Abstract
Chiari malformation type I (CMI) is a brain malformation that is characterized by herniation of the cerebellum into the spinal canal. Chiari malformation type I is highly heterogeneous; therefore, an accurate explanation of the pathogenesis of the disease is often not possible. Although some studies showed the role of genetics in CMI, the involvement of genetic variations in CMI pathogenesis has not been thoroughly elucidated. Therefore, in the current study we aim to reveal CMI-associated genomic variations in familial cases.Four CMI patients and 7 unaffected healthy members of two distinct families were analyzed. A microarray analysis of the affected and unaffected individuals from two Turkish families with CMI was conducted. Analyses of single nucleotide variations (SNVs) and copy number variations (CNVs) were performed by calculation of B allele frequency (BAF) and log R ratio (LRR) values from whole genome SNV data. Two missense variations, OLFML2A (rs7874348) and SLC4A9 (rs6860077), and a 5’UTR variation of COL4A1 (rs9521687) were significantly associated with CMI. Moreover, 12 SNVs in the intronic regions of FAM155A, NR3C1, TRPC7, ASTN2, and TRAF1 were determined to be associated with CMI. The CNV analysis showed that the 11p15.4 chromosome region is inherited in one of the families. The use of familial studies to explain the molecular pathogenesis of complex diseases such as CMI is crucial. It has been suggested that variations in OLFML2A, SLC4A9, and COL4A1 play a role in CMI molecular pathogenesis. The CNV analysis of individuals in both families revealed a potential chromosomal region, 11p15.4, and risk regions that are associated with CMI.
Keywords: Chiari type I malformation, neurogenetics, microarray analysis, molecular karyotyping
1. Introduction
The Chiari malformation (CM) is a condition in which the cerebellar tonsil, which is found at the lower part of the brain right above the foramen magnum, starts extending out through the spinal canal. It blocks spinal fluid flow and can cause fluid accumulation in the canal (syringomyelia) and in the brain. Type I CM (CMI) is the most prevalent type with an incidence rate of approximately 1/1280 (Meadows et al., 2000). Chiari malformation type I can occur due to birth defects and may be detected when the patient is still an infant or during adulthood. However, in some cases—rather than a birth/genetic defect—a traumatic accident or craniocervical tumors may give rise to CMI (Merello et al., 2017). Chiari malformation type I usually comes with accompanying conditions such as syringomyelia, cerebral hydrocephalus, and basilar invagination (Shah et al., 2017). Almost 70%–80% of CMI cases display syringomyelia (Zhao et al., 2016).
The exact pathogenesis of CMI has yet to be discovered. Currently, CMI is diagnosed in clinic when there are more than 5 mm of tonsils protruding from the foramen magnum (Merello et al., 2017). The broad range of explanations for CMI does not aid in formulating a precise diagnosis. Many other conditions may be included in this type of pathogenesis; therefore, there is a need for more accurate parameters. The most classical proposed mechanism for CMI is a mesodermal defect. Neuroectodermal malformation may also be the cause of CM (Schijman, 2004).
Symptoms vary greatly among CM patients; most of the time these symptoms are as simple as a cough, headache, and nausea. Another diagnostic challenge is that decompression of the foramen magnum is handled through surgery as a way of treating CM. However, over the years risks involving the surgery have been observed; taking genetics into consideration may help in determining whether surgery is suitable (Langridge et al., 2017). Along with MRI diagnosis, genetics can be used in diagnosis. Moreover, genetics will help us to determine the genetic basis of CMI, which may elucidate its natural history or serve as a guide to treatment.
For many years, CM was considered a sporadic disease. However, family studies indicate that genetics are involved in the pathogenesis of CM. Familial aggregation shows that there could be a genetic culprit, and identifying the genes/variations responsible for the pathogenesis may aid in accurate diagnosis and treatment (Nagy et al., 2016; Merello et al., 2017).
Here we present the results of a genome-wide analysis of two Turkish families diagnosed with CMI. We present possible candidate genes associated with familial CMI pathogenesis. Moreover, chromosomal regions and CNVs that are potentially related to CMI are reported.
2. Materials and methods
2.1. Subjects
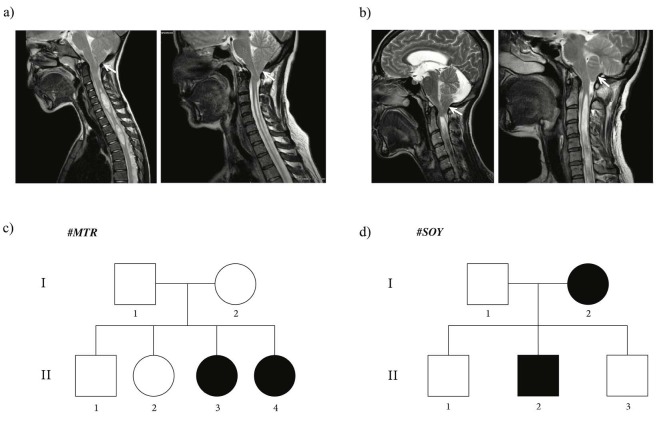
Two families with individuals diagnosed with CMI are included in the study. Families are labelled with suitable letters. The first family (# MTR ) consists of 6 members [2 parents (nonconsanguineous) and 4 children (1 male and 3 female)], and 2 female siblings were operated on for CMI. The second family (# SOY ) consists of 5 members [2 parents (nonconsanguineous) and 3 brothers], and the mother and the second son were operated on for CMI. Herniation of the tonsils through the foramen magnum, which causes brain stem pressure, is seen in Figures 1a and 1b (white arrows). Occipital headache during straining or after coughing, dysphagia, numbness in the extremities, and indifference to discrimination between hot/cold sensations, especially in the lower extremities, constituted the main clinical findings.
The other, unaffected members of both families were checked by cranial MRI, and CMI diagnoses were excluded. This study was approved by the institutional review board (2018-17/02). All procedures performed adhered to ethical guidelines. The pedigrees of both families are shown in Figure 1. In family # MTR , daughters II-3 and II-4 were proband individuals (Figure 1c). Daughter II-3 was operated on when she was 35 years old, and daughter II-4 was operated on when she was 37 years old. Both patients complained of occipital headaches, which were especially aggravated by coughing. In family # SOY , mother (I-2) and son (II-2) were the affected individuals (Figure 1d). The mother (I-2) was 40-years old when she was operated on, and her son (II-2) was 9. The mother’s complaint was untreatable headaches and her son’s was recurrent falling. The surgical procedure performed in all patients was standardized posterior fossa decompression with additional wide duraplasty.
Figure 1.
Pedigrees of two Turkish families with CMI. Squares signify males and circles signify females. Filled symbols signify affected individuals. Blood samples of every individual in the pedigrees were collected and subjected to microarray analysis. a) Pedigree of #MTR family. Individuals II-3 daughter and II-4 daughter were confirmed as affected cases. b) MR images of index patients in #MTR family. Left hand side is individual II-3, right hand side is individual II-4. c) Pedigree of #SOY family. Individuals I-2 mother and II-2 son were confirmed as affected cases. d) MR images of index patients in #SOY family. Left hand side is individual I-2, right hand side is individual II-2.
2.2. Microarray analysis
The isolation of DNA from the blood samples of all individuals in both families shown in the pedigrees (Figure 1) was performed by commercially available kits, according to manufacturer recommendations (NucleoSpin DNA blood, Macherey–Nagel GmbH & Co. KG, Düren, Germany). The DNA samples were studied with an Illumina Infinium CytoSNP-850K v1.2 microarray chip and scanned on Illumina iScan platform (Illumina Inc., San Diego, CA, USA). Around 665,000 single nucleotide polymorphisms (SNPs) were genotyped for each sample. The median range of Illumina microarray chip probes is 2.53 kb. Copy number variation (CNV) analysis was performed by calculation of B allele frequency (BAF) and log R ratio (LRR) values from whole genome SNP data. Copy number variations were determined and visualized via the GenomeStudio (v.2.0.4) and cnvPartition (v.3.2.0) analysis programs provided by Illumina Inc. Human Genome Build 37 was used as a reference to align and report interested positions. The confidence threshold was 50, and GC wave correction was performed for the analysis. In order to determine loss and gain regions, the minimum probe required was set at 8, and copy number loss of heterozygosity (CNLOH) regions greater than 1 Mb were reported.
Microarray results were evaluated by two main approaches. First, SNVs between affected and unaffected members in both families were compared. Mutations in the affected members of both families were reported in Table. Second, chromosomal variations in all affected members and unaffected members were listed for both families in Supplementary Table.
Table .
List of common single nucleotide variations in both families.
| Chr. Location | Gene | Biological Process / Gene Ontologya | n | Variantb | Variant Classc | Enhanced Expressiond |
|---|---|---|---|---|---|---|
| 9q33.3 | OLFML2A | Protein homodimerization activity | 1 | rs7874348 (A/G) | Missense | Low tissue specificity |
| 5q31.3 | SLC4A9 | Anion transmembrane transporter activity | 1 | rs6860077 (T/C/G) | Missense | Kidney, heart |
| 13q.34 | COL4A1 | Basal membrane formation | 1 | rs9521687 (C/A) | 5’UTR | Placenta |
| 13q33.3 | FAM155A | Calcium ion import across plasma membrane | 2 | rs1543002 (C/T), rs1543003 (T/A/C) | Intronic | Brain, pituitary gland |
| 5q31.3 | NR3C1 | Apoptosis, cell cycle, transcription regulation | 2 | rs4607376 (A/G/T), rs6871464 (T/C) | Intronic | Low Tissue Specificity |
| 5q31.1 | TRPC7 | Calcium transport | 2 | rs6893792 (T/A/C), rs10463951 (C/A/G/T) | Intronic | Adrenal gland, brain, intestine, kidney, pituitary gland, testis |
| 9q33.1 | ASTN2 | Protein transport | 5 | rs2274414 (T/G), rs16936575 (G/A/T), rs12554069 (T/C), rs6478300 (C/A), rs10983600 (C/T) | Intronic | Low tissue specificity |
| 9q33.2 | TRAF1 | Apoptosis | 1 | rs3761847 (G/A) | Intronic | Low tissue specificity |
| 3p24.1 | LINC00693 | N/A | 1 | rs9875884 (T/C) | Intronic | Brain |
| 13q33.3 | MYO16 | Motor activity, actin binding | 4 | rs76754941 (G/A), rs2759273 (T/C), rs9514937 (G/A), rs9521082 (A/G) | Intronic | Brain |
| 13q.34 | COL4A2 | Basal membrane formation | 7 | rs73619284 (C/T), rs8001070 (G/A), rs9555689 (G/A/T), rs7317178 (C/A/T), rs12323265 (A/G), rs9515209 (C/T), rs4517640 (C/T) | Intronic | Placenta |
| 5q31.1 | FSTL4 | Calcium ion binding, metal ion binding | 1 | rs55962865 (A/C) | Intronic | Brain |
| 5q31.3 | FGF1 | Angiogenesis, differentiation | 1 | rs2339246 (C/G/T) | Intronic | Brain, heart muscle, kidney |
| 5q31.3 | ARHGAP26 | GTPase activity | 1 | rs77545201 (G/T) | Intronic | Low tissue specificity |
| 5q32 | HTR4 | G protein-coupled receptor activity | 1 | rs1820075 (C/T) | Intronic | Brain, heart muscle, intestine, pituitary gland |
| 5q32 | ADRB2 | G protein-coupled receptor activity | 1 | rs6893517 (G/A) | Intronic | Blood |
| 9q32 | COL27A1 | Extracellular matrix structural constituent | 1 | rs2567714 (T/C) | Intronic | Brain, uterine, cervix |
| 9q33.1 | BRINP1 | Inhibits cell proliferation by negative regulation of the G1/S transition | 1 | rs1332453 (A/C) | Intronic | Brain |
| 9q33.2 | PHF19 | Chromatin regulator | 2 | rs11794516 (A/G), rs2072438 (T/C) | Intronic | Low tissue specificity |
| 9q33.2 | CNTRL | Cell cycle, cell division | 1 | rs746182 (T/A/C) | Intronic | Low tissue specificity |
| 9q33.3 | MVB12B | Protein transport | 1 | rs7047946 (G/A) | Intronic | Brain |
| 9q34.11 | USP20 | Endocytosis, Ubl conjugation pathway | 2 | rs10819567 (A/C/G), rs62583579 (C/T) | Intronic | Low tissue specificity |
| 9q34.11 | LOC101929331 | N/A | 2 | rs10819490 (C/T), rs10988311 (T/G) | Intronic | N/A |
| 5q23.1 | LINC00992 | N/A | 3 | rs1863977 (G/A), rs12332110 (G/A/T), rs62380137 (G/A/T) | Intronic | Pancreas, colon |
| 5q31.1 | LOC100996485 | N/A | 1 | rs2059780 (G/A) | Intronic | N/A |
| 5q31.3 | LOC101926941 | N/A | 3 | rs9324876 (A/G), rs959662 (A/C/G), rs9800206 (C/A/T) | Intronic | N/A |
| 17q21.33 | LOC100288866 | N/A | 1 | rs2537720 (G/A) | Intronic | Low tissue specificity |
| 7q22.2 | LHFPL3 | N/A | 2 | rs2470957 (G/A/T), rs12667640 (T/C) | Intronic | Brain |
| 7q22.3 | LHFPL3 - AS2 | N/A | 1 | rs10953457 (C/T) | Intronic | Kidney |
N/A: no known function or expression, n: number of reported variations in a gene, aAccording to Gene Cards Weizmann Institute of Science (2020). GeneCards: The Human Gene Database [online]. Website https://www.genecards.org [accessed 01 September 2020].and Gene Ontology Gene Ontology Consortium (2020). The Gene Ontology Resource [online]. Website http://geneontology.org/ [accessed 01 September 2020]. databases, bNucleotide changes were determined through dbSNP database National Center for Biotechnology Information (2020). dbSNP database [online]. Website https://www.ncbi.nlm.nih.gov/snp/ [accessed 01 September 2020]., cVariant classes were determined through Genome Browser database University of California Santa Cruz Genomics Institute (2020). Genome Browser database [online]. Website https://genome.ucsc.edu [accessed 01 September 2020., dAccording to Gene Cards database.
3. Results
3.1. Single nucleotide variations
We evaluated single nucleotide variations (SNVs) in both families and listed those that are common between the two families (Table). Most of these variations were intronic; however, there were two missense variations and one 5’UTR variation. None of the variations listed in Table were previously reported as clinically significant variations in the ClinVar database (Landrum et al., 2018).
3.2. Copy number variations
Genotype data of microarray results were used to perform copy number variation (CNV) analysis of the affected individuals in both families. Patient II-3 of family MTR harbored no significant CNV, whereas patient II-4 carried 3 copies of 13q13.1 and homozygote deletion of 22q13.2 variations. In family SOY, patient I-2 carried 1 copy of 7q11.21 and 11p15.4, and 3 copies of 11p12 variations, whereas patient II-2 carried 1 copy of 11p15.4 variation. In addition to the CNVs mentioned, all other nonsignificant CNVs that are common in populations and that were detected in the affected individuals of each family were listed in supplementary Table.
4. Discussion
Several previous studies investigated the genetic underpinnings of CMI by using genome wide analyses. One of the most comprehensive studies of CMI included 23 families with 71 affected individuals and microarray data with 10K SNP Chip analysis. Researchers reported a significant linkage on chromosomes 9 and 15 (Boyles et al., 2006). In another comprehensive study, Markunas et al. conducted the largest genome linkage screen in 66 families and 367 individuals. They reported a maximum logarithm of distance (LOD) score of 3 for chromosomes 8 and 12 and listed candidate associated genes such as SUZ12, NF1, and LR6 (Markunas et al., 2013). A similar exploratory analysis of copy number variations in a Turkish family with 2 members affected by CMI failed to identify any significant CNV or gene candidate (Keser et al., 2019). Various studies reported different results. Therefore, we aim to report potential candidate CNVs and genes for familial CMI.
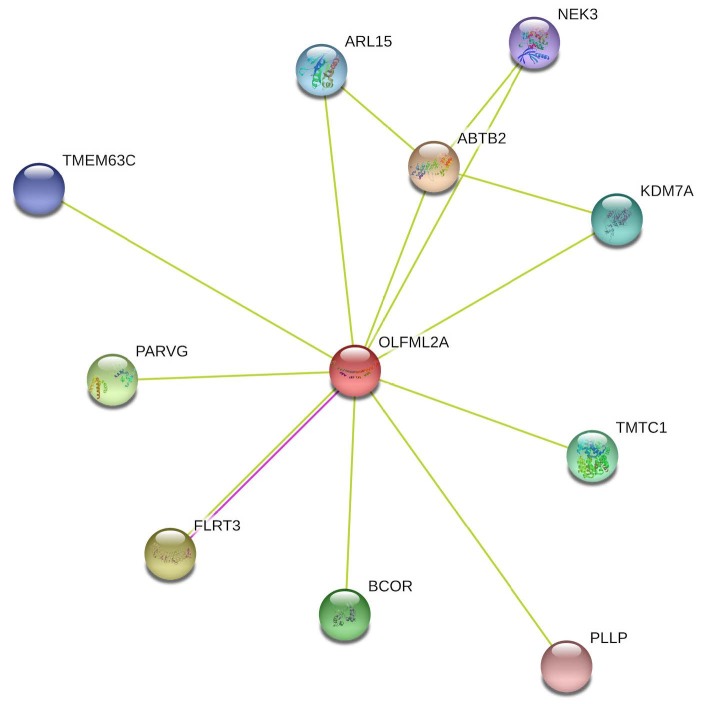
A missense variation was detected in OLFML2A gene (rs7874348) (Table). An SNV was found at exon 7 of the gene and caused Thr to Ala amino acid conversion. OLFML2A (olfactomedin-like 2A) is an olfactomedin-domain–containing protein, and its function in a cell has not been extensively explored (Sistani et al., 2013). Previous studies state that OLFML2A gene has a role in extracellular matrix (ECM) modeling (Furutani et al., 2005; Hong et al., 2019). Since our knowledge about the function of OLFML2A gene is limited, we used the STRING database to determine the functional protein association network for our gene of interest (Szklarczyk et al., 2015). Networks shown in Figure 2 demonstrate functional proteins that are connected with OLFML2A. Some of the genes shown in the network might be helpful for understanding the cellular processes involving OLFML2A (Figure 2). NEK3 gene encodes for a serine/threonine protein kinase which has a role in neuronal morphogenesis through microtubule acetylation (Chang et al., 2009). FLRT3 gene was found to be expressed during craniofacial development (Gong et al., 2009), and it functions in cell-cell adhesion and migration during cortical folding (Lu et al., 2015; Del Toro et al., 2017). Lastly, KDM7A gene in the network encodes for the histone–demethylase required for brain and craniofacial development (Qi et al., 2010; Tsukada et al., 2010). Hence, we propose that OLFML2A gene may be an essential gene in the pathogenesis of CMI due to its role in development of brain structures.
Another missense variation was detected in SLC4A9 gene (rs6860077) and, similarly, this variation has not been previously reported. It is found at exon 12 of the gene and causes Ile to Met amino acid conversion. Solute carrier family 4 member 9 ( SLC4A9 ) belongs to the SCL4A family of anion exchangers. Peña-Münzenmayer et al. reported that SLC4A9 gene has a role in fluid secretion regulation and is specifically responsible for Cl- exchange in the cell (Peña-Münzenmayer et al., 2016). It has been shown that cerebrospinal fluid (CSF) secretion is controlled by Na+, Cl- , and HCO3- transportation (Brown et al., 2004). Disrupted CSF flow in the brain may contribute to many pathologies, including CMI, in which the flow is further disrupted due to cerebellar herniation (Buell et al., 2015).
Figure 2.
STRING functional protein association network of OLFML2A gene (accessed 01 September 2020).
Another significant SNV (rs9521687) was detected in COL4A1 gene as a 5’UTR variation. This particular SNV has not been associated with any condition, and there are no published studies related to rs9521687. Type IV collagens can only be found in basement membranes, and the family consists of 6 distinct a-chains. The earliest discovered chains, a1(IV) and a2(IV), are considered classical chains, and they are encoded by COL4A1 and COL4A2 , respectively. Classical chains are expressed both during development and adulthood; however, four other chains are only expressed during development. The a1a1a2 triple helix is formed and secreted to the extracellular matrix (ECM), which then becomes a molecular scaffold to other ECM components (Khoshnoodi et al., 2008). Type IV collagen family variations have been associated with various sporadic and genetic diseases. COL4A1 gene variations may cause poor speech development, cerebral palsy, epilepsy (Pasternak et al., 1980), porencephaly and adult stroke (Gould et al., 2006; van der Knaap et al., 2006), and encephalopathy (Yaramis et al., 2020). These diseases all suggest that COL4A1 mutations may cause impaired vasculature formation in the brain. Urbizu et al. stated that variations affecting fetal vascular development might cause susceptibility to CMI (Urbizu et al., 2013).
Five of the significantly associated variations (rs1543002, rs10463951, rs4607376, rs6478300, and rs3761847) were reported previously. The SNV (rs1543002) in family with sequence similarity 155 member A gene ( FAM155A ) was reported as a significant SNV for patients diagnosed with bipolar disease and attempted suicide (Willour et al., 2012). Although there is not much information about the function of FAM155 family genes in humans, studies showed that it is associated with calcium channel activities (Gaudet et al., 2011). Calcium channels are known to have a role in neural tube closure during developmental stages (Abdul-Wajid et al., 2015), and CMI can be classified as an neural tube defect (NTD) in some cases. The SNV (rs4607376) in nuclear receptor subfamily 3 group C member 1 gene ( NR3C1 ), which is a glucocorticoid receptor, was previously reported in only one clinical study and it was related with systemic lupus erythematosus (Chen et al., 2017). Although no previous publications have established a relationship between autoimmune diseases and CMI, there have been cases reported suggesting that autoimmune diseases and CM pathogenesis might be related (Ortolani, 2014).
The 3 remaining intronic SNVs (rs10463951) in transient receptor potential cation channel subfamily C member 7 gene ( TRPC7 ), (rs6478300) (Wang et al., 2009) in astrotactin 2 gene ( ASTN2 ), and (rs3761847) (Huang et al., 2019; Plenge et al., 2007) in TNF receptor associated factor 1 gene ( TRAF1 ) are all reportedly related to the autoimmune disease rheumatoid arthritis (RA). TRPC7 gene is a calcium channel that has a role mainly in keratinocyte development (Beck et al., 2006). ASTN2 gene encodes for a protein which regulates surface proteins by endocytic trafficking and protein degradation (Behesti et al., 2018). ASTN2 is reported to form a complex with ASTN1 which plays a role in neuronal migration, and both genes are highly expressed in the cerebellum (Wilson et al., 2010). TRAF1 was first identified as a part of the TNFR2 signaling complex, and it also has relationships with other TNFR superfamily members that play a role in prosurvival signaling. TRAF1 is a well-studied gene, and it has been associated with many different diseases such as B-cell–related cancers, RA, cardiovascular diseases, and many infectious diseases (Edilova et al., 2018).
The CNV analysis provided us with a list of candidate chromosomal regions that are potentially associated with CMI. Furthermore, 11p15.4 one-copy deletion is inherited via mother to son in family SOY.
5. Conclusion
Elucidation of the molecular etiology of CMI by using familial cases is essential for such a complex disease. We conducted molecular karyotyping and SNV analysis for two families and listed important single nucleotide variations with regard to their mutation status and possible roles in CMI etiology. We propose that mutations in OLFML2A, SLC4A9, and COL4A1 genes may have a role in the familial form of CMI. Although a number of chromosomal regions were listed as potential regions associated with CMI, only 11p15.4 chromosome regions were CNVs that were inherited by a son via an affected mother. Our explorative results should be confirmed through further familial CMI case studies and molecular studies.
Informed consent
This study was approved by the ethical committee of Bahçeşehir University, School of Medicine (BAU2018-17/02). All patients and family members provided informed consent. All procedures were performed according to the relevant ethical standards.
Supplementary Materials
Acknowledgements
We offer our sincere thanks to the Bahçeşehir University Technology Transfer Office (BAU TTO) for the project grant they provided and DONE GENETİK Laboratories for their generous support with their infrastructure.
References
- Abdul-Wajid S Morales-Diaz H Khairallah SM Smith WC T-type calcium channel regulation of neural tube closure and EphrinA/EPHA expression. Cell Reports. 2015;13:829–839. doi: 10.1016/j.celrep.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B Zholos A Sydorenko V Roudbaraki M Lehen’kyi V et al. Journal of Investigative Dermatology. 2006;126:1982–1993. doi: 10.1038/sj.jid.5700352. [DOI] [PubMed] [Google Scholar]
- Behesti H Fore TR Wu P Horn Z Leppert M ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E9717–E9726. doi: 10.1073/pnas.1809382115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles AL Enterline DS Hammock PH Siegel DG Slifer SH Phenotypic definition of Chiari type I malformation coupled with high-density SNP genome screen shows significant evidence for linkage to regions on chromosomes 9 and 15. American Journal of Medical Genetics Part A. 2006;140:2776–2785. doi: 10.1002/ajmg.a.31546. [DOI] [PubMed] [Google Scholar]
- Brown PD Davies SL Speake T Millar ID Molecular mechanisms of cerebrospinal fluid production. Neuroscience. 2004;129:957–970. doi: 10.1016/j.neuroscience.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell TJ Heiss JD Oldfield EH Pathogenesis and cerebrospinal fluid hydrodynamics of the Chiari I malformation. Neurosurgery Clinics of North America. 2015;26:495–499. doi: 10.1016/j.nec.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J Baloh RH Milbrandt J The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. Journal of Cell Science. 2009;122:2274–2282. doi: 10.1242/jcs.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ Papp G Póliska S Szabó K Tarr T MicroRNA expression profiles identify disease-specific alterations in systemic lupus erythematosus and primary Sjögren’s syndrome. PLoS One. 2017;12:e0174585–e0174585. doi: 10.1371/journal.pone.0174585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro D Ruff T Cederfjäll E Villalba A Seyit-Bremer G Regulation of cerebral cortex folding by controlling neuronal migration via FLRT adhesion molecules. Cell. 2017;169:621–635. doi: 10.1016/j.cell.2017.04.012. [DOI] [PubMed] [Google Scholar]
- Edilova MI Abdul-Sater AA Watts TH TRAF1 signaling in human health and disease. Frontiers in Immunology. 2018;9:2969–2969. doi: 10.3389/fimmu.2018.02969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani Y Manabe R-I Tsutsui K Yamada T Sugimoto N Identification and characterization of photomedins: novel olfactomedin-domain-containing proteins with chondroitin sulphate-E-binding activity. Biochemical Journal. 2005;389:675–684. doi: 10.1042/BJ20050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P Livstone MS Lewis SE Thomas PD Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Briefings in Bioinformatics. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong SG Mai S Chung K Wei K Flrt2 and Flrt3 have overlapping and non-overlapping expression during craniofacial development. Gene Expression Patterns. 2009;9:497–502. doi: 10.1016/j.gep.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Gould DB Phalan FC Van Mil SE Sundberg JP Vahedi K Role of COL4A1 in small-vessel disease and hemorrhagic stroke. New England Journal of Medicine. 2006;354:1489–1496. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- Hong EP Kim BJ Cho SS Yang JS Choi HJ Genomic variations in susceptibility to intracranial aneurysm in the Korean population. Journal of Clinical Medicine. 2019;8:275–275. doi: 10.3390/jcm8020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-C Hua Sun Q-Q Zhang L-N Cen H Associations of TRAF1/C5 rs10818488 and rs3761847 polymorphisms with genetic susceptibility to rheumatoid arthritis: a case-control study and updated meta-analysis. Central European Journal of Immunology. 2019;44:159–173. doi: 10.5114/ceji.2019.87067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keser N Kuskucu A Is M Celikoglu E Familial Chiari type 1: a molecular karyotyping study in a Turkish family and review of the literature. World Neurosurgery. 2019;121:e852–e857. doi: 10.1016/j.wneu.2018.09.235. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J Pedchenko V Hudson BG Mammalian collagen IV. Microscopy Research and Technique. 2008;71:357–370. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ Lee JM Benson M Brown GR Chao C ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Research. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge B Phillips E Choi D Chiari malformation type 1: a systematic review of natural history and conservative management. World Neurosurgery. 2017;104:213–219. doi: 10.1016/j.wneu.2017.04.082. [DOI] [PubMed] [Google Scholar]
- Lu YC Nazarko OV Sando III R Salzman GS Li N-S Structural basis of latrophilin-FLRT-UNC5 interaction in cell adhesion. Structure. 2015;23:1678–1691. doi: 10.1016/j.str.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markunas CA Soldano K Dunlap K Cope H Asiimwe E Stratified whole genome linkage analysis of Chiari type I malformation implicates known Klippel-Feil syndrome genes as putative disease candidates. PLoS One. 2013;8:e61521–e61521. doi: 10.1371/journal.pone.0061521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J Kraut M Guarnieri M Haroun RI Carson BS Asymptomatic Chiari type I malformations identified on magnetic resonance imaging. Journal of Neurosurgery. 2000;92:920–926. doi: 10.3171/jns.2000.92.6.0920. [DOI] [PubMed] [Google Scholar]
- Merello E Tattini L Magi A Accogli A Piatelli G Exome sequencing of two Italian pedigrees with non-isolated Chiari malformation type I reveals candidate genes for cranio-facial development. European Journal of Human Genetics. 2017;25:952–959. doi: 10.1038/ejhg.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L Mobley J Ray C Familial aggregation of Chiari malformation: presentation, pedigree, and review of the literature. Turkish Neurosurgery. 2016;26:315–320. doi: 10.5137/1019-5149.JTN.10471-14.1. [DOI] [PubMed] [Google Scholar]
- Ortolani F Tummolo A Fedele S Masciopinto M Pesce S Autoimmune disease and Arnold Chiari syndrome: any correlation? Hormone Research in Paediatrics. Hormone Research in Paediatrics. 2014:82. [Google Scholar]
- Pasternak JF Mantovani JF Volpe JJ Porencephaly from periventricular intracerebral hemorrhage in a premature infant. The American Journal of Diseases Children. 1980;134:673–675. doi: 10.1001/archpedi.1980.02130190041010. [DOI] [PubMed] [Google Scholar]
- Peña-Münzenmayer G George AT Shull GE Melvin JE Catalán MA Ae4 (Slc4a9) is an electroneutral monovalent cation-dependent Cl−/HCO3− exchanger. Journal of General Physiology. 2016;147:423–436. doi: 10.1085/jgp.201611571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM Seielstad M Padyukov L Lee AT Remmers EF TRAF1–C5 as a risk locus for rheumatoid arthritis — a genomewide study. New England Journal of Medicine. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH Sarkissian M Hu GQ Wang Z Bhattacharjee A Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijman E History, anatomic forms, and pathogenesis of Chiari I malformations. Child’s Nervous System. 2004;20:323–328. doi: 10.1007/s00381-003-0878-y. [DOI] [PubMed] [Google Scholar]
- Shah A Dhar A Elsanafiry M Goel A Chiari malformation: has the dilemma ended. Journal of Craniovertebral Junction and Spine. 2017;8:297–304. doi: 10.4103/jcvjs.JCVJS_138_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistani L Rodriguez PQ Hultenby K Uhlen M Betsholtz C Neuronal proteins are novel components of podocyte major processes and their expression in glomerular crescents supports their role in crescent formation. Kidney International. 2013;83:63–71. doi: 10.1038/ki.2012.321. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D Franceschini A Wyder S Forslund K Heller D STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y-i Ishitani T Nakayama KI KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes and Development. 2010;24:432–437. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbizu A Toma C Poca MA Sahuquillo J Cuenca-León E Chiari malformation type I: a case-control association study of 58 developmental genes. PLoS One. 2013;8:e57241–e57241. doi: 10.1371/journal.pone.0057241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap MS Smit LM Barkhof F Pijnenburg YA Zweegman S Neonatal porencephaly and adult stroke related to mutations in collagen IV A1. Annals of Neurology. 2006;59:504–511. doi: 10.1002/ana.20715. [DOI] [PubMed] [Google Scholar]
- Wang Y Sha N Fang Y Analysis of genome-wide association data by large-scale Bayesian logistic regression. BMC Proceedings 3 Supplement. 2009;7:S16–S16. doi: 10.1186/1753-6561-3-s7-s16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL Seifuddin F Mahon PB Jancic D Pirooznia M A genome-wide association study of attempted suicide. Molecular Psychiatry. 2012;17:433–444. doi: 10.1038/mp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PM Fryer RH Fang Y Hatten ME Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. Journal of Neuroscience. 2010;30:8529–8540. doi: 10.1523/JNEUROSCI.0032-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaramis A Lochmüller H Töpf A Sonmezler E Yilmaz E COL4A1- related autosomal recessive encephalopathy in 2 Turkish children. Neurology Genetics. 2020;6:e392–e392. doi: 10.1212/NXG.0000000000000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL Li MH Wang CL Meng W A systematic review of Chiari I malformation: techniques and outcomes. World Neurosurgery. 2016;88:7–14. doi: 10.1016/j.wneu.2015.11.087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials