Abstract
Gut microbiota is vital for human health. Shifts in the microbial diversity can affect bacterial function, and dysbiosis is associated with a variety of gastrointestinal disorders, including celiac disease (CD) and irritable bowel syndrome (IBS). The distinction between IBS and non-celiac gluten sensitivity (NCGS) is unclear, and it is conceivable that the gut microbiota profile of these patients may overlap. To our knowledge, no existing literature has evaluated the microbial characteristics in CD, IBS, and NCGS. Hence, this systematic review aims to compare the gut microbiota profile in these three diagnoses. A literature search was conducted in PubMed (Medline) until April 2019. Studies investigating bacterial diversity in the gut of patients with CD, IBS, and NCGS were eligible. Inclusion criteria were observational studies and randomized controlled trials reporting bacterial profile at baseline. Ninety-one articles were identified, of which 13 trials were eligible for inclusion. Overall, the bacterial composition of the gut microbiota of patients with CD and those with IBS shared the many similarities. The microbial richness was correspondingly reduced in these patient-groups compared with healthy controls, but this was not reported for NCGS. Our findings suggest that the bacterial profiles of patients with IBS and CD share certain disease-specific trends. Fewer similarities were observed between the bacterial profiles of patients with IBS and NCGS. Notably, the data are limited; thus, no solid conclusions can be made on the basis of these findings alone. The suggested trends can be a valuable basis for further research.
Keywords: Gastrointestinal microbiota, dysbiosis, celiac disease, gluten sensitivity, irritable bowel syndrome
INTRODUCTION
Microbiota is defined as “the community of microorganisms that lives on the surface and inside the human host” (1). Microorganisms associated with the human host are bacteria, archaea, viruses, and fungi (2). Bacteria along the gastrointestinal (GI) tract are shown to be critical for human health (3, 4), and shifts in microbial diversity can affect bacterial function (4, 5). Dysbiosis refers to a disturbance in both the quantity and composition of the gut microbiota (6) and is associated with a variety of GI-disorders, including celiac disease (CD) and irritable bowel syndrome (IBS) (7, 8).
CD is a chronic immune-mediated response to the ingestion of gluten in genetically susceptible individuals (6). Ingestion of gluten by patients affected by CD induces an inflammatory response causing villous atrophy in the small intestine (9). IBS is a multifactorial functional gastrointestinal disorder (FGID) affecting approximately 10%–20% of the world’s population, characterized by abdominal pain, bloating, and change in the frequency and consistency of stool (10, 11). Subgroups of IBS are characterized by different bowel patterns and are divided into diarrhea-predominant IBS (IBS-D), constipation predominant IBS (IBS-C), and mixed bowel habits (IBS-M) (11, 12).
Non-celiac gluten sensitivity (NCGS) is still an unclear diagnosis, characterized by symptoms related to the ingestion of gluten-containing food in patients who are not affected by either CD or wheat allergy (13). NCGS and IBS share many common features, and current evidence suggests that NCGS might be characterized as a subgroup of IBS (14). Patients with NCGS and IBS suffer from similar symptoms, including classical symptoms such as abdominal pain, bloating, and alterations in bowel habits, as well as a broad range of extra-intestinal complaints (13). There are no reliable diagnostic tests or biomarkers to confirm NCGS and IBS (15). Patients with CD experience many of the same symptoms as patients with NCGS and IBS, but this group of patients can be diagnosed according to positive serology and duodenal biopsies (14). Currently, the distinction between NCGS and IBS has been based on the trigger of symptoms, namely gluten and fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAPs). However, recent studies have questioned gluten as the main trigger of symptoms in NCGS, and suggested fructans and amylase-trypsin inhibitors (ATIs) in wheat as possible symptom triggers (16–18). As the distinction between the diagnoses is unclear, it is conceivable that gut microbiota of these patients may overlap.
Most studies conducted on CD and gut microbiota have confirmed decreased Bifidobacteria and Lactobacilli (8, 19, 20) and increased Gram-negative bacteria compared to with healthy controls (8, 20). Several studies have investigated the composition and diversity of gut microbiota in IBS patients (7, 21). Although the findings are inconsistent, it is suggested an increase in firmicutes to bacteroidetes ratio decreased Lactobacilli and Bifidobacteria (7, 21) and increased Streptococci and Ruminococcus species when compared with healthy individuals (7). In addition, patients with IBS are often characterized by a reduction in bacterial diversity. Differences between IBS subtypes have also been detected (7, 21). As NCGS is a fairly new diagnosis, few studies have investigated gut microbiota in this patient group.
To our knowledge, no previous publications have compared the gut microbiota profile in patients with CD, NCGS, and IBS. Hence, this systematic review aims to compare the gut microbiota profile in these three GI-disorders, focusing on bacterial composition at phylum level.
MATERIALS AND METHODS
The checklist and flowchart of the PRISMA (Preferred Reporting for Systematic Reviews and Meta-Analyses) guidelines were followed for this systematic review (22).
Search strategy and criteria for inclusion
A literature search was conducted in PubMed (Medline) until April 5th, 2019. Three individual searches were conducted, one for each diagnosis. The search terms included “Celiac disease AND (healthy subjects OR healthy controls OR non-celiac controls) AND microbiota,” “Non-celiac gluten sensitivity AND (healthy subjects OR healthy controls) AND microbiota” and “Irritable bowel syndrome AND (healthy subjects OR healthy controls) AND microbiota.”
The criteria for inclusion were observational studies and randomized controlled trials (RCTs) reporting on the gut bacteria profile at baseline. Studies on human individuals that were published within the last five years were included for detailed review. Only patients with IBS fulfilling the Rome III diagnostic criteria, and studies including patients with IBS from all three subgroups (IBS-C, IBS-D, and IBS-M) were selected. Studies including participants with post-infectious IBS were excluded, along with studies including participants with other chronic gastrointestinal (GI) disorders.
RESULTS
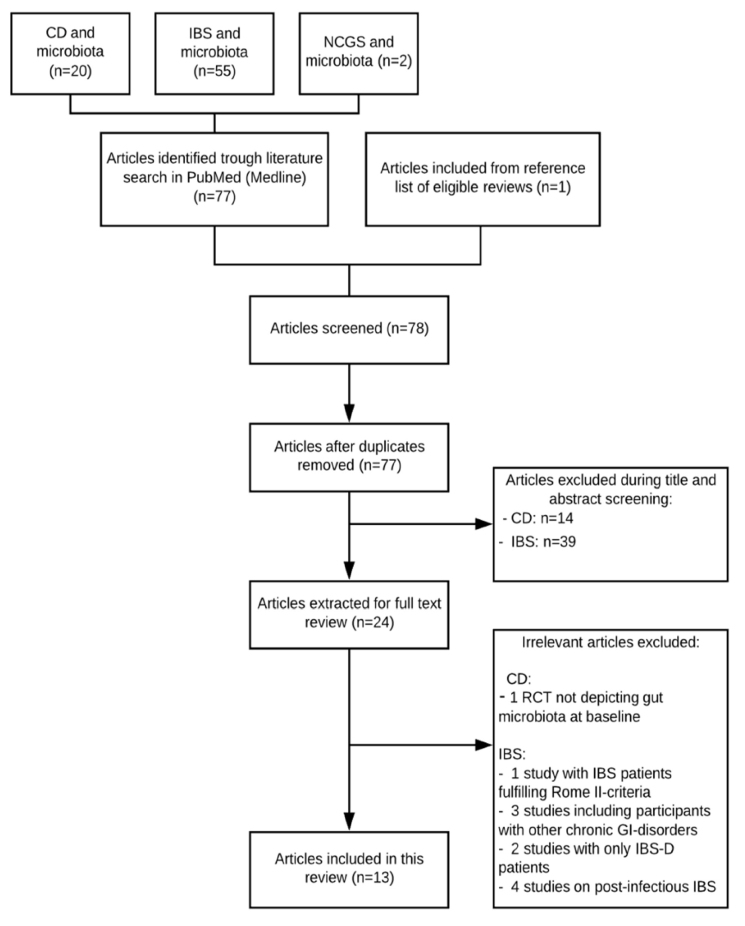
A total of 78 articles were identified, of which 13 trials were included in the systematic review. Figure 1 illustrates the process. Study characteristics and main findings of the 13 studies are summarized in Table 1. Figure 2 illustrates fairly consistent similarities and differences of bacterial strains between the three disorders.
Figure 1.
PRISMA-modified flow diagram depicting the literature search in PubMed (Medline) for this systematic review.
Table 1.
Characteristics and main findings of studies reporting on gut microbiota profile in patients with CD, NCGS, and IBS.
| Diagnosis | First author, year of publication | Study design | Participants (n) | Specimen | Method of detection | Main findings |
|---|---|---|---|---|---|---|
| CD | Golfetto (23) 2014 | Case-control | tCD (14) Control (42) |
Fecal sample | Microscopic analysis | ↓ Bifidobacteria (Actinobacteria) in CD |
| Wacklin (24) 2014 | Cohort | tCD w/symptoms (18) tCD asymptomatic (18) |
Duodenal biopsy | 16s rRNA sequencing PCR |
In symptomatic patients: ↓ Bacteroidetes, Firmicutes, and microbial richness ↑ Proteobacteria |
|
| Nistal (25) 2016 | Case-control | uCD (9) Control (9) |
Duodenal biopsy | 16s rRNA sequencing PCR | Firmicutes and Proteobacteria are the dominant phyla in both CD and HC. No significant differences in bacterial profile ↓ microbial richness in CD |
|
| Quagliarello (26) 2016 | RCT | CD (40) Control (16) |
Fecal sample | 16s rRNA sequencing qPCR | ↑ Bacteroidetes/Firmicutes ratio and ↓ Firmicutes, Actinobacteria, and microbial richness in CD Lower taxonomic levels: ↓ Lactobacillus (Firmicutes) ↑ B. fragilis (Bacteroidetes) |
|
| Herrán (27) 2017 | Case-control | tCD (14) uCD (5) Control (23) |
Duodenal biopsy | 16s rDNA sequencing PCR-DGGE | No significant differences in composition or abundance of bacteria with gluten-hydrolyzing capacity | |
| NCGS | Garcia-Mazcorro (28) 2018 | RCT | NCGS (12) CD (6) Control (12) |
Fecal sample Duodenal biopsy | 16s rDNA sequencing PCR |
Fecal samples Firmicutes (85%) Bacteroidetes (1%) in all ↑ Ruminococcaceae (Firmicutes) in NCGS Duodenal biopsies ↓ Bacteroidetes and Fusobacteria in CD ↑ Saccharibacteria in NCGS |
| Dieterich (29) 2018 | RCT | NCGS (19) Control (10) |
Fecal sample | 16s rRNA sequencing Fluorometric quantitation |
Lower taxonomic levels: ↑ Actinobacillus (Proteobacteria) and Finegoldia (Firmicutes) in NCGS ↑ Sphingobacteria (Bacteroidetes) in HC ↑ Ruminococcaceae, Peptostreptococcaceae (Firmicutes), and ↓ Porphyromonadaceae (Bacteroidetes) in NCGS |
|
| IBS | Halmos (30) 2014 | RCT | IBS (27) Control (6) |
Fecal sample | qPCR | No significant differences in total bacteria count or abundance of bacterial groups |
| Chung (31) 2015 | Case-control | IBS (28) Control (19) |
Fecal sample Jejunal biopsy | 16s rRNA sequencing PCR |
Fecal samples ↑ Firmicutes/Bacteroidetes ratio in IBS-D ↑ Veillonellaceae (Firmicutes) in IBS-D Jejunal biopsies ↑ Firmicutes/Actinobacteria ratio in IBS-M ↑ Prevotellaceae (Bacteroidetes) in IBS ↓ Mycobacteriaceae (Actinobacteria) and Neisseriaceae (Proteobacteria) in IBS |
|
| Pozuelo (32) 2015 | Case-control | IBS (113) Control (66) |
Fecal sample | 16s rRNA sequencing PCR | ↑ Bacteroidetes and Tenericutes in IBS ↓ Firmicutes and microbial richness in IBS Lower taxonomic levels: ↓ Lachnobacterium (Firmicutes) in IBS ↓ Strains within Firmicutes in IBS-D and IBS-M |
|
| Rangel (33) 2015 | Case-control | IBS (35) Control (16) |
Fecal sample Colonic biopsy | 16s rRNA sequencing |
Fecal samples ↑ Actinobacteria and Proteobacteria and ↓ Bacteroidetes in IBS Lower taxonomic levels: ↑ Bacilli and altered composition of species within Clostridia (Firmicutes) Colonic biopsies ↓ Clostridiales (Firmicutes) in IBS |
|
| Shukla (34) 2015 | Case-control | IBS (47) Control (30) |
Fecal sample | 16s rRNA sequencingq PCR | ↑ Gram-negative bacteria in IBS Lower taxonomic levels: ↓ Bifidobacteria (Actinobacteria) ↑ species within Firmicutes and Proteobacteria ↓ Lactobacillus (Firmicutes) and ↑ Bacteroides species (Bacteroidetes) in IBS-D than IBS-C |
|
| Tap (35) 2016 | Case-control | IBS (110) Control (39) |
Fecal sample Colonic biopsy | 16s rRNA sequencing qPCR | No significant differences between fecal and mucosal microbiota in IBS or HC ↑ Bacteroides (Bacteroidetes) in IBS-D and IBS-M ↓ Prevotella (Bacteroidetes) and microbial richness in IBS |
CD: celiac disease; IBS: irritable bowel syndrome; IBS-C: constipation-predominant IBS; IBS-D: diarrhea-predominant IBS; IBS-M: mixed type IBS; NCGS: non-celiac gluten sensitivity; PCR: polymerase chain reaction; PCR-DGGE: PCR denaturing gradient gel electrophoresis; RCT: randomized controlled trial; tCD: treated celiac disease; uCD: untreated celiac disease; qPCR: quantitative PCRNotes: The arrow marks represent increase and decrease.
Figure 2.
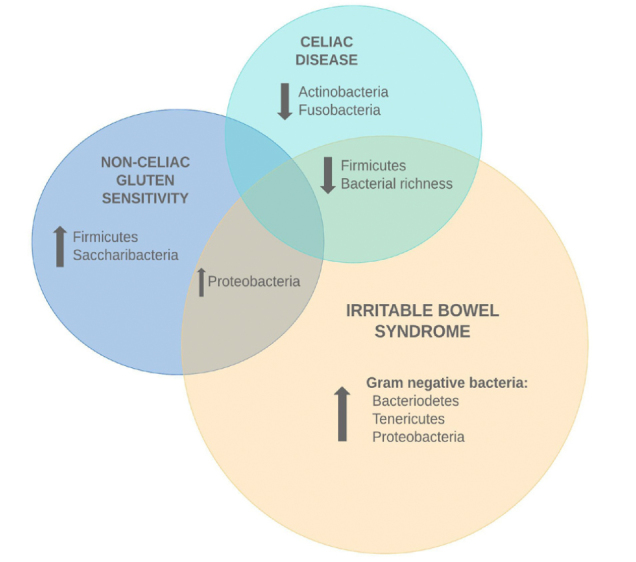
Overview of bacterial strains identified in celiac disease, non-celiac gluten sensitivity, and irritable bowel syndrome.
Gut microbiota profile in patients with celiac disease
Five studies reporting on the gut microbiota of patients with CD were included in this review, of which one was an RCT and four were observational studies. Four of the studies compared patients with CD with healthy controls (HCs), whereas one of the studies compared symptomatic and asymptomatic CD patients. One study included children, whereas the remaining four were conducted in adults.
Golfetto et al. (23) compared the concentration of fecal Bifidobacteria of patients with CD on a gluten-free diet (GFD) (n=14) and HCs (n=42). The samples of patients with CD showed a significant decrease in Bifidobacteria compared with HCs. Wacklin et al. (24) investigated duodenal microbiota composition in treated CD patients with (n=18) and without (n=18) persistent symptoms. At phylum level, the symptomatic patients had a significant decrease in bacteroidetes and firmicutes, and an increase in proteobacteria. Furthermore, symptomatic patients had reduced microbial richness compared with asymptomatic patients. Nistal et al. (25) compared the duodenal microbiota composition in patients with untreated CD (n=9) with HCs (n=9) and reported that bacteria from the upper small intestine mainly belong to the phyla firmicutes and proteobacteria. No significant differences were observed between the two groups of participants, but the microbial composition was specific in each adult. However, the microbial richness was lower in patients with CD compared with the HCs.
Quagliarello et al. (26) conducted an RCT on the effect of Bifidobacterium breve on the intestinal microbiota of children with CD (n=40) and of HCs (n=16). Fecal samples at baseline revealed that patients with CD had a significant decrease in firmicutes and actinobacteria, as well as a higher proportion of bacteroidetes with respect to firmicutes. At lower taxonomic levels, Lactobacillus was significantly reduced in patients with CD, whereas Bacteroides fragilis was increased. In addition, the microbial richness was significantly decreased in patients with CD.
Herrán et al. (27) aimed to characterize whether duodenal microbiota possibly implicated gluten hydrolysis. Bacteria with gluten-hydrolyzing capacity were isolated from all groups enrolled in the study, that is, treated CD (n=14), untreated CD (n=5), first-degree relatives of patients with CD (n=16), and HCs (n=7). Most of the strains were within the phylums firmicutes (88%)—mainly from the genus Lactobacillus (68%)—actinobacteria (8%), proteobacteria (3%), and bacteroidetes (1%). No significant differences were observed between the groups of participants, but the microbial composition was characteristic in each adult. However, some bacterial groups, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Streptococcus anginosus, were only isolated from patients with CD—both treated and untreated.
Gut microbiota profile in non-celiac gluten sensitivity
Two recent RCTs have reported on the gut microbiota profile in patients with NCGS. Both studies compared adult patients with HCs.
Garcia-Mazcorro et al. (28) examined the gut microbiota of patients with NCGS (n = 12) and CD (n = 6) and HCs (n = 12). At baseline, duodenal biopsies were obtained from all participants, whereas only 14 participants (7 NCGS patients; 3 CD patients; 4 HC) provided a fecal sample. At phylum level, the fecal samples showed a high abundance of firmicutes (~85%) and a deficiency of bacteroidetes (~1%), regardless of disease status. At family level, ruminococcaceae was significantly increased in patients with NCGS patients compared with those with CD and HCs. The duodenal microbiota showed significant alterations at phylum level; decreased bacteroidetes and fusobacteria in patients with CD, and increased saccharibacteria in patients with NCGS. Alterations were also observed at genus level; increased Actinobacillus and Finegoldia in patients with NCGS, and increased Sphingobacterium in HCs.
Dieterich et al. (29) evaluated the influence of a GFD and a diet low in FODMAPs on the intestinal microbiota of patients with NCGS (n=19) and HCs (n=10). Fecal samples were collected at baseline while the participants consumed at least two gluten-containing meals per day. At phylum level, no significant differences were observed between patients with NCGS and HCs. However, significant differences were found when comparing bacterial families. Supporting the findings by Garcia-Mazcorro et al., Dietrich et al. also reported an increase in ruminococcaceae. Furthermore, an increase in peptostreptococcaceae and a decrease in porphyromonadaceae were reported in patients with NCGS, compared with HCs.
Gut microbiota profile in irritable bowel syndrome
Six studies reporting on the gut microbiota of patients with IBS were included in this review—one RCT and five observational studies. All studies compared adults with IBS with HCs.
Halmos et al. (30) conducted an RCT investigating how diets with different FODMAP contents alter the colonic microenvironment in patients with IBS (n=27) and HCs (n=6). They reported the absolute and relative abundance of several bacteria within the phylums of firmicutes, actinobacteria, and verrucomicrobia. Neither of these, nor the bacterial count, differed significantly between the two groups.
Chung et al. (31) investigated differences in gut microbiota between subgroups of IBS (n=28) and HCs (n=19). Jejunal biopsies were obtained from 38 of the participants (23 cases and 15 controls), and 44 participants (26 cases and 18 controls) provided a fecal sample. The ratio of firmicutes to actinobacteria in jejenum of patients with IBS-M was increased and so was the ratio of firmicutes to bacteroidetes in fecal samples of patients with IBS-D. Significant differences were also detected at family level; patients with IBS had increased prevotellaceae and decreased mycobacteriaceae and neisseriaceae in the jejenum compared with HCs, regardless of the associated subgroup. Furthermore, patients with IBS-D had increased veillonellaceae in fecal samples compared with those with IBS-C and IBS-M and HCs.
Pozuelo et al. (32) compared fecal samples from 113 IBS patients and 66 HCs. At phylum level, patients with IBS showed increased bacteroidetes and tenericutes and decreased firmicutes compared with HCs. At lower taxonomic levels, Lachnobacterium was significantly decreased in patients with IBS. Overall, patients with IBS had significantly lower bacterial diversity than HCs, and within the subgroups, patients with IBS-D had the lowest diversity. Furthermore, the microbiota of patients with IBS-D and IBS-M had decreased abundance of several strains within the phylum of firmicutes. In contrast, the microbiota of patients with IBS-C did not show significant differences compared with the microbiota of HCs at any taxonomic level.
Rangel et al. (33) investigated the relationship between fecal and mucosal microbiota in patients with IBS (n=35) and HCs (n=16). Colonic biopsies were obtained from 45 of the participants (35 cases and 10 controls), and 29 participants (33 cases and 16 controls) provided a fecal sample. The microbial composition of fecal and mucosal samples differed significantly within both groups of participants, but the separation was much more distinct in patients with IBS. In mucosal microbiota, only one bacterial group within clostridiales differed significantly between HCs and patients with IBS. Regarding fecal microbiota, patients with IBS had increased actinobacteria and proteobacteria and decreased bacteroidetes. Several species of clostridia were altered in feces of patients with IBS, whereas groups of bacilli were increased.
Shukla et al. (34) compared fecal microbiota of patients with IBS (n=47) and HCs (n=30). Three fecal samples were collected from each of the participants. At phylum level, the proportion of Gram-negative bacteria was significantly higher in patients with IBS compared with HCs. At lower taxonomic levels, the abundance of Bifidobacterium species was significantly decreased, whereas several bacteria within firmicutes and proteobacteria were decreased in patients with IBS. Across subgroups of IBS, the microbiota of patients with IBS-D and IBS-M had the most similarities, including a decrease in Lactobacillus and Bacteroides species, compared with those with IBS-C.
Tap et al. (35) aimed to identify a microbiota signature associated with severity of IBS. Fecal samples were collected twice for most patients with IBS (n=110) and once from the HCs (n=39), and colonic biopsies were obtained from 59 of the participants (39 cases and 20 controls). No significant differences were observed between fecal and mucosal microbiota in both HCs and patients with IBS. Regarding microbial composition, Prevotella was significantly increased in HCs, whereas Bacteroides was increased in patients with IBS patients. Further, patients with IBS-D and IBS-M had a significantly higher abundance of Bacteroides compared with those with IBS-C and HCs, whereas the abundance of Prevotella gradually decreased as symptom severity increased. In addition, the microbial richness significantly decreased in line with an increase in Bacteroides.
Comparison of gut microbiota profile in CD, IBS, and NCGS at phylum level
The gut microbiota profile reported in the included trials are compared at phylum level (depicted in Figure 3), and tendencies at lower taxonomic levels are disclosed within the associated phylum.
Figure 3.
Overview of the taxonomic hierarchy of bacteria. Comparison of results in the current review were categorized at phylum level.
Actinobacteria
Two studies reported lower abundance of actinobacteria in CD patients. In fecal samples, Golfetto et al. (23) reported decreased Bifidobacteria, whereas Quagliarello et al. (26) reported decreased abundance at phylum level. Four studies reported altered abundance of actinobacteria in patients with IBS, but the findings were not consistent. Shukla et al. (34) reported decreased Bifidobacteria in fecal samples of patients with IBS in line with the findings of Golfetto et al. (23) on patients with CD (23). A decrease in Bifidobacteria was also detected at a lower taxonomic level by Chung et al (31). In contrast, Rangel et al. (33) reported an increase of actinobacteria in patients with IBS, whereas Halmos et al. (30) found no significant differences.
Bacteroidetes
Two studies reported alterations of bacteroidetes in patients with CD. The results were inconsistent: Quagliarello et al. (26) reported an increased bacteroidetes/firmicutes ratio in the feces of patients with CD compared with HCs. According to Garcia-Mazcorro et al. (28), the abundance of bacteroidetes in feces was similar in patients with CD and NCGS and in HCs. However, the duodenal samples revealed a decrease of bacteroidetes in patients with CD (28). Both studies on NCGS reported on the abundance of bacteroidetes. Dietrich et al. (29) reported a decrease in bacteroidetes in duodenum of patients with NCGS compared with HCs (29). This contrasted with the finding by Garcia-Mazcorro et al. (28), which reported no significant differences between patients with NCGS and CD and HCs (28). Four of the six studies on IBS reported a higher abundance of bacteroidetes among patients with IBS, in feces, jejenum, and in the colonic mucosa (31, 32, 34, 35). However, both Rangel et al. (33) and Tap et al. (35) reported the oppositeat phylum level and within the genus Prevotella, respectively.
Firmicutes
Three studies on CD reported on firmicutes. Quagliarello et al. (26) reported a decrease in firmicutes in feces of patients with CD compared with HCs, including the genus Lactobacillus. However, Garcia-Mazcorro et al. (28) reported that the abundance of firmicutes in fecal samples did not differ between patients with CD and NCGS or HCs. This was supported by Nistal et al. (25) which reported no difference of firmicutes in duodenum of patients with CD and HCs. All though Garcia-Mazcorro et al. (28) reported a similar abundance of firmicutes in feces of patients with CD and NCGS and of HCs, they also reported an increase in firmicutes at lower taxonomic levels in patients with NCGS, both in feces and duodenum. This finding was supported by the results reported by Dietrich et al. (29). Five studies on IBS reported alterations in firmicutes, and the findings were partly contradictory; Pozuelo et al. (32) reported a decreased abundance of firmicutes in feces of patients with IBS compared with HCs. This was not supported by Shukla et al. (34), who reported the opposite. Furthermore, various results were reported at lower taxonomic levels, including altered composition within clostridia and a decrease in clostridiales (33). However, the study conducted by Halmos et al. (30) reported no significant differences.
Proteobacteria
Two studies reported on the abundance of proteobacteria in duodenum of patients with CD, and none of them found significant differences between patients with CD and HCs (25, 27). Interestingly, Herrán et al. (27) reported that P. aeruginosa were only isolated from patients with CD—both treated and untreated. Regarding NCGS, Garcia-Mazcorro et al. (28) reported increased proteobacteria in duodenum of patients with NCGS compared with those with CD and HCs, within the genus actinobacillus.
Three studies reported on proteobacteria in patients with IBS, of which two studies reported increased proteobacteria in feces of patients with IBS (33, 34). One of these studies included an increase of P. aeruginosa, in line with the findings of Herrán et al. in patients with CD (27, 34). In contrast, Chung et al. reported a decrease in proteobacteria at family level in jejenum of patients with IBS (31).
Microbial richness
Regarding microbial diversity and richness, two of the studies on IBS reported that patients with IBS had a reduced microbial diversity compared with HCs (32, 35). This was also reported in two of the studies comparing patients with CD with HCs (25, 26). In addition, the cohort study following treated patients with CD found that patients with symptomatic CD had lower microbial diversity than those with asymptomatic CD (24).
Differences within subgroups of IBS
Across subgroups of IBS, the microbiota of patients with IBS-D and IBS-M had the most similarities. Figure 4 illustrates tendencies within each subtype of IBS. Two studies reported an increase of Bacteroides in patients with IBS-D and IBS-M compared with patients with IBS-C and HCs (34, 35). Furthermore, two studies reported lower abundance of strains within firmicutes in patients with IBS-D and IBS-M compared with patients with IBS-C (32, 34). The latter finding was not supported by Chung et al., who reported an increase in strain within firmicutes in patients with IBS-D compared with HCs (31). Interestingly, Pozuelo et al. reported that the microbiota of patients with IBS-C and HCs did not show significant differences at any taxonomic level (32).
Figure 4.
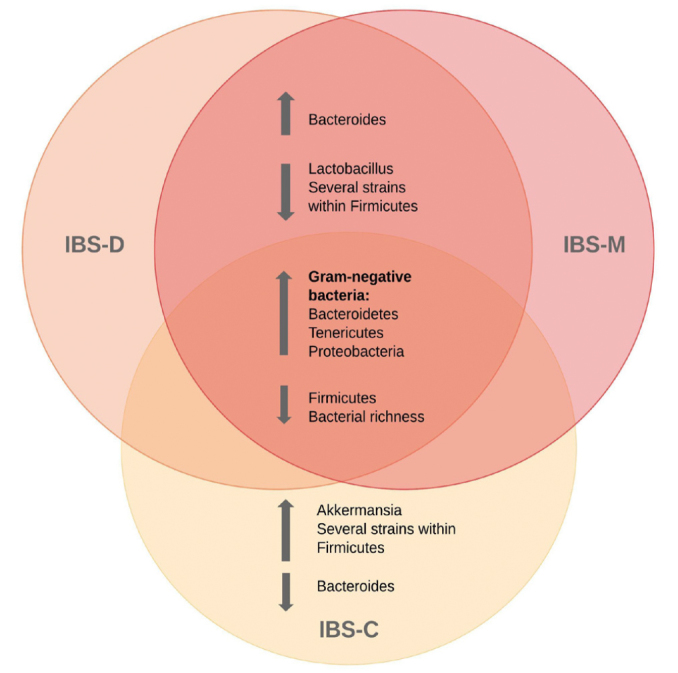
Overview of bacterial strains identified within subgroups of irritable bowel syndrome.
DISCUSSION
To our knowledge, this is the first systematic review aiming to compare the gut microbiota profile in CD, NCGS, and IBS. Although the included studies report inconsistent results, several disease-specific and characteristic trends were identified.
Overall, studies comparing IBS patients to healthy controls suggested higher levels of Gram-negative bacteria, including bacteroidetes, tenericutes, and proteobacteria in IBS (32–35). Furthermore, a decrease within species of firmicutes was reported in three of the included studies (32–34). Previous research has suggested an increase in firmicutes to bacteroidetes ratio in patients with IBS (7, 21), but the results of this review suggests the opposite. However, it is noteworthy that one of the included studies reported increased firmicutes in patients with IBS-D and IBS-M corresponds with previous findings (31). Like the results of previous research, a decrease in Bifidobacteria was reported by one of the studies, but not by the remaining five (34). As for IBS, an increase in proteobacteria was reported in both studies that included patients with NCGS (28). In contrast, this increase was not matched by any other Gram-negative bacteria. Furthermore, an increased abundance of strains within firmicutes was observed in patients with NCGS, which do not overlap with the results of IBS (28, 29). Interestingly, both studies on NCGS reported an increase within the ruminococcaceae family of firmicutes (28, 29). Previous research has reported an increase of ruminococcus, a genus within the ruminococcaceae family, in patients with IBS. Taken together, these findings suggest the ruminococcaceae family as a possible common denominator in NCGS, highlighting an interesting direction for future research (7).
When comparing the microbial profile of patients with CD and IBS, they share the result of a slightly decrease in firmicutes (26). Surprisingly, microbial richness was found to be lower in both patients with CD and those with IBS compared with HCs, but this was not reported in any studies on NCGS (25, 26, 32, 35). Only two studies in patients with NCGS were eligible for inclusion; thus, the comparative basis is somewhat poor. Importantly, some findings indicate that the gut bacterial profile of the participants are unique and independent of disease status (25, 27). The current findings indicate that the gut microbiota profile of patients with CD and IBS share some similarities, however the data are limited.
When interpreting the gut microbiota composition, diet is an important factor. For CD, the only treatment is a strict, life-long GFD (36). A strict GFD is assumed to be necessary for the intestinal microbiota to completely normalize and regain a healthy composition (8). However, it may seem like a GFD allows only a partial recovery of the gut microbiota in CD patients (8). Wacklin et al. have reported significant differences in duodenal microbiota composition between symptomatic and asymptomatic patients with CD. Gut microbiota analysis revealed a significant dysbiosis in the symptomatic patients, which suggests the bacterial dysbiosis does not fully recover in all patients with CD but might still remain after a period of GFD (8, 24). A contradictory theory is that the avoidance of gluten may contribute to further disruption of the intestinal microbiota (37). In summary, there is a need for studies following the same patients’ pre- and post-GFD to evaluate the effect of gluten on the gut microbiota in presence of CD (8, 38). Further, it is hypothesized that NCGS is a dysbiosis-induced disorder. Specific alterations in butyrate-producing bacteria are suggested as a co-causal mechanism for the development of NCGS (37). To date, no specific guidelines are available for the treatment of NCGS. A GFD is most often used for symptom relief (15, 39–41). Since a GFD possibly affect microbiota richness and composition, the avoidance of gluten may contribute to further disruption of the intestinal microbiota in NCGS patients (37). A similar theory applies for IBS, where a low-FODMAP diet is the recommended treatment, and changes in FODMAP intake has been reported to affect the diversity of the gut microbiota (42).
Several issues may explain the lack of consistent findings in the current review. Firstly, the role of gut microbiota in health and disease is fairly undiscovered, and the quality and number of existing articles within the field are limited. Advances in methods of detection over the last years have had a major impact on the field, which make delimitation on year of publication highly relevant (1). Hence, we included only publications from the last five years. Although the results implicate certain similarities in bacterial composition and abundance, the validity of these implications can be discussed. Secondly, the included studies have big differences in design and methodology. The number of participants vary from 18 to 179, which arguably causes differences in the strength of the results. Also, the age of the participants may influence the results. The studies on NCGS and IBS only included adult participants, while one of the five studies on CD was conducted in children. In addition, the review included both observational studies and RCTs. The observational studies do not report on dietary habits. As research have shown similarities in microbiota among subjects of similar age or with common diets (3), such potential confounding factors should be considered. Moreover, methods for assessment of food intake could be a useful tool when investigating gut microbiota.
The included studies analyzed biopsies from the small intestine and colon, as well as fecal samples. When comparing samples from different regions of the gut in healthy individuals, the sites are colonized with different bacterial groups (43). Four of the studies included compared fecal samples and biopsies; while one of them reported no differences between fecal or colonic mucosal microbiota (35), the remaining three reported significant alterations (28, 31, 33). Type of sample material was not specified within the criteria of inclusion for this review. However, this should be an area of comparison in future reviews. Furthermore, the included publications only reported the bacterial proportion of the gut microbiota. As most research performed to date have been focusing mainly on bacteria, the roles of viruses, fungi, and archaea are less known (3). It is possible that important interactions between the different components of the microbiota are somehow missed by such a sharpening of the microbial environment (3, 8, 43).
CONCLUSION
Taken together, the results from the included studies suggest that the bacterial profile of patients with IBS and CD share certain disease-specific trends. Fewer similarities were reported on the bacterial profile of patients with IBS and those with NCGS. The validity of these implications can be discussed because of certain conflicting findings and limited sources of data. Thus, no solid conclusions can be drawn on the basis of these findings alone. However, to our knowledge, no previous publications have compared the gut microbiota profile of patients with CD, NCGS, and IBS. Thus, this review represents an important basis for future research. Future studies should aim to investigate all components of the gut microbiota including bacteria, viruses, fungi, and archaea in order to identify microbial patterns. Considering the potential overlap between NCGS and IBS, an important area of focus should also be to further investigate the existence of NCGS, and potentially establish validated diagnostic biomarkers for the distinction between NCGS and IBS.
MAIN POINTS.
Dysbiosis is associated with a variety of gastrointestinal disorders, including celiac disease (CD) and irritable bowel syndrome (IBS).
The distinction between IBS and non-celiac gluten sensitivity (NCGS) is unclear, and it is conceivable that the gut microbiota profile of these patients may overlap.
We aimed to compare the gut microbiota profile in CD, IBS and NCGS by reviewing the existing literature.
Our findings suggest that the gut microbiota profiles of patients with IBS and CD share certain disease-specific trends differing from those of healthy controls, seen as overall reduced microbial richness.
Few similarities were observed between the gut microbiota profiles of patients with IBS and NCGS.
Footnotes
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - G.A.L., H.F.D.; Design - G.A.L., H.F.D., E.L.T.; Supervision - H.F.D., G.A.L; Data collection and Processing - E.L.T., H.F.D.; Interpretation - G.A.L, H.F.D., E.L.T; Literature search – E.L.T; Writing— E.L.T.; Critical Reviews: G.A.L., H.F.D.
Conflict of Interest: The authors have no conflicts of interest to declare.
Financial Disclosure: The authors declared that this study has received no financial support.
REFERENCES
- 1.D’Argenio V, Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta. 2015;451:97–102. doi: 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Wang ZK, Yang YS. Upper gastrointestinal microbiota and digestive diseases. World J Gastroenterol. 2013;19:1541–50. doi: 10.3748/wjg.v19.i10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017;32:300–13. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schippa S, Conte MP. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6:5786–805. doi: 10.3390/nu6125786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8:143–71. doi: 10.1080/19490976.2016.1278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cukrowska B, Sowinska A, Bierla JB, Czarnowska E, Rybak A, Grzybowska-Chlebowczyk U. Intestinal epithelium, intraepithelial lymphocytes and the gut microbiota - Key players in the pathogenesis of celiac disease. World J Gastroenterol. 2017;23:7505–18. doi: 10.3748/wjg.v23.i42.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointest Liver Physiol. 2017;312:52–62. doi: 10.1152/ajpgi.00338.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marasco G, Di Biase AR, Schiumerini R, et al. Gut Microbiota and Celiac Disease. Dig Dis Sci. 2016;61:1461–72. doi: 10.1007/s10620-015-4020-2. [DOI] [PubMed] [Google Scholar]
- 9.Elli L, Branchi F, Tomba C, et al. Diagnosis of gluten related disorders: Celiac disease, wheat allergy and non-celiac gluten sensitivity. World J Gastroenterol. 2015;21:7110–9. doi: 10.3748/wjg.v21.i23.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Giorgio R, Volta U, Gibson PR. Sensitivity to wheat, gluten and FODMAPs in IBS: facts or fiction? Gut. 2016;65:169–78. doi: 10.1136/gutjnl-2015-309757. [DOI] [PubMed] [Google Scholar]
- 11.Enck P, Aziz Q, Barbara G, et al. Irritable bowel syndrome. Nat Rev Dis Primers. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacy BE, Patel NK. Rome Criteria and a Diagnostic Approach to Irritable Bowel Syndrome. J Clin Med. 2017;6:99. doi: 10.3390/jcm6110099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catassi C, Bai JC, Bonaz B, et al. Non-Celiac Gluten sensitivity: the new frontier of gluten related disorders. Nutrients. 2013;5:3839–53. doi: 10.3390/nu5103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Salhy M, Hatlebakk JG, Gilja OH, Hausken T. The relation between celiac disease, nonceliac gluten sensitivity and irritable bowel syndrome. Nutr J. 2015;14:92. doi: 10.1186/s12937-015-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makharia A, Catassi C, Makharia GK. The Overlap between Irritable Bowel Syndrome and Non-Celiac Gluten Sensitivity: A Clinical Dilemma. Nutrients. 2015;7:10417–26. doi: 10.3390/nu7125541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dale HF, Biesiekierski JR, Lied GA. Non-coeliac gluten sensitivity and the spectrum of gluten-related disorders: an updated overview. Nutr Res Rev. 2019;32:28–37. doi: 10.1017/S095442241800015X. [DOI] [PubMed] [Google Scholar]
- 17.Molina-Infante J, Carroccio A. Suspected Nonceliac Gluten Sensitivity Confirmed in Few Patients After Gluten Challenge in Double-Blind, Placebo-Controlled Trials. Clin Gastroenterol Hepatol. 2017;15:339–48. doi: 10.1016/j.cgh.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Skodje GI, Sarna VK, Minelle IH, et al. Fructan, Rather Than Gluten, Induces Symptoms in Patients With Self-Reported Non-Celiac Gluten Sensitivity. Gastroenterology. 2018;154:529–39. doi: 10.1053/j.gastro.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 19.Chander AM, Yadav H, Jain S, Bhadada SK, Dhawan DK. Cross-Talk Between Gluten, Intestinal Microbiota and Intestinal Mucosa in Celiac Disease: Recent Advances and Basis of Autoimmunity. Front Microbiol. 2018;9:2597. doi: 10.3389/fmicb.2018.02597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losurdo G, Principi M, Iannone A, Ierardi E, Di Leo A. The Interaction Between Celiac Disease and Intestinal Microbiota. J Clin Gastroenterol. 2016;50:145–7. doi: 10.1097/MCG.0000000000000682. [DOI] [PubMed] [Google Scholar]
- 21.Salem AE, Singh R, Ayoub YK, Khairy AM, Mullin GE. The gut microbiome and irritable bowel syndrome: State of art review. Arab J Gastroenterol. 2018;19:136–41. doi: 10.1016/j.ajg.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 23.Golfetto L, de Senna FD, Hermes J, Beserra BT, da Franca FS, Martinello F. Lower bifidobacteria counts in adult patients with celiac disease on a gluten-free diet. Arq Gastroenterol. 2014;51:139–43. doi: 10.1590/S0004-28032014000200013. [DOI] [PubMed] [Google Scholar]
- 24.Wacklin P, Laurikka P, Lindfors K, et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am J Gastroenterol. 2014;109:1933–41. doi: 10.1038/ajg.2014.355. [DOI] [PubMed] [Google Scholar]
- 25.Nistal E, Caminero A, Herran AR, et al. Study of duodenal bacterial communities by 16S rRNA gene analysis in adults with active celiac disease vs non-celiac disease controls. J Appl Microbiol. 2016;120:1691–700. doi: 10.1111/jam.13111. [DOI] [PubMed] [Google Scholar]
- 26.Quagliariello A, Aloisio I, Bozzi Cionci N, et al. Effect of Bifidobacterium breve on the Intestinal Microbiota of Coeliac Children on a Gluten Free Diet: A Pilot Study. Nutrients. 2016;8:E660. doi: 10.3390/nu8100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herran AR, Perez-Andres J, Caminero A, et al. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res Microbiol. 2017;168:673–84. doi: 10.1016/j.resmic.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Mazcorro JF, Rivera-Gutierrez X, Cobos-Quevedo OJ, et al. First Insights into the Gut Microbiota of Mexican Patients with Celiac Disease and Non-Celiac Gluten Sensitivity. Nutrients. 2018;10:1641. doi: 10.3390/nu10111641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dieterich W, Schuppan D, Schink M, et al. Influence of low FODMAP and gluten-free diets on disease activity and intestinal microbiota in patients with non-celiac gluten sensitivity. Clin Nutr. 2019;38:697–707. doi: 10.1016/j.clnu.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Halmos EP, Christophersen CT, Bird AR, Shepherd SJ, Gibson PR, Muir JG. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93–100. doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- 31.Chung CS, Chang PF, Liao CH, et al. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol. 2016;51:410–9. doi: 10.3109/00365521.2015.1116107. [DOI] [PubMed] [Google Scholar]
- 32.Pozuelo M, Panda S, Santiago A, et al. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep. 2015;5:12693. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangel I, Sundin J, Fuentes S, Repsilber D, de Vos WM, Brummer RJ. The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment Pharmacol Ther. 2015;42:1211–21. doi: 10.1111/apt.13399. [DOI] [PubMed] [Google Scholar]
- 34.Shukla R, Ghoshal U, Dhole TN, Ghoshal UC. Fecal Microbiota in Patients with Irritable Bowel Syndrome Compared with Healthy Controls Using Real-Time Polymerase Chain Reaction: An Evidence of Dysbiosis. Dig Dis Sci. 2015;60:2953–62. doi: 10.1007/s10620-015-3607-y. [DOI] [PubMed] [Google Scholar]
- 35.Tap J, Derrien M, Tornblom H, et al. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology. 2017;152:111–23. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 36.Mocan O, Dumitrascu DL. The broad spectrum of celiac disease and gluten sensitive enteropathy. Clujul Med. 2016;89:335–42. doi: 10.15386/cjmed-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leccioli V, Oliveri M, Romeo M, Berretta M, Rossi P. A New Proposal for the Pathogenic Mechanism of Non-Coeliac/Non-Allergic Gluten/Wheat Sensitivity: Piecing Together the Puzzle of Recent Scientific Evidence. Nutrients. 2017;9:1203. doi: 10.3390/nu9111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnareddy S. The Microbiome in Celiac Disease. Gastroenterol Clin North Am. 2019;48:115–26. doi: 10.1016/j.gtc.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Aziz I, Dwivedi K, Sanders DS. From coeliac disease to noncoeliac gluten sensitivity; should everyone be gluten free? Curr Opin Gastroenterol. 2016;32:120–7. doi: 10.1097/MOG.0000000000000248. [DOI] [PubMed] [Google Scholar]
- 40.Catassi C, Elli L, Bonaz B, et al. Diagnosis of Non-Celiac Gluten Sensitivity (NCGS): The Salerno Experts’ Criteria. Nutrients. 2015;7:4966–77. doi: 10.3390/nu7064966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lionetti E, Pulvirenti A, Vallorani M, et al. Re-challenge Studies in Non-celiac Gluten Sensitivity: A Systematic Review and Meta-Analysis. Front Physiol. 2017;8:621. doi: 10.3389/fphys.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi SL, Correa D, Pak SC. Probiotics, prebiotics, and low FODMAP diet for irritable bowel syndrome - What is the current evidence? Complement Ther Med. 2019;43:73–80. doi: 10.1016/j.ctim.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–90. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]