Abstract
Background
The DNAJ family of molecular chaperones maintains protein homeostasis in mitotic and postmeiotic cells, especially germ cells. Recently, we found that the transcription factor SOX30 initiates transcription of Dnajb8 during late meiosis and spermiogenesis in mouse testes.
Methods
We used the CRISPR/Cas9 system to generate Dnajb8 mutant mice and analyze the phenotype of the Dnajb8 mutants.
Results
AlthoughDnajb8 is an evolutionarily conserved gene, it is not essential for spermatogenesis and male fertility. We provide this phenotypic information, which could prevent duplicative work by other groups.
Keywords: Dnajb8, Male fertility, Spermatogenesis
Introduction
Mammalian spermatogenesis is a complicated cellular process during which diploid spermatogonial stem cells (SSCs) generate haploid spermatozoa. The process takes place in seminiferous epithelium of the testis and includes three successive developmental phases: spermatogonial proliferation and differentiation, two meiotic divisions to produce haploid round spermatids, and the differentiation of spermatids into sperm (Nishimura & L’Hernault, 2017). During spermatogenesis, the accurate regulation of protein folding and sorting is fundamental for the production of high-quality spermatozoa (Meccariello et al., 2014).
The DNAJ family is the largest molecular chaperone family and plays a key role in protein folding, trafficking, aggregation, homeostasis and conformation (Zarouchlioti et al., 2018). The human DNAJ proteins are divided into three subgroups based on a highly conserved domain structure containing a J domain that interacts with HSP70 (type III), a Gly/Phe-rich region (type II) and a cysteine-rich region (type I) (Cheetham & Caplan, 1998). Mutations in DNAJ proteins occur frequently in neurodegenerative diseases, such as cerebellar ataxia, distal hereditary motor neuropathy and Parkinson’s disease (Koutras & Braun, 2014). Nevertheless, DNAJ proteins has also been implicated in tumorogenesis and spermatogenesis (Kusumoto et al., 2018; Meccariello et al., 2014; Nishizawa et al., 2012). As there are functional differences among the DNAJ proteins, it is unclear whether deficiencies in each DNAJ protein could lead to male infertility.
DNAJB8, a member of the DNAJB family of proteins, plays a role in suppressing protein expression by interacting with HDACs (Hageman et al., 2010). A recent study also showed that DNAJB8 is a cancer-testis (CT) antigen carrying tumor-initiating ability (Nishizawa et al., 2012). Among mouse tissues, DNAJB8 is highly expressed in the testis, especially in postmeiotic germ cells (Nishizawa et al., 2012). Notably, we previously found that Dnajb8 was directly regulated by the transcription factor SOX30, which controls the expression of a core set of postmeiotic genes (Bai et al., 2018). However, whether DNAJB8 affects spermatogenesis and male fertility is still not clear. Because in vitro spermiogenesis has still not been established, we generated a Dnajb8 knockout (KO) mouse via Cas9/RNA-mediated gene targeting to understand the function of DNAJB8 in spermatogenesis.
Materials and Methods
Animals
Animals were housed in Laboratory Animal Center at University of Science and Technology of China under specific pathogen-free conditions with free access to food and water. All mice were treated humanely and euthanized by cervical dislocation to collected testis and epididymal samples for further analyses. The use of animals and the experimental design were approved by the Institutional Animal Care and Use Committees of the University of Science and Technology of China (No. 2019-N(A)-061), Hefei, China.
Generation of an anti-DNAJB8 polyclonal antibody
DNAJB8 polyclonal antibody was prepared by ABclonal (Wuhan, China) and derived from rabbits. Recombinant fusion protein contained a sequence corresponding to a fragment (amino acids 116-132) of mouse DNAJB8. We have used this antibody for western blot.
Generation of Dnajb8 knockout mice with the CRISPR/Cas9 System
Dnajb8 knockout mice were generated using CRISPR/Cas9 technology. The Dnajb8 exon containing the J-domain was targeted by sgRNAs. The sgRNA was synthesized by Genescript (Nanjing, China) and the sequences were as follows: 5′-ACCTGTCCCTGAGAACGGGG-3′and 5′-CATAGTAGTCGCAACCTAAC-3′. The Cas9 expressed vector pX330 was obtained from Addgene, linearized with NotI (New England Biolabs, USA), transcribed using T7 ULTRA Transcription Kit (Ambion AM1345, USA) and purified using MEGAclear™ Kit (Ambion AM1908, USA). Cas9 mRNA and sgRNAs were coinjected into fertilized eggs for KO mouse production. The pups were genotyped by genomic PCR followed by Sanger sequencing. After genotyping, the F0 mice went through serial mating to generate homozygous mutants.
Genotyping
The genotype identification of offspring was completed by PCR amplification (primers: Dnajb8 _F1: 5′-AGTCAAACAAACAGCCAAACTCAC-3′; R1: 5′-GTGACCGGAATAAATAACCTCCCA-3′; R2: 5′-TCGGAAACCTGCTTAAACTTCTTC-3′) and Sanger sequencing. The results of sequencing were analyzed by SnapGene.
Fertility test
Every 8-week-old Dnajb8+∕+ and Dnajb8−∕− male mouse was caged with two 8-week-old Dnajb8+∕+ female mice for at least 8 weeks. During the breeding test, the number of pups was counted at birth, and the average litter size for each mouse line was recorded.
Histology analysis
Testes and epididymides from 10-week-old Dnajb8+∕+ and Dnajb8−∕− male were fixed in Bouin’s solution overnight, dehydrated with increasing concentrations of ethanol (70%–100%) and embedded in paraffin. The tissue was cut into 5-µm-thick sections and mounted onto glass slides and followed by Hematoxylin and Eosin (H&E; Sigma-Aldrich, USA) staining. For sperm staining, epididymal sperm from the of 8-week-old male mice were extruded, fixed in 4% PFA at 4 °C overnight, and immobilized onto glass slides. The sections were stained with H&E. All sections were analyzed microscopically (LEICA DM2500, Germany).
Sperm parameters analysis
Sperm parameters assays were performed using 10-week-old Dnajb8+∕+ and Dnajb8−∕− male. Cauda epididymal sperm were extracted, incubated in PBS, fixed in 4% of PFA, followed by sperm counting using a hemocytometer. For sperm motility assays, cauda epididymal sperm were released into HTF medium at 37 °C and measured using computer-aided sperm analysis (CASA) system (Hamilton Thorne Biosciences, USA).
Western blotting
Tissues and spermatozoa that separated from seminal plasma were rinsed with PBS and lysed in cold RIPA buffer supplemented with phosphatase inhibitor and protease inhibitor cocktail tablets. Lysates were incubated on ice and centrifuged at 13000 rpm for 15 min at 4 °C. The protein concentration was determined by the Bicinchoninic Acid (BCA) Assay (E11201, Vazyme, China) according to the manufacturer’s instructions. In total, 20 µg of proteins was separated on 10% SDS-PAGE gels. The results were expressed in arbitrary units after normalization for α-TUBULIN protein levels. The primary antibodies used were as follows: anti-DNAJB8 (diluted 1:1000 in TBST, ABclonal, China) and anti-α-TUBULIN (diluted 1:5000 in TBST, 11224-1-AP, Proteintech, China).
Immunofluorescence
For immunofluorescence, the tissue sections were fixed in 4% paraformaldehyde (PFA) and blocked in 10% goat serum. The slides with spermatozoa were incubated with the following primary antibodies: anti-γH2AX (diluted 1:100 in TBST, 16-202A, Merck Millipore, USA), anti-SOX9 (diluted 1:100 in TBST, AB5535, Merck Millipore, USA). Nuclear DNA and acrosome were stained with with 4′,6-diamidino-2-phenylindole (DAPI, F6057, Sigma-Aldrich, USA) and FITC-conjugated peanut agglutinin (PNA, RL-1072, Vector Labs, USA), respectively. All the spermatozoa staining was visualized on a confocal microscope (Carl Zeiss, LSM700, Germany).
Phylogenetic analyses
Multiple amino acid sequence alignments and phylogenetic trees were constructed by the MEGA program. The amino acid sequences were downloaded from the NCBI database.
Statistical analysis
All data are reported as mean ± SD. Significance was tested using a two-tailed unpaired Student’s t test using Prism 7.0 software. A p value < 0.05 was considered statistically significant. NS means not significant.
Results
SOX30 regulates Dnajb8 transcription
In a previous study, we combined bioinformatics analyses of the transcription factor SOX30 RNA-seq and ChIP-seq datasets to reveal that SOX30 directly regulates expression of a core set of postmeiotic genes (Bai et al., 2018). Among these direct targets, most have been reported to be involved in haploid germ cell development, such as Tnp1, Hils1, Ccdc54 and Tsks, while the function of some haploid cell-enriched genes during spermatogenesis, including Dnajb8, remained unclear. We reanalyzed our published SOX30 RNA-seq datasets and found that Dnajb8 was downregulated in both Sox30 null pachytene spermatocytes and round spermatids compared with wild-type cells (Figs. 1A, 1B). In addition, our published SOX30 ChIP-seq datasets also showed that strong binding peaks were observed at the Dnajb8 promoter, indicating that Dnajb8 is a direct downstream target of SOX30 (Fig. 1C).
Figure 1. SOX30 directly regulates Dnajb8 expression.
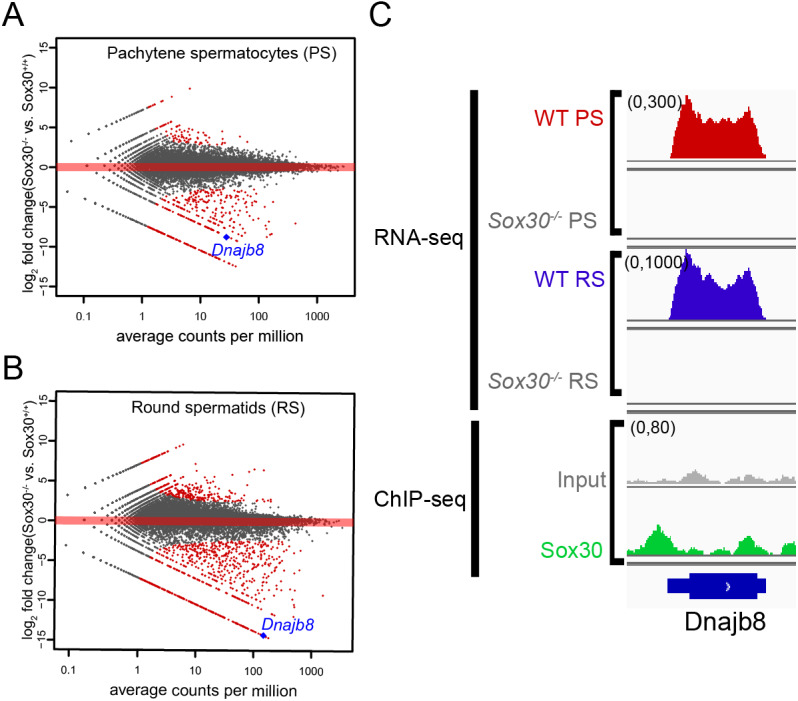
(A, B) Scatter plot of Dnajb8 transcripts in Sox30 null pachytene spermatocytes and round spermatids compared with wild-type cells from published RNA-seq datasets (Bai et al., 2017). (C) Genome browser view of published SOX30 RNA-seq and ChIP-seq data on the Dnajb8 gene loci in isolated germ cells or total testes from wild-type and Sox30−/− mice (Bai et al., 2017).
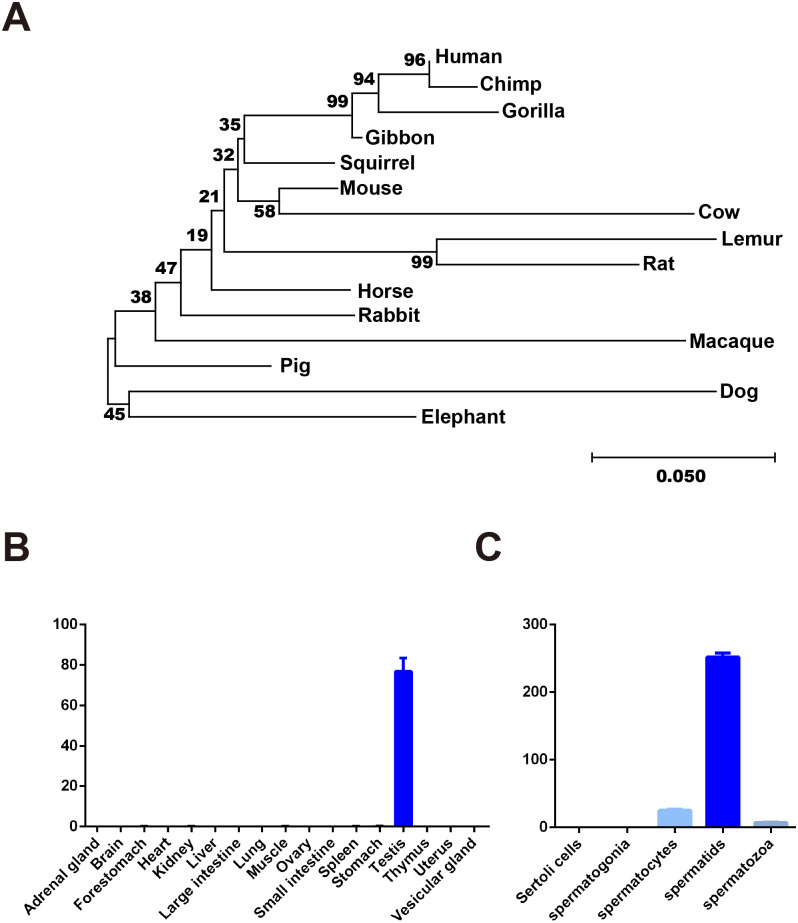
Dnajb8 is a conserved and testis-enriched gene
Phylogenetic analyses demonstrated that DNAJB8 protein is conserved between a variety of mammalian species (Fig. 2A). We further found that Dnajb8 transcripts were highly and exclusively in the testis using published RNA-seq data generated from different mouse tissues (Fig. 2B) (Li et al., 2017). Moreover, Dnajb8 transcripts displayed dynamic expression patterns during spermatogenesis (Soumillon et al., 2013). As shown in Fig. 2C, Dnajb8 transcripts began to increase in pachytene spermatocytes and plateaued in round spermatids, but they were reduced in spermatozoa.
Figure 2. Conserved Dnajb8 is highly expressed in mouse testis.
(A) Phylogenetic trees of DNAJB8 in mammalian species. The numbers in the dendrogram were bootstrap value (%). (B, C) The median-normalized levels of Dnajb8 mRNA expression in different mouse tissues and isolated spermatogenetic cells from published RNA-seq data (Li et al., 2017; Soumillon et al., 2013).
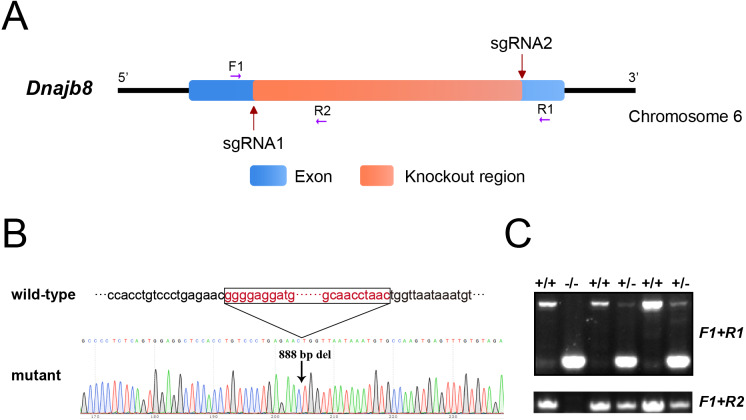
Generation of DNAJB8 knockout mice
To explore the function of DNAJB8, Dnajb8 mutant mice with a Danjb8 allele containing an 888-bp deletion were generated by CRISPR-Cas9 system (Figs. 3A, 3B). Homozygous mutant Dnajb8 alleles were obtained by selective breeding and determined by genotyping PCR (Fig. 3C).
Figure 3. Generation of Dnajb8-null mice by the CRISPR/Cas9 system.
(A) Schematic illustration of the generation of Dnajb8−∕− mice. Two sgRNAs were designed to target the 5′ and the 3′ region respectively of the coding exon. Three primers (F1, R1 and R2) were designed for genotyping. (B) Sanger sequencing from wild-type and Dnajb8−∕− mice. A 888-bp deletion were detected in Dnajb8−∕− mice. (C) Genotype verification of Dnajb8−∕− mice by genomic PCR using primer sets F1-R1 and F1-R2, respectively.
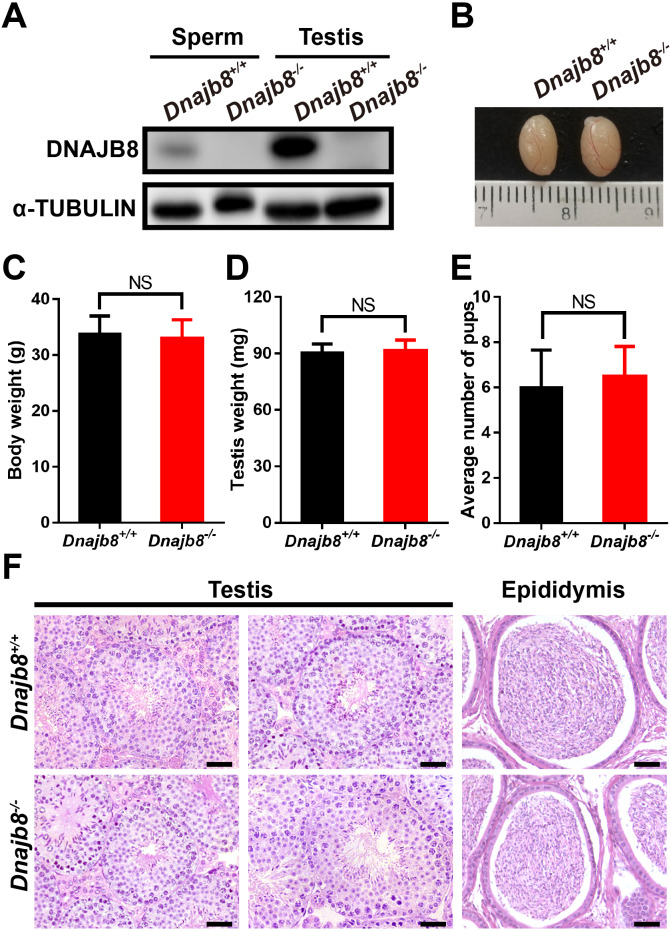
Dnajb8 deficient male mice are fertile
Western blot analysis confirmed the absence of DNAJB8 protein in testis and sperm from adult Dnajb8−∕− mice (Fig. 4A). All Dnajb8−∕− males were viable and phenotypically normal. The body and testicular weight of adult Dnajb8−∕− males (n = 5) was comparable to that of wild-type males (n = 6) at 10-week-old (Figs. 4B–4D). Mating tests showed normal litter sizes from Dnajb8−∕− males (Fig. 4E). Moreover, complete spermatogenesis in seminiferous tubules and normal epididymal structures were observed in Dnajb8−∕− mice by H&E staining (Fig. 4F).
Figure 4. Dnajb8−/− mice are fertile.
(A) Western blot conformed that DNAJB8 protein was absent in adult Dnajb8−∕ − testes and sperm. (B) The morphology of wild-type and Dnajb8-null testes at 10-week-old. (C, D) The body weight (C) and testis weight (D) from Dnajb8+∕+ and Dnajb8−/− mice at 10-week-old. Dnajb8+∕+, n = 6; Dnajb8−/−, n = 5. (E) Number of pups per litter from Dnajb8+∕+ and Dnajb8−/ − males. Each genotype shown was coupled with Dnajb8+∕+ females. n = 3 for each genotype. (F) H&E staining of testes and epididymis from adult Dnajb8+∕+ and Dnajb8−/− mice at 10-week-old. Scale bar: 50 µm. NS, No significant difference.
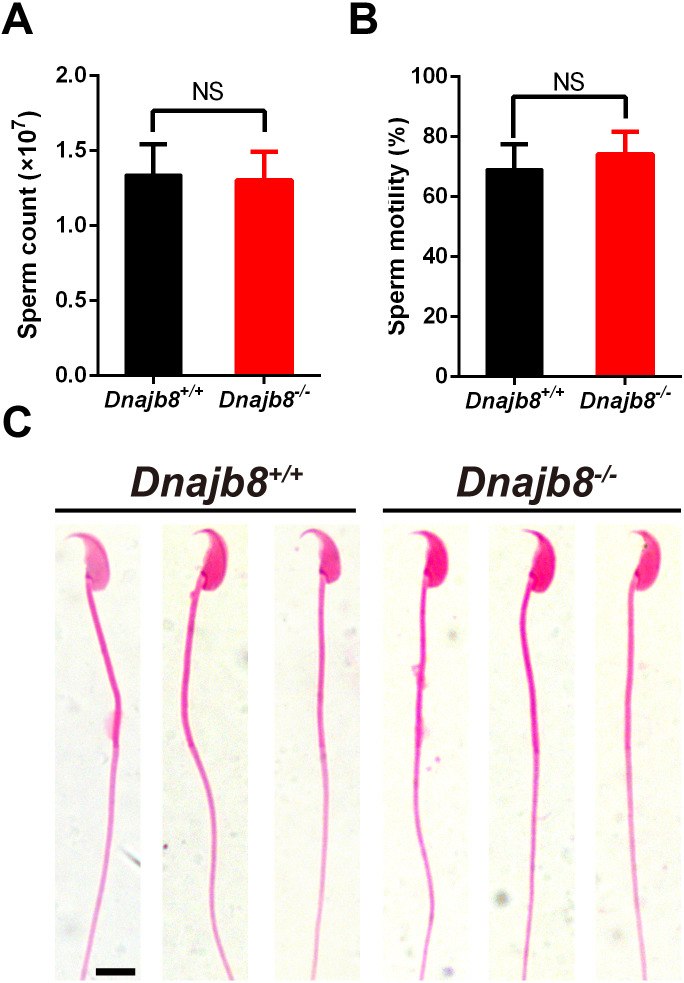
Normal sperm parameters in Dnajb8−∕− mice
The number and motility of mature spermatozoa from the epididymal cauda of Dnajb8−∕− mice (n = 5) were similar with those of Dnajb8+∕+ controls (n = 6) at 10-week-old (Figs. 5A, 5B). In addition, sperm from Dnajb8−∕− males exhibited normal morphology (Fig. 5C).
Figure 5. Normal sperm count, morphology and motility in Dnajb8−/ − mice.
(A, B) Sperm count and motility of the cauda epididymal from Dnajb8+∕+ and Dnajb8−/ − mice at 10-week-old. Dnajb8+∕+, n = 6; Dnajb8−/ −, n = 5. (C) Sperm morphology from Dnajb8 +∕+ and Dnajb8−/ − mice by H&E staining at 10-week-old. n = 3 for each genotype. Scale bar: 10 µm. NS, No significant difference.
DNAJB8 is not essential for germ cell development
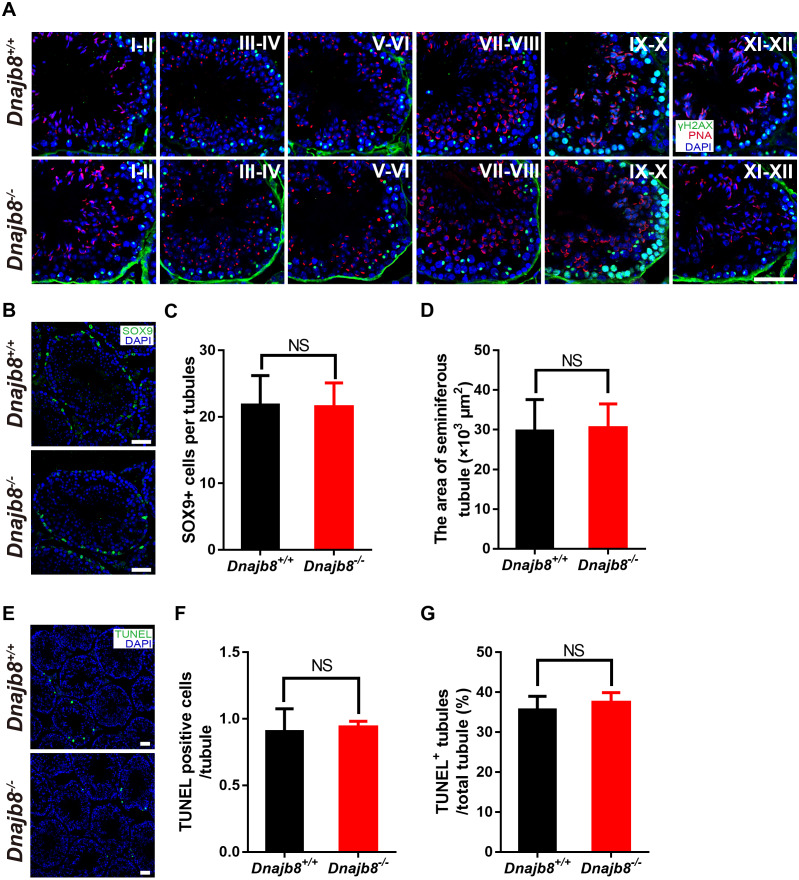
To characterize the spermatogenesis in Dnajb8−∕− mice, we next performed immunostaining for γ-H2AX-positive spermatocytes and PNA-positive acrosomes in spermatids. As shown in Fig. 6A, different stages of spermatocytes and spermatids were both observed in seminiferous tubules from adult wild-type and Dnajb8−∕− mice. We then immunostained testis sections for the Sertoli cell marker SOX9 and found that seminiferous tubules from wild-type and Dnajb8−∕− males (n = 3 for each group) contained similar numbers of Sertoli cells (Figs. 6B, 6C). The area of seminiferous tubules from Dnajb8+∕+ and Dnajb8+∕+testes (n = 3 for each group) were similar (Fig. 6D). Furthermore, TUNEL staining revealed that the numbers of apoptotic cells and tubules did not differ between Dnajb8−∕− and Dnajb8+∕+ testes (n = 3 for each group) (Figs. 6E–6G). These results show that Dnajb8 deficiency does not affect spermatogenesis in Dnajb8−∕−mice.
Figure 6. Dnajb8-/- males show normal spermatogenesis.
(A) Immunodetection of γH2AX and PNA in testis sections from adult Dnajb8+∕+ and Dnajb8−/ − mice. Scale bar: 50 µm. (B) Immunostaining of adult testis sections for SOX9, a marker of Sertoli cells. Scale bar: 50 µm. (C) Quantification of SOX9-positive cells in seminiferous tubule from Dnajb8+∕+ and Dnajb8−/ − mice at 10-week-old. Total 60 tubules from three independent mice per group were counted. (D) Seminiferous tubule area in Dnajb8+∕+ and Dnajb8−/ − testes at 10-week-old. Total 60 tubules from three independent mice per genotype were analyzed. (E) TUNEL assay on testis sections from adult Dnajb8+∕+ and Dnajb8−/ − mice at 10-week-old. Scale bar: 50 µm. (F) Quantification of TUNEL-positive cells per tubule. (G) The ratios of TUNEL-positive tubules to total tubules in Dnajb8+∕+ and Dnajb8−/− testes. Total 157 and 168 from three independent mice were counted per genotype, respectively. NS, No significant difference.
Discussion
Here, we reanalyzed RNA-seq and ChIP-seq datasets from the transcription factor SOX30 and demonstrated that SOX30 directly regulates expression of the postmeiotic gene Dnajb8. As Dnajb8 is a highly conserved and testis-enriched gene, we further generated Dnajb8 KO mice by CRISPR/Cas9 and found that Dnajb8 is not required for male fertility, with no difference in testicular or epididymal histology and average litters sizes compared to wild-type males.
To date, several DNAJB proteins have been identified as being involved in spermatogenesis and male fertility (Meccariello et al., 2014). DNAJB1 is mainly expressed in mouse testis and is localized in the acrosomal region of the sperm head, the middle and end pieces of the sperm tail (Doiguchi et al., 2007). DNAJB3 protein is highly expressed in haploid germ cells and may be involved in vesicle fusion (Berruti & Martegani, 2001). Importantly, it has been reported that homozygous mutations in DNAJB13, a radial spoke protein of the mouse ‘9+2’ axoneme that localized to the sperm flagella, cause male infertility, with severe oligo-astheno-teratozoospermia (El Khouri et al., 2016; Li & Liu, 2014). Moreover, heterozygous variants in DNAJB13 were correlated with male fertility in asthenozoospermia (Li et al., 2020). DNAJB13 interactions with SUN5 play a crucial role in sperm head-tail integration (Shang et al., 2018). Similar to the expression pattern of the DNAJB family, Dnajb8 is a testis-specific gene and is predominantly in spermatids. Notably, Dnajb8 was downregulated in spermatozoa of infertile men by 12-fold compared to that of normospermic individuals (Montjean et al., 2012). Hence, DNAJB8 was thought to play a role in germ cell development. However, our study showed that all stages of spermatogenic cells were detected in Dnajb8−∕− seminiferous tubules. Meanwhile, immunofluorescence staining of γH2AX-positive spermatocytes and PNA-positive spermatids, as well as SOX9-positive Sertoli cells, were not different between Dnajb8−∕− and Dnajb8+∕+ mice. These data indicate that DNAJB8 protein is not essential for spermatogenesis.
Previous studies have found that more than 54 conserved testis-enriched proteins were not essential for fertility (Miyata et al., 2016). This may be because these genes are highly covered by the redundancy of other genes. Functional redundancy of DNAJ family proteins has been shown to exist in yeast (Sahi & Craig, 2007). Interestingly, most Dnajb transcripts were mainly detected in the testis, especially the haploid germ cell-enriched genes Dnajb3 and Dnajb7 (Li et al., 2017; Soumillon et al., 2013), indicating that redundancy may be found in human DNAJs. Thus, DNAJ proteins work in parallel, and this redundancy could explain the normal male fertility of Dnajb8 KO mice.
Although we investigated litter size using co-housing assay, which provides unrestricted access of males to females, the protocol may be not detect subtle changes in Dnajb8 KO males. Nevertheless, we have shown that Dnajb8 is not essential for male fertility under normal laboratory mating conditions in this study.
Taken together, our findings demonstrate that mouse DNAJB8 is dispensable for spermatogenesis and male fertility. Even though Dnajb8 is a postmeiotic gene directly regulated by SOX30, we did not observe any defects in germ cell development in Dnajb8−∕− males. One possible explanation for this is the redundant mechanisms of DNAJ proteins that control male fertility.
Supplemental Information
Acknowledgments
We thank Jianqiang Bao and Wenxiang Fang for support with mice feeding.
Funding Statement
This study was supported by National Natural Science Foundation of China (No. 81901543, 81972641, 81901545, 81971333), the Fundamental Research Funds for the Central Universities (No. WK9110000063), State Key Laboratory of Reproductive Medicine (No. SKLRM-K201904) and the National Key Research and Development Project (2019YFA0802600). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Fengsong Wang, Shuai Kong and Shun Bai conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Xuechun Hu, Xin Li, Bo Xu, Qiuling Yue and Kaiqiang Fu performed the experiments, prepared figures and/or tables, and approved the final draft.
Lan Ye analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The Institutional Animal Care and Use Committee (IACUC) of the University of Science and Technology of China approved this study (Approval No. 2019-N(A)-061).
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
SOX30 RNA-seq and ChIP-seq data are available at NCBI SRA: PRJNA433934 (accession: SRP143508).
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files.
References
- Bai et al. (2018).Bai S, Fu K, Yin H, Cui Y, Yue Q, Li W, Cheng L, Tan H, Liu X, Guo Y, Zhang Y, Xie J, He W, Wang Y, Feng H, Xin C, Zhang J, Lin M, Shen B, Sun Z, Guo X, Zheng K, Ye L. Sox30 initiates transcription of haploid genes during late meiosis and spermiogenesis in mouse testes. Development. 2018;145 doi: 10.1242/dev.164855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti & Martegani (2001).Berruti G, Martegani E. MSJ-1, a mouse testis-specific DnaJ protein, is highly expressed in haploid male germ cells and interacts with the testis-specific heat shock protein Hsp70-2. Biology of Reproduction. 2001;65:488–495. doi: 10.1095/biolreprod65.2.488. [DOI] [PubMed] [Google Scholar]
- Cheetham & Caplan (1998).Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doiguchi et al. (2007).Doiguchi M, Kaneko T, Urasoko A, Nishitani H, Iida H. Identification of a heat-shock protein Hsp40, DjB1, as an acrosome- and a tail-associated component in rodent spermatozoa. Molecular Reproduction and Development. 2007;74:223–232. doi: 10.1002/mrd.20609. [DOI] [PubMed] [Google Scholar]
- El Khouri et al. (2016).El Khouri E, Thomas L, Jeanson L, Bequignon E, Vallette B, Duquesnoy P, Montantin G, Copin B, Dastot-Le Moal F, Blanchon S, Papon JF, Lores P, Yuan L, Collot N, Tissier S, Faucon C, Gacon G, Patrat C, Wolf JP, Dulioust E, Crestani B, Escudier E, Coste A, Legendre M, Toure A, Amselem S. Mutations in DNAJB13, encoding an HSP40 family member, cause primary ciliary dyskinesia and male infertility. American Journal of Human Genetics. 2016;99:489–500. doi: 10.1016/j.ajhg.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman et al. (2010).Hageman J, Rujano MA, Van Waarde MA, Kakkar V, Dirks RP, Govorukhina N, Oosterveld-Hut HM, Lubsen NH, Kampinga HH. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Molecular Cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Koutras & Braun (2014).Koutras C, Braun JE. J protein mutations and resulting proteostasis collapse. Frontiers in Cellular Neuroscience. 2014;8:191. doi: 10.3389/fncel.2014.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto et al. (2018).Kusumoto H, Hirohashi Y, Nishizawa S, Yamashita M, Yasuda K, Murai A, Takaya A, Mori T, Kubo T, Nakatsugawa M, Kanaseki T, Tsukahara T, Kondo T, Sato N, Hara I, Torigoe T. Cellular stress induces cancer stem-like cells through expression of DNAJB8 by activation of heat shock factor 1. Cancer Science. 2018;109:741–750. doi: 10.1111/cas.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2017).Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L. A Comprehensive Mouse Transcriptomic BodyMap across 17 Tissues by RNA-seq. Scientific Reports. 2017;7:4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Liu (2014).Li W, Liu G. DNAJB13, a type II HSP40 family member, localizes to the spermatids and spermatozoa during mouse spermatogenesis. BMC Developmental Biology. 2014;14:38. doi: 10.1186/s12861-014-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2020).Li WN, Zhu L, Jia MM, Yin SL, Lu GX, Liu G. Missense mutation in DNAJB13 gene correlated with male fertility in asthenozoospermia. Andrology. 2020;8:299–306. doi: 10.1111/andr.12685. [DOI] [PubMed] [Google Scholar]
- Meccariello et al. (2014).Meccariello R, Chianese R, Ciaramella V, Fasano S, Pierantoni R. Molecular chaperones, cochaperones, and ubiquitination/deubiquitination system: involvement in the production of high quality spermatozoa. BioMed Research International. 2014;2014:561426. doi: 10.1155/2014/561426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata et al. (2016).Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR, Isotani A, Kiyozumi D, Kriseman ML, Mashiko D, Matsumura T, Matzuk RM, Mori M, Noda T, Oji A, Okabe M, Prunskaite-Hyyrylainen R, Ramirez-Solis R, Satouh Y, Zhang Q, Ikawa M, Matzuk MM. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7704–7710. doi: 10.1073/pnas.1608458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montjean et al. (2012).Montjean D, De La Grange P, Gentien D, Rapinat A, Belloc S, Cohen-Bacrie P, Menezo Y, Benkhalifa M. Sperm transcriptome profiling in oligozoospermia. Journal of Assisted Reproduction and Genetics. 2012;29:3–10. doi: 10.1007/s10815-011-9644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura & L’Hernault (2017).Nishimura H, L’Hernault SW. Spermatogenesis. Current Biology. 2017;27:R988–R994. doi: 10.1016/j.cub.2017.07.067. [DOI] [PubMed] [Google Scholar]
- Nishizawa et al. (2012).Nishizawa S, Hirohashi Y, Torigoe T, Takahashi A, Tamura Y, Mori T, Kanaseki T, Kamiguchi K, Asanuma H, Morita R, Sokolovskaya A, Matsuzaki J, Yamada R, Fujii R, Kampinga HH, Kondo T, Hasegawa T, Hara I, Sato N. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem-like cells. Cancer Research. 2012;72:2844–2854. doi: 10.1158/0008-5472.CAN-11-3062. [DOI] [PubMed] [Google Scholar]
- Sahi & Craig (2007).Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang et al. (2018).Shang Y, Yan J, Tang W, Liu C, Xiao S, Guo Y, Yuan L, Chen L, Jiang H, Guo X, Qiao J, Li W. Mechanistic insights into acephalic spermatozoa syndrome-associated mutations in the human SUN5 gene. Journal of Biological Chemistry. 2018;293:2395–2407. doi: 10.1074/jbc.RA117.000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon et al. (2013).Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, Dym M, De Massy B, Mikkelsen TS, Kaessmann H. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Reports. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Zarouchlioti et al. (2018).Zarouchlioti C, Parfitt DA, Li W, Gittings LM, Cheetham ME. DNAJ Proteins in neurodegeneration: essential and protective factors. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2018;373 doi: 10.1098/rstb.2016.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files.