Abstract
Dojo loach (Misgurnus anguillicaudatus) that inhabit Japan are composed of two genetically divergent groups (A and B). Although most individual loach reproduce bisexually, clone lineages exist in certain populations that reproduce gynogenetically. To investigate the molecular phylogenetic relationships among the M. anguillicaudatus groups and clone lineages, complete mitogenomes of members from groups A and B and a clone lineage were sequenced using long range PCR and primer walking methods. The three groups of mitogenomes shared the same gene order and had similar base compositions and codon usage patterns. Phylogenetic analysis indicated group A and the clone lineage were genetically close with group B being genetically divergent.
Keywords: Dojo loach, Misgurnus anguillicaudatus, mitochondrial genome
Dojo loach (Misgurnus anguillicaudatus) are small freshwater fish widely distributed along the eastern coasts of Asia from the Amur River to the northern regions of Vietnam (Global Biodiversity Information Facility, 2020). Previous studies using the mitochondrial DNA control region (mtDNA-CR) and two nuclear genes, recombination activating gene 1 (RAG1) and interphotoreceptor retinoid-binding protein 2 (IRBP2) revealed M. anguillicaudatus in Japan can be genetically discriminated into three groups, A, B1, and B2 (Morishima et al. 2008, Yamada et al. 2015). Although most M. anguillicaudatus in nature commonly reproduces bisexually, gynogenetically reproducing clone lineages are distributed in certain districts in Japan (Morishima et al. 2002). Based on phylogenetic and cytogenetic studies, the clone lineages are thought to be hybrids originating between group A females and group B1 males (Yamada et al. 2015, Kuroda et al. 2018).
Complete mitogenomes of M. anguillicaudatus, independent of nuclear genomes, have been reported in previous studies (He et al 2008, Zhang et al 2018, Zhang et al 2019). Here, we analyzed the mitogenomes of group A, group B1, and a clone lineage of M. anguillicaudatus and report their molecular phylogenetic relationship among the Misgurnus genus. Specimens of M. anguillicaudatus group A, group B1, and a clone lineage were collected from Hokkaido prefecture. Specifically, the group A and clone lineage specimens were collected from Ozora, Japan and the group B1 specimen were collected from Nanae, Japan. Genotypes of the nuclear genomes were previously confirmed by Fujimoto et al. (2017). All specimens have been deposited in the DNA collection of the Hokkaido University Museum, Hakodate, Japan (catalog number: HUMZ 231155–231157).
Mitogenomes of the three groups were sequenced using long range PCR and primer walking methods (Miya and Nishida 1999). The complete genomes were 16,566 bp for group A (accession number LC532166), 16,641 bp for group B1 (accession number LC532167), and 16,568 bp for the clone lineage (accession number LC532168). The mitogenomes of all three groups included 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, one control region, and the origin of light strand replication (OriL), consistent with what is found in other teleost fish. The NADH-ubiquinone oxidoreductase chain 6 protein gene (ND6) and eight of the tRNA genes were encoded on the L-strand with the other genes encoded on the H-strand. The group A and clone lineage specimens shared the same gene directions and sequence lengths. In comparison, these genes in group B1 also had the same direction, but two PCGs (COII and ND3) and two rRNA genes had sequence lengths that differed from those of group A and the clone lineage. The initiation codons of the PCGs, order of the tRNAs, and location of the two of the rRNA genes were consistent with mitogenomes of other M. anguillicaudatus isolates. Based on pairwise comparisons, mitogenome sequence similarity was 86.22% between groups A and B1, 95.71% between group A and the clone lineage, and 86.17% between group B1 and the clone lineage.This was consistent with the findings of Morishima et al. (2008), who showed that the clone lineage had inherited their mitochondria from group A strains.
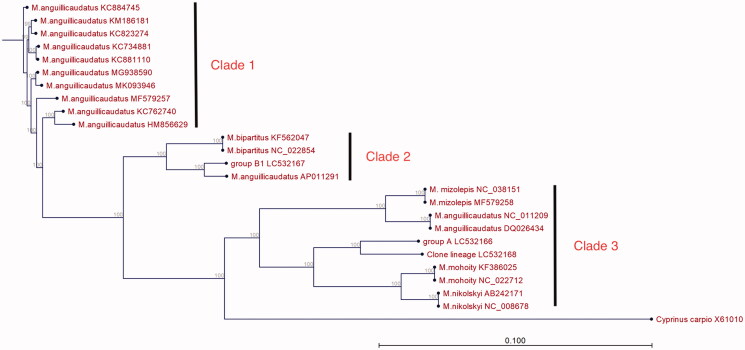
A phylogenetic relationship of Misgurnus spp. was reconstructed based on their mitogenomes using CLC Genomics Workbench (ver. 9.5.3) and the neighbor-joining method. The resulting phylogenetic tree consisted of three major clades (Figure 1). The first clade contained only M. anguillicaudatus strains for which the sequences were previously deposited by Zeng et al. (2012), Yu et al. (2014a, 2014b, 2014c), Zhou et al. (2014), Lee et al. (2018), Zhang et al. (2018), and Zhang et al. (2019). The second clade contained a M. anguillicaudatus isolate and another isolate of group B1, as well as two M. bipartitus isolates. The sequences for these isolates were previously deposited by Huang et al. (2015) and Miya et al. (2015). The third clade contained M. anguillicaudatus isolates, another isolate of group A, and the clone lineage isolated from the current study, in addition to other Misgurnus species including M. mizolepis, M. mohoity, and M. nikolskyi for which the sequences were previously deposited by He et al. (2008) and Saitoh et al. (2006). The results support the previous report that M. anguillicaudatus consists of genetically diverse groups (Zhang et al. 2018). The results also suggest that group B1 M. anguillicaudatus strains, which are distributed mainly in Japan, are closely related to M. bipartitus. Meanwhile, group A and the clone lineage were genetically closer related to M. mohoity and M. nikolskyi than to group B1. Further research regarding the mitogenome and nuclear genome with respect to geographical and morphological data will help solve the taxonomical complexity of the genus Misgurnus.
Figure 1.
Phylogenetic relationship of Misgurnus spp. based on their mitogenomes. The phylogenetic tree was constructed using CLC Genomics Workbench (ver. 9.5.3) and the neighbor-joining method. Numbers above branches indicate bootstrap values for 1000 replicates. The phylogenetic tree consists of three major clades.
Acknowledgments
We thank Dr. Fumihito Tashiro, (Hokkaido University Museum, Fisheries Science Center) for assistance with archival of the M. anguillicaudatus specimen DNAs at HUMZ.
Funding Statement
Duong Thuy Yen participated in this work during her visit to the Laboratory of Aquaculture Genetics and Genomics under the Can Tho University Improvement Project VN14-P6, supported by a Japanese Official Development Assistance (ODA) loan.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in ‘DNA Date Bank of Japan’ at http://getentry.ddbj.nig.ac.jp/, accession number [LC532166, LC532167, LC532168].
References
- Fujimoto T, Yamada A, Kodo Y, Nakaya K, Okubo-Murata M, Saito T, Ninomiya K, Inaba M, Kuroda M, Arai K, et al. 2017. Development of nuclear DNA markers to characterize genetically diverse groups of Misgurnus anguillicaudatus and its closely related species. Fish Sci. 83(5):743–756. [Google Scholar]
- Global Biodiversity Information Facility (GBIF), 2020. https://www.gbif.org/species/2367919 [Google Scholar]
- He S, Gu X, Mayden RL, Chen WJ, Conway KW, Chen Y.. 2008. Phylogenetic position of the enigmatic genus Psilorhynchus (Ostariophysi: Cypriniformes): evidence from the mitochondrial genome. Mol Phylogenet Evol. 47(1):419–425. [DOI] [PubMed] [Google Scholar]
- Huang S, Tian X, Wang W, Song W, Zhang X, Bai X, Cao X.. 2015.The complete mitochondrial genome of natural Misgurnus bipartitus (Cypriniformes: Cobitidae). Mitochondr DNA. 26(5):680–681. [DOI] [PubMed] [Google Scholar]
- Lee SY, Bang I, Nam YK.. 2018. Complete mitochondrial genome of albino cyprinid loach, Misgurnus anguillicaudatus (Cypriniformes: Cobitidae). Conserv Genet Resour. 10:507–510. [Google Scholar]
- Miya M, Nishida M.. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol (NY)). 1(5):416–426. [DOI] [PubMed] [Google Scholar]
- Miya M, Sato Y, Fukunaga T, Sado T, Poulsen JY, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, et al. 2015. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc Open Sci. 2(7):150088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima K, Horie S, Yamaha E, Arai K.. 2002. A Cryptic clonal line of the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae) evidenced by induced gynogenesis, interspecific hybridization, microsatellite genotyping and multilocus DNA fingerprinting. Zoolog Sci. 19(5):565–575. [DOI] [PubMed] [Google Scholar]
- Morishima K, Nakamura-Shiokawa Y, Bando E, Li YJ, Boroń A, Khan MMR, Arai K.. 2008. Cryptic clonal lineages and genetic diversity in the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae) inferred from nuclear and mitochondrial DNA analyses. Genetica. 132(2):159–171. [DOI] [PubMed] [Google Scholar]
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M.. 2006. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63(6):826–841. [DOI] [PubMed] [Google Scholar]
- Yamada A, Kodo Y, Murakami M, Kuroda M, Aoki T, Fujimoto T, Arai K.. 2015. Hybrid origin of gynogenetic clones and the introgression of their mitochondrial genome into sexual diploids through meiotic hybridogenesis in the loach, Misgurnus anguillicuadatus. J Exp Zool. 323A(9):593–606. [DOI] [PubMed] [Google Scholar]
- Yu YY, Li HY, Li RW, Wang WM, Zhou XY.. 2014. a. Complete mitochondrial genome of the natural hexaploid loach, Misgurnus anguillicaudatus (Teleostei: Cypriniformes: Cobitididae). Mitochondr DNA. 25(2):100–101. [DOI] [PubMed] [Google Scholar]
- Yu YY, Li HY, Li RW, Wang WM, Zhou XY.. 2014. b. Complete mitochondrial genome of the natural tetraploid loach Misgurnus anguillicaudatus. Mitochondrial DNA. 25(2):115–116. [DOI] [PubMed] [Google Scholar]
- Yu YY, Li HY, Li RW, Wang WM, Zhou XY.. 2014. c. Complete mitochondrial genome of the natural triploid loach, Misgurnus anguillicaudatus (Teleostei: Cypriniformes: Cobitididae). Mitochondr DNA. 25(5):353–354. [DOI] [PubMed] [Google Scholar]
- Zeng L, Wang J, Sheng J, Gu Q, Hong Y.. 2012. Molecular characteristics of mitochondrial DNA and phylogenetic analysis of the loach (Misgurnus anguillicaudatus) from the Poyang Lake. Mitochondr DNA. 23(3):187–200. [DOI] [PubMed] [Google Scholar]
- Zhang G, Zhu D, Li X, Liang X, Cai K, Zhang H, Zhang G.. 2018. Complete mitochondrial genome of natural diploid loaches Misgurnus anguillicaudatus from the Taihu Lake. Mitochondr DNA B Resour. 3(2):566–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chu D, Cai K, Zhang G, Zhu D, Deng Z, Jiang H.. 2019. Complete mitochondrial genome of Misgurnus anguillicaudatus, natural diploid loach from Nansi Lake. Mitochondr DNA B Resour. 4(1):399–400. [Google Scholar]
- Zhou X, Yu Y, Li Y, Wu J, Zhang X, Guo X, Wang W.. 2014. Comparative analysis of mitochondrial genomes in distinct nuclear ploidy loach Misgurnus anguillicaudatus and its implications for polyploidy evolution. PLOS One. 9(3):e92033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in ‘DNA Date Bank of Japan’ at http://getentry.ddbj.nig.ac.jp/, accession number [LC532166, LC532167, LC532168].