Scholar]
- Gao J.; He L.; Shi Y.; Cai M.; Xu H.; Jiang J.; Zhang L.; Wang H. Cell contact and pressure control of YAP localization and clustering revealed by super-resolution imaging. Nanoscale 2017, 9, 16993–17003. 10.1039/c7nr05818g. [DOI] [PubMed] [Google Scholar]
- Chen J.; Liu T.; Gao J.; Gao L.; Zhou L.; Cai M.; Shi Y.; Xiong W.; Jiang J.; Tong T.; Wang H. Variation in Carbohydrates between Cancer and Normal Cell Membranes Revealed by Super-Resolution Fluorescence Imaging. Adv. Sci. 2016, 3, 1600270. 10.1002/advs.201600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.; He L.; Zhou L.; Jing Y.; Wang F.; Shi Y.; Cai M.; Sun J.; Xu H.; Jiang J.; Zhang L.; Wang H. Mechanical force regulation of YAP by F-actin and GPCR revealed by super-resolution imaging. Nanoscale 2020, 12, 2703–2714. 10.1039/c9nr09452k. [DOI] [PubMed] [Google Scholar]
- Elson E. L.; Fried E.; Dolbow J. E.; Genin G. M. Phase separation in biological membranes: integration of theory and experiment. Annu. Rev. Biophys. 2010, 39, 207–226. 10.1146/annurev.biophys.093008.131238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle F. A.; Feigenson G. W. Phase separation in lipid membranes. Cold Spring Harbor Perspect. Biol. 2011, 3, a004630. 10.1101/cshperspect.a004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levet F.; Hosy E.; Kechkar A.; Butler C.; Beghin A.; Choquet D.; Sibarita J.-B. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 2015, 12, 1065–1071. 10.1038/nmeth.3579. [DOI] [PubMed] [Google Scholar]
- Harder T.; Scheiffele P.; Verkade P.; Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 1998, 141, 929–942. 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R. A. Influence of increased membrane cholesterol on membrane fluidity and cell function in human red blood cells. J. Supramol. Struct. 1978, 8, 413–430. 10.1002/jss.400080404. [DOI] [PubMed] [Google Scholar]
- Yeagle P. L. Cholesterol and the cell membrane. Biochim. Biophys. Acta 1985, 822, 267–287. 10.1016/0304-4157(85)90011-5. [DOI] [PubMed] [Google Scholar]
- Gaus K.; Rodriguez M.; Ruberu K. R.; Gelissen I.; Sloane T. M.; Kritharides L.; Jessup W. Domain-specific lipid distribution in macrophage plasma membranes. J. Lipid Res. 2005, 46, 1526–1538. 10.1194/jlr.m500103-jlr200. [DOI] [PubMed] [Google Scholar]
- Hansen G. H.; Niels-Christiansen L.-L.; Thorsen E.; Immerdal L.; Danielsen E. M. Cholesterol depletion of enterocytes. Effect on the Golgi complex and apical membrane trafficking. J. Biol. Chem. 2000, 275, 5136–5142. 10.1074/jbc.275.7.5136. [DOI] [PubMed] [Google Scholar]
- Lin J. C.; Wu Y. Y.; Wu J. Y.; Lin T. C.; Wu C. T.; Chang Y. L.; Jou Y. S.; Hong T. M.; Yang P. C. TROP2 is epigenetically inactivated and modulates IGF-1R signalling in lung adenocarcinoma. EMBO Mol. Med. 2012, 4, 472–485. 10.1002/emmm.201200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karey K. P.; Sirbasku D. A. Differential responsiveness of human breast cancer cell lines MCF-7 and T47D to growth factors and 17 beta-estradiol. Cancer Res. 1988, 48, 4083–4092. [PubMed] [Google Scholar]
- Zwettler F. U.; Reinhard S.; Gambarotto D.; Bell T. D. M.; Hamel V.; Guichard P.; Sauer M. Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nat. Commun. 2020, 11, 3388. 10.1038/s41467-020-17086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R.; Lelek M.; Fornasiero E. F.; Valtorta F.; Zimmer C.; Mhlanga M. M. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 2010, 7, 339–340. 10.1038/nmeth0510-339. [DOI] [PubMed] [Google Scholar]
- Laine R. F.; Tosheva K. L.; Gustafsson N.; Gray R. D. M.; Almada P.; Albrecht D.; Risa G. T.; Hurtig F.; LindÅs A.-C.; Baum B.; Mercer J.; Leterrier C.; Pereira P. M.; Culley S.; Henriques R. NanoJ: a high-performance open-source super-resolution microscopy toolbox. J. Phys. D: Appl. Phys. 2019, 52, 163001. 10.1088/1361-6463/ab0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.