Abstract
Background:
Although the implementation of systematic review and evidence mapping methods stands to improve the transparency and accuracy of chemical assessments, they also accentuate the challenges that assessors face in ensuring they have located and included all the evidence that is relevant to evaluating the potential health effects an exposure might be causing. This challenge of information retrieval can be characterized in terms of “semantic” and “conceptual” factors that render chemical assessments vulnerable to the streetlight effect.
Objectives:
This commentary presents how controlled vocabularies, thesauruses, and ontologies contribute to overcoming the streetlight effect in information retrieval, making up the key components of Knowledge Organization Systems (KOSs) that enable more systematic access to assessment-relevant information than is currently achievable. The concept of Adverse Outcome Pathways is used to illustrate what a general KOS for use in chemical assessment could look like.
Discussion:
Ontologies are an underexploited element of effective knowledge organization in the environmental health sciences. Agreeing on and implementing ontologies in chemical assessment is a complex but tractable process with four fundamental steps. Successful implementation of ontologies would not only make currently fragmented information about health risks from chemical exposures vastly more accessible, it could ultimately enable computational methods for chemical assessment that can take advantage of the full richness of data described in natural language in primary studies. https://doi.org/10.1289/EHP6994
Introduction
Chemical assessment has seen significant improvement in the validity and utility of its outputs over the last few decades, in parallel with the introduction of an increasing variety of open-source and online tools and resources that facilitate communication, flexibility, access to information, and inclusiveness of scope (NRC 2007). However, further gains in the quality and inclusivity of chemical assessment are being challenged by exponential growth in the volume of risk-relevant research being published and a burgeoning array of innovative study designs being developed by scientists for investigating health risks from chemical exposures. All this data has to be found, assembled into logical cause–effect frameworks, and evaluated as to what it all means for health risks from chemical exposures. Continued improvement of chemical assessment outputs therefore hinges on the development of new methods for data acquisition, and the rapid, reproducible, and reusable identification of old and new scientific information (Watford et al. 2019).
In parallel to the increasing diversity, volume, and complexity of toxicological research has been the development of systematic methods for reviewing (Woodruff et al. 2011; Whaley et al. 2016; Hoffmann et al. 2017) and mapping (Walker et al. 2018; Wolffe et al. 2019) evidence relevant to assessing health risks posed by exposure to chemical substances. Although systematic methods improve the transparency and accuracy of chemical assessment products, they also accentuate the challenge of locating, evaluating, and integrating the many types of study design that provide evidence for the health effects that an exposure might be causing. This commentary provides an overview of systematic methods for literature-based chemical assessments, presents the authors’ views of the challenges that current approaches to reporting and organizing toxicological research present to its systematic aggregation and analysis, and makes a series of recommendations for developing general “knowledge organization systems” for environmental health, all of which would enable more comprehensive application of systematic methods for assessing health risks posed by exposure to chemical substances.
Systematic Methods in Chemical Assessments
One of the major methodological innovations in chemical assessment over the last decade has been the introduction of systematic methods for exploring and synthesizing evidence. Systematic methods componentize the evidence assessment workflow, dividing it into a modular sequence of steps (Figure 1). The approaches fall into two broad categories: systematic reviews and systematic evidence maps. Systematic approaches are considered an advance on traditional, expert-based narrative approaches to summarizing evidence because they use explicit, discussable methods in each component, allowing the validity of decisions to be scrutinized, assessed, and improved on (Garg et al. 2008).
Figure 1.
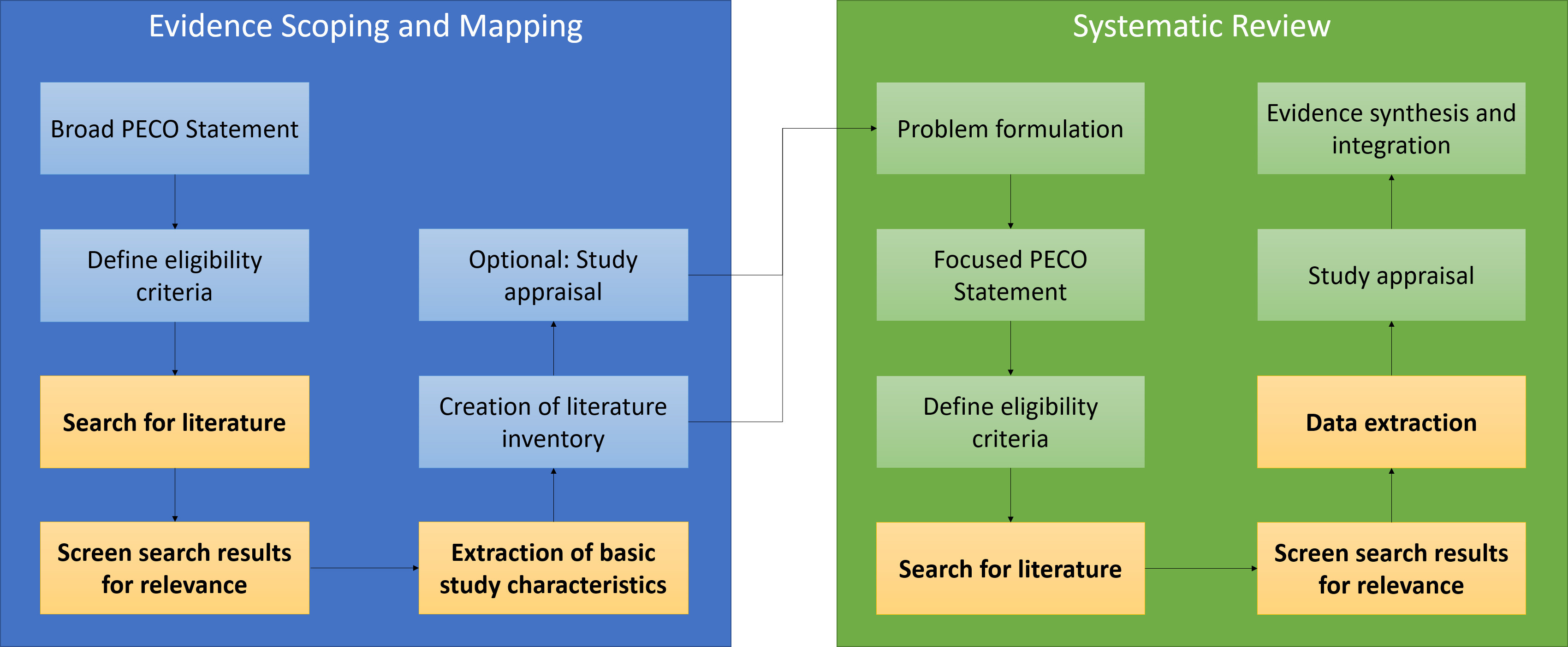
The relationship between the processes involved in systematically mapping and systematically reviewing evidence. The elements where what we call the “information retrieval challenge” comes into play are highlighted in bold and yellow. Comprehensive evidence maps, if they represent complete inventories of the literature, should ultimately obviate the need for additional literature searches in systematic reviews conducted in response to the findings of a systematic evidence mapping exercise.
Systematic Reviews
Systematic review (SR) has traditionally been defined as “attempts to identify, appraise and synthesize all the empirical evidence that meets prespecified eligibility criteria to answer a specific research question” and use “explicit, systematic methods that are selected with a view aimed at minimizing bias” (Higgins et al. 2019). We favor defining it as a methodology for testing a research hypothesis using existing evidence that employs techniques intended to minimize random and systematic error and maximize transparency of decision-making. Either way, SR breaks the evidence assessment process down into discrete steps of specifying objectives, defining search strategies and eligibility criteria, appraising the validity of each individual included study, synthesizing the evidence using quantitative and narrative techniques as appropriate, and assessing certainty in the results of the synthesis (Institute of Medicine 2011; Higgins et al. 2019; Whaley et al. 2020). Each step is thoroughly documented so the reader can assess the validity of each judgement being made by the reviewers as they move from stating their research objective through to providing their final conclusions.
Although there have been several historical precursors to the approach, SR methods as currently recognized were first formally introduced in the health care and social sciences in the late 1980s and early 1990s (Chalmers et al. 2002). Since then, SR has become a fundamental technique for evaluating existing evidence of the efficacy of interventions in health care, education, criminal justice, and other fields (Farrington and Ttofi 2009; Braga et al. 2012; Roberts et al. 2017).
The potential value of SR methods for similarly advancing toxicology and chemical risk assessment was first mooted in the published literature around the mid-2000s (Guzelian et al. 2005; Hoffmann and Hartung 2006). By 2014, the first SR frameworks for chemical risk assessment had been published (European Food Safety Authority 2010; Rooney et al. 2014; Woodruff and Sutton 2014), with subsequent rapid uptake from regional (Schaefer and Myers 2017), national (Yost et al. 2019), and international agencies (Descatha et al. 2020; Orellano et al. 2020).
Systematic Evidence Maps (SEMs)
SR methods function best when responding to focused questions posed in “confirmatory mode” research contexts (Nosek et al. 2018), where researchers are testing a hypothesis or quantifying a specific exposure–outcome relationship using existing evidence in lieu of conducting an experiment. However, many research contexts are not confirmatory but exploratory, generating new hypotheses that might need to be tested and identifying novel issues that may warrant further investigation. In these contexts the methods of SR, developed for narrowly defined questions, rapidly become unwieldy and demand interrogation of evidence at a level of detail at odds with the broader objectives of an exploratory research exercise (Leenaars et al. 2020; Radke et al. 2020. In response to the limitations of SR methods for exploratory research, systematic evidence maps (SEMs), also known as “evidence maps” or “systematic maps” have been developed.
SEMs are designed to apply the same principles of comprehensiveness and transparency as SR; however, instead of answering specific research questions, they produce queryable databases of evidence that catalog research of relevance to an open question, theme, or policy area, which has been developed to support a broad range of decision-making contexts (James et al. 2016, Wolffe et al. 2020). In a chemical assessment, the characteristics summarized in an SEM will vary depending on decision-making context but will usually consist of study type, chemical or test substance, population, outcome, summary results, and (potentially) indicators of the validity of a study. This is much less information than required for an SR, with the bare minimum of information required for priority-setting being extracted and stored in the map database. The resulting inventory of studies and findings allows a user to make screening-level decisions based on regulatory needs, outcomes of regulatory concern, research questions, and so forth.
In essence, SEMs are the application of systematic methods to scoping reviews (Wolffe et al. 2019), providing an evidence-based approach to deciding when to conduct new SRs (e.g., when a confluence of sufficiently high-quality evidence suggests a need for a regulatory exposure limit to be revised), new primary studies (e.g., when sufficiently high-quality data required for a decision may be absent), or not do anything at all (e.g., when a new confluence of data would not lead to a change in exposure values). Although SEMs are one of the newest innovations in evidence synthesis methods, they are already seeing uptake in the environmental and social sciences (Cheng et al. 2019), environmental economics (Fagerholm et al. 2016), and health care (El Idrissi et al. 2019), among others. Examples from environmental health include SEMs of evidence for transgenerational inheritance of health effects from environmental exposures (Walker et al. 2018), health effects of exposure to acrolein (Keshava et al. 2020), protocols for health effects of per- and polyfluoroalkyl substances (PFAS) exposure (Pelch et al. 2019), and interventions to reduce traffic-related air pollution (Sanchez et al. 2020).
The Information Retrieval Challenge
Systematic methods are a natural fit for many chemical assessments, providing a mechanism for meeting the expectation that an assessment fully and transparently uses all relevant evidence in the course of analyzing health risks posed by exposure to chemical substances (NRC 2014). In our experience, however, in spite of the contribution made by the various online research platforms, databases, and indexing systems that have emerged over the last three decades, the extent to which evidence relevant to chemical assessments can be systematically accessed remains heavily constrained by current approaches to storing and cataloging scientific knowledge. These issues are systemic, constraining what can be retrieved even by the best search strategies and most expert information specialists.
The formal record of scientific research is almost exclusively the written study report. Researchers report their methods and findings in manuscripts that are published in scientific journals. These documents are stored in multiple siloed databases and are retrieved using complex and sensitive queries that require detailed understanding of the varying data schemas and search interfaces employed by each database. Because each database is siloed, covers different areas of the total literature, and stores documents in its own unique manner, these searches have to be redesigned and reconducted multiple times to ensure all relevant documents are retrieved. The searches also return a large proportion of false-positive results that have to be screened out to identify the documents of true relevance to the objectives of the reviewing or mapping exercise. Then the data in the relevant documents has to be manually read and extracted into an appropriate format for analysis in the SR or SEM.
The result is a lengthy location and extraction process that may still inadvertently exclude potentially large numbers of relevant records because of the “streetlight effect.” This is the phenomenon by which research tends to be conducted in established areas of understanding, rather than around novel ideas (Kaplan 1973; Battaglia and Atkinson 2015). Although there are multiple causes of the streetlight effect, database queries are affected in two principal ways that are relevant to our discussion. First, in most databases, the content of stored documents is represented using only a relatively limited selection of keywords in comparison to the full set of concepts actually discussed in the documents in question, plus the words in the title and abstract. This means that queries can only retrieve certain results: a) records where the search terms happen to be concepts deemed by the database designers as important enough to be cataloged in the database’s keywords and b) records where the search terms happen to match the words used by the authors in the manuscript’s title, abstract, and author keywords. Second, only information known by the searchers or coded into the database as conceptually related to the research problem can be retrieved.
Overcoming the streetlight effect and quickly and accurately locating and extracting the relevant data in scientific documents is what we refer to as the “information retrieval challenge.” In setting out this challenge and how it can be addressed, we first describe the difficulties that retrieving information in written documents presents to developers of databases. We explain this in terms of two root factors that we designate as the “semantic” and “conceptual” factors in information retrieval. The “semantic factor” concerns how natural variation in language presents an obstacle to identifying relevant research, whereas the “conceptual factor” concerns how limits in knowledge of the conceptual relations between research topics make it difficult to access research documents that are related to, but not directly about, the immediate topic of interest.
The Semantic Factor
The language that scientists use to describe their work can be quite varied, with researchers using different words for the same things (synonyms) and the same words for different things (homographs and polysemes). Because meaning is a function of the relationships between words and the context in which they are presented (Gasparri and Marconi 2019), scientists can even use incorrect words to describe their activities and still successfully get their meaning across to a sufficiently fluent reader.
The flexibility of language allows it to evolve over time and enables us to use familiar words to talk about new things in our changing physical and intellectual worlds (Sorensen 2018). However, in our experience this variation and evolution in natural language also presents significant challenges to the information retrieval process: Not only do databases have to be engineered to accommodate such variation, but because approaches to accommodating the variation differ from one database to the next, to design searches that maximize the amount of relevant literature being retrieved, a database user has to be aware of both the variation in the way language is being employed by authors of the documents in which they are interested and how this variation is handled by the database itself.
This is why complex search strings are typically used in querying research databases to cover the many different ways of expressing the same concepts. It is also why the strings are different for each database: There is no one correct way of solving the problem of variation in language, just different optimizations; hence, the designers of each database end up implementing different solutions fashioned according to different priorities depending on the database’s intended use.
If an information retrieval strategy does not include all the words that have been or are being used for the concepts of interest in a way which responds to the individual characteristics of the database being searched, then relevant documents will be overlooked (Salvador-Oliván et al. 2019). This is one of the reasons information specialists are needed for SR projects (Rethlefsen et al. 2015). An example of how linguistic variation can affect the number of results retrieved for a search concept is illustrated in Table 1, where different terms for the same concept can return different results within and across databases.
Table 1.
Demonstration of how variation in language used by study authors in title, abstract, and author keywords fields affects search results in PubMed. Database syntax is used to ensure the phrase entered is the exact one being searched for. Date of searches: 15 July 2020.
| PubMed Query | Results |
|---|---|
| “PAHs” [Title/Abstract] OR “PAHs” [Other Term] | 15,912 |
| “PAH” [Title/Abstract] OR “PAH” [Other Term] | 22,605 |
| “polycyclic aromatic hydrocarbon” [Title/Abstract] OR “polycyclic aromatic hydrocarbon” [Other Term] |
4,545 |
| “aromatic polycyclic hydrocarbons” [Title/Abstract] OR “aromatic polycyclic hydrocarbons” [Other Term] |
59 |
| “polycyclic aromatic hydrocarbons” [Title/Abstract] OR “polycyclic aromatic hydrocarbons” [Other Term] |
19,311 |
The Conceptual Factor
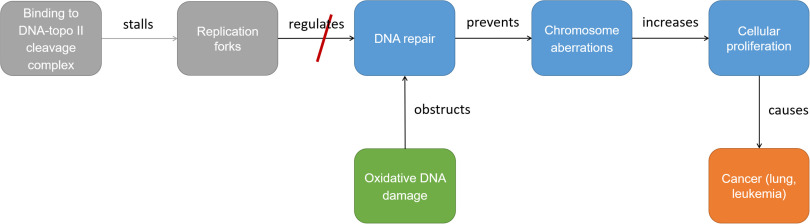
For any given domain of interest, there will be an expansive network of related concepts and subconcepts of relevance to a SR or SEM exercise. Having a complete map of the relationships among these concepts is necessary if the full body of assessment-relevant information is to be retrieved (Figure 2); however, expert knowledge is finite, which means that important relationships outside the knowledge sphere of the expert conducting a review are always at risk of being missed. This risk is illustrated in Figure 2. Here, an expert might be aware that DNA strand breaks are related to inadequate DNA repair, and the expert is consequently able to include exposures that increase oxidative DNA damage in a cancer assessment. However, if the expert is not aware that DNA strand breaks are also related to the collapse of stalled replication forks, then research into this event and others that are related to it may be overlooked in the assessment.
Figure 2.
Illustration of how lack of knowledge of relations between concepts relevant to a research topic can result in evidence of potential importance to a given question being overlooked. In this example, awareness that DNA repair is obstructed by oxidative DNA damage allows lung cancer and leukemia to be connected to stressors that cause oxidative DNA damage to be incorporated into a cancer assessment. However, lack of awareness that replication forks regulate DNA repair may result in studies of stressors that stall replication forks by binding to cleavage complexes being excluded from cancer assessments.
Other examples of conceptual relationships that are often of relevance in synthesizing evidence to answer a research question but might not be known to researchers conducting a SR or SEM exercise include a) comparable chemicals, where the known effects of exposure to one substance can be informative of the potential effects of another; b) biologically comparable species, where an animal might serve as a better model for a disease process in humans than another animal model; and c) surrogate outcomes, where an upstream biomarker of health effects might be a strong predictor of a final health outcome. Although this evidence is indirectly relevant to an assessment, it is nonetheless conceptually related to it and therefore potentially informative of its conclusions. Some of these conceptual relationships will be known and some speculative; however, unless they are accounted for in an information retrieval strategy, evidence of potential importance for answering a given question may be overlooked.
The conceptual factor is fundamental to the streetlight problem in information retrieval: Without measures to augment search terms with related concepts, search strategies can only find information on concepts that the searcher already knows to be related to those specified in the research question. Addressing the challenge of finding what is relevant, but not necessarily known by the searcher as relevant, is a central element of modern information retrieval strategies.
The Conceptual and Semantic Factors in SRs and SEMs
One strategy for addressing the conceptual factor in the information retrieval challenge, which at least ensures saturation of concepts in relation to a research question, is simply to narrow the topic of the review. This is fundamental to current practice in SR, whereby a tightly focused research objective is a common recommendation (Institute of Medicine 2011; Morgan et al. 2018). Such focus means fewer concepts need to be covered in an information-retrieval strategy, thereby helping to ensure that a finitely resourced research project provides comprehensive coverage of the topics that must be included to answer its question.
The problem with topic-narrowing as a strategy is that it deliberately excludes evidence that may be relevant to the review question on the assumption that the excluded evidence is going to be insufficiently informative to materially alter the conclusions of the review. This assumption may be reasonable for SRs where the knowledge objective is very specific. However, it is much less available as a strategy for SEM exercises, where the purpose is to map domain topics and the evidence associated with them in a broad thematic or policy area rather than in relation to a specific question (Miake-Lye et al. 2016; Saran and White 2018).
Whether narrow or broad, the same structural issue is confronted in SRs and SEMs. Researchers need to access a universe of information, but because they only know or are able to recall a certain proportion of terms for and linkages between concepts, they have only partial access to the full universe of information they might need. This situation can be improved by groups of experts working together using effective knowledge elicitation strategies; however, their view of the evidence will still be biased by what they can collectively access. The streetlight might be larger, but it still offers only partial illumination. To allow movement across conceptual linkages that are unknown to particular individuals or groups requires systems that make those linkages accessible without the end user having to be aware of them.
Following work by Hodge and Digital Library Federation (2000), we call these overarching technologies and structures for providing access to information “Knowledge Organization Systems” (KOS) and discuss how their evolution, particularly the introduction of ontologies, is fundamental to the ongoing modernization of chemical assessments.
Knowledge Organization Systems
Here, we discuss three KOS technologies: controlled vocabularies, thesauruses, and ontologies. Although controlled vocabularies and thesauruses are KOS technologies already well-established in chemical assessment, the value of a broader adoption of ontologies is highlighted.
Controlled Vocabularies
A controlled vocabulary (CV) is a defined list of words and phrases used to tag content in a database to make that content retrievable via navigation or search (Pomerantz 2015). It is a type of metadata (data about data) that provides an interpretive layer between the user of a database and the content in the database. CVs can be used in tools that expand, translate, or map user queries to the terminology used to classify content in the database, and sometimes to map additional entry terms (synonyms) that the user may not have applied but the CV defines as being semantically equivalent to the terms in the user query (Ashburner et al. 2000; Stearns et al. 2001; Fragoso et al. 2004).
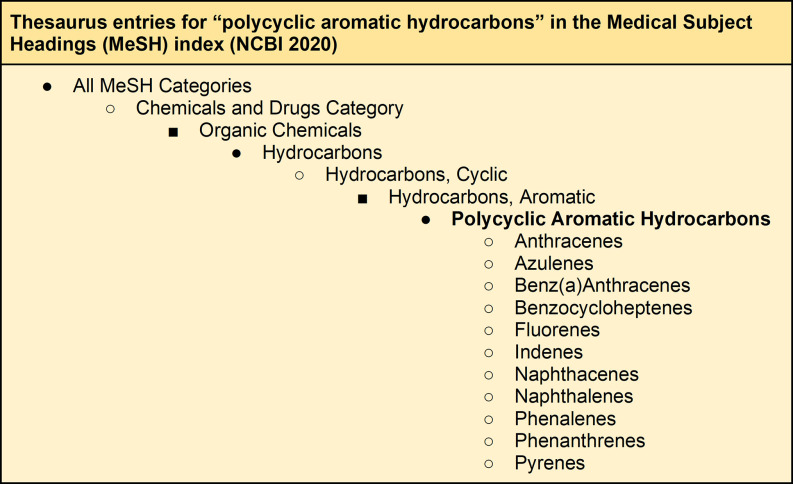
In its simplest form, a CV is a consistent labeling system in which the same concept is always given the same name (e.g., “PAH,” “polycyclic aromatic hydrocarbons,” “polycyclic aromatic hydrocarbon,” “polycyclic aromatic hydrocarbons,” “aromatic polycyclic hydrocarbons,” “polycyclic aromatic hydrocarbons” can be understood to mean the same chemical). In a database that tags all records about a concept with the same CV label, the user is able to retrieve all documents known to the system as discussing that concept, independent of the author’s terminology, simply by searching for the CV label (“polycyclic aromatic hydrocarbons”). The CV allows the user to do this without needing to specify each individual synonymous term, the full range of which the user may not have access to. This utility is illustrated by the CV terms of the Medical Subject Headings (MeSH) used to index research in the Medline database (see Figure 3). We have ourselves applied a controlled Environmental Health Vocabulary (EHV) in the EPA Health Assessment Workplace Collaborative (HAWC) that is freely accessible at the following url: https://hawcprd.epa.gov/assessment/100000039/.
Figure 3.
The MeSH CV entry for “polycyclic aromatic hydrocarbons,” 21 July 2020.
CVs are one approach to addressing the semantic factor in information retrieval, increasing the recall of queries by augmenting users’ search terms with a set of synonyms. They can also improve the precision of a query by disambiguating word senses (e.g., “bank” as a mound of earth, rather than as a place to deposit money) and reducing false positives (a paper about the use of pesticides in the home will not be indexed as occupational exposure). CVs can, however, reduce recall if the user is expecting to find a concept not included in the CV, if indexers (human or machine) fail to assign relevant terms, or if some records are not tagged with CV terms at all.
The main limitation of CVs, in terms of their function as part of a KOS, is that they capture only one type of logical relationship between concepts, i.e., an equivalence relation where one thing is defined as being the same as another thing (e.g., PAH means polycyclic aromatic hydrocarbons) (W3C 2020). Although capture of synonyms that are unknown to a system user is valuable, there are other types of relationship that, if they can be built into a KOS, go further in overcoming the semantic and conceptual factors in information retrieval.
Thesauruses
Thesauruses expand beyond the equivalence relation of synonymy by introducing an overarching conceptual hierarchy in which the CV terms are organized and related. Such hierarchies are valuable for a KOS because they allow information consisting of related but nonequivalent concepts to be defined as relevant to a user’s search term (Pomerantz 2015). By organizing concepts in terms of how they are related, rather than simply in terms of when two words or strings refer to the same concept, the introduction of a thesaurus begins to address the conceptual factor in information retrieval.
An illustration is the MeSH thesaurus, which organizes MeSH CV terms in a parent-child hierarchy (see Figure 4). This “is a class of” type of logical relationship can be exploited for greater recall in search results than is allowed for by an equivalence relation. For example, a PubMed search for “polycyclic aromatic hydrocarbons” using MeSH headings will return citations that have not only been indexed with terms synonymous with polycyclic aromatic hydrocarbons but also that contain terms that are subclasses thereof, such as anthracenes, fluorenes, and pyrenes.
Figure 4.
The MeSH thesaurus entries for “polycyclic aromatic hydrocarbons,” 21 July 2020. For brevity, only first-level entries are shown.
This search result is not possible in a CV alone because although a pyrene is a type of polycyclic aromatic hydrocarbon, it is not equivalent to one: It is false to state “pyrene means the same as polycyclic aromatic hydrocarbon.” Because CVs are restricted to the equivalence relation, they have no mechanism to describe the relationship between pyrenes and polycyclic aromatic hydrocarbons and therefore have to treat them as unrelated entities. When being queried, a system employing only a CV thus requires the user to enter terms for each subclass of polycyclic aromatic hydrocarbons. Continuing with the example of polycyclic aromatic hydrocarbon and pyrenes, if the user does not know all the subclasses, then citations that are about pyrenes but do not use the term “polycyclic aromatic hydrocarbon” would be missing from the search results, even though they are relevant to the user’s information needs.
In developing a more comprehensive taxonomy of the concepts that have been labeled by the CV, simply by adding the “is a class of” relationship via a thesaurus, MeSH greatly increases the conceptual coverage of a user’s search for PAHs without the user needing to account for related, but not synonymous, terms in their search.
Ontologies
Thesauruses, as hierarchical taxonomies, are a powerful strategy in KOS development. When implemented comprehensively and fully exploited by a user, they make a significant contribution to addressing the semantic and conceptual factors in the information retrieval challenge. However, being able to codify more information about the relationships between the concepts of the CV than simple hierarchies can further increase the information retrieval capacity of a KOS. After all, there are many more types of relationships than “is a class of,” however powerful that relationship is as a general organizing principle.
When a taxonomy moves beyond a hierarchy toward a representation of the properties of and the relations between concepts, it becomes an ontology. An ontology is a formal method for representing knowledge, usually within a particular knowledge domain, that relates terms or concepts to one another in a format that supports reading and searching, not only for the terms themselves but also for the relationships between those terms (Whetzel et al. 2011). Using an ontology allows knowledge to be stored in a mathematical graph, which is a well-studied structure that has many useful properties in terms of searching and/or querying.
Returning to our example of cancer and DNA damage, Figure 2 provides a visual representation of the richer way in which an ontology can relate concepts to each other in a graphical schema, with concepts (nodes) related to each other via edges. The ontology is not restricted to being hierarchical, because both nodes (the things in the database) and the edges between nodes (the relationships between them) can be the object of a controlled vocabulary and carry semantic value. This lack of restriction allows highly specific relationships such as “stalls” and “regulates” to be represented in the KOS, enabling information about those relationships, or things related by those relationships, to be retrieved. Queries can be written that trace a path through the graph, in principle returning information about, e.g., replication forks and oxidative damage in relation to DNA repair, whether or not the user is aware of any relationships between the concepts.
Building an Ontologized KOS
A KOS that incorporates ontologies can be used for much more complex information retrieval tasks than one that only incorporates thesauruses, because the ontologized KOS is able to represent complex connections between units of information. This is particularly valuable for making systematically accessible information that is indirectly related to an exposure–outcome relationship of concern but nonetheless informative for a chemical assessment.
The challenge with the development and implementation of ontologies is how they present a dilemma in that, although they provide a formal way of representing knowledge in a domain, they rely on the existing knowledge within that domain to determine how that knowledge is organized: The system needs to be known in order to be described yet needs to be described in order to be known. We now use the development of Adverse Outcome Pathways (AOPs) as an example to illustrate this challenge of building ontologies and indicate how it can be solved.
Adverse Outcome Pathways as an Example of an Ontologized KOS
AOPs are a way of formalizing the steps by which a disease or injury progresses from exposure through to final adverse outcome via increasing levels of biological complexity (Knapen et al. 2018). The AOP framework was designed to provide a consistent, generic, and chemical-independent description of toxicological mechanisms across differing levels of biological organization and to make clear the gaps in our knowledge concerning these mechanisms. They are of interest in chemical assessments because they provide a means of integrating data across different assays targeting varied components of a biological system and allow a user to organize the available evidence to more fully interrogate potential cause–effect relationships and identify data gaps. As such, they provide a means for incorporating mechanistic data into chemical assessments (Vinken 2013).
Specific interpretations vary because the concept is still under development, but AOPs are essentially logic models that connect an initial exposure to an outcome via a sequence of biological events (Villeneuve et al. 2018). As illustrated in Figure 5, the sequence of events begins with a Molecular Initiating Event (MIE), where a stressor initiates a biological change at the molecular level in a cell in an organism. Activation of the MIE initiates progression through a sequence of Key Events (KEs) occurring at increasing levels of biological complexity—from subcellular to cellular to organ, to whole organism, to population. The final event in the chain of Key Events is the Adverse Outcome (AO).
Figure 5.
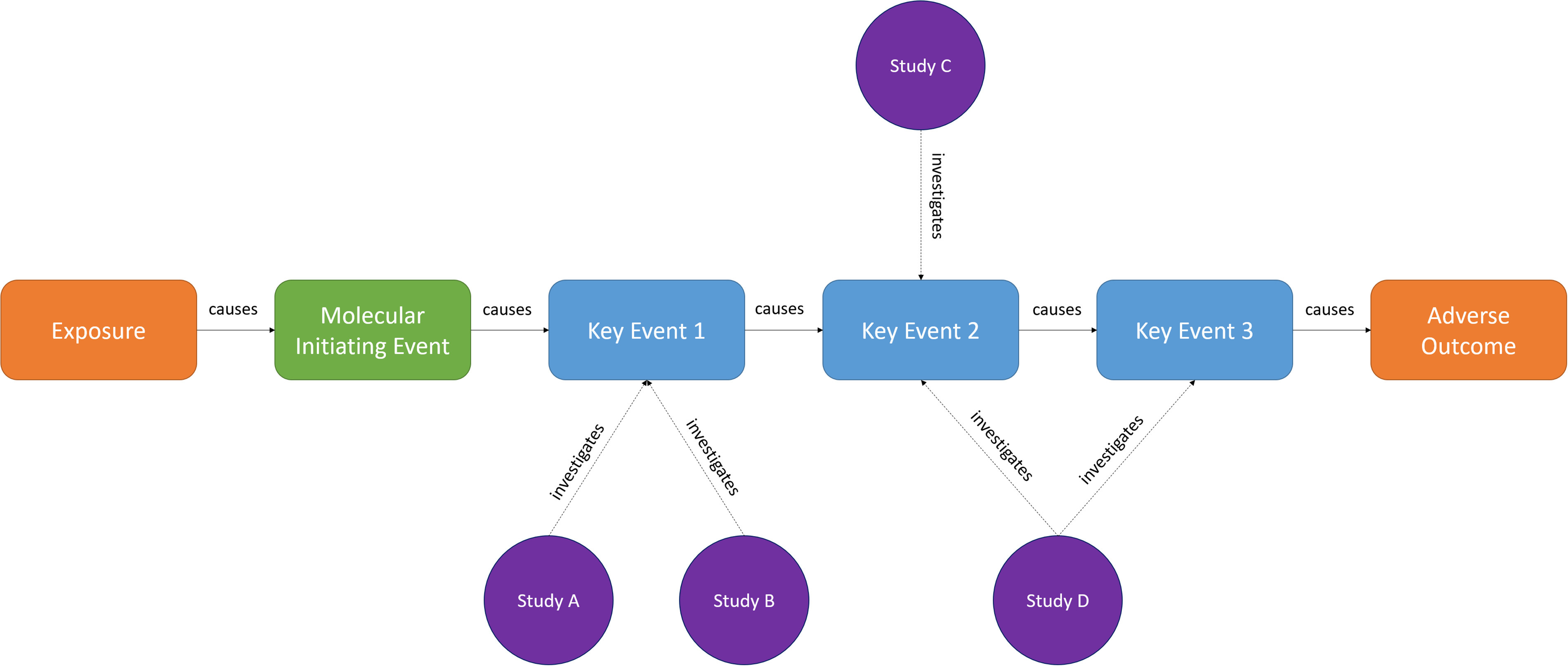
The elements of an Adverse Outcome Pathway, whereby an exposure causes a Molecular Initiating Event, initiating a biological sequence of causally related Key Events that result in a final Adverse Outcome being manifest. Experimental research can target how a challenge might affect a Key Event (Studies A, B, and C) or how one Key Event might cause another Key Event in a Key Event Relationship (Study D). Arranging biological events, exposures, and the evidence around them in these sorts of AOP chains can be very valuable for integrating mechanistic evidence into chemical assessments but requires knowledge organization systems that reflect the complexity and heterogeneity of the relationships and event types.
Although nascent, the AOP framework is an example of an ontologized KOS. By connecting relevant literature to KEs and connecting KEs to each other using logical relationships (known as “Key Event Relationships” or KERs), an AOP allows the full evidence space around an exposure–outcome relationship to be accessed from any single entry-point by traversing across KERs (either upstream or downstream). This allows system users who lack any prior knowledge of the AOP to access connected evidence within that space. For example, as shown in Figure 3, it is possible to move upstream from the AO to Assays A and B via KEs 3, 2, and 1—thereby incorporating information in the chemical assessment that might otherwise have been excluded by searches or inclusion criteria focusing on the AO alone (NRC 2007; Schwarzman et al. 2015).
How the Conceptual and Semantic Factors Challenge the Building of AOPs
Contemporary methods for development of AOPs rely exclusively on human expert knowledge of the mechanisms and biological pathways from which the AOP is ultimately derived. As far as we are aware, every registered AOP has been developed manually. As such, AOPs lack transparency and are highly vulnerable to both the semantic and conceptual factors in information retrieval, and therefore unlikely to be based on an evaluation of the complete evidence base that is relevant to their development.
Currently, an AOP author will define the key events associated with an AOP based on their expert knowledge of the mechanisms by which one or more prototypical stressors causes an adverse outcome. The author ties assays and biomarkers that are associated with each of the steps leading toward the adverse outcome of interest to the underlying biological events they represent. The AOP author then assembles the literature that supports the linkages between each pair of events and evaluates the overall strength of the evidence supporting each linkage, based on guidance provided by the OECD AOP Development Program (OECD 2018).
In theory, it should be possible to develop AOPs using systematic methods. We are currently involved in an effort under the Extended Advisory Group on Molecular Screening and Toxicogenomics (EAGMST) Handbook Guidance, Gardening, and Internal Review (HGGIR) scoping the practical application and feasibility of doing so. We envision a process that involves systematically mapping the scientific literature to develop a model of the current known biology and to identify candidate KEs, then using SR methods to evaluate the relationship between each pair of candidate KEs, considering the upstream key event as the “exposure” and the downstream key event as the “outcome.” Those candidate KEs that attain a sufficiently high level of certainty as being causally related would be elevated to formal KEs and become part of the approved AOP.
However, this process is severely challenged by the breadth of knowledge required to fully understand an entire toxicological pathway, covering literature from molecular, physiological, clinical, and epidemiological domains. Given the overwhelming number of publications in the scientific literature, with over 800,000 citations being added per year to MEDLINE (National Library of Medicine 2020), it seems impossible for a small group of experts to be fully aware of the complete evidence base and, therefore, the entire universe of biological concepts relevant to an AOP from across all related knowledge domains.
The problem is that this map-and-review approach is not practically feasible. Literature databases currently represent only a minority of AOP concepts in their controlled vocabularies, whereas representation of the relationships between the concepts is more limited still. Although these issues can to some degree be mitigated by running large numbers of complex, iterated searches that spider out to related concepts and terms for those concepts, such searches are challenging and time-consuming to develop, and their completeness is difficult to validate. The building of such queries is still dependent on expert knowledge and painstaking analysis of the literature to map the relevant components of the biology when developing AOPs.
In response to the challenges of mapping and reviewing such a complex evidence base, AOPs have generally been developed in the publicly accessible AOP Wiki (https://aopwiki.org/), a resource that facilitates crowdsourcing while also implementing some controlled vocabularies and descriptors of AOP components. However, the number of experts who can realistically contribute tends to in fact be quite small, and the system is still vulnerable to the streetlight effect. According to the AOP Wiki, only 16 AOPs of the 306 in development have been endorsed. The 306 in development represent only a fraction of the thousands of biological processes we know we could be evaluating.
Escaping the Streetlight
Recent developments in AOPs illustrate what we perceive as four fundamental steps in a general strategy for addressing the streetlight problem and overcoming the semantic and conceptual factors in retrieving information about health risks posed by exposure to chemical substances. We argue that these steps require further development in the AOP sphere, and we believe they can be applied in general to the development and implementation of ontologized KOSs in toxicology and environmental health. The steps are derived from the experience of the authors and generally accepted principles of ontology development, as described in, e.g., Arp et al. (2015).
Step 1. Enumerate AOP-relevant entities, how they are related, and specify the vocabulary for labeling them.
The first step in developing an ontologized KOS is to define the things that are to be covered by the ontology (known in the technical vocabulary of ontology development as “entities”), the ways in which those things are related (the relationships between the entities), and the terms that will be used to label the entities and relationships (the controlled vocabulary). An AOP ontology would include many entities, examples of which include MIEs, KEs, and many types of relationships denoting how an upstream biological event affects a downstream one. Although some examples of potential entities and relationships are illustrated in Figure 2, the entities and relationships would ultimately be characterized in whatever formalism best serves a universalized approach to describing biological processes. Developing an ontology is a bootstrapping exercise of iteratively defining, mapping, and refreshing the conceptual framework that constitutes the ontology. It is based on expert knowledge and active surveillance of the literature. In at least its initial phase it is conducted manually before computationally assisted approaches can be applied later.
An AOP ontology has already been developed within the international AOP KnowledgeBase (https://aopkb.oecd.org/) and incorporates terms from existing biological ontologies into the AOP descriptions within the AOP KnowledgeBase (Ives et al. 2017). Some existing ontologies and how they relate to levels of cellular organization in an AOP are shown in Figure 6, indicating options for how the AOP ontology might be extended in the future. There has also been work on semantically defining AOPs (Wang et al. 2019; Wang 2020), which may also inform these efforts in the future. Finally, the Gene Ontology Causal Activity Model (Thomas et al. 2019) is suggestive of an approach to defining the relationships between events in an AOP.
Figure 6.
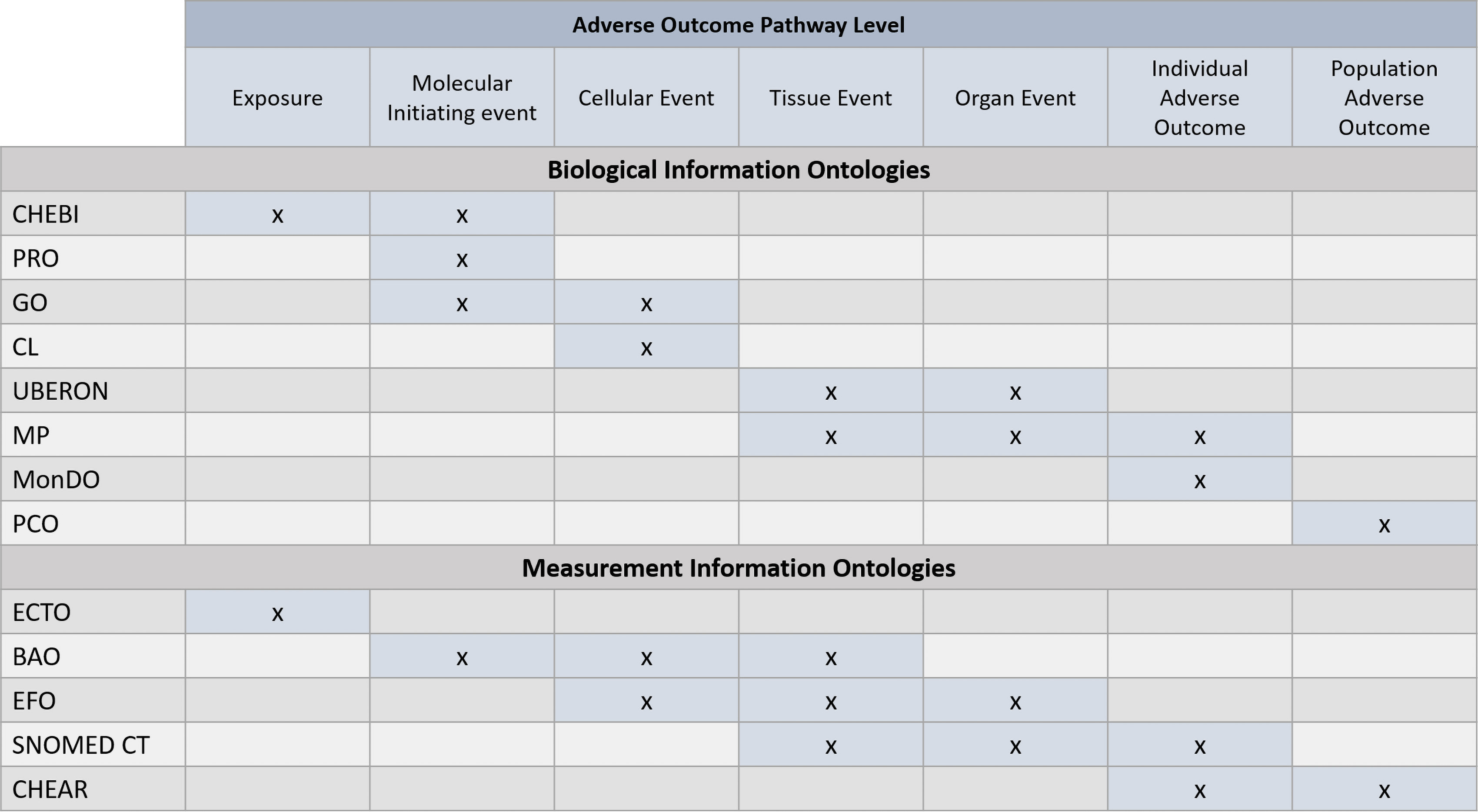
Existing biological ontologies can be used to define key events in computable terms and thereby make AOP information more interoperable with other toxicological data sources. The same can be done when describing the assays and biomarkers used to measure the key events. Note: BAO, BioAssay Ontology; CHEAR, Children’s Health Exposure Analysis Resource; CHEBI, Chemical Entities of Biological Interest; CL: Cell Ontology; ECTO, Environment Exposure Ontology; EFO, Experimental Factor Ontology; GO, Gene Ontology; MonDO, Mondo Disease Ontology; MP, Mammalian Phenotype Ontology; PCO, Population and Community Ontology; PRO, Protein Ontology; SNOMED CT, SNOMED Clinical Terms; UBERON, Uber Anatomy Ontology.
Step 2. Catalog the evidence for hypothetical relationships.
We propose that in developing an AOP, at least some minimum evidence for the existence of an entity or a relationship needs to be identified for something to be put forward as a candidate Key Event or Key Event Relationship. The purpose of this is to enable meaningless relations between completely unrelated entities to be excluded from the ontology. This evidence can be as little as a speculatively hypothesized relationship in a single document (even if the relationship proves false, this is still part of the knowledge that the ontology is being used to map and would need to be cataloged). If sufficient evidence with the appropriate agreed-upon characteristics accumulates, candidate events and relationships can be elevated for evaluation to being characterized as “Key.”
Evidence can be put behind relationships by tagging natural language expressions in relevant research documents with authorized terms from controlled vocabularies, a process illustrated in Figure 7. This would ensure the ontology developed via the expert process of Step 1 is associated with the real-world knowledge that the ontology is intended to describe. It also allows spurious relationships and factually nonexistent entities to be discarded. Both manual and automated methods are required to effectively tag the literature with concepts from the ontology. In the early stages, the process is almost exclusively manual, with an essential role for editors and biocurators in annotating documents. This is well documented in, for example, the Gene Ontology (Poux and Gaudet 2017).
Figure 7.
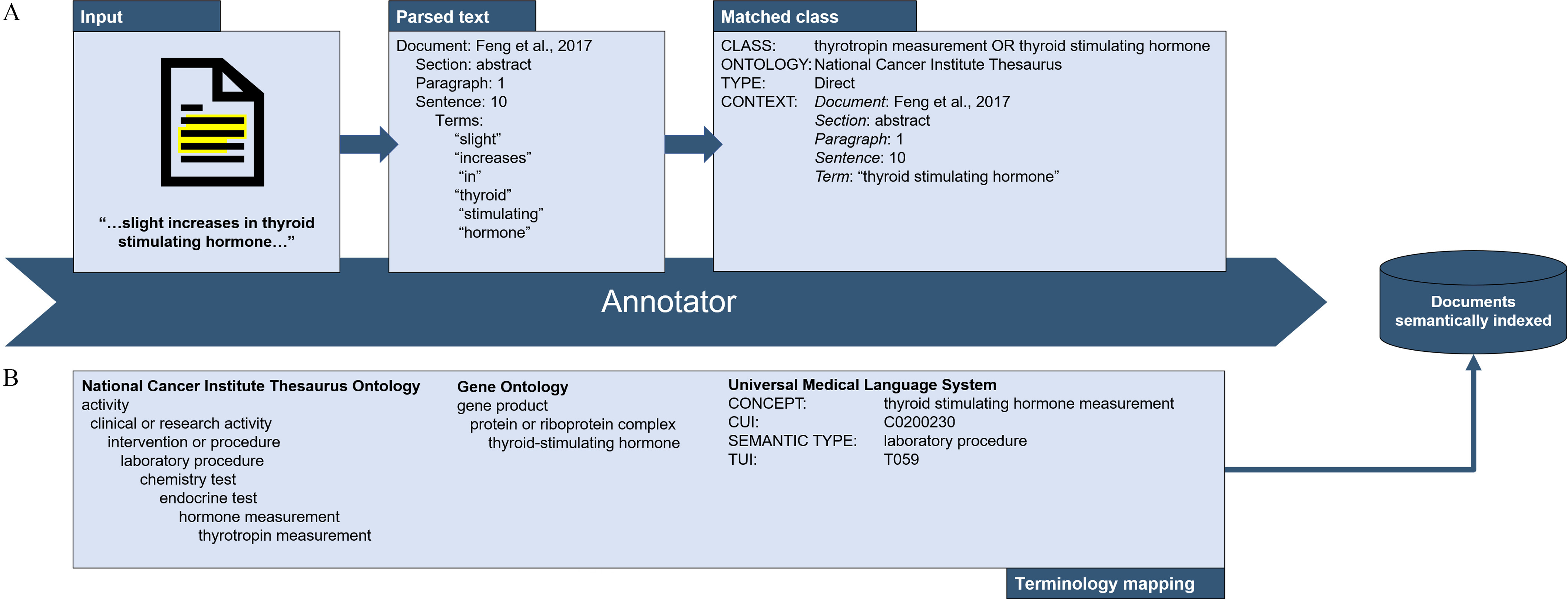
The workflow for matching natural language strings in research reports to a hierarchy of concepts in an ontology. Natural language information is extracted from included studies (e.g., phrases such as “increase in thyroid stimulating hormone”) into an evidence inventory (A). The terms “increase,” “thyroid,” “stimulating,” and “hormone” are cleaned and mapped to ontological classes in preparation for integration with other data sets. The inventory can then be connected to other data models by mapping terminology between CVs (B). Done enough times, a large data inventory begins to accumulate.
Because the rate-limiting step in creating annotations is the physical process of reading and tagging the scientific literature, it is necessary to automate the annotation of documents to scale the application of the ontology to the growing volume of new research (Thomas et al. 2019). The results of manual annotation exercises should, in theory, be usable as training data for automated methods for tagging free text with controlled vocabularies. We believe that conducting SEMs and SRs provides an opportunity to do this: With the right tools and training, data extractors should in principle be able to annotate the documents included in their map or review. Natural Language Processing (NLP) techniques, including Named Entity Recognition for tagging entities and sentiment analysis for identifying relationships, will be central to automation (Marshall and Wallace 2019; O’Connor et al. 2020). Various other machine learning applications could drastically reduce the time needed to review and vet evidence (Wittwehr et al. 2020). The use of semantic authoring tools that would render new studies machine-readable (Eldesouky et al. 2016; Oliveira et al. 2017; Oldman and Tanase 2018) would obviate many of the challenges in annotating research documents and should be explored for toxicology and environmental health contexts.
Step 3. Integrate different systems.
In the case of the biological mechanisms that underlie the indirect evidence supporting a chemical assessment, there is not one but many domains of knowledge. In addition, there are gaps in that knowledge even for the most well-studied toxicological mechanisms. Consequently, a framework is needed that can incorporate and represent biological knowledge in an interoperable (the ability for systems to exchange and use information) network of resources including visualizations, workflows, and computational pipelines that are online, interactive, and automatically updated. To be usable as an information resource, we believe, they must not only make intelligible to the user the knowledge from the many domains that they cover, but they must also explicitly account for missing information, so users are not led to overestimate the domain coverage of these systems.
Illustrative examples of such systems include the Health Assessment Workplace Collaborative (HAWC, https://hawcprd.epa.gov/portal/), the U.S. Environmental Protection Agency (U.S. EPA) Chemicals Dashboard (Williams et al. 2017), the U.S. EPA ChemView Portal (https://chemview.epa.gov/chemview), and the AOP KnowledgeBase (https://aopkb.oecd.org/); however, although these are functional and interactive depots for aggregating toxicological information, they are not yet interoperable. Achieving interoperability will require data management and stewardship that promotes the FAIR principles of information findability, accessibility, interoperability, and reusability (Wilkinson et al. 2016; Watford et al. 2019).
Step 4. Apply and evaluate.
The payoff in annotating free text in scientific documents with controlled vocabulary terms from ontologies is that it makes research machine-readable, enabling the use of computational intelligence in manipulating data that is still intelligible to humans. This facility is critical for increasing the speed at which evidence can be analyzed to a level commensurate with the rate at which new studies are being produced, meeting the information requirements of chemical assessment stakeholders, while still preserving a “white box” computational approach to analyzing chemical safety. “White box” systems are human-understandable computational approaches to solving analysis problems, just conducted at a speed and scale not achievable by humans working manually (Rudin 2019).
The final step of the process for strategic development of KOS development is in its evaluation. Although computational approaches to chemical assessment are still in very early development, and none that we are aware of use semantic data, a nascent predictive toxicology application based on AOPs has already been attempted (Burgoon 2017), and a comparison has been made between in silico approaches to in vivo assays and human data for identifying skin sensitizers (Luechtefeld et al. 2018; Golden 2020). These are suggestive of the future direction of computational toxicology and would be well supported by KOSs that make computationally accessible the human knowledge written up in scientific documents.
Conclusion
Chemical assessment can often involve the analysis of evidence that is only indirectly related to the target populations, exposures, and outcomes of concern. Surrogate populations are used because experimental toxicology is unethical in humans, so animal and in vitro models are relied on instead. For many chemicals (and by definition for novel substances) few studies have been conducted, requiring their potential toxicity to be inferred from suitably similar chemicals whose characteristics are better understood. Evidence of health outcomes may also be sparse. This is especially the case for diseases with long latency periods, such as certain brain cancers, or those that cannot be observed in a test system, such as when an in vitro model is being used for an apical outcome.
Systematically incorporating such indirect evidence is essential to all but the simplest of chemical assessments; however, systematic approaches are greatly challenged by the semantic and conceptual factors we have discussed in this commentary, cutting researchers off from relevant evidence. AOPs are illustrative of a certain way of overcoming the semantic and conceptual challenges of the streetlight effect, applying ontologies to the task of mapping the biological pathways by which exposures may cause adverse health outcomes and organizing the empirical evidence that underpins their development and evaluation. We believe that by following the four strategic steps of enumerating entities and their relationships, cataloguing evidence, systems integration, and evaluating effectiveness, the development of generalized KOSs for environmental health research can be achieved. Through such systems, broad-scale application of systematic methods that incorporate the full range of evidence relevant to a chemical assessment will become a reality.
Acknowledgments
The authors would like to thank G. Woodall, S. Bell, J. Lee, and K. Thayer for their technical review. The authors would also like to thank K. Markey for conceptual and intellectual knowledge contributions. The work described in this article has been reviewed by the Center for Environmental and Public Health Assessment of the U.S. EPA and approved for publication. The views expressed in this commentary are those of the authors and do not necessarily reflect the views or policies of the U.S. EPA. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- Arp R, Smith B, Spear AD. 2015. Building Ontologies with Basic Formal Ontology. Cambridge, MA: MIT Press; https://play.google.com/store/books/details?id=AUxQCgAAQBAJ [accessed 19 August 2020]. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25(1):p 25–29, PMID: 10802651, 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Atkinson MA. 2015. The streetlight effect in type 1 diabetes. Diabetes 64(4):p 1081–1090, PMID: 25805758, 10.2337/db14-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga A, Papachristos A, Hureau D. 2012. Hot spots policing effects on crime. Campbell Systematic Rev 8(1):1–96, 10.4073/csr.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoon LD. 2017. The AOPOntology: a semantic artificial intelligence tool for predictive toxicology. Appl in Vitro Toxicol 3(3):278–281, 10.1089/aivt.2017.0012. [DOI] [Google Scholar]
- Chalmers I, Hedges LV, Cooper H. 2002. A brief history of research synthesis. Eval Health Prof 25(1):12–37, PMID: 11868442, 10.1177/0163278702025001003. [DOI] [PubMed] [Google Scholar]
- Cheng SH, MacLeod K, Ahlroth S, Onder S, Perge E, Shyamsundar P, et al. 2019. A systematic map of evidence on the contribution of forests to poverty alleviation. Environ Evid 8(1):221, 10.1186/s13750-019-0148-4. [DOI] [Google Scholar]
- Descatha A, Sembajwe G, Pega F, Ujita Y, Baer M, Boccuni F, et al. 2020. The effect of exposure to long working hours on stroke: a systematic review and Meta-analysis from the WHO/ILO joint estimates of the work-related burden of disease and injury. Environ Int 142:105746, PMID: 32505015, 10.1016/j.envint.2020.105746. [DOI] [PubMed] [Google Scholar]
- El Idrissi T, Idri A, Bakkoury Z. 2019. Systematic map and review of predictive techniques in diabetes self-management. Int J Inform Manag 46:263–277, 10.1016/j.ijinfomgt.2018.09.011. [DOI] [Google Scholar]
- Eldesouky B, Bakry M, Maus H, Dengel A. 2016. Seed, an end-user text composition tool for the semantic web. In Proceedings of the 15th International Semantic Web Conference, Part I: The Semantic Web – ISWC 2016 P Groth, et al., eds. 17–21 October, 2016. Kobe, Japan. Cham, Switzerland: Springer International Publishing (Lecture Notes in Computer Science), 218–233, 10.1007/978-3-319-46523-4_14. [DOI] [Google Scholar]
- European Food Safety Authority. 2010. Application of systematic review methodology to food and feed safety assessments to support decision making. EFSA Journal 8(6):1637, PMID: 32625630, 10.2903/j.efsa.2010.1637.32625630 [DOI] [Google Scholar]
- Fagerholm N, Torralba M, Burgess PJ, Plieninger T. 2016. A systematic map of ecosystem services assessments around European agroforestry. Ecolog Ind 62:p 47–65, 10.1016/j.ecolind.2015.11.016. [DOI] [Google Scholar]
- Farrington DP, Ttofi MM. 2009. School-based programs to reduce bullying and victimization. Campbell Systematic Rev 5(1): i–148, 10.4073/csr.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G, de Coronado S, Haber M, Hartel F, Wright L. 2004. Overview and utilization of the NCI thesaurus. Comp Funct Genomics 5(8):648–654, PMID: 18629178, 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg AX, Hackam D, Tonelli M. 2008. Systematic review and meta-analysis: when one study is just not enough. Clin J Am Soc Nephrol 3(1):253–260, PMID: 18178786, 10.2215/CJN.01430307. [DOI] [PubMed] [Google Scholar]
- Gasparri L, Marconi D. 2019. Word meaning. In: Edward NZ, ed. Stanford Encyclopedia of Philosophy. Stanford, CA: Metaphysics Research Lab, Center for the Study of Language and Information (CSLI), Stanford University. [Google Scholar]
- Golden E. 2020. Evaluation of the global performance of eight in silico skin sensitization models using human data. ALTEX, 10.14573/altex.1911261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzelian PS, Victoroff MS, Halmes NC, James RC, Guzelian CP. 2005. Evidence-based toxicology: a comprehensive framework for causation. Hum Exp Toxicol 24(4):161–201, PMID: 15957536, 10.1191/0960327105ht517oa. [DOI] [PubMed] [Google Scholar]
- Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al. , eds. 2019. Cochrane Handbook for Systematic Reviews of Interventions version 6.0 (updated July 2019). London, UK: Cochrane; www.training.cochrane.org/handbook. [Google Scholar]
- Higgins JPT, et al. 2019. Methodological Expectations of Cochrane Intervention Reviews (MECIR). London, UK: Cochrane; https://community.cochrane.org/mecir-manual. [Google Scholar]
- Hodge GM, Digital Library Federation. 2000. Systems of Knowledge Organization for Digital Libraries: beyond Traditional Authority Files. Digital Library Federation, Council on Library and Information Resources. https://play.google.com/store/books/details?id=FpB8S5uT3xkC.
- Hoffmann S, Hartung T. 2006. Toward an evidence-based toxicology. Hum Exp Toxicol 25(9):p 497–513, PMID: 17017003, 10.1191/0960327106het648oa. [DOI] [PubMed] [Google Scholar]
- Hoffmann S, de Vries RBM, Stephens ML, Beck NB, Dirven HAAM, Fowle JR 3rd, et al. 2017. A primer on systematic reviews in toxicology. Arch Toxicol 91(7): 2551–2575, PMID: 28501917, 10.1007/s00204-017-1980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine. 2011. Finding What Works in Health Care: Standards for Systematic Reviews. Washington, DC: National Academies Press, 10.17226/13059. [DOI] [PubMed] [Google Scholar]
- Ives C, Campia I, Wang R-L, Wittwehr C, Edwards S. 2017. Creating a structured AOP knowledgebase via ontology-based annotations. Appl in Vitro Toxicol 3(4):298–311, PMID: 30057931, 10.1089/aivt.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James KL, Randall NP, Haddaway NR. 2016. A methodology for systematic mapping in environmental sciences. Environ Evid 5(1):7, 10.1186/s13750-016-0059-6. [DOI] [Google Scholar]
- Kaplan A. 1973. The Conduct of Inquiry. New Brunswick, NJ: Transaction Publishers; https://play.google.com/store/books/details?id=ks8wuZHSKs8C. [Google Scholar]
- Keshava C, Davis JA, Stanek J, Thayer KA, Galizia A, Keshava N, et al. 2020. Application of systematic evidence mapping to assess the impact of new research when updating health reference values: a case example using acrolein. Environ Int 143:105956, PMID: 32702594, 10.1016/j.envint.2020.105956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D, Angrish MM, Fortin MC, Katsiadaki I, Leonard M, Margiotta-Casaluci L, et al. 2018. Adverse outcome pathway networks I: development and applications. Environ Toxicol Chem 36(6):1723–1733, 10.1002/etc.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenaars C, et al. 2020. Reviewing the animal literature: how to describe and choose between different types of literature reviews. Lab Anim. Preprint posted online November 1, 2020, PMID: 33135562, 10.1177/0023677220968599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T, Marsh D, Rowlands C, Hartung T. 2018. Machine learning of toxicological big data enables read-across structure activity relationships (RASAR) outperforming animal test reproducibility. Toxicol Sci 165(1):198–212, PMID: 30007363, 10.1093/toxsci/kfy152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall IJ, Wallace BC. 2019. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Syst Rev 8(1): 163, PMID: 31296265, 10.1186/s13643-019-1074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miake-Lye IM, Hempel S, Shanman R, Shekelle PG. 2016. What is an evidence map? A systematic review of published evidence maps and their definitions, methods, and products. Syst Rev 5:28, PMID: 26864942, 10.1186/s13643-016-0204-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RL, Whaley P, Thayer KA, Schünemann HJ. 2018. Identifying the PECO: a framework for formulating good questions to explore the association of environmental and other exposures with health outcomes. Environ Int 121(Pt 1):1027–1031, PMID: 30166065, 10.1016/j.envint.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Library of Medicine. 2020. MEDLINE Citation Counts by Year of Publication. Bethesda, MD: National Library of Medicine; https://www.nlm.nih.gov/bsd/medline_cit_counts_yr_pub.html [accessed 9 November 2020]. [Google Scholar]
- Nosek BA, Ebersole CR, DeHaven AC, Mellor DT. 2018. The preregistration revolution. Proc Natl Acad Sci USA 115(11):2600–2606, PMID: 29531091, 10.1073/pnas.1708274114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC (National Research Council). 2007. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press, 216, 10.17226/11970. [DOI] [Google Scholar]
- NRC. 2014. Review of EPA’s Integrated Risk Information System (IRIS) Process. Washington, DC: National Academies Press; http://www.nap.edu/openbook.php?record_id=18764 [PubMed] [Google Scholar]
- O’Connor AM, et al. 2020. A focus on cross-purpose tools, automated recognition of study design in multiple disciplines, and evaluation of automation tools: a summary of significant discussions at the fourth meeting of The International Collaboration for Automation of Systematic Reviews (ICASR). Syst Rev 9(1): 100, 10.1186/s13643-020-01351-4, PMID: 32366302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development). 2018. Users’ Handbook supplement to the Guidance Document for developing and assessing Adverse Outcome Pathways. OECD Series on Adverse Outcome Pathways, No. 1. Paris, France: OECD Publishing, 63, 10.1787/5jlv1m9d1g32-en. [DOI]
- Oldman D, Tanase D, et al. 2018. Reshaping the knowledge graph by connecting researchers, data and practices in ResearchSpace. In: 17th International Semantic Web Conference, Proceedings, Part II. Vrandečić D, ed. 8–12 October 2018. Monterey, CA, USA: The Semantic Web – ISWC 2018. Cham, Switzerland: Springer International Publishing (Lecture Notes in Computer Science), 325–340, 10.1007/978-3-030-00668-6_20. [DOI] [Google Scholar]
- Oliveira EC, Thabata HG, Hironouchi LH, Ishikawa E, de A. Nunes MV, Gois L. et al. 2017. Ontology-based CMS news authoring environment. In: 2017 IEEE 11th International Conference on Semantic Computing (ICSC). January 30–February 1, 2017. San Diego, CA. IEEE, 264–265, 10.1109/ICSC.2017.91. [DOI]
- Orellano P, Reynoso J, Quaranta N, Bardach A, Ciapponi A. 2020. Short-term exposure to particulate matter (PM10 and PM2.5), nitrogen dioxide (NO2), and ozone (O3) and all-cause and cause-specific mortality: systematic review and meta-analysis. Environ Int 142:105876, PMID: 32590284, 10.1016/j.envint.2020.105876. [DOI] [PubMed] [Google Scholar]
- Pelch KE, Reade A, Wolffe TAM, Kwiatkowski CF. 2019. PFAS health effects database: protocol for a systematic evidence map. Environ Int 130:104851, PMID: 31284092, 10.1016/j.envint.2019.05.045. [DOI] [PubMed] [Google Scholar]
- Pomerantz J. 2015. Metadata. Cambridge, MA: MIT Press; https://play.google.com/store/books/details?id=j0X7CgAAQBAJ [Google Scholar]
- Poux S, Gaudet P. 2017. Best practices in manual annotation with the gene ontology. Methods Mol Biol 1446: 41–54, PMID: 27812934, 10.1007/978-1-4939-3743-1_4. [DOI] [PubMed] [Google Scholar]
- Radke EG, Yost EE, Roth N, Sathyanarayana S, Whaley P. 2020. Application of US EPA IRIS systematic review methods to the health effects of phthalates: lessons learned and path forward. Environ Int 145:105820, PMID: 33081976, 10.1016/j.envint.2020.105820. [DOI] [PubMed] [Google Scholar]
- Rethlefsen ML, Farrell AM, Osterhaus Trzasko LC, Brigham TJ. 2015. Librarian co-authors correlated with higher quality reported search strategies in general internal medicine systematic reviews. J Clin Epidemiol 68(6):617–626, PMID: 25766056, 10.1016/j.jclinepi.2014.11.025. [DOI] [PubMed] [Google Scholar]
- Roberts D, Brown J, Medley N, Dalziel SR, et al. 2017. Antenatal corticosteroids for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst Rev 3(3):CD004454, 10.1002/14651858.CD004454.pub3, PMID: 28321847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney AA, Boyles AL, Wolfe MS, Bucher JR, Thayer KA. 2014. Systematic review and evidence integration for literature-based environmental health science assessments. Environ Health Perspect 122(7):711–718, PMID: 24755067, 10.1289/ehp.1307972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin RS, Fischer SH, Damberg CL, Shi Y, Shekelle PG, Xenakis L. 2020. Optimizing health IT to improve health system performance: a work in progress. Healthc (Amst) 8(4):100483, PMID: 33068915, 10.1016/j.hjdsi.2020.100483. [DOI] [PubMed] [Google Scholar]
- Salvador-Oliván JA, Marco-Cuenca G, Arquero-Avilés R. 2019. Errors in search strategies used in systematic reviews and their effects on information retrieval. JMLA 107(2): 210–221, PMID: 31019390, 10.5195/jmla.2019.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez KA, Foster M, Nieuwenhuijsen MJ, May AD, Ramani T, Zietsman J, et al. 2020. Urban policy interventions to reduce traffic emissions and traffic-related air pollution: protocol for a systematic evidence map. Environ Int 142:105826, PMID: 32505921, 10.1016/j.envint.2020.105826. [DOI] [PubMed] [Google Scholar]
- Saran A, White H. 2018. Evidence and gap maps: a comparison of different approaches. Campbell Syst Rev 14(1): 1–38, 10.4073/cmdp.2018.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer HR, Myers JL. 2017. Guidelines for performing systematic reviews in the development of toxicity factors. Regul Toxicol Pharmacol 91:124–141, PMID: 29080853, 10.1016/j.yrtph.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Schwarzman MR, Ackerman JM, Dairkee SH, Fenton SE, Johnson D, Navarro KM, et al. 2015. Screening for chemical contributions to breast cancer risk: a case study for chemical safety evaluation. Environ Health Perspect 123(12):1255–1264, PMID: 26032647, 10.1289/ehp.1408337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen R. 2018. Vagueness. In: Stanford Encyclopedia of Philosophy. Summer 2018. Edward NZ, ed. Stanford, CA: Metaphysics Research Lab, Center for the Study of Language and Information (CSLI), Stanford University. [Google Scholar]
- Stearns MQ, Price C, Spackman KA, Wang AY. 2001. SNOMED clinical terms: overview of the development process and project status. Proc AMIA Symp. February 1, 2001, 662–666, PMID: 11825268. [PMC free article] [PubMed]
- Thomas PD, Hill DP, Mi H, Osumi-Sutherland D, Van Auken K, Carbon S, et al. 2019. Gene ontology causal activity modeling (GO-CAM) moves beyond GO annotations to structured descriptions of biological functions and systems. Nat Genet 51(10):1429–1433, PMID: 31548717, 10.1038/s41588-019-0500-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Angrish MM, Fortin MC, Katsiadaki I, Leonard M, Margiotta-Casaluci L, et al. 2018. Adverse outcome pathway networks II: Network analytics. Environ Toxicol Chem 37(6):1734–1748, 10.1002/etc.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, et al. 2014. Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol Sci 142(2):312–320, PMID: 25466378, 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M. 2013. The adverse outcome pathway concept: a pragmatic tool in toxicology. Toxicology 312:158–165, PMID: 23978457, 10.1016/j.tox.2013.08.011. [DOI] [PubMed] [Google Scholar]
- W3C (World Wide Web Consortium). 2020. Vocabularies. https://www.w3.org/standards/semanticweb/ontology [accessed 23 November 2020].
- Walker VR, Boyles AL, Pelch KE, Holmgren SD, Shapiro AJ, Blystone CR, et al. 2018. Human and animal evidence of potential transgenerational inheritance of health effects: an evidence map and state-of-the-science evaluation. Environ Int 115:48–69, PMID: 29549716, 10.1016/j.envint.2017.12.032. [DOI] [PubMed] [Google Scholar]
- Wang R-L. 2020. Semantic characterization of adverse outcome pathways. Aquat Toxicol 222:105478, PMID: 32278258, 10.1016/j.aquatox.2020.105478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R-L, Edwards S, Ives C. 2019. Ontology-based semantic mapping of chemical toxicities. Toxicology 412: 89–100, PMID: 30468866, 10.1016/j.tox.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watford S, Edwards S, Angrish M, Judson RS, Paul Friedman K. 2019. Progress in data interoperability to support computational toxicology and chemical safety evaluation. Toxicol Appl Pharmacol 380:114707, PMID: 31404555, 10.1016/j.taap.2019.114707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley P, Halsall C, Ågerstrand M, Aiassa E, Benford D, Bilotta G, et al. 2016. Implementing systematic review techniques in chemical risk assessment: challenges, opportunities and recommendations. Environ Int 92-93:556–564, PMID: 26687863, 10.1016/j.envint.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley P, Aiassa E, Beausoleil C, Beronius A, Bilotta G, Boobis A, et al. 2020. Recommendations for the conduct of systematic reviews in toxicology and environmental health research (COSTER). Environ Int 143:105926, PMID: 32653802, 10.1016/j.envint.2020.105926. [DOI] [PubMed] [Google Scholar]
- Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. 2011. BioPortal: enhanced functionality via new web services from the national center for biomedical ontology to access and use ontologies in software applications. Nucleic Acids Res 39(Web Server issue):W541–5, PMID: 21672956, 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. 2016. The FAIR guiding principles for scientific data management and stewardship. Sci Data 3(1):160018, PMID: 26978244, 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, et al. 2017. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform 9(1): 61–61, PMID: 29185060, 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwehr C, Blomstedt P, Gosling JP, Peltola T, Raffael B, Richarz A-N, et al. 2020. Artificial intelligence for chemical risk assessment. Comput Toxicol 13:100114, PMID: 32140631, 10.1016/j.comtox.2019.100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe TAM, Whaley P, Halsall C, Rooney AA, Walker VR. 2019. Systematic evidence maps as a novel tool to support evidence-based decision-making in chemicals policy and risk management. Environ Int 130:104871, PMID: 31254867, 10.1016/j.envint.2019.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe TAM, Vidler J, Halsall C, Hunt N, Whaley P. 2020. A survey of systematic evidence mapping practice and the case for knowledge graphs in environmental health and toxicology. Toxicol Sci 175(1):35–49, 10.1093/toxsci/kfaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff TJ, Sutton P. 2014. The navigation guide systematic review methodology: a rigorous and transparent method for translating environmental health science into better health outcomes. Environ Health Perspect 122(10):1007–1014, PMID: 24968373, 10.1289/ehp.1307175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff TJ, Sutton P, Navigation Guide Work Group. 2011. An evidence-based medicine methodology to bridge the gap between clinical and environmental health sciences. Health Aff (Millwood) 30(5):931–937, PMID: 21555477, 10.1377/hlthaff.2010.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost EE, Euling SY, Weaver JA, Beverly BEJ, Keshava N, Mudipalli A, et al. 2019. Hazards of diisobutyl phthalate (DIBP) exposure: a systematic review of animal toxicology studies. Environ Int 125:579–594, PMID: 30591249, 10.1016/j.envint.2018.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]